Abstract
Double-stranded RNAs that are complementary to non-coding transcripts at gene promoters can activate or inhibit gene expression in mammalian cells. Understanding the mechanism for modulating gene expression by promoter-targeted antigene RNAs (agRNAs) will require identification of the proteins involved in recognition. Previous reports have implicated argonaute (AGO) proteins, but identifications have differed with involvement of AGO1, AGO2, or both AGO1 and AGO2 being reported by different studies. The roles of AGO3 and AGO4 have not been investigated. Here, we examine the role of AGO 1–4 in gene silencing and activation of the progesterone receptor (PR) gene. Expression of AGO2 is necessary for efficient gene silencing or activation and AGO2 is recruited to the non-coding transcript that overlaps the promoter during both gene silencing and activation. Expression of AGO1, AGO3 and AGO4 are not necessary for gene silencing or activation nor are AGO1, AGO3, or AGO4 recruited to the target non-coding transcript during gene activation. These data indicate that AGO2 is the primary AGO variant involved in modulating expression of PR by agRNAs.
INTRODUCTION
RNA interference (RNAi) involves silencing gene expression through recognition of mRNA by small duplex RNAs (1). Some recent reports have suggested that RNAs complementary to gene promoters can inhibit (2–9) or activate (10–14) gene expression in mammalian cells. In contrast to duplex RNAs that recognize mRNA and act post-transcriptionally, RNAs that target gene promoters modulate gene transcription. We describe RNAs that target gene promoters as antigene RNAs (agRNAs) to distinguish them from traditional siRNAs that target and cleave mRNA.
There is no evidence that promoter-targeted RNAs directly interact with chromosomal DNA. Instead, they have been reported to bind to non-coding RNA transcripts that overlap gene promoters (8,9,14–17). Three studies have proposed that small duplex RNAs associate with non-coding RNAs that are transcribed in the sense orientation (i.e. the same direction as mRNA) (8,9,15). Our laboratory identified an antisense transcript as the molecular target for agRNAs that modulate expression of the PR gene (16). This PR antisense transcript initiates within the coding region of the gene and spans ∼70 000 bases upstream from the transcription start site.
Our approach for further understanding how agRNAs bind to non-coding transcripts and alter transcription from gene promoters involves examining the potential role of RNA-binding proteins that facilitate RNA/RNA interactions. We reasoned that studying the function of the argonaute (AGO) family of proteins provided a logical starting point since members of this family are critical components in the RNAi pathway.
There are four AGO proteins (AGO1–4) in humans. AGO2 is the ‘catalytic engine’ of RNAi, responsible for recognition of mRNA and subsequent cleavage of the transcript (18–21). AGO2 has also been suggested to be involved in miRNA biogenesis (22). Using a minimal in vitro system AGO1 and AGO2 have been shown to possess the ability to dissociate miRNA duplexes, while AGO3 and AGO4 do not (23). In another report, reintroduction of any AGO variant into embryonic stem (ES) cells deficient for expression of all four AGO variants rescues miRNA silencing defects and reduces apoptosis, suggesting that AGO3 and AGO4 can assist RNAi (24). Functional redundancy of AGO has also been inferred from mRNA or miRNA pull-down experiments showing detection of similar bound transcripts regardless of which AGO variant is being isolated (20,25). Finally, all four human AGO proteins exhibit similar preferences for binding to duplex RNA with mismatches at different positions, although only AGO2 efficiently unwound fully complementary duplexes (26). Taken together, these data demonstrate a role for AGO2 in these RNA-mediated processes, but also suggest that AGO1, AGO3 and AGO4 proteins may be involved in these mechanisms.
For AGO proteins to alter promoter activity, they must be located within the cell nucleus. Although AGO proteins primarily reside in the cytoplasm, studies have indicated that they are also found in the nucleus (27–31). In Caenorhabditis elegans an AGO protein NRDE-3 was found to be required for nuclear siRNA import (27). In mammalian cells, nuclear activity of AGO was first inferred from the observation of potent gene silencing of small nuclear RNA 7SK (28). A highly specific anti-AGO2 antibody was subsequently used to identify AGO2 in nuclear lysate (29) and fluorescence correlation and cross-correlation spectroscopy also revealed nuclear AGO2 (30). Most recently, importin-8 has been reported to be involved in the translocation of AGO2 from cytoplasm to nucleus (31).
There have been multiple reports on the role of AGO proteins in the mechanism of promoter-targeted RNAs. One laboratory has implicated AGO2 in RNA-mediated gene activation (10). Our laboratory reported that either AGO1 or AGO2 might be necessary for gene silencing (32), while other reports determined AGO1, along with other non-AGO proteins, as critical using multiple experimental approaches (9,17,33–36). However, few reports investigated a role for AGO3 or AGO4 in either gene silencing or activation (10). Here we investigate involvement of human AGO1–4 in agRNA-mediated gene silencing or activation of PR expression. Using multiple experimental strategies we find that AGO2 protein is the best candidate for mediating both gene silencing and activation (Table 1).
Table 1.
Summary of data on participation of AGO variants in modulation of PR gene expression by agRNAs
| Located in nucleus? | Silencing reverses agRNA-mediated silencing | Silencing reverses agRNA-mediated activation | RIP shows binding to antisense transcript during gene silencing | RIP shows binding to antisense transcript during gene activation | HA-Tag RIP shows binding to antisense transcript during gene activation | |
|---|---|---|---|---|---|---|
| AGO1 | Yes | No | No | Yes | No | No |
| AGO2 | Yes | Yes | Yes | Yes | Yes | Yes |
| AGO3 | Yes | No | No | No | No | No |
| AGO4 | Yes | No | No | No | No | No |
MATERIALS AND METHODS
Double-stranded RNAs
RNAs were synthesized by Integrated DNA Technologies. The siRNAs for AGO1, AGO2, AGO3 and AGO4 were provided by Dharmacon, Inc. The sequences are listed in Supplementary Table S1. Multiple siRNAs were tested for their ability to specifically decrease AGO gene expression. We were able to identify a single efficient siRNA duplex for AGO1, AGO3 and AGO4. For AGO2 the Smartpool siRNAs gave us the best knockdown efficiency. Three negative control RNAs were used: MM (a mismatched RNA duplex based on PR-9 sequence), SCR1 (a scrambled RNA duplex based on PR-9 sequence) and siGL2 (a luciferase based siRNA).
