Abstract
Geobacter species often play an important role in bioremediation of environments contaminated with metals or organics and show promise for harvesting electricity from waste organic matter in microbial fuel cells. The ability of Geobacter species to fix atmospheric nitrogen is an important metabolic feature for these applications. We identified novel regulatory cascades controlling nitrogen-fixation gene expression in Geobacter sulfurreducens. Unlike the regulatory mechanisms known in other nitrogen-fixing microorganisms, nitrogen-fixation gene regulation in G. sulfurreducens is controlled by two two-component His–Asp phosphorelay systems. One of these systems appears to be the master regulatory system that activates transcription of the majority of nitrogen-fixation genes and represses a gene encoding glutamate dehydrogenase during nitrogen fixation. The other system whose expression is directly activated by the master regulatory system appears to control by antitermination the expression of a subset of the nitrogen-fixation genes whose transcription is activated by the master regulatory system and whose promoter contains transcription termination signals. This study provides a new paradigm for nitrogen-fixation gene regulation.
INTRODUCTION
Geobacter species are predominant microorganisms in subsurface environments in which organic compounds such as acetate are oxidized with the reduction of metals including Fe(III) (1,2). Geobacter species play a pivotal role in the bioremediation of organic or metal contaminants in subsurface environments (3–9). Geobacter species are also considered to play an important part in the biogeochemical cycles in subsurface environments (1,10). One of the factors contributing to the capability of Geobacter species to be such effective competitors in a diversity of subsurface environments may be their ability to fix atmospheric nitrogen (11–15). Analyses of transcript abundance demonstrated that Geobacter species expressed genes for nitrogen fixation in petroleum-contaminated subsurface sediments and uranium-contaminated groundwater (14,16,17). This suggests that nitrogen fixation was an important process in these environments. Moreover, Geobacter species show promise for harvesting electricity from waste organic matter in microbial fuel cells (18–20). Geobacter species are often the predominant microorganisms colonizing anodes of microbial fuel cells harvesting electricity from sediments or simulated wastewater (21–25) and the ability to fix nitrogen might aid in growth deep within the thick anode biofilms (26,27) where nutrient access might be limiting. Nitrogen fixation was also demonstrated with Geobacter isolates, Geobacter metallireducens (12) and Geobacter sulfurreducens (13). These Geobacter species fix nitrogen gas present in the headspace of the pressure tubes containing liquid medium devoid of fixed nitrogen. The disruption of the nifD gene encoding a subunit of nitrogenase eliminated the ability of G. sulfurreducens to grow in this medium (13).
Microbial nitrogen fixation is an important component of the global nitrogen cycle. Molecular mechanisms regulating nitrogen-fixation gene expression have been well characterized in some nitrogen-fixing microorganisms (diazotrophs) (28). In the Proteobacteria characterized, nitrogen-fixation genes are invariably subject to transcriptional activation by the master regulator of nitrogen fixation, NifA, a member of the enhancer-binding protein (EBP) family, together with the RNA polymerase sigma factor, RpoN (28,29). Transcription initiation by RNA polymerase containing the sigma factor RpoN (RNAP/RpoN) requires a transcription factor EBP (30–33). EBPs typically consist of three domains: an N-terminal sensory (regulatory) domain; a central ATPase domain; and a C-terminal DNA-binding domain. NifA has a GAF domain at the N-terminus. The GAF domain is a ubiquitous signaling module known to bind small molecules such as cyclic nucleotide monophosphates, tetrapyrroles and formate (34,35). The expression of the nifA gene is controlled by the two-component His–Asp phosphorelay system in these proteobacterial diazotrophs. For instance, Klebsiella pneumonia utilizes the histidine kinase NtrB and the response regulator NtrC, which is also a member of the EBP family. The two-component His–Asp phosphorelay system is a major scheme in bacterial signal transduction pathways during responses to environmental changes. The two-component system typically consists of a sensor histidine kinase, which senses environmental signals, and a response regulator, which often exerts gene expression necessary for the adaptation (36). The sensor histidine kinase autophosphorylates its own histidine residue upon signal sensing and subsequently transfers the phosphoryl group to the aspartate residue in the receiver domain of the response regulator. Phosphorylation of the receiver domain results in the activation of the effector (output) domain of the response regulator.
Although the ability of Geobacter species to fix nitrogen appears to be a key to growth in subsurface environments, as described above, little is known about regulatory molecular mechanisms for nitrogen fixation in Geobacter species. The genome sequence analyses of Geobacter species, which belong to the δ-subdivision of Proteobacteria, revealed no NifA homologue in Geobacter species, unlike other proteobacterial diazotrophs. Here we report novel regulatory cascades controlling gene expression during nitrogen fixation in G. sulfurreducens that consist of two two-component His–Asp phosphorelay systems, which regulate transcriptional activation, repression and antitermination of genes involved in nitrogen metabolism.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Geobacter sulfurreducens DL1 (37) was used for genetic and biochemical studies. Geobacter sulfurreducens strains were grown anaerobically in NBAF medium (acetate and fumarate as the electron donor and the acceptor, respectively) (11), unless otherwise described. Proper antibiotics were supplemented when necessary. Nitrogen fixation was examined with 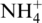 -free NBAF media as described previously (15). Cell growth was monitored by measuring the optical density at 600 nm (OD600). Escherichia coli DH5α (38) was used for plasmid preparation and grown in LB medium (39) supplemented with proper antibiotics when necessary.
-free NBAF media as described previously (15). Cell growth was monitored by measuring the optical density at 600 nm (OD600). Escherichia coli DH5α (38) was used for plasmid preparation and grown in LB medium (39) supplemented with proper antibiotics when necessary.
Primer extension assay
Total RNA was prepared from the wild type and the mutants grown in the media indicated in the figure legends by the hot-phenol method. The primers used in the assays are listed in Supplementary Table S1.
Construction of EBP expression vectors and purification of EBPs
DNA fragments encoding EPBs were amplified by PCR with primers listed in Supplementary Table S1. PCR products were digested with restriction enzymes and cloned in pET24b (Novagen). EBPs were overexpressed by Autoinduction system (Novagen) as instructed by the manufacturer and prepared as histidine-tagged proteins at the C-terminus as described previously (40).
DNA-binding assay
Promoter regions of genes gnfK, nifH, glnB, gdhA and GSU3409, were used as probes in DNA-binding assays. The DNA fragments containing their promoter regions were prepared by PCR with primers listed in Supplementary Table S1. DNA fragments were labeled with [γ-32P]ATP by T4 polynucleotide kinase. DNA-binding assays were carried out as described previously (41).
Footprint assay
DNA fragments containing the region from nt −243 to −3 of the gnfK promoter or from nt −223 to +66 of the gdhA promoter were amplified with primers listed in Supplementary Table S1 and cloned in the plasmid vector. DNA fragments for the probe were prepared by digesting the plasmid with SacII and XhoI located in the cloning vector and isolating the fragment with gel electrophoresis. The isolated DNA fragments were then labeled with [α-32P]dATP by Klenow fragment of DNA polymerase I. Thus, only the top strand was labeled. DNA-binding reactions were conducted with different amounts of GnfM and DNase I footprint assays were conducted as described previously (41).
lacZ fusion assay
For the WT construct of gnfK, the promoter region from nt −243 to +32 of gnfK was amplified by PCR with primers −243T and +32B. The PCR product was digested with restriction enzymes and cloned into the transcriptional lacZ fusion vector, pCMZKT (42). To examine the effects of the binding sites determined by the footprint assays on the promoter activity, the regions from nt −128 to −112 (BS1) and from −105 to −89 (BS2) were deleted. For the ΔBS1 construct, the promoter regions from nt −243 to −129 and from nt −111 to +32 were amplified by primers −243T and −129B, and −111T and +32B, respectively, and digested with XbaI and EcoRI, and EcoRI and BamHI, respectively. The XbaI-EcoRI fragment and the EcoRI-BamHI fragment were ligated and cloned into pCMZKT. For the ΔBS2 construct, the promoter regions from nt −243 to −106 and from nt −88 to +32 were amplified by primers −243T and −106B, and −88T and +32B, respectively, and cloned as described for the ΔBS1 construct. For the ΔBS1ΔBS2 construct, the promoter regions from nt −243 to −129 and from nt −88 to +32 were amplified by primers −243T and −129B, and −88T and +32B, respectively, and cloned as described above. The plasmids thus constructed were introduced into G. sulfurreducens DL1 by electroporation (13).
