Abstract
Decapping enzymes are required for the removal of the 5′-end m7GpppN cap of mRNAs to allow their decay in cells. While many cap-binding proteins recognize the cap structure via the stacking of the methylated guanosine ring between two aromatic residues, the precise mechanism of cap recognition by decapping enzymes has yet to be determined. In order to get insights into the interaction of decapping enzymes with the cap structure, we studied the vaccinia virus D10 decapping enzyme as a model to investigate the important features for substrate recognition by the enzyme. We demonstrate that a number of chemically modified purines can competitively inhibit the decapping reaction, highlighting the molecular features of the cap structure that are required for recognition by the enzyme, such as the nature of the moiety at positions 2 and 6 of the guanine base. A 3D structural model of the D10 protein was generated which suggests amino acids implicated in cap binding. Consequently, we expressed 17 mutant proteins with amino acid substitutions in the active site of D10 and found that eight are critical for the decapping activity. These data underscore the functional features involved in the non-canonical cap-recognition by the vaccinia virus D10 decapping enzyme.
INTRODUCTION
The synthesis, translation and decay of messenger RNAs (mRNAs) are highly regulated processes in cells (1–3). In order to allow mRNA turnover, the protective structures found on both the 5′- and 3′-ends of mRNAs have to be removed. mRNA decay occurs mainly in the cytoplasm by the recruitment of a plethora of enzymes involved in degradation. While mRNA decay has previously been associated with P-bodies, recent evidences suggest that the initiation of decapping is independent of P-body formation, and that the latter would rather be a consequence of mRNA decay (4–6). The m7GpppN cap structure found at the 5′-end of mRNAs is degraded by decapping enzymes that have now been identified in mammalians, plants, and yeasts (Dcp2 proteins), as well as in some viruses (vaccinia virus and African swine fever virus) (7–13). The Dcp2 and viral decapping enzymes exhibit a specific hydrolase activity resulting in cleavage between the α- and β-phosphates of the m7GpγpβpαN cap to generate both m7GDP and monophosphorylated RNA products. The remaining monophosphorylated mRNAs are then further subjected to 5′→3′ exonucleolytic degradation (14,15). Decapping enzymes possess a conserved NUDIX motif common to Dcp2 and viral decapping enzymes as well as other hydrolases members of the NUDIX hydrolase superfamily (7–9,16,17). The conserved 23 amino acids NUDIX motif GX5EX7REUXEEXGU (where U is a bulky hydrophobic amino acid and X is any amino acid) has previously been demonstrated to be required for catalysis during the decapping reaction (8,18,19).
The cellular mRNA 5′ cap structure is recognized by a number of cap-binding proteins other than the decapping enzymes. Structural studies of the interaction of cap derivatives with the human cap-binding complex (CBC), the mouse eIF4E translation initiation factor, and the vaccinia virus VP39 mRNA 2′-O-methyltransferase, revealed that cap-recognition is primarily achieved by the stacking of the methylated guanosine ring between two aromatic residues within the catalytic center of these proteins (20–23). These evolutionarily unrelated proteins have evolved to share a canonical cap-recognition mode. In contrast, the recently published crystal structures of the yeast Schizosaccharomyces pombe Dcp2 enzyme (SpDcp2) indicated that no aromatic residues are found in the vicinity of the cap-binding site of the enzyme (10,24). So far, no mechanism has been proposed for the cap-recognition mode of metazoan or viral decapping enzymes. However, structural data and mutational analysis of the yeast decapping enzyme highlighted the presence of amino acids located in the catalytic pocket, such as Lys129 and Gln169, that could potentially be involved in recognition of the cap structure (10).
The discovery that mammalian viruses can harbor decapping enzymes has important implications for the control of viral and cellular gene expression. The vaccinia virus D10 decapping enzyme (D10) is expressed during late viral transcription, and deletion of the D10 gene was shown to produce a 10-fold reduction of the specific infectivity of the vaccinia virions (25). The presence of decapping enzymes in vaccinia virus is suggested to be required to eliminate competing host mRNAs and to allow stage specific synthesis of viral proteins (7). Previous studies have further demonstrated the ability of the D10 decapping enzyme to hydrolyze capped RNAs of at least 24 nt in length in the presence of divalent cations. Notably, the decapping reaction catalyzed by D10 was also shown to be inhibited by methylated cap analogs such as m7GpppG and m7GTP (7,26). With no available crystal structure for D10, we previously generated a structural model of the tertiary structure of the active site of the D10 protein based on the SpDcp2 3D structure (26). Our biochemical assays also demonstrated that the D10 protein utilizes a two-metal ion mechanism for catalysis. The importance of Glu132 and Glu145 in metal ion coordination, as well as Glu141 in the positioning of a water molecule in the catalytic center was also highlighted by this study (26).
In order to gain additional insights into the molecular determinants involved in substrate recognition by the vaccinia virus D10, we now perform a thorough probing of the active site of the enzyme using a collection of nucleotide analogs. Moreover, mutagenesis studies underline the importance of specific amino acids located within the catalytic pocket of D10 for the decapping activity. Using this combination of nucleotide analogs and site-directed mutagenesis, this study illustrates the specific functional groups that are required for the non-canonical cap-recognition mode of the D10 decapping enzyme.
MATERIALS AND METHODS
Homology modeling
The crystal structure of the S. pombe decapping enzyme [SpDcp2, amino acids 1–266 (10)] was used as a template to model the vaccinia virus D10 protein (amino acids 1–226). The atomic coordinates were obtained from the Protein Data Bank file 2A6T. The predicted 3D structure of the D10 protein was generated with the Deep View program (27). The coordinates of the amino acids from the D10 protein sequence that were not folded by the program were deleted from the generated PDB file to produce a workable file for the modelized D10 structure. The modified D10 PDB file was then uploaded in the Visual Molecular Dynamics program (28) to generate the presented structures.