Plasmids and antibodies
The pIRESneo FLAG/HA AGO plasmids was purified according to manufacturer’s instructions (Addgene Plasmid 10820, 10822, 10823, 10824) (20). An empty vector plasmid pUC was used as a control plasmid. The following antibodies were used in this study: monoclonal anti-HA antibody 16B12 (MMS-101P, Covance, for IP and western blotting (WB); monoclonal anti-PR antibody 6A1 (3172, Cell Signaling, for WB); monoclonal anti-AGO2 4G8 (011-22033, Wako, for IP and WB); monoclonal anti-AGO1 (4B8, for WB and IP), anti-AGO2 (11A9, for IP), anti-AGO3 (5A3, for IP) and anti-AGO4 (6C10, for IP) (gifts from Dr G. Meister) (31); monoclonal anti-AGO3 (3H8-A5 or 4B1-F6, gift from Dr M.C. Soimi, for WB) (37), monoclonal anti-β-tubulin (T5201, Sigma, for WB), monoclonal anti-GAPDH (ab9484, Abcam, for WB), monoclonal anti-Dicer [ab14601, chromatin immunoprecipitation (ChIP) grade, for IP], monoclonal anti-lamin A/C (ab8984, Abcam, for WB), rabbit polyclonal anti-TRBP (HIV TAR-binding protein) (gift from Dr Q. Liu, for IP) (38), normal rabbit IgG (2729, Cell Signaling, for IP), normal mouse IgG (12-371, Millipore, for IP), monoclonal anti-RNA polymerase II (05-623, Millipore, for IP) and anti-β-actin (Sigma).
Cell culture and transfection
T47D and MCF7 cells (American Type Culture Collection) were maintained in RPMI-1640 media supplemented with 10% (v/v) FBS, 0.5% (w/v) non-essential amino acids, 0.4 units/ml bovine insulin (all reagents from Sigma). Cells were cultured at 37°C and 5% (v/v) CO2. Cells were plated in six-well plates (Costar) 2 days before transfection without antibiotics so that the cells are 30–50% confluent by the time of transfection.
Oligofectamine RNAiMAX (Invitrogen) was used as lipid for all RNA duplex transfection. For single transient transfection experiments (unless otherwise noted) 25 nM duplex RNA (1.2 μl lipid) in OPTI-MEM (Invitrogen) was used in a final volume of 250 μl. Media was added to the duplex-lipid mixture for a final volume of 1.25 ml, then added on cells. Media was changed 24 h later, and cells were harvested after 72 h of transfection for mRNA analysis and 96 h for protein analysis. For double transient transfection experiments, the first transfection was performed as described earlier. The second transfection utilizing a reverse transfecting method was carried out 72 h after the first one. The cells were first detached using trypsin and then transferred to another plate, to which appropriate RNA duplex/lipid/OPTI-MEM mixture were added (final concentration 25 nM for each RNA duplex). Media was changed 2 days later and the cells were harvested 72 h after the second transfection for mRNA analysis and 96 h for protein analysis.
The plasmid transient transfection in MCF7 and T47D cells was carried out using Oligofectamine 2000 (Invitrogen) as the lipid according to the manufacturer’s instruction in a six-well plate fashion (Costar). The transfected quantities of each AGO plasmid in Supplementary Figure S13 were 0.5 µg for AGO1, AGO2 and AGO3 plasmids, and 1.0 µg for AGO4 plasmid. The plasmid/lipid ratio was kept at 1:2 (µg/µl). For anti-HA RIP experiments in Figure 7, a double transfection assay was carried out on 150 cm2 large dishes. During the first transfection, 4 µg of AGO1 or AGO3 plasmids, or 12 µg of AGO2 plasmid, or 8 µg of AGO4 plasmids were added on 90% confluent MCF7 cells. After 2 days the transfected cells were split (1:3) and PR-11 (25 nM final) were added at the same time. The cells were harvested 72 h later (after the second transfection) for mRNA analysis or RNA immunoprecipitation assay, and 96 h for protein analysis.
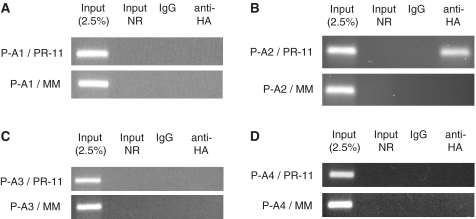
Figure 7.
RIP examining the association of Flag/HA tagged AGO proteins with the PR antisense transcript. MCF7 cells were transfected with Flag/HA-Ago plasmid first and then PR-11 in 48 h (final 25 nM). (A) Flag/HA-AGO1, (B) Flag/HA-AGO2, (C) Flag/HA-AGO3 and (D) Flag/HA-AGO4, were examined. The transfection protocol is described in the ‘Materials and Methods’ section. P-A1: Flag/HA-AGO1 plasmid, etc. MM: negative control RNA duplex. IgG: negative control antibody. Input: nuclear extract prior to treatment with antibody. Input NR: input sample with no reverse transcriptase treated. The above data is the representative data set from three independent experiments.
Western blotting
Western blots were performed on protein lysates (30 or 40 µg per sample). Primary antibodies included monoclonal anti-PR, anti-HA, anti-AGO1 and anti-AGO2, anti-AGO3 and anti-AGO4. Anti-β-actin was used as an internal control and for quantitation. Protein was visualized using secondary anti-mouse, anti-rabbit or anti-rat (Jackson Immunolabs) and Supersignal developing solution (Pierce).