For the WT construct of gdhA, the promoter region from nt −84 to +6 of the gdhA gene was prepared by annealing oligonucleotides WT12T and WT12B and oligonucleotides WT34T and WT34B, and by ligating these annealed oligonucleotides. For the M_BS3/BS4 construct, oligonucleotides Mut34T and Mut34B were used instead of WT34T and WT34B. For the ΔBS1/BS2 construct, oligonucleotides WT34T and WT34B were used. These fragments were cloned into pCMZKT. The plasmids thus constructed were introduced into G. sulfurreducens DL1 by electroporation (13).
β-Galactosidase activity was measured by the method described and calculated by the formula given by Miller (39). The strains carrying the plasmids were grown in NBAF media supplemented with kanamycin in the presence or absence of NH4Cl. Presented values are averages of two independent experiments.
Promoter regions for testing the transcription termination signals were prepared by oligonucleotides containing an RpoD-dependent like −35/−10 elements without or with the wild type or mutated transcription termination signals from nifH (Supplementary Figure S5A). Oligonucleotides were phosphorylated with T4 polynucleotide kinase, annealed and ligated with pCMZKT (42). The resultant plasmids were transformed into E. coli and β-galactosidase activity was examined by plating on LB agar media containing kanamycin and X-gal.
In vitro phosphorylation
The DNA fragments encoding GnfLK from 103Met to 363Gln of GnfL (GSU1004), GnfM, GnfK (GSU0941) or GnfR (GSU2822) were amplified by PCR with primers listed in Supplementary Table S1. The PCR products were cloned into pET24b (Novagen). The cloned genes were overexpressed by Autoinduction system (Novagen) as instructed by the manufacturer and the proteins were prepared as a histidine-tagged protein at the C-terminus. Purification of these proteins was performed as described previously (40).
In vitro phosphorylation assays were performed as described previously (40) with modifications. Reactions were performed in the kinase buffer [25 mM Tris–HCl (pH 7.4); 10 mM MgCl2; 5 mM β-mercaptoethanol; 20 µM ATP; 2 µCi [γ-32P]ATP]. GnfLK (20 pmol) was incubated at room temperature for 5 min for the autophosphorylation reaction. GnfM (20 pmol) was then added and further incubated at room temperature for 5 min for the phosphotransfer reaction. GnfK (10 pmol) was incubated at a room temperature for 5 min for the autophosphorylation reaction. GnfR (10 pmol) was then added and further incubated at a room temperature for 5 min for the phosphotransfer reaction. The reactions were stopped by adding the stop solution (10% SDS; 0.4 M Tris–HCl (pH 6.8); 50% glycerol; 0.1 M β-mercaptoethanol; 0.02 % bromphenol blue). The samples were subjected to 10% SDS–PAGE and analyzed by autoradiography.
Overproduction of GnfLK and GnfM in G. sulfurreducens
An expression vector was constructed to overproduce GnfLK and GnfM in G. sulfurreducens. Oligonucleotides listed in Supplementary Table S1 were phosphorylated, annealed, and ligated with the plasmid pCD341 (43) at the EcoRI site. These oligonucleotides contain a ribosome binding site and an NdeI site for the initiation codon preceded by three stop codons in three open reading frames. After confirmation of proper sequence and orientation of the oligonucleotides in pCD341, the expression vector was designated pCDNdeII.
The DNA fragments encoding GnfLK or GnfM were amplified by PCR with primers listed in Supplementary Table S1. The PCR products were digested with BglII and XbaI or NdeI and XhoI, respectively, and ligated with pET24b (Novagen). For overexpression of GnfLK and GnfM together, after sequence confirmation of the PCR products, the DNA fragments encoding GnfLK or GnfM (with the ribosome binding site from pET24b) were isolated from the plasmids by digestion with EcoRI and XbaI or XbaI and HindIII, respectively, and ligated with pCDNdeII in one step. The plasmid containing the DNA fragments encoding GnfLK and GnfM was designated pCDNdeII/GnfLM. For overexpression of GnfM, the DNA fragment encoding GnfM was isolated from the plasmid by digestion with NdeI and HindIII, and ligated with pCDNdeII. The plasmid containing the DNA fragment encoding GnfM was designated pCDNdeII/GnfM.
Plasmids pCDNdeII, pCDNdeII/GnfLM, and pCDNdeII/GnfM were introduced into G. sulfurreducens DL1 by electroporation (13). Overproduction of GnfLK and GnfM was achieved by growing in NBAF media (in the presence of ammonium) supplemented with kanamycin and adding IPTG at a final concentration of 1 mM at a mid-log phase.
Construction of gnfK or gnfR deletion mutants
The gnfK gene was replaced with a kanamycin resistance gene, such that the coding region for amino acid residues from 13Asp to 164Leu was deleted. Double-crossover homologous recombination was carried out by electroporation (13) with the linear DNA fragment consisting of the kanamycin resistance gene flanked by 0.7-kb DNA fragments containing the upstream and the downstream regions of gnfK. These flanking DNA fragments were amplified by PCR with primers listed in Supplementary Table S1. The DNA fragment of the kanamycin resistance gene was amplified by PCR with primers listed in Supplementary Table S1, and pBBR1MCS-2 (44) as a template. The replacement was confirmed by PCR amplification.
The gnfR gene was replaced with the kanamycin resistance gene, such that the coding region for amino acid residues from 12Asp to 139Thr was deleted, as described above. The flanking DNA fragments were amplified by PCR with primers listed in Supplementary Table S1. The proper replacement was confirmed by PCR amplification.
RNA-binding assay
RNA fragments CGGAGUAUUCGCUGCUGAACGCAGGCAAGGGCGCCACCAUUAAACGGA (wild type, the putative GnfR-binding site is indicated by bold letters) and CGGAGUAUUCGCUGCUGAACGCAUCACUGCUCAUAACCAUUAAACGGA (mutant, mutated positions are underlined) were synthesized by INTEGRATED DNA TECHNOLOGIES. RNA fragments were labeled with [γ-32P]ATP by T4 polynucleotide kinase. After the labeling, the fragments were boiled and kept on ice until used. RNA-binding assays were performed in the binding buffer [50 mM Tris–HCl (pH 7.9); 10 mM MgCl2; 50 mM KCl; 0.1 mM DTT; 0.1 mg/ml BSA; 20 µM ATP]. GnfK (1 pmol) was incubated at room temperature for 5 min for the autophosphorylation reaction. GnfR (1 pmol) was then added and further incubated at room temperature for 5 min for the phosphotransfer reaction. The 32P-labeled RNA fragments (40 fmol) and yeast tRNA (1µg) were added and incubated at room temperature for 15 min. The reaction mixtures were subjected to 5% PAGE and analyzed by autoradiography.
Computational tools
Homology search was conducted by NCBI/BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi/). The Genbank accession numbers for the genome sequences of G. sulfurreducens, G. metallireducens and G. uraniireducens are AE017180, CP000148 and CP000698, respectively. Sequence alignments were constructed by MAFFT version 6 (http://align.bmr.kyusyu-u.ac.jp/mafft/online/server/). Sequence logos were generated by WebLogo (http://weblogo.Berkeley.edu).