Molecular docking
Docking calculations were carried out using the Docking Server software and the Dreiding force field was used for energy minimization of N7-methyl diguanosine triphosphate (m7GpppG) using built-in Chemaxon tools in Docking Server (29). PM6 semi-empirical charges calculated by MOPAC2007 were added to the ligand atoms. Non-polar hydrogen atoms were merged and rotatable bonds were defined (30). Docking calculations were carried out using the SpDcp2 crystal structure (Protein Data Bank 2A6T). Essential hydrogen atoms, Kollman united atom type charges and solvation parameters were added with the aid of AutoDock tools (31). Affinity (grid) maps of 20 × 20 × 20 Å grid points and 0.375 Å spacing were generated using the Autogrid program (31). AutoDock parameter set- and distance-dependent dielectric functions were used in the calculation of the van der Waals and the electrostatic terms, respectively. Docking simulations were performed using the Lamarckian genetic algorithm and the Solis and Wets local search method (32). Initial position, orientation, and torsions of the ligand molecules were set randomly. Each docking experiment was derived from two different runs that were set to terminate after a maximum of 2 500 000 energy evaluations. The population size was set to 150. During the search, a translational step of 0.2 Å, and quaternion and torsion steps of 5 Å were applied.
D10 expression, purification and decapping activity
The wild-type D10 protein and the substitution mutants were expressed and purified as described earlier (26). Reaction mixtures (10 μl) containing 10 mM Tris–HCl (pH 7.5), 100 mM KOAc, 10 mM MnCl2, 10 mM MgCl2, 1 mM DTT, 0.9 μM γ-phosphate-labeled 7-methyl-capped RNA substrate and 1 μM purified D10 WT or mutant protein were incubated for 15 min at 25°C. The reactions were quenched by adding 1 μl of 50 mM EDTA. Aliquots of the mixtures were applied to a polyethyleneimine–cellulose TLC plate, which was developed with 0.5 mM LiCl and 1 M methanoic (formic) acid. The release of γ-phosphate-labeled m7GDP was revealed by autoradiography with a Storm phosphoimager (Amersham Biosciences) and quantified using ImageQuant 5.0 (Molecular Dynamics). The average of at least two single independent experiments is presented.
N7-Methyl-capped RNA synthesis
A radiolabeled 81-mer RNA substrate was synthesized and capped as described earlier (26).
Inhibition assays
Reaction mixtures (10 μl) containing 10 mM Tris–HCl (pH 7.5), 100 mM KOAc, 10 mM MnCl2, 10 mM MgCl2, 1 mM DTT, 0.9 μM γ-phosphate-radiolabeled N7-methyl-capped RNA substrate and 2.5 μM purified D10 WT protein were incubated for 15 min at 25°C in the presence 100 μM of various GTP analogs. The reactions were quenched by adding 1 μl of 50 mM EDTA and applied to a polyethyleneimine–cellulose TLC plate, which was developed with 0.5 mM LiCl and 1 M methanoic (formic) acid. The average of at least two single independent experiments is presented. Inhibition assays were performed in the presence of increasing concentrations of nucleotide analogs, while competitive inhibition was assessed by performing decapping assays with methylated-capped RNA concentrations ranging from 0 to 1 μM in the presence of 0, 50, 100 or 200 μM of GTP. The Ki values were determined using the following equation:
where [S] represents the concentration of N7-methyl-cap RNA in the reaction mixture.
Circular dichroism spectroscopy measurements
Circular dichroism measurements were performed with a Jasco J-810 spectropolarimeter. The samples were analyzed in quartz cells with path lengths of 1 mm. Far-UV wavelength scans were recorded from 200 to 260. The average of three wavelength scans is presented. The ellipticity results were expressed as mean residue ellipticity, in millidegrees (mdeg).
RESULTS
Probing of the D10 enzyme active site with nucleotide analogs
With the intention of probing the active site of the vaccinia virus D10 decapping enzyme with nucleotide analogs, we first verified if the activity of a recombinant His-tagged version of the enzyme could be inhibited in vitro by GTP. All the decapping assays were performed at sub-optimal conditions in order to account for the possible increase in decapping activity that could potentially result from the addition of nucleotide or nucleotide analogs in solution. The addition of increasing GTP concentrations to the reaction resulted in a decrease in the generation of the m7GDP decapping reaction product (Figure 1A and B). This concentration-dependent inhibition was also observed in the presence of ATP, however to a lower extent (Figure 1B). Further characterization demonstrated that both the GTP and ATP inhibitions are competitive, suggesting that the purine base binds to the active site of the viral decapping enzyme. A lineweaver–burk representation of the competitive inhibition data obtained for GTP is presented in Figure 1C. Furthermore, derivation of binding data of GTP to the D10 protein with the Scatchard equation yielded a linear plot consistent with a single binding site for the GTP molecule on the enzyme (data not shown). It should be noted that no inhibition was observed in the presence of either CTP or UTP (data not shown).
Figure 1.
Competitive inhibition of the RNA decapping activity of D10 by GTP. (A) The D10 protein (2.5 μM) and N7-methyl-cap RNA substrate (0.9 μM) were incubated in 10 mM Tris–HCl (pH 7.5), 100 mM KOAc, 1 mM DTT, 10 mM MgCl2 and 10 mM MnCl2 (decapping buffer) for 15 min at 25°C with increasing concentrations of GTP (10, 20, 50, 100, 200, 500 and 1000 μM). The reactions were stopped by the addition of EDTA to 50 mM. The products were analyzed by TLC on a polyethyleneimine-cellulose plate and developed with 0.5 M LiCl and 1 M formic acid. An autoradiogram of the plate is shown. The positions of the unlabeled products, visualized under ultraviolet light, are indicated. (B) Dose–response inhibition of the decapping activity. The mean values of three replicates with standard errors are presented for the inhibition of the decapping reaction observed in the presence of both GTP (filled square) and ATP (unfilled square). (C) Competitive inhibition of the decapping reaction by GTP. Double-reciprocal plot of the inhibition of cap hydrolysis by D10 in the presence of 0 μM (filled square), 50 μM (filled triangle), 100 μM (unfilled circle), 200 μM (unfilled diamond) of GTP in solution. A close-up view of the data is shown in inset.