RNA analysis
Expression of AGO1–4, PR and antisense PR transcript was evaluated by real-time quantitative PCR. Total RNA from treated T47D or MCF7 cells was extracted using Trizol (Invitrogen). Samples of 2 μg were treated with DNase 1 (6355, Worthington) first, followed by reverse transcription using random primers (Applied Biosystems) with the High Capacity cDNA Archive kit (Applied Biosystems). Products were detected using TaqMan Gene Expression Assays (EIF2C1, EIF2C2, EIF2C3, EIF2C4 and PGR, Applied Biosystems) with 50 ng of complementary DNA. Data were normalized relative to measured levels of GAPDH (Applied Biosystems). Error is expressed as standard deviation from the mean (SD).
RNA immunoprecipitation and ChIP
ChIP assays were performed essentially as described (16). The following antibodies were used for immunoprecipitations: normal mouse IgG and monoclonal anti-RNA polymerase II. Four micrograms of each of the appropriate antibody were used for each ChIP. Primers used for ChIP are described in Supplementary Table S2.
For RNA immunoprecipitation (RIP), MCF7 or T47D cells were grown in 150 cm2 dishes and transfected with either duplex RNAs or plasmids. Cells (∼40 × 106) were harvested in 72 h and nuclear fraction was isolated (28). A nuclear lysis buffer [150 mM KCl, 20 mM Tris–HCl 7.4, 3 mM MgCl2, 0.5% NP-40, 1 × Roche protease inhibitors cocktail, RNAse•in (50 U/ml final)] was added to the nuclei (Note: no formaldehyde cross-linking is used in this protocol). The mixture was left on ice for 10 min. After vigorous vortexing and pipetting, nuclei were freeze-thawed three times in liquid nitrogen and a 22°C water bath. Insoluble material was removed by centrifugation at maximum speed for 15 min at 4°C. Nuclear extracts were quickly frozen in liquid nitrogen and stored at −80°C. An amount of 60 µl Protein A/G agarose Plus was washed with phosphate-buffered saline (1 × PBS, pH 7.4) and incubated with 0.5 ml anti-AGO1–4B8, anti-AGO2-11A9, anti-AGO3-5A3 or anti-AGO4-6C10 at 4°C with gentle agitation overnight. After washes with 1 × PBS twice, beads were incubated with the above nuclear cell lysate for 3 h at 4°C. For purified antibodies, the beads, antibody and nuclear cell lysate were mixed and under constant rotation for a total of 3 h. The beads were extensively washed with nuclear lysis buffer once, IP wash buffer twice [300 mM NaCl, 3 mM MgCl2, 0.1% NP-40 and 20 mM Tris–HCl (pH 7.4) and finally 1 × PBS once (31)]. The beads were then eluted with elution buffer (1% SDS, 0.1 M NaHCO3 and RNase inhibitor). Following proteinase K treatment, RNA extraction and precipitation, samples were treated with recombinant DNase I, followed by reverse transcription. Corresponding cDNA was amplified using primers complementary to antisense PR transcripts (Supplementary Table S2).
RESULTS AND DISCUSSION
Experimental design
We chose to investigate the role of AGO proteins in agRNA-mediated modulation of progesterone receptor (PR) gene expression. PR has two major isoforms, PR-B and PR-A. The isoforms are expressed from different promoters, with the PR-B transcription start site upstream from the start site of PR-A within the gene (Figure 1A). Expression of PR-B and PR-A is linked and we have found that levels of the two isoforms vary proportionally regardless of whether silencing is performed using agRNAs, siRNAs complementary to PR mRNA or single-stranded locked nucleic acids (LNAs) or peptide nucleic acids (PNAs) (39–42).
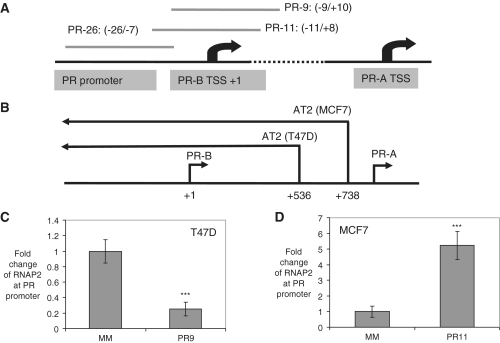
Figure 1.
Scheme showing PR gene promoter structure and the effect of promoter targeted agRNAs on the recruitment of RNAP2 to the transcription start site. (A) The regions targeted by representative agRNA duplexes. Relative to PR-B transcription starting site (TSS), PR-26, PR-11 and PR-9 target −26 to −7, −11 to +8 and −9 to +10 regions, respectively. (B) The non-coding 5′-antisense transcript AT2 starts within the coding region of PR in both T47D and MCF7 cells. ChIP assay evaluates the recruitment of RNAP2 in (C) T47D cells treated by agRNA PR-9; (D) MCF7 cells treated by agRNA PR-11. MM: negative control RNA duplex. Mouse IgG was used as a negative control antibody. The experiments were repeated at least three times. ***P < 0.005 as compared to cells treated with a mismatched RNA (MM). P-values were calculated using the two tailed unpaired Student’s t-test with equal variances. All error bars represent standard deviation.
PR is a productive model for studying agRNA-mediated regulation of cellular gene expression for several reasons: (i) The PR transcription start site has been well defined (43,44); (ii) cell lines are available that express PR at different levels; (iii) we have previously characterized the involvement of antisense transcripts in the mechanism of agRNAs at the PR locus (16); and (iv) PR provides a model for both agRNA-mediated activation and inhibition. agRNAs inhibit gene expression in T47D cells (where PR expression is high) and activate gene expression in MCF7 cells (where PR expression is low).
The target antisense transcript originates downstream from the PR transcription start site (at +536 in T47D cells and +738 in MCF7 cells), and extends 70 000 bases upstream beyond the PR promoter (Figure 1B). Evidence of involvement of the antisense transcript includes: (i) detection of the antisense transcript by 5′- and 3′-RACE and failure to detect a sense transcript overlapping the promoter; (ii) when biotin labeled agRNA duplexes are added binding is observed with biotin-labeled strands complementary to the antisense (but not the sense) transcript; and (iii) reduced expression of the antisense transcript upon addition of a complementary oligonucleotide reverses agRNA-mediated gene modulation (16). We had also used RNA immunoprecipitation (RIP) to show that AGO protein was recruited to the antisense transcript, although these experiments used a non-selective anti-AGO antibody and did not distinguish between AGO 1–4 (16).