RESULTS
Promoter region of the nitrogen-fixation genes
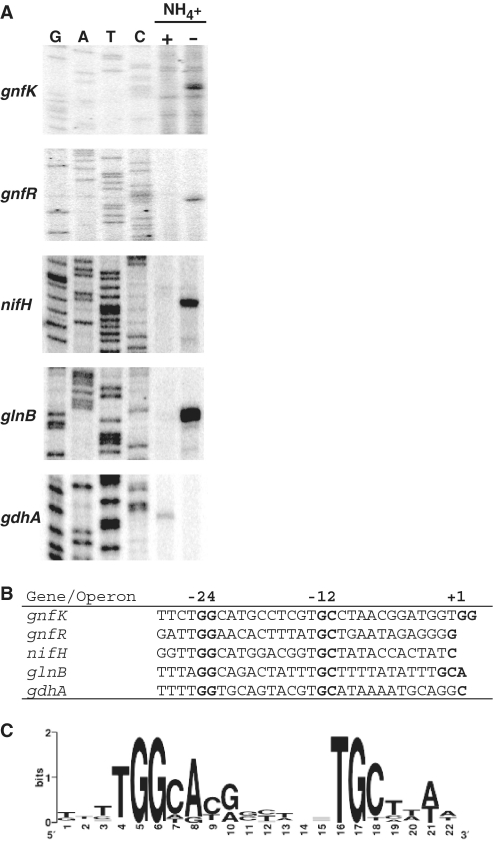
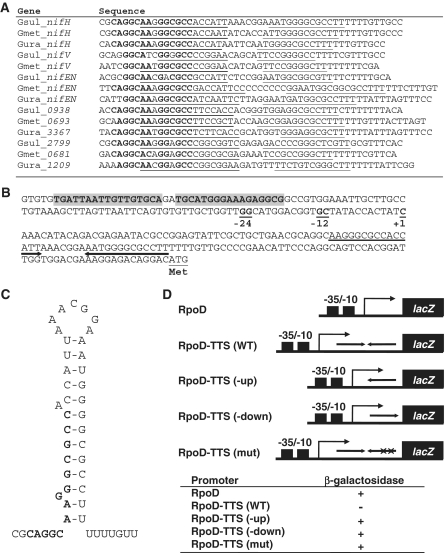
In order to elucidate the regulation of the genes involved in nitrogen fixation, it is necessary to define the promoter region of the genes. Therefore, transcription initiation sites for genes whose expression was previously shown to be significantly changed during nitrogen fixation in G. sulfurreducens (15) were determined by primer extension assays. Among the genes up-regulated during nitrogen fixation, gnfK (Geobacter nitrogen fixation histidine kinase) and gnfR (Geobacter nitrogen fixation response regulator) encoding a histidine kinase and a response regulator in the two-component system were examined, because they likely play a role in nitrogen-fixation gene regulation (see below). In addition, nifH, which encodes a nitrogenase iron protein, and glnB, which encodes a nitrogen regulatory protein PII, were tested, as they are well known to be involved in nitrogen fixation in other bacteria. Moreover, gdhA encoding glutamate dehydrogenase was investigated, because it was highly down-regulated during nitrogen fixation (15). The expression patterns of these genes were consistent with those found by the transcriptome analysis in that the transcription of gnfK, gnfR, nifH and glnB was activated during nitrogen fixation and gdhA expression was repressed during nitrogen fixation (Figure 1A).
Figure 1.
RpoN-dependent promoter in nitrogen-fixation genes. (A) Expression of nitrogen-fixation genes and identification of their transcription initiation site. The expression of gnfK, gnfR, nifH, glnB and gdhA was examined by primer extension assays. Total RNA was prepared from cells grown in the presence (+) or absence (−) of  . G, A, T and C represent sequence ladders generated by the same primer used in the primer extension assays. (B) RpoN-dependent −24/−12 promoter elements of the nitrogen-fixation genes. The −24/−12 promoter elements for nifH, glnB, gnfK, gnfR and gdhA of G. sulfurreducens were assigned from the transcription initiation sites (+1) determined by the primer extension assays (A). (C) The consensus sequence of the −24/−12 promoter elements. The consensus sequence was obtained from the alignment shown in Supplementary Table S2 and is presented as a logo.
. G, A, T and C represent sequence ladders generated by the same primer used in the primer extension assays. (B) RpoN-dependent −24/−12 promoter elements of the nitrogen-fixation genes. The −24/−12 promoter elements for nifH, glnB, gnfK, gnfR and gdhA of G. sulfurreducens were assigned from the transcription initiation sites (+1) determined by the primer extension assays (A). (C) The consensus sequence of the −24/−12 promoter elements. The consensus sequence was obtained from the alignment shown in Supplementary Table S2 and is presented as a logo.
Based on a region upstream of the transcription initiation site, it appears likely that these genes including gdhA have an RpoN-dependent promoter (Figure 1B). RpoN, a subunit of RNA polymerase, recognizes −24 and −12 promoter elements, which contain highly conserved sequences, GG and GC, respectively (45,46). In addition to these genes from G. sulfurreducens, homologues of these genes in other Geobacter species were predicted to have RpoN-dependent promoter elements (Supplementary Table S2). Furthermore, the sequence analysis of upstream regions of other up-regulated genes during nitrogen fixation identified by the transcriptome analysis in G. sulfurreducens as well as homologues in other Geobacter species revealed that many of them also have RpoN-dependent promoter elements (Supplementary Table S2). In contrast, gdhA in G. sulfurreducens was the only gene containing −24/−12 promoter elements that was among the genes down-regulated during nitrogen fixation (15). By using these RpoN-dependent promoter elements, the consensus sequence for the −24/−12 promoter elements was determined (Figure 1C). The consensus sequence is similar to other −24/−12 promoter elements in G. sulfurreducens (47) as well as other bacteria (45,46). Therefore, it is likely that the majority of the genes involved in nitrogen fixation are controlled by RpoN in G. sulfurreducens.
Master regulator for the nitrogen-fixation genes
As shown above, the majority of the genes up-regulated during nitrogen fixation appear to have an RpoN-dependent promoter in Geobacter species. Transcription initiation by RNAP/RpoN generally requires an EBP. However, the transcriptome analysis failed to identify a gene encoding an EBP (15). This might be due to the fact that many transcription factors including EBPs are regulated at the level of activity by modulation of the proteins such as phosphorylation and ligand binding. EBPs typically are able to bind specific DNA without modulation, but are incapable of activating transcription initiation with RNAP/RpoN without modulation (30–33).
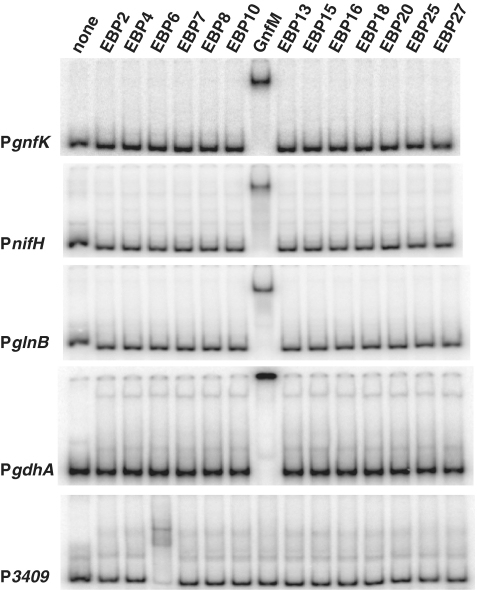
Because the genes involved in nitrogen fixation are highly conserved in Geobacter species, it is reasonable to speculate that the EBP responsible for the transcription initiation of the nitrogen-fixation genes with an RpoN-dependent promoter is also conserved in Geobacter species. In order to identify the EBP, Geobacter genome sequences were analyzed for EBPs (Supplementary Table S3). The genome sequence of G. sulfurreducens encodes 28 EBPs and 14 of them are highly conserved in both G. metallireducens and G. uraniireducens. The 14 EBPs conserved in Geobacter species were then tested for DNA-binding activity to promoters of gnfK, nifH, glnB and gdhA (Figure 2). Only EBP11, termed GnfM (Geobacter nitrogen fixation master regulator), exhibited DNA-binding activity to the promoter region of gnfK, nifH, and glnB as well as to the promoter region of gdhA, which was repressed during nitrogen fixation. To evaluate the specificity of the GnfM binding, the upstream promoter of the GSU3409 gene, which was recently identified as a direct target of Grr1 (EBP6), another EBP in G. sulfurreducens (Ueki and Lovley, a manuscript in preparation), was tested. Only EBP6 (Grr1) showed DNA-binding activity to the GSU3409 promoter (Figure 2). Therefore, these results suggest that GnfM is involved in the regulation of these genes. GnfM is a typical EBP and contains a receiver domain at the N-terminus, a AAA/Sigma54_activation domain, and a DNA-binding domain at the C-terminus (Supplementary Figure S1).
Figure 2.
Identification of the master regulator for the nitrogen-fixation genes by DNA-binding assay. DNA fragments (40 fmol) containing upstream promoter regions of gnfK, nifH, glnB and gdhA were tested for DNA-binding activity of the EBPs (0.5 pmol) conserved in Geobacter species (Supplementary Table S3). The upstream promoter region of the GSU3409 gene was used as a control.
