Summary
The dikaryons of basidiomycete fungi represent an unusual cell type required for complete sexual development. Dikaryon formation occurs via the activities of cell type-specific homeodomain transcription factors, which form regulatory complexes to establish the dikaryotic state. Decades of classical genetic and cell biological studies in mushrooms have provided a foundation for more recent molecular studies in the pathogenic species Ustilago maydis and Cryptococcus neoformans. Studies in these systems have revealed novel mechanisms of regulation that function downstream of classic homeodomain complexes to ensure that dikaryons are established and propagated. Comparisons of these dikaryon-specific networks promise to reveal the nature of regulatory network evolution and the adaptations responsible for driving complex eukaryotic development.
Introduction
Sexual modes of reproduction have been maintained throughout eukaryotic life over very long spans of evolutionary time, indicating the apparent advantage that sex confers on fitness [1]. The advantage of sex, however, can only be realized through the mixing of nuclear material from genetically distinct parents. In most cases, this occurs during gamete fusion, where the processes of cellular fusion (plasmogamy) and nuclear fusion (karyogamy) are coupled and occur in concert [2]. For example, during metazoan zygote formation, sperm and egg cell membranes fuse, and the two nuclei fuse immediately thereafter. In some systems, however, karyogamy does not coincide with cellular fusion. Following cell fusion in these systems, the two parent nuclei remain distinct. In many fungi, when two mating partners undergo cellular fusion, the resulting bi-nucleate cell grows filamentously, with each filament cell maintaining two independent nuclei that are replicated in a coordinate fashion (Figure 1) [3,4]. This growth stage is referred to as the dikaryon or dikaryotic filament. The largest and most studied group of organisms with a prolonged dikaryotic stage is the basidiomycete fungi. This phylum includes mushrooms, bracket fungi, and many plant pathogens, including the corn smut Ustilago maydis [5]. The basidiomycetes also include the globally distributed human pathogenic Cryptoccocus species, which are among the leading causes of fungal meningitis [6].
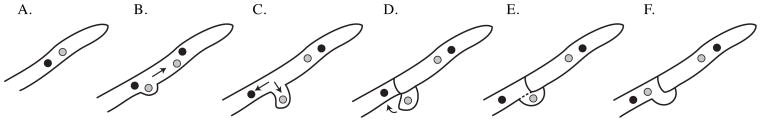
Figure 1. Model of dikaryotic growth.
(A) Each cell of the multicellular dikaryotic filament contains two distinct nuclei. (B) During cell division, the two nuclei are synchronously replicated via mitosis. (C) One daughter from each parent nucleus is maintained near the hyphal tip to generate the nascent filament cell. Simultaneously, one of the remaining parent nuclei moves into a clamp cell (hook-shaped cells required for proper nuclear distribution along the filament), while the other parent nucleus moves further away from the hyphal tip. (D) Septa emerge to produce a dikaryotic apical cell, monokaryotic clamp cell, and a monokaryotic subapical cell. (E, F) The clamp cell then fuses with the subapical cell to deliver its nucleus and restore dikaryosis along the filament.
Despite the complexities of managing two nuclei per cell throughout very long multicellular filaments, the dikaryon predominates as the vegetative growth form for many basidiomycete fungi and most mushroom species [7]. The advantages conferred by dikaryotic growth are currently unknown, but there is evidence that dikaryotic cells may have fitness advantages over their monokaryotic counterparts. When grown in culture, dikaryons of the mushroom Schizophyllum commune have been observed to diverge phenotypically to a much greater extent than monokaryons [7]. These data suggest that dikaryotic growth provides an increased potential for genetic and phenotypic variation. It is not known if these findings apply to dikaryotic cells of Cryptococcus neoformans and U. maydis, where the dikaryon is a developmental rather than vegetative cell type [8,9]. Despite their transitional natures, the dikaryotic phases in these systems are of particular interest because of their apparent importance to pathogenesis. Dikaryons are the infectious cell type of U. maydis and other plant pathogens [10] and dikaryotic growth of C. neoformans is required for the formation of spores, potential infectious particles in human cryptococcosis [11]. Characterization of the dikaryotic cell type is providing insights into the evolution, ecology, and pathogenesis of the basidiomycete fungi.