For gene silencing we used PR-9, a duplex RNA that targets the −9 to +10 region surrounding the PR-B transcription start site. For gene activation we used PR-11, a duplex that targets the region −11 to +8, relative to the +1 start site. Using ChIP we observed that introduction of PR-9 into cells leads to decreased recruitment of RNA polymerase 2 (RNAP2) at the PR promoter, while addition of PR-11 leads to increased RNAP2 (Figure 1C and D) (16). Unless otherwise noted all experiments in this study were performed in duplicate or triplicate independent determinations and results are reported as averages (for qPCR) or by showing representative primary data (for western analysis).
Inhibition of AGO expression
All four AGO proteins are expressed in both T47D and MCF7 cells, although AGO1 and AGO2 are expressed at higher levels than AGO3 or AGO4 (Figure 2A and B). We tested at least five duplex siRNAs designed to suppress each AGO expression, including several used previously in the literature (20) and identified siRNAs that potently and specifically inhibited expression of each AGO variant in T47D (Figure 2C–F) or MCF7 cells (Supplementary Figure S1, S2 and S3). We chose to perform transfections using 25 nM anti-AGO duplex RNA because, while silencing of AGO was not optimally potent, that concentration had minimal effects on cell growth rates.
Figure 2.
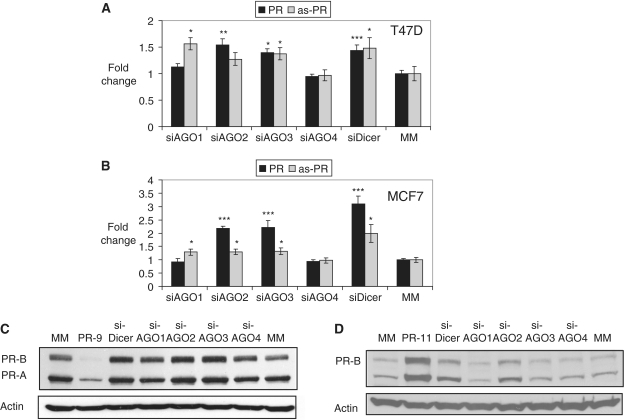
AGO expression and silencing. qPCR analysis of relative expression levels of AGO1–4 in human breast cancer cell lines, (A) T47D and (B) MCF7. Effect of silencing (C) AGO1, (D) AGO2, (E) AGO3 and (F) AGO4 on expression of other AGO variants (using 25 nM siRNA). All data were normalized to GAPDH. Error bars represent standard deviations, calculated from four independent experiments. ***P < 0.005, **P < 0.01, *P < 0.05 as compared to cells treated with a mismatched RNA (MM). P-values were calculated using the two tailed unpaired Student’s t-test with equal variances.
Silencing AGO proteins can have unexpected consequences because RNAi pathways affect the expression of many genes inside cells. Observed effects might be direct, indirect, or a mixture of both. We examined whether silencing AGO protein expression would affect PR expression or alter levels of the PR antisense transcript that acts as a direct target for agRNAs. In T47D cells, inhibition of AGO1–4 expression had relatively modest effects on levels of PR mRNA (<1.5-fold), the PR antisense transcript, (<1.5-fold) (Figure 3A) or PR protein (<1.5-fold) (Figure 3C; Supplementary Figure S4A). Inhibition of dicer, an enzyme involved in processing miRNAs also yielded only modest changes for PR expression in T47D cells (Figure 3A, C and Supplementary Figure S4B and S5). Given that AGO and dicer proteins affect the expression of many cellular proteins, the small changes in PR expression are not surprising.
Figure 3.
Effect of inhibiting AGO1–4 or dicer on expression of PR mRNA, PR antisense transcript (as-PR), and PR protein. (A) qPCR analysis of PR and as-PR after silencing expression of AGO1–4 or dicer by siRNA in T47D cells. (B) qRT-PCR analysis of PR and as-PR after a repression of AGO1–4 or dicer by siRNA in MCF7 cells. (C) Western analysis of PR protein after inhibiting expression of AGO1–4 or dicer in T47D cells. (D) Western analysis of PR protein following inhibition of AGO1–4 or dicer in MCF7 cells. PR-9 and PR-11: positive controls. MM: negative control. The final concentration for all used RNAs was 25 nM. qPCR data was normalized relative to GAPDH. Error bars represent standard deviations, calculated from four independent experiments. ***P < 0.005, **P < 0.01, *P < 0.05 as compared to cells treated with a mismatched RNA (MM). P-values were calculated using the two tailed unpaired Student’s t-test with equal variances.
In MCF7 cells inhibition of AGO2 or AGO3 expression caused a 2-fold increase in PR mRNA expression (Figure 3B) and, for inhibition of AGO2, similar increases in levels of PR protein (Figure 3D; Supplementary Figure S4). Inhibition of dicer expression also led to increases in PR mRNA and protein expression (Figure 3B and D; Supplementary Figure S4). Inhibition of AGO1 or AGO4 expression did not affect PR expression. The activation of PR expression upon silencing AGO2, AGO3, or dicer indicates that PR expression may be affected by endogenous AGO-mediated pathways. The mechanism of this modulation may be through direct miRNA interactions at the PR locus or may be indirect involving other genes.
Effect of AGO expression on agRNA-mediated silencing
To test whether expression of AGO proteins was necessary for potent silencing by agRNAs we used a two step transfection method. In the first transfection (transfection on Day 0), expression of AGO1–4 was reduced using siRNAs (Figure 2; Supplementary Figure S6). In the second transfection silencing agRNA, PR-9 was added (Day 3). The effect of reduced AGO expression on the activity of PR-9 was evaluated by quantitative PCR (Day 6) (Figure 4A–D) or western analysis (Day 7) (Figure 4E–H).