GnfM-binding sites
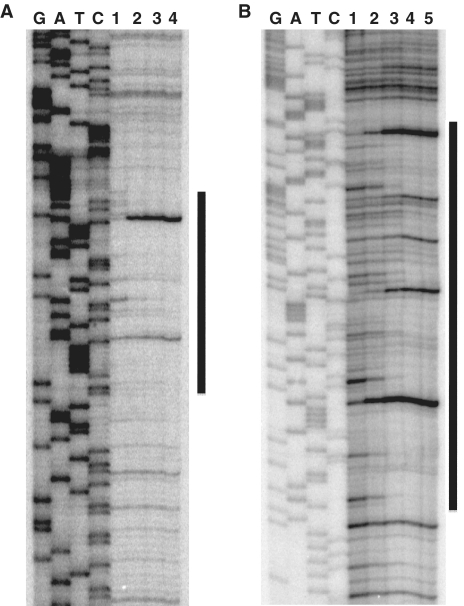
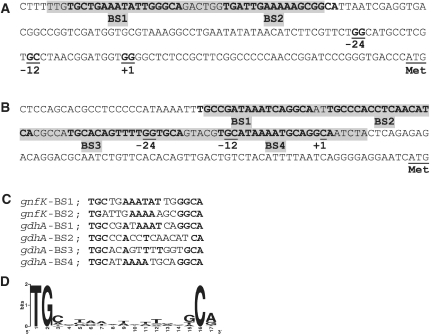
To determine the binding sites of GnfM, footprint assays were performed, and the region from nt −131 to −91 in the gnfK promoter with respect to the transcription initiation sites was protected from or modified by DNase I in the presence of GnfM (Figure 3A). In addition, the region from nt −77 to +7 in the gdhA promoter with respect to the transcription initiation site was protected from or modified by DNase I in the presence of GnfM (Figure 3B). It is likely that there are two and four GnfM-binding sites in gnfK (Figure 4A) and gdhA (Figure 4B), respectively, as highly similar sequences are found in these identified regions (Figure 4C). The presence of more GnfM-binding sites in gdhA than in gnfK is consistent with the results of the DNA-binding assays in that the GnfM/DNA complex migrated slower with gdhA than with gnfK (Figure 2). The GnfM-binding sites are located upstream of the RpoN-dependent promoter in gnfK, suggesting that GnfM functions as a transcriptional activator. In gdhA, the GnfM-binding sites are located upstream of the RpoN-dependent promoter as well as at the region overlapping the RpoN-dependent promoter, suggesting that GnfM functions as a transcriptional repressor for gdhA. These results are consistent with their expression patterns (Figure 1A). A sequence analysis of upstream regions of the nitrogen-fixation genes with an RpoN-dependent promoter identified similar sequences with those in gnfK and gdhA, which may function as a binding site of GnfM (Supplementary Table S4). A consensus sequence for the GnfM-binding sites was determined (Figure 4D).
Figure 3.
GnfM-binding site. Footprint assays were carried out with GnfM and the upstream region of gnfK (A) or gdhA (B). DNA-binding reactions were conducted with GnfM (lane 1, 0 pmol; lane 2, 0.7 pmol; lane 3, 1.3 pmol; lane 4, 2.5 pmol for gnfK, lane 1, 0 pmol; lane 2, 0.3 pmol; lane 3, 0.7 pmol; lane 4, 1.3 pmol; lane 5, 2.5 pmol for gdhA) and probes (0.2 pmol). G, A, T and C represent sequence ladders. The region protected from or modified by DNase I in the presence of GnfM is indicated by a vertical bar.
Figure 4.
Promoter regions. (A) gnfK. (B) gdhA. Regions modified in the presence of GnfM in the footprint assays (Figure 3) are highlighted in gray. Similar sequences found in the GnfM-binding sites are indicated by bold letters. Highly conserved dinucleotides GG and GC in RpoN-dependent −24/−12 promoter elements are underlined. Transcription initiation site is indicated by +1. Translation initiation codon is indicated by Met. (C) Alignment of similar sequences found in the GnfM-binding sites. (D) The consensus sequence of the GnfM-binding sites. The consensus sequence was obtained from the alignment of the putative GnfM-binding sites in the nitrogen-fixation genes (Supplementary Table S4) and is presented as a logo.
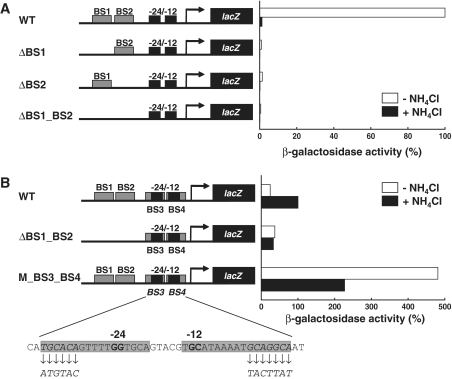
To evaluate the effects of the identified GnfM-binding sites on the expression of gnfK and gdhA, lacZ fusion assays were conducted (Figure 5). When one or both of the GnfM-binding sites were deleted, the gnfK promoter was not activated during nitrogen fixation (Figure 5A). These results indicate that the identified GnfM-binding sites were essential for the induction of gnfK expression during nitrogen fixation. When both the GnfM-binding sites (BS1 and BS2) located upstream of the RpoN-dependent promoter were deleted, the activity of the gdhA promoter in the presence of ammonium was similar to that in the absence of ammonium (Figure 5B), indicating that BS1 and BS2 were required for the induction of gdhA expression in the presence of ammonium. In contrast, when the GnfM-binding sites (BS3 and BS4) located at the region overlapping the RpoN-dependent promoter were mutated, the activity of the gdhA promoter was higher both in the presence and absence of ammonium than the activity of the wild-type gdhA promoter (Figure 5B), indicating that BS3 and BS4 were necessary for the repression of gdhA expression. The RpoN-dependent promoter elements were not mutated in this assay (Figure 5B). These results suggest that GnfM functions as a repressor in the absence of ammonium and an activator in the presence of ammonium for gdhA.
Figure 5.
Effect of the GnfM-binding sites on the expression of gnfK and gdhA. The promoter activity of gnfK (A) and gdhA (B) was analyzed by lacZ fusion assays. The activity is shown as a percentage of the activity of the wild-type promoter in the absence of  for gnfK and in the presence of
for gnfK and in the presence of 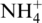 for gdhA. The location of GnfM-binding sites (BS) is indicated in Figure 4A and B. The binding sites were deleted (Δ) or mutated (M) in the assays. Diagrams of promoter constructs are shown and changes in the binding sites (M_BS3_BS4) are presented.
for gdhA. The location of GnfM-binding sites (BS) is indicated in Figure 4A and B. The binding sites were deleted (Δ) or mutated (M) in the assays. Diagrams of promoter constructs are shown and changes in the binding sites (M_BS3_BS4) are presented.
The two-component system GnfL/GnfM
In order to assess the physiological function of GnfM, construction of a deletion mutant of the gene encoding GnfM was attempted. However, such a mutant could not be isolated even in the presence of ammonium (data not shown). Therefore, the gnfM gene appears to be essential for growth even in the presence of ammonium. An apparent reason is that GnfM regulates genes involved in nitrogen metabolism not only in the absence of ammonium but also in its presence, such as gdhA as described above. A deletion mutant of the gnfM gene could not be isolated even when the mutant selection medium was supplemented with glutamine and glutamate (data not shown). The results suggest that GnfM regulates other pathway(s) in nitrogen metabolism and/or that the simultaneous inactivation of the genes regulated by GnfM resulting from the deletion of the gnfM gene is deleterious to a cell.