Genetic Determinants of Dikaryotic Growth
To date, much of what is known about dikaryons of basidiomycete fungi has been garnered using cell biological and genetic approaches in the mushrooms Coprinopsis cinerea [12] and S. commune [3]. Observations in these and other systems have been fundamental in developing the current understanding of dikaryon formation and maintenance [9]. Dikaryons are formed following the fusion of two cells of compatible mating types. Mating types in fungi are specified by information at Mating Type (MAT) loci, specialized regions of fungal genomes akin to the sex chromosomes of larger eukaryotes. MAT alleles must differ between mating partners for complete sexual development to take place. Most basidiomycete fungi contain two unlinked MAT loci: one encoding pheromones and pheromone receptors and the other encoding homeodomain transcription factors. These components control the initiation and continued development of the dikaryon [9] (Figure 1). In many mushrooms, cellular fusion occurs among cells of all mating types. Post-fusion, compatibility at the MAT locus encoding pheromones and pheromone receptors is required for proper nuclear migration as well as the fusion of specialized cells called clamps. The locus encoding the homeodomain transcription factors governs nuclear pairing and clamp formation. A different mechanism is utilized by C. neoformans and U. maydis, where compatibility of the pheromone and pheromone receptor genes is required for the initial cellular fusion event [13,14]. Following plasmogamy, a filamentous dikaryon grows in the presence of compatible homeodomain transcription factors in apparent accordance with the model shown in Figure 1 [11,15]. Although the specific nature of MAT locus contributions to dikaryotic growth vary among species characterized thus far, in all cases described, one feature is common: establishment of the dikaryon is mediated by a heterodimeric transcription factor complex comprised of a pair of MAT-encoded homeodomain proteins with one member contributed by each parent nucleus [9,16]. While studies in mushrooms have been instrumental in determining the genetic features of dikaryotic growth, the molecular and biochemical tools now available in the pathogenic basidiomycetes U. maydis and C. neoformans provide ideal opportunities to characterize the molecular mechanisms by which heterodimeric homeodomain transcription factors control establishment of the dikaryotic state.
Heterodimer Control of Fungal Sexual Development
The paradigm of homeodomain control of cell identity and sexual development comes from the budding yeast Saccharomyce cerevisiae, where the a1 and α2 homeodomain proteins heterodimerize following the mating of an a cell with an α cell [17]. The a1/α2 mechanism of diploid specification has been characterized in detail; the heterodimer transcriptionally represses 19 target genes that contain the a1/α2 binding site GATGN9ACA in their promoter regions [18,19] (Figure 2A). The target genes function directly to confer the diploid cell phenotype in a simple one-to-many regulatory mechanism. The resulting diploid cell is incapable of mating but is competent to undergo meiosis and sporulation in response to appropriate nutritional signals [17].
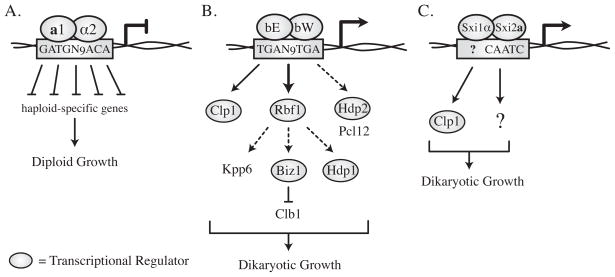
Figure 2. Mechanisms of molecular control of sexual development.
(A) The a1/α2 heterodimer of S. cerevisiae established the paradigm for cell-identification following the fusion of mating partners. 19 targets are repressed directly by the heterodimer, resulting in the stable diploid cell type. (B) The bE/bW complex of U. maydis operates at the top of a transcriptional cascade responsible for establishing the dikaryon. It acts directly to induce a number of regulators, including rbf1 and clp1. The transcription factor Rbf1 then induces the expression of biz1, hdp1, and kpp6. hdp2 and pcl12 show b-dependent expression, although this regulation has yet to be verified as direct. (C) The Sxi1α/Sxi2a complex in C. neoformans is known to act directly to induce the expression of CLP1. It is as yet unclear what pattern of transcriptional control will be utilized downstream of Sxi1α/Sxi2a activity to establish the dikaryotic state. Solid arrows indicate direct regulation; dashed arrows indicate indirect (or not yet confirmed as direct) regulation.