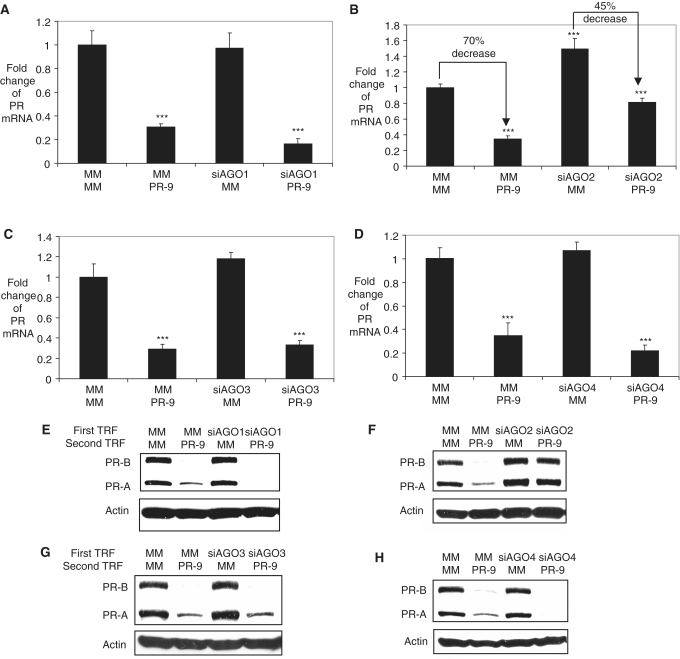
Figure 4.
Effect of reduced expression of AGO1, AGO2, AGO3, or AGO4 (siRNAs, 25 nM) on silencing by PR-9 (25 nM) in T47D cells. qPCR analysis of PR after a double transfection assay with (A) siAGO1, (B) siAGO2, (C) siAGO3, (D) siAGO4 being added first and PR9 added second (in 72 h). Western analysis of PR protein levels after a double transfection assay with (E) siAGO1, (F) siAGO2, (G) siAGO3, (H) siAGO4 being added first and PR-9 added second (in 72 h). Transfection combinations: MM/PR-9 represents that the first transfection is MM, a control RNA duplex and the second is PR-9, etc. All data were normalized to GAPDH. Error bars represent standard deviations, calculated from three independent experiments. ***P < 0.005 as compared to cells treated with a mismatched RNA (MM). P-values were calculated using the two tailed unpaired Student’s t-test with equal variances.
Inhibition of AGO2 expression partially reversed gene silencing by agRNA PR-9. In the presence of wild-type levels of AGO2, addition of PR-9 led to a 75% decrease of PR mRNA expression, while under conditions of reduced AGO2 expression PR-9 was a less efficient silencing agent, reducing expression by only 45% (Figure 4B). We also observed a similar reversal of agRNA-mediated inhibition of PR protein expression (Figure 4F). Inhibition of AGO1, AGO3, or AGO4 expression had little effect on gene silencing by agRNA PR-9 as measured by RNA levels (Figure 4A, C and D) or protein levels (Figure 4E, G and H). These data suggest that AGO2 is the best candidate for involvement in agRNA-meditated silencing of PR gene expression. A similar reversal of gene silencing was observed when knockdown of AGO2 was followed by addition of PR-26 (an agRNA targeting −26 to −7) that was also known to silence gene expression (Supplementary Figures S7 and S8) (6).
We had previously reported that both AGO1 and AGO2 proteins were necessary for agRNA-mediated silencing of PR (32). The siRNA used in the previous study for inhibiting expression of AGO1 was at least as potent as the siRNA used in this study. However, while comparing various anti-AGO siRNAs we found that the previously-used anti-AGO siRNA caused a 4-fold increase in expression of AGO4, suggesting that the previous siRNA was prone to off-target effects and was likely to be less reliable. It is also possible that the previously used siRNA and transfection conditions yielded more potent silencing of AGO proteins. Such potent silencing may have revealed a role of AGO1, but the potential for greater off-target effects render this conclusion uncertain and we have adopted the more conservative interpretation of our data.
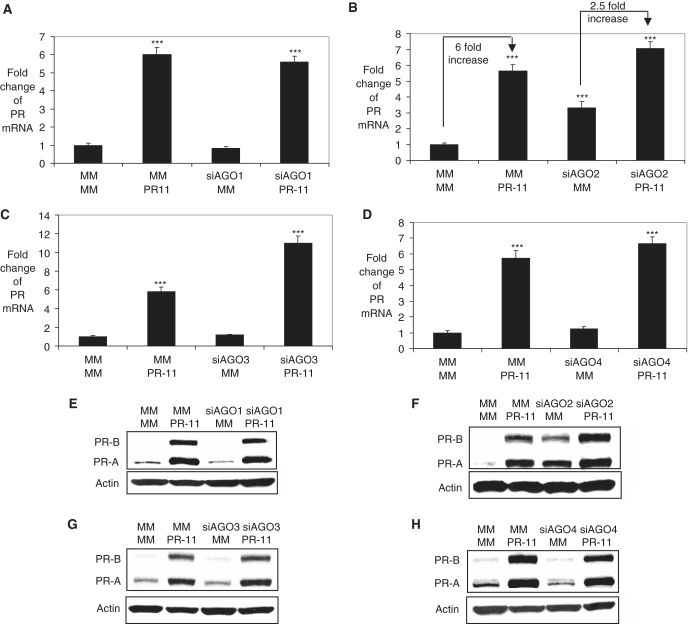
Effect of AGO expression on agRNA-mediated gene activation
We used the double transfection strategy to study the effect of inhibiting expression of AGO1–4 on activation of PR expression by agRNA PR-11 (Figure 5A–H; Supplementary Figure S9). Studying the role of AGO2 was complicated by the fact that silencing AGO2 alone increased PR mRNA and protein expression by ∼2- to 3-fold (Figure 3). Addition of PR-11 to MCF7 cells yielded a ∼6-fold activation of gene expression at the level of RNA (Figure 5B). When AGO2 expression was repressed the addition of PR-11 only produced a 2.5-fold increase in PR expression (relative to cells treated with siAGO2/MM). Inhibiting expression of AGO1, AGO3 and AGO4 did not reverse gene activation by PR-11 (Figure 5A, C–E, G and H; Supplementary Figure S9). These data suggest that AGO2 is the best candidate for involvement in RNA-mediated gene activation by PR-11.