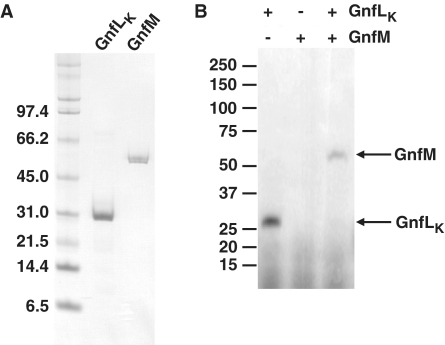
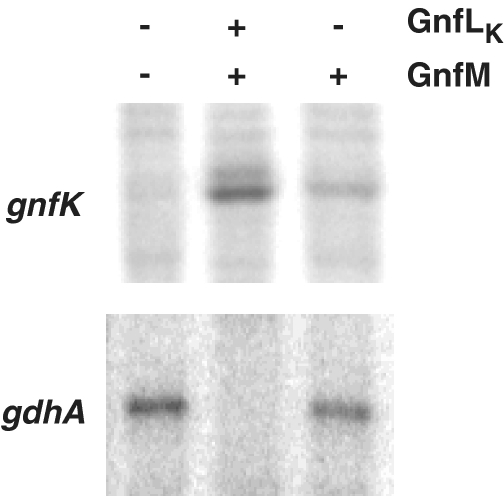
As an alternative to elucidating GnfM function, a strain harboring an expression vector for GnfM was constructed to examine the effects of the overproduction of GnfM on the expression of the nitrogen-fixation genes. GnfM contains a receiver domain at the N-terminus (Supplementary Figure S1) and thus is likely a response regulator of the two-component His–Asp phosphorelay system. In addition, a gene termed gnfL encoding a histidine kinase is located immediately upstream of the gnfM gene on the genome. GnfL appears to have a PAS domain as a sensor with some homology to NtrB homologues (Supplementary Figure S2). A gene encoding the kinase domain of GnfL (GnfLK) was included in the expression vector because the histidine kinase, which becomes autophosphorylated upon sensing a signal, activates the cognate response regulator by phosphorylation, but histidine kinases without the sensor domain are often capable of autophosphorylation independent of their signals and subsequently of phosphorylation of the cognate response regulators (48). In fact, GnfLK had autophosphorylation activity and phosphorylated GnfM in vitro (Figure 6). The short duration (5 min) of this reaction strongly suggests that GnfLK displays specificity to GnfM in this assay. It has been shown that cross-talk observed in vitro likely arises from excesses in reaction time (49,50). Taken together, it is likely that GnfM and GnfL function as a two-component system involved in sensing and responding to nitrogen availability. The expression of gnfK, which is induced during nitrogen fixation, and gdhA, which is repressed during nitrogen fixation, was examined (Figure 7). The expression of gnfK was induced in the presence of ammonium when the two-component system GnfL/GnfM was overexpressed, whereas the expression of gdhA was repressed under the same conditions. In contrast, when only GnfM was overexpressed, the induction of gnfK expression drastically decreased, suggesting that the phosphorylated form of GnfM is more active in the transcription initiation of gnfK. Moreover, gdhA expression was only slightly repressed by the overexpression of GnfM alone, suggesting that the phosphorylation enhances the activity of GnfM as a transcriptional repressor for gdhA. These results suggest that GnfM functions as a transcription activator for gnfK and a repressor for gdhA and these activities of GnfM are modulated by phosphorylation via GnfL.
Figure 6.
The two-component GnfL/GnfM system. (A) Purification of GnfLK and GnfM. The sizes of molecular mass standards are shown in kilodaltons. (B) In vitro phosphorylation assay. GnfLK and GnfM were tested for phosphotransfer activity in vitro. GnfLK was incubated at room temperature for 5 min for the autophosphorylation reaction. GnfM was then added and further incubated at room temperature for 5 min for the phosphotransfer reaction. The sizes of molecular mass standards are shown in kilodaltons.
Figure 7.
Effect of overproduction of GnfL/GnfM on the expression of gnfK and gdhA. Total RNA was prepared from strains harboring the vector only, the GnfL/GnfM expression vector or the GnfM expression vector grown in the presence of 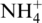 . The expression of gnfK and gdhA was analyzed by primer extension assays.
. The expression of gnfK and gdhA was analyzed by primer extension assays.
Another two-component system in the nitrogen-fixation gene regulation
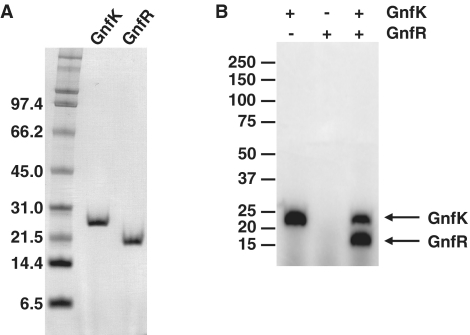
A potential additional mechanism for the regulation of the genes involved in nitrogen fixation in G. sulfurreducens is regulation via another two-component system. The previous transcriptome analysis for nitrogen fixation indicated that only two genes, gnfK and gnfR, predicted to encode a histidine kinase and a response regulator, respectively, had higher transcript abundance during nitrogen fixation in G. sulfurreducens (15). As shown above, these genes were activated by the master two-component system GnfL/GnfM during nitrogen fixation. This is the first demonstration that the regulatory mechanisms for nitrogen-fixation gene expression consist of two two-component His–Asp phosphorelay systems. gnfK and gnfR are not closely located on the Geobacter genomes. However, purified GnfK exhibited autophosphorylation activity as well as phosphotransfer activity to purified GnfR in in vitro phosphorylation assays (Figure 8A and B). It is likely that the phosphotransfer from GnfK to GnfR is a specific reaction because the reaction time was short (5 min), as described above. These results suggest that GnfK and GnfR function as a two-component system during nitrogen fixation.
Figure 8.
The two-component GnfK/GnfR system. (A) Purification of GnfK and GnfR. The sizes of molecular mass standards are shown in kilodaltons. (B) In vitro phosphorylation assay. The histidine kinase GnfK and the response regulator GnfR were tested for phosphotransfer activity in vitro. GnfK was incubated at a room temperature for 5 min for the autophosphorylation reaction. GnfR was then added and further incubated at a room temperature for 5 min for the phosphotransfer reaction. The sizes of molecular mass standards are shown in kilodaltons.
Homologues of GnfK and GnfR were found in G. metallireducens and G. uraniireducens as well as Pelobacter propionicus. Pelobacter species, which like Geobacter species belong to the δ-subdivision of Proteobacteria, are phylogenetically intertwined with Geobacter species (51,52). Therefore, Pelobacter species may also regulate nitrogen-fixation gene expression by the homologues of the two-component GnfK/GnfR system.
Physiological function of GnfK/GnfR
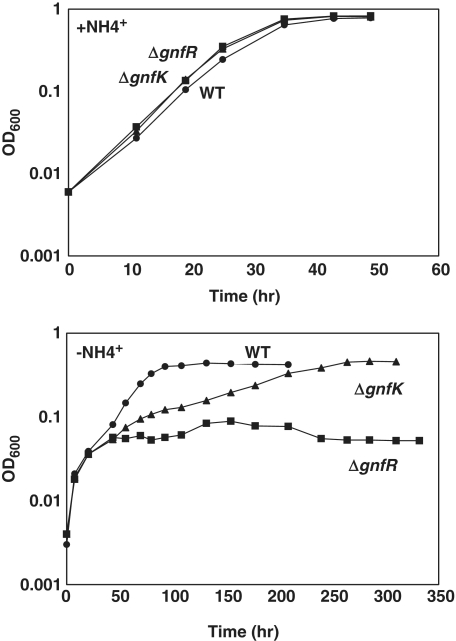
In order to assess the physiological function of the two-component GnfK/GnfR system, deletion mutants of gnfK or gnfR were constructed. The mutant cells grew as well as the wild-type cells when ammonium was provided (Figure 9). In contrast, the deletion mutants exhibited growth defects in the absence of ammonium (Figure 9). The mutants grew similarly with the wild-type cells until OD600 ≈ 0.04 after being transferred from the ammonium-containing medium to the ammonium-free medium, presumably by utilizing the ammonium transferred with the inoculum or stored inside the cells during growth in the ammonium-containing medium. Subsequent growth of the gnfK deletion mutant was slower than the wild-type, and the gnfR deletion mutant stopped growing. These results suggest that the GnfK/GnfR system plays an important role during nitrogen fixation in G. sulfurreducens.
Figure 9.
Growth under nitrogen-fixation condition. The wild-type (circles) and gnfK (triangles) or gnfR (squares) mutants were grown in the presence or absence of 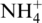 . Growth was monitored by measuring the optical density at 600 nm (OD600). Data are a representative of three replicate cultures.
. Growth was monitored by measuring the optical density at 600 nm (OD600). Data are a representative of three replicate cultures.
Premature transcription termination
An analysis of the amino acid sequence of GnfR suggested that GnfR is a transcription antiterminator, as it has an RNA-binding domain or the ANTAR domain (53) at the C-terminus (Supplementary Figure S3). Furthermore, an analysis of promoter regions of the genes up-regulated during nitrogen fixation identified transcription termination-like sequences in the promoter region of some of the genes such as nifH, nifEN, nifX, glnK and amtB (Figure 10A, B and Supplementary Figure S4), which are similar to the Rho-independent transcription termination signals that typically consist of a hairpin followed by a T-rich region (Figure 10A, B and C; 54). Therefore, it is likely that premature transcription termination and its antitermination are one of the regulatory mechanisms controlling a subset of the nitrogen-fixation genes that includes the genes regulated by the GnfL/GnfM two-component system and containing a transcription termination signal in their promoter regions (Figure 10A).