In basidiomycetes, homeodomain heterodimer activity obeys the basic paradigm established in S. cerevisiae; two differentially encoded transcription factors dimerize to form a new transcriptional regulatory complex that specifies the fused cell type [19–21]. However, until recently, the molecular mechanisms underlying the MAT-encoded homeodomain regulation of dikaryotic growth were completely unknown. Recent work on the bE/bW complex of U. maydis and Sxi1α/Sxi2a complex of C. neoformans reveals novel regulatory networks critical for establishment of the dikaryon.
Establishing a Dikaryon: Transcriptional Regulatory Cascade
The bE/bW complex of U. maydis is necessary for pathogenic growth, a process that includes dikaryon formation, a cell cycle arrest, appressorium formation, and plant penetration [10]. These events require the homeodomains of both proteins, and it is likely that both contribute to DNA binding in vivo [20]. Molecular studies have identified the bE/bW DNA binding site (bbs) in a small number of promoters. It spans approximately 20 nucleotides, and among the instances described, contains the core sequence TGAN9TGA [22–26]. bE/bW binding to the bbs results in transcriptional activation of the direct targets identified thus far, which contrasts with a1/α2 regulation in which binding to direct targets results only in transcriptional repression [18] (Figure 2A, B).
Microarray studies of b-inducible strains filamenting in the absence of host tissue have identified hundreds of genes with bE/bW-dependent expression patterns, likely including both direct and indirect targets [23,27]. In contrast, S. cerevisiae a1/α2 influences the levels of fewer than 25 genes, and nearly all appear to be regulated directly [18]. To identify genes regulated directly by the bE/bW, Heimel et al. analyzed 39 candidates whose transcription was induced by b-complex activity [28]. Of these, four were of particular interest because, when deleted, they all showed phenotypes related to dikaryotic growth and they encode putative transcription factors: rbf1, biz1, hdp1, and hdp2. rbf1 contains identifiable bbs sequences in its promoter, suggesting that rbf1 is a direct target of bE/bW. However, biz1, hdp1, and hdp2 do not appear to be direct targets. These findings suggest that bE/bW initiates a transcriptional cascade with numerous tiers of activation controlling processes crucial to dikaryotic growth (Figure 2B).
Of the morphological and transcriptional changes induced by bE/bW, the majority are controlled by Rbf1, a putative C2H2 Zinc finger transcription factor [28]. rbf1 mutants show defects in very early steps of pathogenic growth and do not form appressoria or undergo the cell cycle arrest required for plant invasion. Expression of rbf1 in b-deletion strains is sufficient to promote the normally b-induced cell cycle arrest and initiate filamentous growth. Additionally, combinatorial microarray experiments found that Rbf1 is necessary for >90% and sufficient for >50% of b-induced transcriptional changes, including the activation of biz1 and hdp1. Direct evidence for the cascading regulatory model was recently attained via chromatin-immunoprecipitation (ChIP) experiments, which localized bE to a region of the rbf1 promoter that contains a site very similar to the previously described bbs. These findings indicate that Rbf1 is a master regulator of dikaryotic growth downstream of bE/bW [28] (Figure 2B).
A possible direct target of rbf1 is biz1, another C2H2 zinc finger transcription factor-encoding gene. biz1 is required for pathogenic filamentation, and its expression is dependent on both Rbf1 and bE/bW activity, although no bbs has been identified in its promoter [28]. biz1 has also been shown to directly regulate clb1, a cyclin whose repression is required for plant invasion. This ordering of biz1 further supports the model of a multi-tier transcriptional cascade downstream of bE/bW.
Other potential members of the bE/bW regulatory cascade are the homeodomain-encoding genes hdp1 and hdp2. These genes have yet to be characterized in detail; however it is known that hdp1 expression is dependent on Rbf1 and is potentially a direct target of Rbf1. In contrast, hdp2 shows b-dependent but not rbf1-dependent expression, and thus, it may be functioning in parallel to Rbf1 during dikaryotic growth [28] (Figure 2B). Overall, the transcriptional regulators identified and characterized to date comprise a cascade in which early signals through bE/bW are amplified to effect dikaryotic growth.