In order to gain insights on the molecular determinants involved in substrate binding, we then tested a collection of 26 analogs of ATP and GTP for their ability to inhibit the decapping reaction catalyzed by the D10 protein. The purine analogs selected display various modifications on both the ribose and the guanine or adenine base (Figure 2A). The modifications ranged from relatively minor (modification of a single functional group) to severe (modification of multiple functional groups). The informative finding is that a number of analogs were able to efficiently inhibit the reaction at a concentration of 100 μM (Figure 2B, Table 1). For instance, the presence of 100 μM 2-methyl-oxo-GTP inhibited the decapping activity of D10 by 75%, while the addition of xanthosine triphosphate (XTP) to the decapping reaction did not inhibit the activity of the enzyme (Figure 2B). All analogs tested in the current study were competitive inhibitors of the decapping reaction, indicating that they bind to the active site of the enzyme. A typical inhibition example using N1-methyl-ATP is presented in Figure 2C. Both the IC50 and Ki values were systematically evaluated for the various analogs that were able to reduce the decapping activity by at least 25% (Table 1). It should be noted that although UTP and CTP are not potent inhibitors of the decapping reaction, various CTP and UTP analogs (e.g. 5-methyl-UTP, 5-methyl-CTP, UDP, CDP) were also tested for their potential ability to inhibit the decapping reaction by the vaccinia virus D10. Our results demonstrate that these UTP and CTP analogs were unable to affect the activity of the enzyme (data not shown).
Figure 2.
Inhibition of the RNA decapping activity by nucleotide analogs. (A) Representation of the purine analogs tested. (B) Inhibition of the decapping reaction in the presence of 100 μM of selected nucleotide analogs (0.9 μM capped RNA, 1 μM D10, 50 mM Tris–HCl, 10 mM MgCl, 10 mM MnCl and 5 mM DTT). Inhibition parameters are compiled in Table 1. (C) Competitive inhibition with N1-methyl ATP. Double-reciprocal plot of the inhibition of cap hydrolysis by D10 in the presence of 0 μM (filled square), 50 μM (filled triangle), 100 μM (unfilled circle) of N1-methyl-ATP in solution. (D) Representation of the important functional features of the GTP molecule required for the inhibition of the decapping reaction.
Table 1.
Parameters for the inhibition of the decapping activity of D10 by nucleotide analogs
| Nucleotide analog | Relative decapping activity (%) (100 µM analog)a | IC50 (µM) | Ki (µM) |
|---|---|---|---|
| None | 100 | ||
| GTP | 35 ± 8 | 22 | 0.09 |
| Guanine | 92 ± 4 | >200 | >0.80 |
| GMP | 94 ± 4 | >200 | >0.80 |
| GDP | 69 ± 11 | 171 | 0.68 |
| N7-methyl-GDP | 24 ± 7 | 21 | 0.08 |
| N7-methyl-GTP | 3 ± 1 | 3 | 0.01 |
| N7-methyl-GpppG | 36 ± 5 | 69 | 0.28 |
| 6-thio-GTP | 83 ± 5 | >200 | >0.80 |
| 6-methyl-thio-GTP | 95 ± 2 | >200 | >0.80 |
| 6-methyl-oxo-GTP | 78 ± 5 | >200 | >0.80 |
| 8-bromo-GTP | 76 ± 5 | >200 | >0.80 |
| 8-iodo-GTP | 95 ± 2 | >200 | >0.80 |
| 8-oxo-GTP | 80 ± 2 | >200 | >0.80 |
| 2′-deoxy-GTP | 22 ± 7 | 27 | 0.11 |
| 2′-methyl-oxo-GTP | 25 ± 5 | 27 | 0.11 |
| 3′-methyl-oxo-GTP | 41 ± 12 | 65 | 0.26 |
| XTP | 97 ± 1 | >200 | >0.80 |
| Ribavirin triphosphate | 96 ± 3 | >200 | >0.80 |
| ATP | 65 ± 6 | 84 | 0.34 |
| ADP | 74 ± 1 | 188 | 0.75 |
| 2′-deoxy-ATP | 63 ± 5 | 129 | 0.51 |
| 2′-3′-dideoxy-ATP | 67 ± 8 | 166 | 0.66 |
| 2′-fluoro-ATP | 76 ± 5 | >200 | >0.80 |
| N6-methyl-ATP | 92 ± 4 | >200 | >0.80 |
| N1-methyl-ATP | 60 ± 10 | 175 | 0.7 |
| 6-methyl-thio-ITP | 95 ± 1 | >200 | >0.80 |
aThe decapping assays were performed a minimum of three times separately. Results are shown as the means of these independent experiments and normalized to the specific activity observed in the absence of nucleotide analog.
These data indicate that the active site of D10 is flexible and can accommodate a number of chemically modified purines. The analogs harboring chemical modifications on the ribose moiety and the purine rings (Figure 2A) helped to illuminate the flexibility of the D10 active site. As suggested by a previous study (7), the presence of a methyl group on the N7 position greatly increased the inhibitory potential of GTP and GDP (Table 1), highlighting the importance of this position for recognition by D10 (Figure 2D). In fact, the decapping activity was completely abolished in the presence of 100 μM m7GTP. Moreover, we initially suspected that the ability of the 6-oxo group of GTP (which is substituted by a 6-amino group in ATP) to act as a hydrogen-bond acceptor might be a key determinant promoting an increase in the inhibitory potential of GTP over ATP. The inhibitory properties of three GTP analogs harboring chemical modifications at this position provided clues on the importance of this functional group. Replacement of the 6-oxo group of GTP by SCH3 (6-methyl-thio-GTP) or OCH3 (6-methyl-oxo-GTP), that both maintain the capacity to act as hydrogen-bond acceptors, had a strong negative effect on the inhibition of the decapping activity in comparison to GTP, with IC50 and Ki values >200 and 0.80 µM, respectively (Table 1). These initial results suggest that the addition of the bulkier methyl group at position 6 weakens the ability of the analog to bind to the D10 protein. However, we observed that the substitution of the 6-oxo group of GTP by a thiol group (6-thio GTP), which lacks hydrogen bond acceptor properties, also severely decreased the inhibitory potential of the molecule (Table 1). We therefore conclude that the presence of a functional group which can display hydrogen-bond acceptor properties is important at the C6 position of the substrate (Figure 2D).