Figure 10.
Premature transcription termination in the nitrogen-fixation genes. (A) Alignment of transcription termination-like signals in the nitrogen-fixation genes. It is likely that the nifEN operon contains nifEN and nifX in this order, the Gsul_0938 homologue operon contains a gene encoding a hypothetical protein, glnK and amtB in this order, and the Gsul_2799 homologue operon contains genes encoding a putative radical SAM domain protein and a putative acetyltransferase. The inverted-repeat sequences are underlined. Highly conserved nucleotides are indicated in bold letters. (B) Promoter region of nifH. Predicted GnfM-binding sites (Supplementary Table S4) are highlighted in gray. Highly conserved dinucleotides GG and GC in RpoN-dependent −24/−12 promoter elements are underlined. Transcription initiation site is indicated by +1. Transcription termination-like signals are indicated by arrows. Translation initiation codon is indicated by Met. (C) Putative RNA secondary structure of nifH. The putative stem-loop structure was predicted on the basis of base-pairing. A predicted binding site for GnfR is indicated in bold letters. (D) lacZ fusion assay. The assays were conducted with artificial promoters. Diagrams of promoter constructs are shown (see Supplementary Figure S5A for sequences). RpoD, the RpoD-dependent like promoter; RpoD-TTS (WT), the RpoD-dependent like promoter with the wild-type transcription termination signals from nifH; RpoD-TTS (-up), the RpoD-dependent like promoter with the transcription termination signals lacking the upstream half of the stem structure; RpoD-TTS (-down), the RpoD-dependent like promoter with the transcription termination signals lacking the downstream half of the stem structure; RpoD-TTS (mut), the RpoD-dependent like promoter with the transcription termination signals mutated in the stem structure (Supplementary Figure S5A). Mutations in RpoD-TTS (mut) were made by changing GGGGCGCCTT to ACATACAACA. β-Galactosidase activity was examined with the X-gal plate (Supplementary Figure S5B).
To examine the function of these identified transcription termination signals, lacZ fusion assays were conducted with artificial promoters that contain an RpoD-dependent like −35/−10 elements with or without the wild-type or mutated sequences from the identified transcription termination signals in nifH (Figure 10D and Supplementary Figure S5). β-Galactosidase activity by the X-gal plate assays indicated that the RpoD promoter was active. In contrast, RpoD-TTS (WT) promoter in which the RpoD promoter was placed upstream of the wild-type transcription termination signals from nifH was inactive under the same condition. When the upstream or downstream half of the transcription termination signals from nifH was deleted and thus the stem structure was disrupted, RpoD-TTS (-up) and RpoD-TTS (-down) promoters became active. In addition, when the region predicted to form the stem structure was mutated and thus a stem structure was also disrupted, RpoD-TTS (mut) promoter was active as well. These results suggest that the identified sequences function as transcription termination signals.
Transcription antitermination
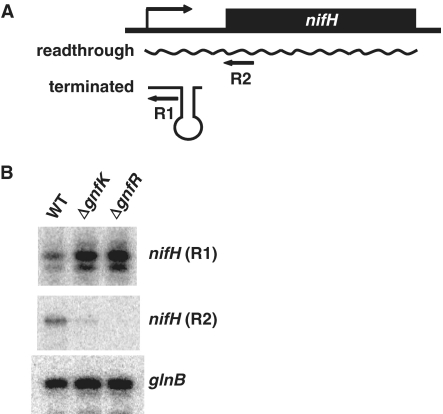
To assess whether or not the GnfK/GnfR system is involved in transcription antitermination during nitrogen fixation, transcripts of nifH, which has transcription termination signals, and of glnB, which does not, were examined by primer extension assays (Figure 11). Both nifH and glnB appear to be under the control of the RNA polymerase RpoN together with the master two-component GnfL/GnfM system and up-regulated during nitrogen fixation. For nifH transcripts, two different primers were used to distinguish prematurely terminated transcripts and readthrough transcripts (Figure 11A). Under nitrogen-fixing conditions, the transcripts of nifH were prematurely terminated in the mutant strains in which gnfK or gnfR was deleted, but not in the wild-type strain (Figure 11B). The expression of glnB was not affected by the deletion of gnfK or gnfR (Figure 11B). These results suggest that premature transcription termination and its antitermination are an important regulatory mechanism controlling the subset of the genes containing transcription termination signals in their promoter among the nitrogen-fixation genes regulated by GnfL/GnfM and that the two-component GnfK/GnfR system is involved in the antitermination of prematurely terminated transcription. It is also possible that the nifH mRNA is destabilized by the slow growth conditions imposed by the gnfK and gnfR deletions. It is noteworthy that primer extension products by the R1 primer increase in intensity in these mutants.
Figure 11.
Transcription antitermination. (A) Diagrams of two transcripts of nifH mRNA. The locations of regions where primers (R1 and R2) used in the primer extension assay (B) hybridize are presented by arrows. (B) Expression of nifH and glnB. Total RNA was prepared from the wild-type and gnfK or gnfR mutants after they were grown to the early log phase in the media containing 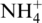 , harvested, resuspended and grown in the
, harvested, resuspended and grown in the 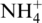 -free media. Equal amounts of total RNA for each strain were used to examine the expression of nifH and glnB by primer extension assays.
-free media. Equal amounts of total RNA for each strain were used to examine the expression of nifH and glnB by primer extension assays.
RNA binding of GnfR activated by GnfK
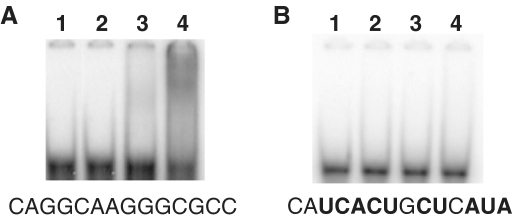
As the upstream regions of the sequences forming a hairpin in the premature transcription termination signals contain highly conserved sequences, CAGGCAANGGCGCC, in the subset of the nitrogen-fixation genes in G. sulfurreducens (Figure 10A), it is possible that these conserved sequences serve as a recognition site for GnfR to bind their mRNA and to inhibit the premature transcription termination of these genes. The results shown above suggest that the activity of GnfR is regulated by GnfK via phosphorylation. To test these, RNA-binding assays were conducted in the presence of ATP (Figure 12). GnfR bound the RNA fragment containing the predicted binding site, CAGGCAAGGGCGCC, from the nifH promoter in the presence of GnfK, whereas GnfR was unable to bind the same RNA fragment in the absence of GnfK (Figure 12A). In contrast, GnfR was unable to bind the RNA fragment lacking the predicted binding site (Figure 12B). These results indicate that GnfR is an RNA-binding protein and that its RNA-binding ability is activated by phosphorylation via GnfK.
Figure 12.
RNA binding of GnfR activated by GnfK. RNA-binding activity of GnfR was tested with RNA fragments containing the putative GnfR-binding site. (A) The wild-type RNA sequence. (B) A mutated RNA sequence. The RNA sequences of the putative GnfR-binding site are shown below the images of the gels. Mutated nucleotides are indicated with bold letters (see also Figure 10A). Lane 1, no protein; lane 2, GnfK; lane3, GnfR; lane 4, GnfR and GnfK.
DISCUSSION
Nitrogen fixation is a key physiological feature in Geobacter species during growth in subsurface environments and bioremediation of contaminated environments. The results presented in this study show that the majority of nitrogen-fixation genes appear to be controlled by the sigma factor RpoN and the two-component system consisting of the histidine kinase GnfL and the response regulator GnfM, which also belongs to the EBP family. Unlike most other bacterial rpoN genes, the G. sulfurreducens rpoN is essential under all conditions tested, including ammonium-sufficient conditions (47). In addition to nitrogen metabolism, RpoN controls genes involved in a wide range of cellular processes such as fumarate respiration, Fe(III) reduction, and pili and flagella biosyntheses in G. sulfurreducens. Although most of the up-regulated genes during nitrogen fixation appear to be controlled by RpoN in G. sulfurreducens, there are some genes and operons that are up-regulated during nitrogen fixation and appear to be controlled by other sigma factors. For instance, the operon for a putative molybdenum transporter is up-regulated during nitrogen fixation in G. sulfurreducens (15). Molybdenum is known to be an important co-factor for nitrogenase as well as other enzymes such as formate dehydrogenase. Genes encoding the molybdenum transporter are regulated by the molybdenum-responsive transcription factor ModE in other bacteria (55). This operon in G. sulfurreducens appears to lack an RpoN-dependent promoter and to be regulated by a homologue of ModE, which is encoded by the gene located immediately upstream of the operon on the G. sulfurreducens genome (56). In addition, several genes encoding a hypothetical protein are up-regulated during nitrogen fixation and appear to lack an RpoN-dependent promoter. Therefore, it may be possible that genes mainly involved in nitrogen metabolism such as nitrogen fixation genes and gdhA are controlled by both RpoN and the GnfL/GnfM system. In contrast, it is likely that genes required for other cellular processes in addition to nitrogen fixation such as molybdenum transporter genes are regulated by other sigma factors and transcription factors in G. sulfurreducens.