Establishing a Dikaryon: Network Configuration Unknown
In C. neoformans, the regulatory network controlling dikaryon formation is not yet known. Central to dikaryotic growth in this system, however, is the Sxi1α/Sxi2a homeodomain regulatory complex. Strains lacking either protein cannot form dikaryons [29,30], and while few direct targets are known in vivo, the Sxi proteins have been characterized in vitro. Studies of the DNA-binding properties of Sxi1α and Sxi2a reveal that both are capable of binding DNA and that Sxi2a binding is specific and of high affinity, even in the absence of Sxi1α [31]. This contrasts with many homeodomain heterodimers in which both partners are required for efficient DNA binding [19,32]. Protein binding microarray studies show that Sxi2a binds preferentially to the core DNA consensus CAATC, which differs somewhat from the half-site bound by its S. cerevisiae homolog a1 (CATC) [19] (Figure 2A, C). It is not yet known what sequence(s) are bound by the Sxi1α/Sxi2a complex in vivo or in vitro, although the homeodomains of both Sxi1α and Sxi2a are required for heterodimer function in vivo [31].
Microarray studies of sxi1α and sxi2a mutant strains have identified many genes with Sxi-dependent expression patterns (Stanton & Hull, unpublished data); however, no functional equivalent of Rbf1 has been identified. Although there are candidate homologs exhibiting sequence similarity to Rbf1, none has been observed to be regulated by Sxi1α/Sxi2a (Kruzel and Hull, personal observation). Given the available information on the Sxi regulatory pathway, it is as yet unclear which molecular regulatory paradigm (multiple direct targets vs. transcriptional cascade) the Sxi1α/Sxi2a transcriptional network will follow to establish the dikaryon and control sexual development in C. neoformans (Figure 2C). Continuing analyses of likely direct targets will facilitate comparisons between the U. maydis and C. neoformans systems and elucidate both common and disparate features of these heterodimer-controlled transcriptional networks.
Basidiomycete-specific regulator: CLP1
A common feature of heterodimer-controlled networks among basidiomycetes characterized thus far is the regulator Clp1. The CLP1 gene was first identified in a screen in C. cinerea for mutants that block homeodomain-driven dikaryon formation [33]. clp1 mutants do not form clamp cells. Homologs of CLP1 have subsequently been identified and functionally characterized in both U. maydis and C. neoformans [24,34,35].
In U. maydis, clp1 transcription is activated by bE/bW, and its promoter contains multiple instances of the bbs, making it a likely direct target in vivo [24]. clp1 mutants invade plant tissues but fail to replicate and propagate the dikaryon, and mutant phenotypes are consistent with a prolonged cell cycle arrest and an inability to resume cell division. Clp1 has been shown to interact directly with bW and Rbf1 in vivo, and Clp1 overexpression negatively regulates many bE/bW and Rbf1 target genes [35]. This overexpression blocks b- and rbf1-dependent filamentation [24,35]. Working models suggest that Clp1 acts as a negative feedback regulator to relieve the bE/bW and Rbf1-dependent gene expression changes that induce cell cycle arrest. This feedback then permits re-initiation of the cell cycle and propagation of the dikaryon in planta.
In C. neoformans, clp1 mutant strains are unable to form dikaryons following cellular fusion [34]. This defect resembles that of U. maydis clp1 mutants, and supports the hypothesis that Clp1 activity facilitates escape from the cell cycle arrest that occurs preceding dikaryotic growth [24]. C. neoformans CLP1 shows Sxi-dependent transcriptional induction, and a 19 base-pair sequence within its promoter (GTTATTGTTTTTCATTCAA) is bound by Sxi2a in vitro. This makes CLP1 a likely direct target of Sxi1α/Sxi2a in vivo (Figure 2C). These data indicate that CLP1 is a critical regulator of transcription whose activity is required for establishing the dikaryotic state in basidiomycete fungi.
In addition to transcriptional regulators, other b-dependent genes, and potential effectors of dikaryotic growth, have been identified in U. maydis (Figure 2B). For example, kpp6, encoding a MAP kinase, is required for appressoria formation and shows b- and Rbf1 dependent expression [27]. pcl12, encoding a regulatory cyclin that interacts with Cdk5, is expressed in a b-dependent manner, although appears to be an indirect target of bE/bW as no bbs sites have been identified in its promoter. pcl12 plays a role in polarized growth occurring in concert with cell cycle arrest [36,37]. Of the other numerous putative bE/bW targets identified, the vast majority are not required for dikaryotic growth [23]. However, the characterization of homeodomain targets to date has facilitated an understanding of the molecular mechanisms by which a gene regulatory network establishes the dikaryotic state.