Another key difference between GTP and ATP is the nature of the functional group located at the C2 position of the purine rings. GTP displays a 2-amino group with hydrogen-bond donor properties while this functional group is absent from ATP. The importance of the hydrogen-bond at this position is elegantly highlighted by inhibition studies performed with XTP, which only differs with GTP by harboring a 2-oxo group at this position (instead of the 2-amino group). The replacement of the hydrogen-bond donor group by a hydrogen-bond acceptor group drastically reduced the inhibitory potential of this analog, demonstrating its importance for substrate recognition. These results clearly indicate the importance of a hydrogen-bond donor group at this position of the substrate. This finding is supported by inhibition studies performed with ribavrin triphosphate, a GTP analog that completely lacks the C2 and C3 positions of the purine rings. This analog had a weak ability to inhibit the decapping reaction with IC50 and Ki values >200 and 0.80 µM, respectively (Table 1).
Analogs harboring substituents at the C8 position of the guanine base (8-bromo-GTP, 8-iodo-GTP and 8-oxo-GTP) did not efficiently inhibit the decapping reaction, as reflected by their high IC50 and Ki values (Table 1). We postulate that the addition of these substituents likely results in steric hindrance with active site residues, thereby limiting the access of these analogs to the active site of the D10 protein. This suggests that the C8 position likely resides in close proximity to amino acis located in the D10 active site. In contrast, the addition of a methyl group to the N1 position of adenine had no significant effect on the inhibition of the decapping activity. The analog harboring such a modification (N1-methyl ATP) was not a very potent inhibitor of the enzyme (Table 1), indicating that the enzyme does not make critical interaction with the substrate through this position.
Although the current analysis largely focuses on the importance of specific molecular determinants of the purine rings required for substrate recognition, some of the analogs contained modifications on the ribose moiety of the nucleotide analogs. Surprisingly, all GTP and ATP analogs harboring various modifications/substitutions on the ribose moieties were potent inhibitors of the decapping reaction. As seen from the inhibition parameters, the removal of the 2′-OH group (2′-dGTP and 2′-dATP) had no significant effect on the inhibition of the decapping activity. Most strikingly, 2′,3′-dATP which lacks both ribose hydroxyls was still able to significantly inhibit the decapping reaction. Similarly, the addition of substituents on the ribose hydroxyls did not alter the ability of analogs to inhibit the decapping reaction. 2′-methyl-oxo-GTP and 3′-methyl-oxo-GTP inhibited the reaction with Ki values of 0.11 and 0.26 µM, respectively. We conclude that the binding of the substrate to the vaccinia virus D10 protein does not involve significant interactions with the ribose hydroxyls.
Moreover, we evaluated the ability of guanine and GMP to inhibit the decapping reaction. These two molecules did not affect the decapping reaction, as evidenced by their high IC50 and Ki values (Table 1). In addition, pyrophosphate and tripolyphosphate were also unable to inhibit the decapping activity of D10 (data not shown). From these data, we infer that a minimum of two phosphates linked to a purine base is required for interaction with the vaccinia virus D10 protein (Figure 2D).
Docking of m7GDP to SpDcp2
While no crystal structure is yet available for the vaccinia virus D10 protein, the crystal structure of the S. pombe Dcp2 (SpDcp2) decapping enzyme has been resolved (10,24). We previously utilized this latter structure to generate a model of the active site of D10 using the Deep View bioinformatics software (26,27). Since the predicted D10 catalytic center structure was shown to be similar to that of SpDcp2, we now utilized the SpDcp2 crystal structure for molecular docking to provide information on the interaction between the decapping enzyme and the m7GpppG moiety. Extensive computational docking and structure optimizations were used to generate a model of the m7GpppG-enzyme complex. More than 2 000 000 energy evaluations were performed in order to provide an accurate description of the enzyme–substrate interactions. The model underwent 150 rounds of steepest descent energy minimization and did not contain energetically unfavorable bonds, angles or torsions. The molecular docking model predicted that the most energetically favorable position for the m7GpppG molecule is located within the central cavity between the two main domains of the yeast enzyme (Figure 3A and B). The m7GpppG-SpDcp2 docking model highlighted amino acids that could be involved in the interaction of the cap structure in the catalytic pocket of SpDcp2 (Figure 3C). Four of the eight residues highlighted by molecular docking (Lys129, Glu147, Gln169, Tyr220) were previously mutated in SpDcp2 and shown to be important for the activity of the decapping enzyme (10,24).
Figure 3.
Molecular docking model for the binding of m7GpppG to a decapping enzyme. (A) Molecular docking model for the binding of m7GpppG to the S. pombe decapping enzyme (SpDcp2, amino acids 1–266). Ribbon diagram looking into the catalytic center of the enzyme with bound m7GpppG. (B) Side view of the enzyme following a 90° rotation to the right depicting the loop—α helix—loop NUDIX motif to the front. (C) The numerous amino acids interacting with the m7GpppG moiety are presented in a close-up view. The atoms of the cap analog are represented in spheres.
According to the similarity between the predicted D10 structure and the SpDcp2 tertiary structure, we hypothesized that corresponding amino acids could be found in D10. We further proceeded with alignment of the amino acid sequences of SpDcp2 and the D10 protein to search for analogous residues in the viral decapping enzyme (Figure 4A). Using the information from the SpDcp2 molecular docking and the sequence alignment, 17 amino acids of the viral decapping enzyme that could potentially be involved in the interaction with the cap structure were selected. In order to get further insights into the global architecture of the active site of D10 and the position of the 17 residues selected for mutational analysis, we generated a model for the tertiary structure of the complete viral enzyme (Figure 4B and C). Amino acids 1–226 of D10 were aligned with the corresponding amino acids of SpDcp2 and modeled onto the SpDcp2 crystal structure to generate the 3D D10 protein model. The D10 structure obtained is similar to the SpDcp2 structure, presenting a beta-sheet cap-binding cavity bordered by the NUDIX motif alpha-helix (Figure 4B and C). It should be noted that the 17 amino acids selected for mutational analysis are located in the potential active site of the D10 decapping enzyme, as well as in the two alpha-helical sequences closely surrounding the cavity (Figure 4B and C).