The expression of gdhA was shown to be activated by RpoN and the GnfL/GnfM system in the ammonium-sufficient conditions and repressed during nitrogen fixation in G. sulfurreducens. The repression of gdhA expression was observed in some bacteria (57,58). However, gdhA expression is controlled by the major sigma factor RpoD in other known bacteria (52). In Klebsiella aerogenes the NAC (nitrogen assimilation control) protein, which is a member of a LysR transcriptional regulator, represses transcription of the gdhA gene under nitrogen-limiting conditions (59). NtrC is known to act as both a repressor and an activator in other bacteria such as E. coli (60). Although gdhA homologues are conserved in Geobacter species and among the EBPs conserved in Geobacter species only GnfM bound the promoter region of the gdhA gene, which has an RpoN-dependent promoter, it is possible that another EBP is involved in the regulation of the gdhA gene. Furthermore, it is also possible that another transcription factor plays a role in modulating the activity of GnfM as a repressor and an activator. For instance, integration host factor (IHF) is known to be involved in the transcription mediated by the EBP such as NifA (61). Moreover, phosphorylation of GnfM by GnfL affects the activity of GnfM as a repressor and an activator. Although it is generally thought that phosphorylation is not required for the DNA-binding activity of the EBP family transcription factor (30–33), it has been shown that phosphorylation affects DNA-binding activity of NtrC (62,63). In addition, it is known that the expression of ompC and ompF, encoding outer membrane porins, is differentially regulated by the phosphorylation levels of the response regulator OmpR, which is not a member of the EBP family, in the E. coli osmoregulation system (64). Therefore, it is possible that during growth in the ammonium-sufficient conditions, under which the phosphorylated form of GnfM is less abundant, most of GnfM binds the upstream binding sites (BS1 and BS2) in the gdhA promoter that may have higher affinity with GnfM than the downstream binding sites in gdhA (BS3 and BS4) and the binding sites in the nitrogen-fixation genes, resulting in the activation of gdhA expression. In contrast, during growth dependent on nitrogen fixation, under which the phosphorylated form of GnfM is more abundant, GnfM may be able to bind the downstream binding sites in gdhA (BS3 and BS4) and the binding sites in the nitrogen-fixation genes, resulting in the repression of gdhA expression as well as the activation of the nitrogen-fixation gene expression. Known EBPs including NtrC bind sites located far upstream of an RpoN-dependent promoter when compared with other types of bacterial transcription factors that activate other sigma factors (30,31,33). However, GnfM-binding sites (BS1 and BS2) in the gdhA promoter that are essential for the activation of gdhA are located closer to the RpoN-dependent promoter than typical EBP-binding sites including the GnfM-binding sites in the gnfK promoter (Figure 4A and B). This difference may also contribute to different activation mechanisms in the expression of gnfK and gdhA.
Another two-component system GnfK/GnfR controls the subset of nitrogen-fixation genes by the transcription antitermination. Transcription of these genes is activated by the GnfL/GnfM system. Although antitermination is found in gene regulation during a variety of cellular processes (65), this is the first demonstration of the involvement of antitermination in nitrogen-fixation gene regulation. Genome sequence analysis revealed that the ANTAR domain is found in other response regulators of the two-component system. However, to our knowledge, GnfR is the first response regulator with the ANTAR domain to be characterized in detail. AmiR, an antiterminator involved in regulating the expression of the aliphatic amidase operon of Pseudomonas aeruginosa (66,67), has an N-terminal domain whose structure is similar to that of the receiver domain of the response regulator (68). However, AmiR is not likely to be a response regulator, because residues typically conserved in the receiver domain are absent from the N-terminal domain of AmiR, most notably a phosphorylation site. Another well characterized antiterminator, NasR, which has an NIT domain at the N-terminus, is involved in nitrate-responsive transcription antitermination of the nasF operon in Klebsiella oxytoca that is required for nitrate assimilation (69,70). The NIT domain is found in nitrate- and nitrite-sensing receptor components (71). The untranslated region or the leader region containing transcription termination signals has been shown to fold into two different secondary structures in many cases as shown for the NasR-mediated nitrate-responsive transcription antitermination (69,70). Binding of the antiterminator protein to the leader RNA inhibits formation of the transcription terminator structure. It is possible that GnfR functions as an antiterminator in a similar fashion during nitrogen fixation in G. sulfurreducens. GnfR inhibits formation of the transcription terminator structure by binding to the highly conserved sequences, CAGGCAANGGCGCC, located upstream of and at the region overlapping transcription termination signals in leader RNA of the subset of nitrogen-fixation genes.
Although GnfK has a typical histidine kinase domain, it does not have an apparent sensor domain. The kinase activity of the sensor histidine kinase of the two-component system is normally regulated by an environmental signal or stimulus, which is detected by the sensor domain of the histidine kinase (36,48). Therefore, it is likely that GnfK is constitutively active, as suggested by the in vitro phosphorylation assays, and that its expression is regulated at the level of transcription by the GnfL/GnfM system. Therefore, when expressed, GnfK might function as the activator for GnfR regardless of an environmental signal. It is, however, possible that an intracellular signal could regulate the activity of GnfK. For example, the activity of the histidine kinases is dependent on divalent cations (Ueki and Lovley, manuscript in preparation; 48). On the other hand, the response regulator GnfR appears to be regulated at levels of both the expression by transcription via the GnfL/GnfM system and the activity by phosphorylation via GnfK.
The phenotypes of mutants of GnfK and GnfR were somewhat different during growth dependent on nitrogen fixation. This difference may be explained by the fact that some response regulators have been shown to utilize small phosphate-containing molecules such as acetylphosphate as a phospho-donor in the absence of their cognate histidine kinases (72,73). Thus, GnfR might still be active with such small phosphate-containing molecules in the absence of GnfK, but not as efficient as with GnfK. Different phenotypes between the kinase mutant and the response regulator mutant have also been observed in other two-component systems. For instance, in the two-component RegB/RegA system in Rhodobacter capsulatus, disruption of regB encoding a kinase affected photosystem gene expression less dramatically than did disruption of regA encoding a response regulator (74).
In contrast to GnfK, GnfL, the histidine kinase of the master two-component system for the nitrogen-fixation gene regulation in G. sulfurreducens, has a PAS domain as the sensor at the N-terminus and shows some similarity to NtrB homologues (Supplementary Figure S2). The PAS domain is a signaling module known to sense light, oxygen, redox and small molecules (75,76). Oxygen is known to be an important regulatory factor for nitrogen-fixation gene expression due to the oxygen sensitivity of nitrogenase in diazotrophs. As Geobacter species are strictly anaerobic and oxygen has a negative impact on anaerobically growing Geobacter cells (77; Leang, personal communication), critical cellular processes are thought to be sensitive to oxygen in Geobacter species. The transcriptome analysis of G. uraniireducens growing in uranium-contaminated subsurface sediments showed higher levels of transcripts for genes for nitrogen fixation as well as oxidative stress response (13), suggesting that nitrogen fixation is among the critical cellular processes sensitive to oxygen in Geobacter species. NtrB homologues are known to be regulated by the PII regulatory protein GlnB in nitrogen regulation in some bacteria such as Escherichia coli and Klebsiella pneumoniae (16). Geobacter sulfurreducens contains homologues of the PII protein, and genes encoding GlnB and GlnK are up-regulated during nitrogen fixation (15). Therefore, it is possible that GnfL is regulated by GlnB as found in nitrogen regulation in other bacteria.