Conclusions and Future Directions
Future work in the field will certainly focus on identifying additional key effectors and targets required to establish and maintain dikaryotic growth. While the C. neoformans and U. maydis systems are both robust for molecular studies, they confer distinct advantages. In C. neoformans, wild type sexual development occurs under nutrient-controlled conditions in the laboratory in the absence of a plant or animal host. This is in contrast to U. maydis where native dikaryotic growth occurs only in the context of host plant tissue, making molecular studies of wild-type dikaryon formation and subsequent development technically challenging. Features required for host interactions may not be readily distinguished from plant-independent components of dikaryotic growth.
In contrast, U. maydis has been an exceptional system for studying dikaryon interactions with the plant host and for gene expression studies to determine the heterodimer-controlled transcriptional network. Further studies in both systems will provide insights into eukaryotic control of sexual development. Preliminary studies in C. neoformans indicate that unique modes of cell-type specification will emerge. Gene expression microarray studies show that approximately 70% of genes induced during the dikaryotic stage have no recognizable protein domains or functionally characterized homologs (Kruzel & Hull, unpublished data). Thus, the basic cellular processes underlying dikaryotic growth have yet to be described. Identifying targets downstream of Sxi1α/Sxi2a that are required for wild-type dikaryosis in C. neoformans will provide a solid foundation for developing a model of dikaryon formation in a human fungal pathogen and provide context for discerning dikaryon vs. host-derived regulatory events in the U. maydis model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Otto S, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- 2.Chen E, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 3.Iwasa M, Tanabe S, Kamada T. The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet Biol. 1998;23:110–116. doi: 10.1006/fgbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 4.Raper JR. Genetics of sexuality in higher fungi. New York: Ronald Press; 1966. [Google Scholar]
- 5.James T, Kauff F, Schoch C, Matheny P, Hofstetter V, Cox C, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall A, Perfect JR. Cryptococcus neoformans. Washington DC, USA: ASM Press; 1998. p. 541. [Google Scholar]
- 7.Clark T, Anderson J. Dikaryons of the basidiomycete fungus Schizophyllum commune: evolution in long-term culture. Genetics. 2004;167:1663–1675. doi: 10.1534/genetics.104.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alspaugh J, Davidson R, Heitman J. Morphogenesis of Cryptococcus neoformans. Contrib Microbiol. 2000;5:217–238. doi: 10.1159/000060352. [DOI] [PubMed] [Google Scholar]
- 9.Casselton L, Olesnicky N. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 11.Hull C, Heitman J. Genetics of Cryptococcus neoformans. Annu Rev Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- 12.Raper CA. Controls for development and differentiation of the dikaryon of basidiomycetes. In: JB, AC, editors. Secondary Metabolism and Differentiation in Fungi. Dekker; 1983. pp. 195–238. [Google Scholar]
- 13.Stanton B, Giles S, Staudt M, Kruzel E, Hull C. Allelic exchange of pheromones and their receptors reprograms sexual identity in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000860. doi: 10.1371/journal.pgen.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bölker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- 15.Wahl R, Zahiri A, Kämper J. The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol Microbiol. 2010;75:208–220. doi: 10.1111/j.1365-2958.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- 16.Kronstad J, Staben C. Mating type in filamentous fungi. Annu Rev Genet. 1997;31:245–276. doi: 10.1146/annurev.genet.31.1.245. [DOI] [PubMed] [Google Scholar]
- 17.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galgoczy D, Cassidy-Stone A, Llinás M, O’Rourke S, Herskowitz I, DeRisi J, Johnson A. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goutte C, Johnson A. a1 protein alters the DNA binding specificity of α2 repressor. Cell. 1988;52:875–882. doi: 10.1016/0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- 20.Schlesinger R, Kahmann R, Kämper J. The homeodomains of the heterodimeric bE and bW proteins of Ustilago maydis are both critical for function. Mol Gen Genet. 1997;254:514–519. doi: 10.1007/pl00008609. [DOI] [PubMed] [Google Scholar]
- 21.Asante-Owusu R, Banham A, Böhnert H, Mellor E, Casselton L. Heterodimerization between two classes of homeodomain proteins in the mushroom Coprinus cinereus brings together potential DNA-binding and activation domains. Gene. 1996;172:25–31. doi: 10.1016/0378-1119(96)00177-1. [DOI] [PubMed] [Google Scholar]