Figure 4.
D10 amino acids targeted for mutational analysis. (A) Alignment of amino acids 81–99, 120–150, 166–171 and 217–223 of the S. pombe decapping enzyme (SpDcp2) with the corresponding sequences of the vaccinia virus D10 protein. The NUDIX motif sequence is presented in bold. The positions of the SpDcp2 amino acids interacting with m7GpppG in the docking model are presented in red, while the black dots underlining the residues indicate residues that were previously shown to affect decapping activity. The amino acids in the vaccinia virus D10 protein selected for mutational analysis are indicated in blue, while amino acids previously mutated and shown to be important for D10 activity are shown in green. (B) Ribbon diagram of the predicted 3D structure of the vaccinia virus D10 protein (1–226). The alpha-helices are shown in red, while beta-sheets appear in yellow. The amino acids selected for mutational analysis are shown in blue, and the previously mutated amino acids in green. (C) Top view representation of the enzyme and amino acids following a 90° rotation towards the apex.
Mutational analysis of the D10 catalytic pocket
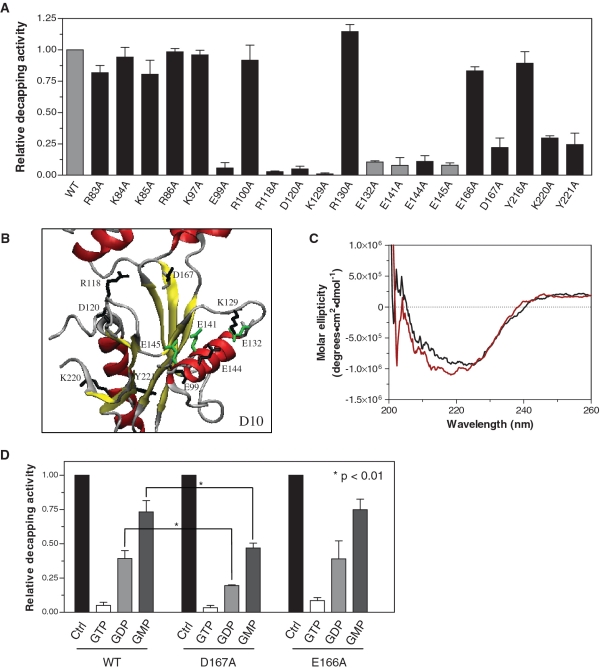
The importance of the D10 amino acids that potentially interact with the cap structure was investigated by generating a series of enzymatic mutants. Each of the 17 amino acids presented in Figure 4 was individually substituted by alanine, and the mutant polypeptides were expressed and purified in parallel with the wild-type D10 enzyme. Three other mutants (E132A, E141A, E145A) previously shown to drastically reduce the decapping activity were used as controls (Figures 4A and 5A) (7,26). The relative decapping activies of the mutants were tested on a N7-methyl-cap RNA substrate and compared with the activity of an equimolar concentration of the wild-type D10. Eight mutations (E99A, R118A, D120A, K129A, E144A, D167A, K220A, Y221A) resulted in a strong reduction of the decapping activity, thereby demonstrating the importance of these amino acids for the catalytic activity of D10 (Figure 5A). Specifically, the substitution of residues Glu99, Arg118, Asp120, Lys 129 and Glu144 by alanine resulted in a 90% decrease in the decapping activity. The substitution of the three other amino acids (Asp167, Lys220 and Tyr221) produced a 75% reduction of the decapping activity of D10.
Figure 5.
Decapping activity of D10 mutants. (A) The wild-type and D10 mutants (1 μM) were incubated with a N7-methyl-cap RNA substrate (0.9 μM) in decapping buffer for 15 min at 25°C. The decapping activity of each mutant is presented relative to the WT activity. The activity of the WT enzyme and E132A, E141A and E145A control mutations are presented in gray. (B) Ribbon diagram of the predicted 3D structure of D10 with the amino acids demonstrated to be important for the catalytic activity of the enzyme presented in black. Control mutations are shown in green. (C) Circular dichroism spectra were recorded for the D10 wild-type protein (black line) and E99A (red line). The average of three wavelength scans is presented. (D) Inhibition of the activity of the WT, D167A and E166A D10 in the presence of GTP, GDP and GMP.
Strikingly, five of the eight mutations significantly affecting the decapping activity (Arg118, Asp120, Lys129, Glu144, Asp167) are positioned in the D10 tertiary structure model with their side chains pointing towards the catalytic center of the enzyme (Figure 5B). This observation led to the premise that some of these amino acids could be involved in the interaction with specific components of the cap structure substrate. Unfortunately, the experiments to evaluate the respective roles of these amino acid residues are limited because of the lack of decapping activity of the mutant enzymes. Nevertheless, analysis of the position of the amino acids within the predicted tertiary structure of D10 and the decapping assay results allowed us to propose hypotheses for the role of some of the important amino acids of D10.