Figure 5.
Effect of inhibiting AGO1, AGO2, AGO3 or AGO4 gene expression on activation of PR expression by PR-11 (25 nM) in MCF7 cells. qPCR analysis of PR after a double transfection assay with (A) siAGO1, (B) siAGO2, (C) siAGO3, (D) siAGO4 being added first (Day 0) and PR-11 added second (after 72 h). Western analysis of PR after a double transfection assay with (E) siAGO1, (F) siAGO2, (G) siAGO3, (H) siAGO4 being added first (Day 0) and PR-11 added second (after 72 h). Transfection combinations: MM/PR-11 represents that the first transfection is MM, a control RNA duplex and the second is PR-11, etc. All data were normalized to GAPDH for q-PCR. Error bars represent standard deviations, calculated from three independent experiments. ***P < 0.005 as compared to cells treated with a mismatched RNA (MM). P-values were calculated using the two tailed unpaired Student’s t-test with equal variances.
Nuclear localization of AGO protein and the PR antisense transcript
Our proposed mechanism of action for agRNAs requires that the AGO/agRNA complex bind the antisense transcript in the nucleus. To determine whether AGO proteins and the antisense transcript were available to form nuclear interactions, we obtained nuclear and whole cell extracts and compared their localization.
We detected all four AGO variants in the nucleus of T47D (Figure 6A). In the nucleus of MCF7, AGO1, AGO2 and AGO4 are easily seen, but AGO3 is barely detectable (Supplementary Figure S10). Cytoplasmic markers β-tubulin or GAPDH were detected only in the whole-cell fraction, not in the nuclear fraction, suggesting that the nuclear fraction was not contaminated with cytoplasmic proteins. Conversely, the nuclear envelope marker lamin A/C is enriched in the nuclear fraction, suggesting that nuclear isolation was efficient.
Figure 6.
Association of AGO1–4 with the PR antisense transcript upon addition of PR-9 or PR-11 determined by RIP. (A) Western analysis of AGO1, AGO2, AGO3 and AGO4 proteins in whole cell and nuclear fractions from T47D cells. 40 µg of protein was loaded in each lane. WC: whole cell fraction; Nuc: nuclear fraction. (B) The nuclear distribution of PR mRNA, GAPDH mRNA and PR pre-mRNA in MCF7 cells. Data is represented as percent ratio of amplified transcript in nuclear to whole cell extracts. SN 7SK was used as a nuclear internal control. (C) RIP examining association of AGO1, AGO2, AGO3 and AGO4 with the PR antisense transcript in T47D cells treated with PR-9. (D) RIP examining association of AGO1, AGO2, AGO3 and AGO4 with the PR antisense transcript in MCF7 cells treated with PR-11. MM: negative control RNA duplex. IgG: negative control antibody. Input: nuclear extract prior to treatment with antibody. Input NR: input sample with no reverse transcriptase treated. The above data is the representative data set from three independent experiments.
We also examined the nuclear extract for the presence of the antisense transcript that is presumed to be the target for agRNAs PR-9 and PR-11. We observed that the antisense transcript was predominantly localized in the nucleus (Figure 6B). GAPDH mRNA and PR mRNA were distributed between cytoplasm and nucleus. SN7SK, known to exclusively localize in the nucleus (28), was used as an internal control. PR pre-mRNA was localized exclusively to the nuclear fraction. These data indicated that the PR antisense transcript is present with the four AGO proteins in the nucleus.
Recruitment of AGO proteins to non-coding RNA during gene silencing
To further evaluate involvement of AGO proteins in agRNA-mediated gene silencing we used nuclear RNA immunoprecipitation (RIP) (16) to examine recruitment of AGO protein to the non-coding transcript at the PR promoter. T47D cells were transfected with agRNA PR-9 and nuclear extract was obtained. RIP using anti-AGO1 (4B8) or two different anti-AGO2 (4G8 and 11A9) antibodies revealed an association between AGO1 or AGO2 and the antisense transcript after transfection of PR-9 (Figure 6C). Sequencing confirmed that the RIP PCR product was derived from amplification of the antisense transcript (Supplementary Figure S11). No RIP product was observed after transfection with mismatch-containing duplex RNA. No association with antisense transcript was apparent after testing with anti-AGO3 or anti-AGO4 antibodies. Similar to PR-9, addition of inhibitory agRNA PR-26 to cells also revealed association with AGO1 and AGO2 (Supplementary Figure S12).
These data are consistent with the results from our AGO knockdown experiments (Figure 4) indicating that AGO2 is responsible for agRNA-mediated gene silencing of PR-9 but also suggest that AGO1 can bind the antisense transcript. Binding of AGO1 protein may not induce silencing activity either because association of AGO1 may be insufficient or AGO1 may not form interactions necessary for gene silencing.
While AGO2 is a known critical component of RISC involved in siRNA/miRNA mediated gene regulation pathway, dicer and TRBP have also been reported as essential RISC members (38,45,46). We used RIP to examine if Dicer or TRBP were associated with the antisense transcript in PR-9 treated cells. No association with antisense transcript was apparent after testing with anti-Dicer or anti-TRBP antibodies (Supplementary Figure S12).
Recruitment of AGO proteins to non-coding RNA during gene activation
We also used RIP to examine involvement of AGO proteins in agRNA-mediated gene activation. RIP using two different anti-AGO2 antibodies (4G8 or 11A9) revealed association with the PR antisense transcript upon transfection with PR-11 but not a mismatch-containing RNA duplex (Figure 6D). Sequencing confirmed that the RIP PCR product was derived from amplification of the antisense transcript (Supplementary Figure S11). No association with antisense transcript was apparent after testing with anti-AGO1, anti-AGO3 or anti-AGO4 antibodies. We also examined the association of dicer or TRBP with the antisense transcript in PR-11 treated cells and we failed to detect any signal in both cases (Supplementary Figure S12).