- 22.Romeis T, Brachmann A, Kahmann R, Kämper J. Identification of a target gene for the bE-bW homeodomain protein complex in Ustilago maydis. Mol Microbiol. 2000;37:54–66. doi: 10.1046/j.1365-2958.2000.01978.x. [DOI] [PubMed] [Google Scholar]
- 23.Brachmann A, Weinzierl G, Kämper J, Kahmann R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol. 2001;42:1047–1063. doi: 10.1046/j.1365-2958.2001.02699.x. [DOI] [PubMed] [Google Scholar]
- 24.Scherer M, Heimel K, Starke V, Kämper J. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell. 2006;18:2388–2401. doi: 10.1105/tpc.106.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doehlemann G, Wahl R, Vranes M, de Vries R, Kämper J, Kahmann R. Establishment of compatibility in the Ustilago maydis/maize pathosystem. J Plant Physiol. 2008;165:29–40. doi: 10.1016/j.jplph.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Brefort T, Doehlemann G, Mendoza-Mendoza A, Reissmann S, Djamei A, Kahmann R. Ustilago maydis as a Pathogen. Annu Rev Phytopathol. 2009;47:423–445. doi: 10.1146/annurev-phyto-080508-081923. [DOI] [PubMed] [Google Scholar]
- 27.Brachmann A, Schirawski J, Müller P, Kahmann R. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 2003;22:2199–2210. doi: 10.1093/emboj/cdg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Heimel K, Scherer M, Vranes M, Wahl R, Pothiratana C, Schuler D, Vincon V, Finkernagel F, Flor-Parra I, Kämper J. The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001035. Authors demonstrate that bE/bW initates a transcriptional regulatory cascade during dikaryotic growth of U. maydis that operates through Rbf1. Additionally, bE/bW is localized to the rbf1 promoter in vivo via the first published chromatin-immunoprecipitation experiments on a basidiomycete homeodomain heterodimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hull C, Boily M, Heitman J. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell. 2005;4:526–535. doi: 10.1128/EC.4.3.526-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hull C, Davidson R, Heitman J. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 2002;16:3046–3060. doi: 10.1101/gad.1041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Stanton B, Giles S, Kruzel E, Warren C, Ansari A, Hull C. Cognate Site Identifier analysis reveals novel binding properties of the Sex Inducer homeodomain proteins of Cryptococcus neoformans. Mol Microbiol. 2009;72:1334–1347. doi: 10.1111/j.1365-2958.2009.06719.x. Authors use a novel technique to characterize comprehensively the DNA-binding properties of Sxi2a and demonstrate that the homeodomains of both Sxi1a and Sxi2a are required for establishment of the dikaryon in C. neoformans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark M, Escher D, Johnson A. A trans-acting peptide activates the yeast a1 repressor by raising its DNA-binding affinity. EMBO J. 1999;18:1621–1629. doi: 10.1093/emboj/18.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inada K, Morimoto Y, Arima T, Murata Y, Kamada T. The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics. 2001;157:133–140. doi: 10.1093/genetics/157.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Ekena J, Stanton B, Schiebe-Owens J, Hull C. Sexual development in Cryptococcus neoformans requires CLP1, a target of the homeodomain transcription factors Sxi1alpha and Sxi2a. Eukaryot Cell. 2008;7:49–57. doi: 10.1128/EC.00377-07. This study provides the first biochemical characterization of Sxi1a and Sxi2a as DNA-binding proteins, and both are shown to bind the CLP1 promoter in vitro. This work also demonstrates that CLP1, a likely Sxi1a/Sxi2a direct target, is required for dikaryon formation in C. neoformans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Heimel K, Scherer M, Schuler D, Kämper J. The Ustilago maydis Clp1 protein orchestrates pheromone and b-dependent signaling pathways to coordinate the cell cycle and pathogenic development. Plant Cell. 2010 doi: 10.1105/tpc.110.076265. This study demonstrates the role of Clp1 during U. maydis dikaryotic growth as a modulator of gene expression via direct interactions with specific transcription factors. Authors map Clp1 activity in a negative feedback loop that is induced by bE/bW and required for pathogenic filamentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flor-Parra I, Castillo-Lluva S, Pérez-Martín J. Polar growth in the infectious hyphae of the phytopathogen Ustilago maydis depends on a virulence-specific cyclin. Plant Cell. 2007;19:3280–3296. doi: 10.1105/tpc.107.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castillo-Lluva S, Alvarez-Tabarés I, Weber I, Steinberg G, Pérez-Martín J. Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J Cell Sci. 2007;120:1584–1595. doi: 10.1242/jcs.005314. [DOI] [PubMed] [Google Scholar]