Glu144 is located within the alpha-helix of the NUDIX motif while Lys129 is pointing towards the latter helix (Figure 5B), suggesting that both these amino acids could be involved in direct contact with the methylated guanosine of the cap structure. In contrast, according to their position in the catalytic pocket, we theorize that amino acids Arg118 and Asp120 are likely to be involved in direct interactions with the first nucleotide of the RNA substrate (Figure 5B). Moreover, amino acid residues Lys220 and Tyr221 appeared to be part of the alpha-helix running down the back of the D10 tertiary structure model and could be required for interaction with the RNA body (Figure 5B). However, binding experiments of a non-capped RNA with the K220A and Y221A mutants did not show any reduced affinity for RNA compared to the wild-type enzyme (data not shown). Additionally, Glu99 is situated at the base of the active site cavity, pointing away from the substrate binding site (Figure 5B). It seemed reasonable to imagine that this residue may be important for the structural integrity of the protein or the positioning of the alpha-helix of the NUDIX motif. Circular dichroism experiments were performed to assess these possibilities. The typical alpha-helical (222 nm) and beta-sheet (208 nm) peaks of the far-UV spectra are significantly shifted in the case of the E99A mutation (Figure 5C), compared to the spectra obtained for the wild-type D10 protein as well as R100A, R130A and E166A mutants (data not shown). These results supported the idea that the E99A mutation alters the structure of the enzyme, which could result in the observed decrease in decapping activity of the mutant polypeptide. The last amino acid shown to be important for the catalytic activity of D10, Asp167, is positioned in the middle of the binding cavity in the 3D model and could potentially be involved in the interaction with the α- or β-phosphates of the cap structure (Figure 5B). To evaluate this latter hypothesis, we evaluated whether the activity of the D167A mutant enzyme could be inhibited more potently than the wild-type enzyme by nucleotide analogs with one, two or three phosphates. Interestingly, a 50% increased inhibition was produced by GDP and GMP on the decapping activity of the D167A mutant compared to the wild-type enzyme and the E166A mutant (Figure 5D). This increased inhibition could be explained by the presence of contacts between the Asp167 side chain and the phosphates of the cap in the wild-type enzyme that are abolished upon substitution of Asp167 for alanine in the D167A mutant. This loss of interaction between the phosphates of the cap structure and the enzyme would weaken the binding of the substrate to the catalytic site and allow an easier competition for GDP and GMP. These results support the suggestion that Asp167 is involved in the coordination of the phosphates of the RNA cap structure. In order to demonstrate that this effect is selective for Asp167, the E166A substitution mutant was utilized as a control for this experiment and the levels of inhibition observed in the presence of GTP, GDP and GMP were comparable to that of the wild-type D10 (Figure 5D). Overall, these results revealed a number of amino acids that are required for both substrate interaction and structural integrity of the vaccinia virus D10 decapping enzyme.
DISCUSSION
The RNA cap structure has been shown to be recognized by many enzymes via a stacking interaction between aromatic residues located within their respective active sites and the methylated guanosine of the cap structure. While this canonical cap-recognition mode is shared between the human eIF4E translation initiation factor, the mouse CBC and the vaccinia virus 2′-O-methyltransferase VP39, other cap-binding proteins, such as the NUDIX decapping enzymes, do not harbor any aromatic residues in the vicinity of their cap-binding site (10). This observation suggests that decapping enzymes employ a different mechanism for cap-recognition. The present study illustrates the non-canonical cap-recognition mode of the vaccinia virus D10 decapping enzyme using a combination of nucleotide analogs, molecular modeling, and site-directed mutagenesis.
The competitive inhibition of the decapping activity of the D10 protein by various purine analogs led to the identification of specific features that are important for the recognition of the cap structure by the viral enzyme. The requirement for specific chemical groups on positions 2 and 6 of the purine rings, as well as the importance of the triphosphate moiety were revealed. However, the ribose of the methylated guanosine does not seem to be critical for substrate recognition. Notably, cap-recognition by both eIF4E and VP39 also occurs without contacts with the ribose of the cap (33,34). The 6-oxo/6-amino group of purines has previously been demonstrated to be important for ATP/GTP recognition by other nucleotide interacting proteins. Specifically, ATP hydrolysis by the West Nile virus NS3 helicase is dependent on the presence of a hydrogen-bond donor in position 6, while incorporation of larger substituents at this position results in steric hindrance for the interaction between the yeast RNA 5′-triphosphatase and its substrate (34,35). Interestingly, we now demonstrate that a hydrogen-bond acceptor at position 6 is important for the interaction of GTP with D10. These results support previous evidences that proteins have evolved in a varieties of way to recognize the same or very similar substrates (33,36). Moreover, the ability of GTP to inhibit the decapping reaction was not affected by modifications at the 2′ and 3′ of the ribose, corresponding to the ribose of the methylated guanosine of the cap structure. The hydrogen-bond networks surrounding the cap structure in the catalytic pocket of the two latter enzymes reveals interactions with both the methylated-guanosine rings and the phosphate groups, but none with the ribose (33), suggesting that some similarities are still present between the non-canonical cap-recognition by D10 and the canonical cap-binding by eIF4E and VP39.
A previous study demonstrated that the activity of a D10 recombinant protein fused to the maltose-binding protein (MBP) was inhibited by methylated nucleotide derivatives (m7GpppG, m7GTP, m7GDP), but no significant inhibition could be observed with non-methylated derivatives (GTP, GDP) (7). In contrast, the recombinant his-tagged D10 protein used in the current present can clearly be inhibited by GTP and GDP. The discrepancy might arise from the presence of the MBP tag which may affect the flexibility of the active site of the MBP recombinant viral enzyme utilized in the previous study. Moreover, decapping experiments were performed here in multiple turnover conditions, while previous studies were done in single turnover, which might also account for the observed divergences.
By performing alanine mutations of 17 amino acids of the vaccinia virus D10 decapping enzyme, we identified eight novel amino acids that are important for the decapping activity (Glu99, Arg118, Asp120, Lys129, Glu144, Asp167, Lys220, Tyr221). The effects of mutations K129A, D167A and Y221A in D10 correlate with previous mutational analysis of corresponding amino acids in the yeast S. pombe Dcp2 enzyme. The crystal structure of SpDcp2 with ATP suggested the interaction of Lys129, Gln169 and Tyr220 with the bound substrate, and the individual mutation of these amino acids to alanine also resulted in a significant decrease in decapping activity for this fungal enzyme (24). We also observed that the D10 Arg118 and Asp120 residues were required for the decapping activity of the viral enzyme. However, corresponding amino acids are not found in the SpDcp2 enzyme. These results underline both similarities and discrepancies between the amino acids required for cap recognition in viral and yeast decapping enzymes.