To further investigate the association of AGO proteins with the non-coding antisense transcript during gene activation we introduced vectors encoding HA epitope-tagged variants of AGO1–4 (20) into MCF7 cells (Figure 7) (Supplementary Figure S13). We observed that HA-tagged AGO2 associated with the antisense transcript after addition of PR-11. No association with antisense transcript was detected in the presence of HA-tagged AGO1, AGO3, or AGO4. Expression levels of the FLAG-HA tagged AGO variants were much lower in transfected T47D cells than in MCF7 cells, preventing similar experiments examining recruitment of AGO variants by silencing agRNA PR-9.
AGO2 is known to act to promote cleavage of mRNA during RNAi. For transcriptional activation, Morris reported that duplex RNAs act by relieving the suppressive effect of antisense RNAs by a mechanism that may involve cutting the antisense transcript (14). Using 5′-RACE we readily detected cleavage of PR mRNA by a duplex complementary to PR mRNA. By contrast, we did not detect any RACE products after addition of PR-9, PR-26, or PR-11 when analyses were done in parallel (Supplementary Figure S14). We also note that our RIP protocol employs primers on either side of agRNA target site and can only detect intact antisense transcript. Therefore, while cleavage of non-coding transcripts may be involved in regulating expression of other genes, we do not observe evidence that cleavage is involved in agRNA-mediated modulation of PR expression.
AGO proteins and agRNAs
The majority of studies with small duplex RNAs to modulate gene expression have used duplexes that are complementary to mRNA. However, there has also been significant work on RNAs that target sequences outside of mRNA and some of these studies have characterized the role of AGO proteins during transcriptional silencing in plants, yeast, worms and flies (47–59). To date, relatively few papers have appeared describing the action of promoter-targeted agRNAs in human cells (2–12,14). A better understanding of agRNAs in human cells is needed because gene promoters might also be targets for miRNAs (60–62). It is also possible that agRNAs might have therapeutic applications (7), especially for activating expression of beneficial genes, such as p21 (12). This topic has been recently reviewed (17).
A more complete picture of the mechanism of action of agRNAs will require an inventory of proteins at the promoters of target genes and how that inventory changes upon agRNA recruitment. For such studies to be successful, it is essential to base them on knowledge of involvement of a core protein. AGO is an ideal protein to build studies of mechanism around because of its central role in mediating recognition by small RNAs not only in the cell cytoplasm (63–65), but also in the nucleus of mammalian cells (20,28,30). When considering such studies it is essential to recall that the nucleic acid target of agRNAs are relatively lowly expressed non-coding transcripts associated with promoter DNA. The abundance of target sites is low and creating inventories of associated proteins is likely to face challenges more similar to those associated with studying transcription factors than the challenges encountered during the study of proteins that interact with mRNA.
Studies have suggested that the four AGO proteins have both unique and overlapping functions in human cells (23,25,26). These are not simple experiments. Experiments that investigate the role of AGO by decreasing AGO expression run the risk of off-target effects caused by global disruption of AGO-mediated expression pathways. Therefore, we have used an approach that couples silencing of AGO variants with direct examination of recruitment of AGO proteins using RIP.
For agRNA-mediated gene silencing, our data point to a primary role for AGO2 (Table 1). Our ChIP results also show recruitment of AGO1 during agRNA-mediated gene silencing (32). Involvement of AGO1, therefore, remains possible and we note that other laboratories previously observed involvement of AGO1 in silencing of CCR5 (33). Thus, while our data support a primary role for AGO2 in transcriptional silencing at the PR promoter, it is possible that AGO1 might also be involved or might even be the primary AGO variant involved in silencing other genes. In previous studies showing involvement of AGO1 (9,15), association with a non-coding sense transcript in the promoter region was observed. However, another recent report described that AGO2, not AGO1, is required for promoter-directed siRNA silencing of c-Myc and the direct target of this siRNA is the non-coding sense transcript in the promoter region (8).
We also identify AGO2 as the primary candidate for involvement in RNA-mediated activation (Table 1). Silencing AGO2 reduces the expected level of gene activation. RIP data using native anti-AGO and epitope tagged antibodies both supported that AGO2 was the most likely candidate for agRNA-mediated recruitment to the antisense transcript (Figures 6 and 7). Involvement of AGO2 in agRNA-mediated gene activation is consistent with previous findings by Li et al. (10), who evaluated the effect of silencing AGO2 on gene expression. Morris et al. have also reported that AGO2 is involved in small RNA mediated gene activation (14).
AGO2 is known to cleave target mRNA sequences. However, we do not observe cleavage of the targeted non-coding antisense transcript after addition of activating or inhibitory agRNAs. Our RIP protocol would not have detected any transcript if it had been cleaved, and the fact that it does detect product upon addition of anti-AGO2 antibody demonstrates that AGO2 can bind to the non-coding transcript without cleavage. One potential explanation is that, in the context of nuclear recognition, AGO2 forms interactions with nucleic acids or proteins that inhibit cleavage of the non-coding transcript. Our data suggests that investigators should keep an open mind about the capabilities of AGO2 during interactions with target nucleic acids inside cells.
AGO3 and AGO4 have received much less attention than AGO1 and AGO2 even though they are expressed at significant levels relative to their better studied counterparts and have been shown to interact with a similar spectrum of RNA transcripts (20,25,37). The involvement of AGO3 and AGO4 in the action of agRNAs had not been investigated previously. We find no evidence to support involvement of AGO3 or AGO4 in agRNA-mediated modulation of PR expression. Thus, of the four AGO variants, AGO3 and AGO4 are the least likely candidates for involvement in the mechanism of agRNA action.
Our proposed mechanism for agRNA action at the PR promoter involves recognition of an antisense transcript (16). We now localize this transcript within the nucleus. We also find that AGO proteins reside in the nuclear fraction and the presence of the PR antisense transcript in the nucleus is consistent with its participation in transcriptional silencing and recognition by nuclear AGO2 protein.