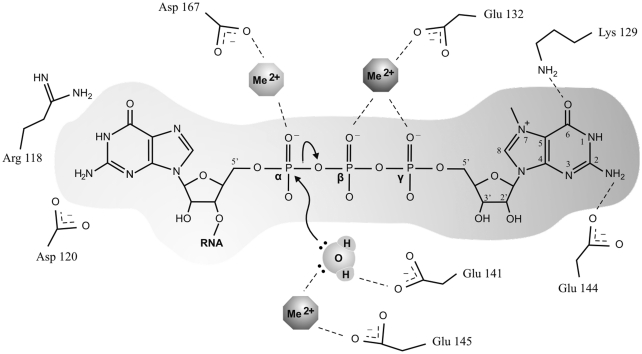
The results from the mutational analysis of the catalytic center of D10 as well as the data from the probing of the active site with nucleotide analogs provided us with a plethora of new informations on important features for cap-recognition by the viral decapping enzyme. In the absence of crystallographic data for D10, our generated structural model and the crystal structures of other decapping enzymes lead us to postulate the roles in cap-recognition for some of the residues observed to be critical for decapping. Previous crystallographic studies on NUDIX decapping enzymes have revealed that the bound cap structure stretches through the catalytic center, with the methylated guanosine positioned over the alpha-helix of the NUDIX motif (13,24). Accordingly, amino acids within the NUDIX motif of D10 are likely to be involved in catalysis by coordination of a water molecule or metal ions, as demonstrated by previous biochemical characterization of D10 (26), or in the interaction with the methylated guanosine of the cap structure. Moreover, the mutation of either Lys129 or Glu144, residues of the NUDIX motif, strongly affected the decapping activity of D10 and these amino acids could be involved in direct contacts with the cap structure according to their relative position in the structural model. Based on the probing results with nucleotide analogs and their position in the structural model, it seems reasonable that the amino group of Lys129 participates in hydrogen bonding with the 6-oxo group of the methylated guanosine of the cap structure, while Glu144 could be involved in a hydrogen-bond with the 2-amino position (Figure 6). Additionally, the crystal structure of the yeast Dcp1–Dcp2 complex resolved in the presence of ATP highlights contacts between the side chain of Gln169 and the α-phosphate of the ATP molecule. In the current work, the sequence alignment, tertiary structure modelization and catalytic activity assays suggest that Asp167 in D10 could play an homologous role to that of Gln169 in SpDcp2 (Figures 4A and 5A). Moreover, it was demonstrated that a D10 protein harboring the D167A mutation is 50% more readily inhibited by nucleotides with one or two phosphates than the wild-type enzyme (Figure 5D). According to these data, we postulate that Asp167 could be involved in interactions with the α-phosphate of the cap structure. However, since the Asp167 harbors a negatively charged side chain, a metal ion is likely to be making direct contacts with both the amino acids and the oxygen groups of the phosphate to allow proper coordination (Figure 6). Furthermore, while Arg118 and Asp120 present no evident counterparts in SpDcp2, their relative position in the structural model of D10 is consistent with a participation in direct interactions with the first base of the RNA. However, this study did not focus of the characterization of these interactions and the results could not specify the precise interplay between these amino acids and the capped RNA. Finally, to complete the suggested model, the results of our previous biochemical analysis of the catalytic mechanism of D10, with the amino acids involved in the coordination of two metal ions and a water molecule for the hydrolysis of the cap structure (Glu132, Glu141, Glu145) were also incorporated (26).
Figure 6.
Model for N7-methyl-cap RNA interaction with D10. (A) Mechanistic model for the cap-recognition and decapping mechanism of the D10 protein. During cap-recognition, Asp167 is engaged in the coordination of a divalent cation, Lys129 and Glu144 are involved in direct contacts with the methylated guanosine of the cap, while Arg118 and Asp120 interact with the first base of the RNA. In the chemical step, Glu132 and Glu145 also engage in the coordination of divalent cations, and Glu141 would be required for the polarization of the attacking water molecule involved in catalysis.
In Dcp2 enzymes, the RNA-binding channel is located on the dorsal surface of the NUDIX domain (24,37,38). Even though the D10 protein does not possess the conserved RNA-binding motif B found in Dcp2 enzymes (26), the decapping activity of the viral enzyme is still efficiently inhibited by non-capped RNA [data not shown, (7)] suggesting a strong contribution by the RNA body in the interaction. While mutations of amino acids Lys220 and Tyr221, located on the dorsal alpha-helix of the NUDIX domain in our 3D model of D10, resulted in a decrease in the decapping activity of D10, binding assays did not support their direct implication in the interaction with RNA. One could argue that this C-terminal segment of D10 may be incorrectly folded in our predicted structure (amino acids 1–226). Furthermore, amino acids Lys227, Lys229, Lys242 and Lys243 of D10 are not represented in the generated tertiary structure model and could be constituents of an RNA-binding channel for the D10 decapping enzyme. Future mutational analysis of these amino acids will address this hypothesis.
Although the current study largely focused on the important features for cap-recognition by the vaccinia virus D10 protein, inhibition of a viral decapping enzyme conserved throughout the poxvirus family, including variola virus, is also of medical relevance (25,39). The importance of D10 for vaccinia virus replication and virion infectivity suggests that targeting this enzyme with antiviral drugs could lead to the inhibition of viral replication (25,39). Dcp2 enzymes have been shown to be poorly inhibited by nucleotide or cap analogs (9,40), and this is also the case for the second vaccinia virus decapping enzyme, D9, and the newly discovered African swine fever virus g5R (11,12). Therefore, inhibitors designed specifically to prevent the non-canonical cap binding by D10 would have low probabilities of also inhibiting other decapping enzymes or cap-interacting proteins in a cellular context. Some of the analogs utilized in this work appear as interesting starting molecules for the design of specific inhibitors against the viral decapping enzyme. For example, 2′-methyl-oxo GTP potently inhibited the decapping reaction, while addition of a methyl group in N1 position of ATP did modify the inhibitory potency compared to ATP itself. These two modifications could be combined to specifically target D10 and reduce the off-target effect. The potential of guanosine analogs against viruses has already been demonstrated by the utilization of Ribavirin, Acyclovir and Entecavir respectively against hepatitis C, herpes virus and hepatitis B infections (41–44). The type of catalytic pocket probing and structural modelization as presented in this study opens the door to the potential development of other effective viral inhibitors.
FUNDING
Canadian Institutes of Health Research (CIHR). CIHR and by the Université de Sherbrooke (to the RNA group); Canada Research Chair in Genomics and Catalytic RNA (to J.P.P.); Doctoral training award from the Fonds de Recherche en Santé du Québec (to M.F.S.). Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Guy Lemay (Université de Montréal) for the generous gift of the vaccinia virus DNA. M.B. is a Chercheur Boursier Junior 2 from the Fonds de Recherche en Santé du Québec. Both M.B. and J.P.P. are members of the Infectious diseases group of the Centre de Recherche Clinique Étienne-Lebel.
REFERENCES
- 1.Jackson DA, Pombo A, Iborra F. The balance sheet for transcription: an analysis of nuclear RNA metabolism in mammalian cells. FASEB J. 2000;14:242–254. [PubMed] [Google Scholar]
- 2.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Wickens M, Goldstrohm A. A place to die, a place to sleep. Science. 2003;300:753–755. doi: 10.1126/science.1084512. [DOI] [PubMed] [Google Scholar]
- 4.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell. Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc. Natl Acad. Sci. USA. 2007;104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunawardana D, Cheng HC, Gayler KR. Identification of functional domains in Arabidopsis thaliana mRNA decapping enzyme (AtDcp2) Nucleic Acids Res. 2008;36:203–216. doi: 10.1093/nar/gkm1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She M, Decker CJ, Chen N, Tumati S, Parker R, Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrish S, Moss B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J. Virol. 2007;81:12973–12978. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parrish S, Hurchalla M, Liu SW, Moss B. The African swine fever virus g5R protein possesses mRNA decapping activity. Virology. 2009;393:177–182. doi: 10.1016/j.virol.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarsdale JN, Peculis BA, Wright HT. Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes. Structure. 2006;14:331–343. doi: 10.1016/j.str.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 15.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 21.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, et al. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 2003;328:365–383. doi: 10.1016/s0022-2836(03)00314-0. [DOI] [PubMed] [Google Scholar]
- 22.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 23.Mazza C, Segref A, Mattaj IW, Cusack S. Co-crystallization of the human nuclear cap-binding complex with a m7GpppG cap analogue using protein engineering. Acta Crystallogr. D Biol. Crystallogr. 2002;58:2194–2197. doi: 10.1107/s0907444902015445. [DOI] [PubMed] [Google Scholar]
- 24.She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish S, Moss B. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J. Virol. 2006;80:553–561. doi: 10.1128/JVI.80.2.553-561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Souliere MF, Perreault JP, Bisaillon M. Characterization of the vaccinia virus D10 decapping enzyme provides evidence for a two-metal-ion mechanism. Biochem. J. 2009;420:27–35. doi: 10.1042/BJ20082296. [DOI] [PubMed] [Google Scholar]
- 27.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Mayo SL, Olafson BD, Goddard WA. DREIDING: a generic force field for molecular simulations. J. Phys. Chem. 1990;94:8897–8909. [Google Scholar]
- 30.Stewart JJ. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1661. [Google Scholar]
- 32.Solis FJ, Wets RJB. Minimization by random search techniques. Math. Oper. Res. 1981;6:19–30. [Google Scholar]
- 33.Quiocho FA, Hu G, Gershon PD. Structural basis of mRNA cap recognition by proteins. Curr. Opin. Struct. Biol. 2000;10:78–86. doi: 10.1016/s0959-440x(99)00053-6. [DOI] [PubMed] [Google Scholar]
- 34.Despins S, Issur M, Bougie I, Bisaillon M. Deciphering the molecular basis for nucleotide selection by the West Nile virus RNA helicase. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq276. [Epub ahead of print 25 April 2010] 10.1093/nar/gkq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Issur M, Despins S, Bougie I, Bisaillon M. Nucleotide analogs and molecular modeling studies reveal key interactions involved in substrate recognition by the yeast RNA triphosphatase. Nucleic Acids Res. 2009;37:3714–3722. doi: 10.1093/nar/gkp227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nobeli I, Laskowski RA, Valdar WS, Thornton JM. On the molecular discrimination between adenine and guanine by proteins. Nucleic Acids Res. 2001;29:4294–4309. doi: 10.1093/nar/29.21.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 38.Souliere MF, Bisaillon M. Structures and mechanisms of Dcp2-like decapping enzymes. Curr. Top. Biochem. Res. 2008;10:55–62. [Google Scholar]
- 39.Shors T, Keck JG, Moss B. Down regulation of gene expression by the vaccinia virus D10 protein. J. Virol. 1999;73:791–796. doi: 10.1128/jvi.73.1.791-796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen LS, Mikhli C, Jiao X, Kiledjian M, Kunkel G, Davis RE. Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent. Mol. Cell. Biol. 2005;25:8779–8791. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Bisceglie AM, Conjeevaram HS, Fried MW, Sallie R, Park Y, Yurdaydin C, Swain M, Kleiner DE, Mahaney K, Hoofnagle JH. Ribavirin as therapy for chronic hepatitis C. A randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 1995;123:897–903. doi: 10.7326/0003-4819-123-12-199512150-00001. [DOI] [PubMed] [Google Scholar]
- 42.Leyssen P, Balzarini J, De Clercq E, Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 2005;79:1943–1947. doi: 10.1128/JVI.79.3.1943-1947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reardon JE, Spector T. Herpes simplex virus type 1 DNA polymerase. Mechanism of inhibition by acyclovir triphosphate. J. Biol. Chem. 1989;264:7405–7411. [PubMed] [Google Scholar]
- 44.Kamiya N. The mechanisms of action of antivirals against hepatitis B virus infection. J. Antimicrob. Chemother. 2003;51:1085–1089. doi: 10.1093/jac/dkg236. [DOI] [PubMed] [Google Scholar]