Non-coding transcripts overlap the promoters of many genes and may play a substantial role in regulating gene expression (66–73). At this early stage of studying recognition of non-coding RNAs, we recognize that other AGO variants may be involved in recognition at other gene loci in different cell lines. There may be a multiplicity of mechanisms for small RNA:non-coding transcript interactions and their regulation of expression. For example, recognition of an antisense transcript has recently been shown to affect gene splicing, a process that clearly will differ in significant ways for regulation of gene transcription (74). In addition, we have recently observed that agRNAs complementary to a sense transcript expressed beyond the 3′ terminus of PR mRNA is also a target for agRNAs that modulate PR expression (75).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
FUNDING
National Institutes of Health (NIGMS 77253 to D.R.C. and NIGMS 85080 to B.A.J.); Alnylam Pharmaceutical; Robert A. Welch Foundation (I-1244). Funding for open access charge: National Institutes of Health (NIGMS 77253).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Amritha Bhat for technical assistance and Dr Mikikio C. Siomi, Dr Gunter Meister and Dr Qinghua Liu for generously providing antibodies.
REFERENCES
- 1.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J. RNAi Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M-X, Ou H, Shen YH, Wang J, Coselli J, Wang XL. Regulation of endothelial nitric oxide synthase by small RNA. Proc. Natl Acad. Sci. USA. 2005;102:16967–19972. doi: 10.1073/pnas.0503853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Gen. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA by antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 7.Pulukuri SMK, Rao JS. Small-interfering RNA-directed reversal of urokinase plasminogen activator dimethylation inhibits prostate tumor growth and metastasis. Cancer Res. 2007;67:6637–6646. doi: 10.1158/0008-5472.CAN-07-0751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L-C, Okino ST, Zhao H, Place RF, Pookot D, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li L-C. Antitumor effect of dsRNA-induced p21WAF1/CIP1 gene activation in human bladder cancer cells. Mol. Cancer Ther. 2008;7:698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- 13.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li L-C. RNAa is conserved in mammalian cells. PLoS ONE. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris KV, Santoso S, Turner A-M, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl Acad. Sci. USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz JC, Younger ST, Nguyen N, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009;19:299–305. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 19.Rand TA, Ginalski K, Grishin NV, Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl Acad. Sci. USA. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–187. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. Binding and cleavage specificities of human argonaute2. J. Biol. Chem. 2009;284:26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diederichs S, Haber DA. Dual role for argonautes in MicroRNA processing and posttranscriptional regulation of MicroRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human argonaute proteins. Nat. Struct. Mol. Biol. 2009;16:1259–1267. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 24.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian argonautes in microRNAs silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landthater M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human argonaute-containing ribonucleoprotein complexes and their bound target RNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoda M, Kawamata T, Paro Z, Xuecheng Y, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 29.Rüdel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohrt T, Mütze J, Staroske W, Weinmann L, Höck J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of Ago1 and Ago2 link mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 33.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 34.Kim DH, RossiJ J. Transcriptional gene silencing using small RNAs. Methods Mol. Biol. 2009;555:119–125. doi: 10.1007/978-1-60327-295-7_9. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher A. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J. Biol. Chem. 2008;283:23353–23363. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner AMW, Cruz J, De La, Morris KV. Mobilization-competent lentiviral vector-mediated sustained transcriptional modulation of HIV-1 expression. Mol. Ther. 2009;17:360–368. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl Acad. Sci. USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, CoreyD R. Inhibiting transcription of chromosomal DNA using antigene peptide nucleic acids. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, Corey DR. Inhibiting gene expression with PNA-peptide conjugates that target chromosomal DNA. Biochemistry. 2007;46:7581–7589. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beane R, Ram R, Gabillet S, Arar K, Monia B, Corey DR. Inhibiting gene expression with locked nucleic acids (LNAs) that target chromosomal DNA. Biochemistry. 2007;46:7572–7580. doi: 10.1021/bi700227g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beane R, Gabillet S, Montallier C, Arar K, Corey DR. Recognition of chromosomal DNA by locked nucleic acids. Biochemistry. 2008;47:13147–13149. doi: 10.1021/bi801930p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misrahi M, Atger M, d’Auriol L, Loosfelt H, Meriel C, Fridlansky F, Guiochon-Mantel A, Galibert F, Milgrom E. Complete amino acid sequence of the human progesterone receptor deduced from cloned cDNA. Biochem. Biophys. Res. Comm. 1987;143:740–748. doi: 10.1016/0006-291x(87)91416-1. [DOI] [PubMed] [Google Scholar]
- 45.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 48.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 50.Grewal SIS, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis argonaute4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 52.Zilberman D, Cao X, Jacobsen SE. Argonaute4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2004;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 53.Qi Y, He X, Wang X-J, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and noncatalytic roles of argonaute4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Zhu J, Kapoor A, Zhu J-K. Role of Arabodopsis AGO6 in siRNA accumulation, DNA methylation, and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baumberger N, Baulcombe DC. Arabidopsis argonaute1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sigova A, Rhind N, Zamore PD. A single argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18:2359–2367. doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buker SM, Iida T, Buhler M, Villen J, Gygi SP, Nakayama J-I, Moazed D. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- 58.Yigit E, Batista PJ, Bei Y, Pang KM, Chen C-C, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 59.Claycomb JM, Batista PJ, Pang K, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting PF, et al. The argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim DH, Saetrom P, Snøve O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 63.Hutvangner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 64.Hock J, Meister G. The argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jinek M, Doudna JA. A three dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 66.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 67.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 68.Goodrich JA, Kugel JF. From Bacteria to humans, chromatin to elongation, and activation to repression: the expanding roles of noncoding RNAs in regulating transcription. Crit. Rev. Biochem. Mol. Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 71.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 72.Wahlstedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov. Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 73.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotech. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 74.Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 75.Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Regulation of transcription by small RNAs complementary to sequences downstream from the 3′ termini of genes. Nat. Chem. Biol. 2010;6:621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.