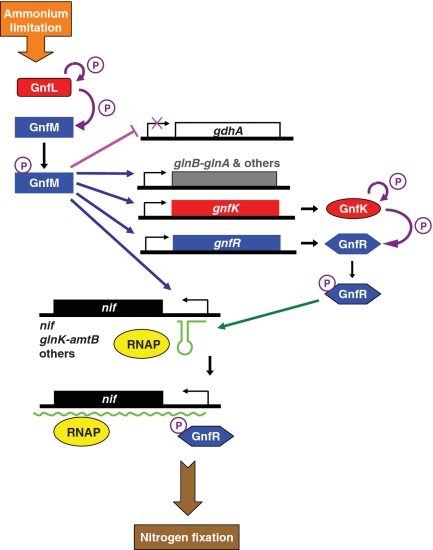
In summary, G. sulfurreducens appears to utilize novel regulatory mechanisms for the nitrogen-fixation gene expression (Figure 13). The master regulator, the two-component GnfL/GnfM system, appears to activate the transcription of the majority of the nitrogen-fixation genes and repress the transcription of gdhA encoding glutamate dehydrogenase under nitrogen-fixing conditions. In addition, it is likely that the GnfL/GnfM system activates the transcription of gdhA under ammonium-sufficient conditions. The other two-component system, GnfK/GnfR, whose expression is directly activated by the GnfL/GnfM system during nitrogen fixation, appears to control the subset of the nitrogen-fixation genes by the transcription antitermination mechanism. These genes contain transcription termination signals in their promoter and are regulated by the GnfL/GnfM system. These regulatory cascades appear to play a key role in fine-tuning the expression of the nitrogen-fixation genes in G. sulfurreducens to ensure an adequate response to nitrogen availability, as nitrogen fixation is a costly cellular process. These findings provide new insights into regulatory molecular mechanisms in nitrogen-fixation gene expression and important information on how Geobacter species respond to growth conditions in subsurface environments and during in situ bioremediation.
Figure 13.
Model for the regulation of gene expression during nitrogen fixation in G. sulfurreducens. Histidine kinases are presented in red. Response regulators are presented in blue. Phosphorylation is indicated in purple arrows. Transcription activation and repression are indicated in blue arrows and a pink line, respectively. Transcription antitermination is indicated by a green arrow. RNAP represents RNA polymerase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genomics: GTL program of the Office of Science (BER); US Department of Energy [DE-FC02-02ER63446]. Funding for open access charge: US Department of Energy.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank E. Blunt, J. Ward, and A. Murchie for technical support. They are grateful to R. Glaven, C. Leang, and M. Coppi for helpful discussion. They also thank T. Woodard for critical reading of this manuscript.
REFERENCES
- 1.Lovley DR. Microbial Fe(III) reduction in subsurface environments. FEMS Microbiol. Rev. 1997;20:305–313. [Google Scholar]
- 2.Lovley DR, Holmes DE, Nevin KP. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 2004;49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- 3.Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, Marutzky S, Metzler DR, Karp K, Lowe M, et al. Stimulated in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botton S, van Harmelen M, Braster M, Parsons JR, Röling WFM. Dominance of Geobacteraceae in BTX-degrading enrichments from an iron-reducing aquifer. FEMS Microbiol. Ecol. 2007;62:118–130. doi: 10.1111/j.1574-6941.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- 5.Holmes DE, O'Neil RA, Vrionis HA, N'Guessan LA, Ortiz-Bernad I, Larrahondo MJ, Adams LA, Ward JA, Nicoll JS, Nevin KP, et al. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 2007;1:663–677. doi: 10.1038/ismej.2007.85. [DOI] [PubMed] [Google Scholar]
- 6.Lovley DR. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 2003;1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- 7.Lovley DR, Baedecker MJ, Lonrgan DJ, Cozzarelli IM, Phillips EJP, Siegel DI. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature. 1989;339:297–299. [Google Scholar]
- 8.Lovley DR, Phillips EJ, Gorby YA, Landa ER. Microbial reduction of uranium. Nature. 1991;350:413–416. [Google Scholar]
- 9.Rooney-Varga JN, Anderson RT, Fraga JL, Ringelberg D, Lovley DR. Microbial communities associated with anaerobic benzene mineralization in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 1999;65:3056–3063. doi: 10.1128/aem.65.7.3056-3063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson RT, Lovley DR. Ecology and biogeochemistry of in situ groundwater bioremediation. Adv. Microbial. Ecol. 1997;15:289–350. [Google Scholar]
- 11.Aklujkar M, Krushkal J, Dibartolo G, Lapidus A, Land ML, Lovley DR. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 2009;9:109. doi: 10.1186/1471-2180-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazylinski DA, Dean AJ, Schüler D, Phillips EJ, Lovley DR. N2-dependent growth and nitrogenase activity in the metal-metabolizing bacteria, Geobacter and Magnetospirillum species. Environ. Microbiol. 2000;2:266–273. doi: 10.1046/j.1462-2920.2000.00096.x. [DOI] [PubMed] [Google Scholar]
- 13.Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 2001;67:3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes DE, Nevin KP, Lovley DR. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 2004;70:7251–7259. doi: 10.1128/AEM.70.12.7251-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Methé BA, Webster K, Nevin KP, Butler JE, Lovley DR. DNA Microarray Analysis of Nitrogen Fixation and Fe(III) Reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 2005;71:2530–2538. doi: 10.1128/AEM.71.5.2530-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes DE, O'Neil RA, Chavan MA, N'Guessan LA, Vrionis HA, Perpetua LA, Larrahondo MJ, Didonato R, Liu A, Lovley DR. Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J. 2009;3:216–230. doi: 10.1038/ismej.2008.89. [DOI] [PubMed] [Google Scholar]
- 17.Mouser PJ, N’Guessan AL, Elifantz H, Holmes DE, Williams KH, Wilkins MJ, Long PE, Lovley DR. Influence of heterogeneous ammonium availability on bacterial community structure. Environ. Sci. Technol. 2009;43:4386–4392. doi: 10.1021/es8031055. [DOI] [PubMed] [Google Scholar]
- 18.Lovley DR. Microbial fuel cells: novel microbial physiologies and engineering approaches. Curr. Opin. Biotechnol. 2006;17:327–332. doi: 10.1016/j.copbio.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Lovley DR. Bug juice: harvesting electricity with microorganisms. Nat. Rev. Microbiol. 2006;4:497–508. doi: 10.1038/nrmicro1442. [DOI] [PubMed] [Google Scholar]
- 20.Lovley DR. The microbe electric: conversion of organic matter to electricity. Curr. Opin. Biotechnol. 2008;19:564–571. doi: 10.1016/j.copbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Bond DR, Holmes DE, Tender LM, Lovley DR. Electrode-reducing microorganisms harvesting energy from marine sediments. Science. 2002;295:483–485. doi: 10.1126/science.1066771. [DOI] [PubMed] [Google Scholar]
- 22.Holmes DE, Bond DR, O'Neil RA, Reimers CE, Tender LR, Lovley DR. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb. Ecol. 2004;48:178–190. doi: 10.1007/s00248-003-0004-4. [DOI] [PubMed] [Google Scholar]
- 23.Ishii S, Watanabe K, Yabuki S, Logan BE, Sekiguchi Y. Bacterial electrode reducing activities of Geobacter sulfurreducens compared to an enriched consortium in an air-cathode microbial fuel cell. Appl. Environ. Microbiol. 2008;74:7345–7355. doi: 10.1128/AEM.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung S, Regan JM. Comparison of anode bacterial communities and performance in microbial fuel cells with different electron donors. Appl. Microbiol. Biotechnol. 2007;77:393–402. doi: 10.1007/s00253-007-1162-y. [DOI] [PubMed] [Google Scholar]
- 25.Ren Z, Steinberg LM, Regan JM. Electricity production and microbial biofilm characterization in cellulose-fed microbial fuel cells. Water Sci. Technol. 2008;58:617–622. doi: 10.2166/wst.2008.431. [DOI] [PubMed] [Google Scholar]
- 26.Nevin KP, Richter H, Covalla SF, Johnson JP, Woodard TL, Orloff AL, Jia H, Zhang M, Lovley DR. Power output and columbic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ. Microbiol. 2008;10:2505–2514. doi: 10.1111/j.1462-2920.2008.01675.x. [DOI] [PubMed] [Google Scholar]
- 27.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 2006;72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. The NifL-NifA System: a multidomain transcriptional regulatory complex that integrates environmental signals. J. Bacteriol. 2004;186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck M, Bose D, Burrows P, Cannon W, Joly N, Pape T, Rappas M, Schumacher J, Wigneshweraraj S, Zhang X. A second paradigm for gene activation in bacteria. Biochem. Soc. Trans. 2006;34:1067–1071. doi: 10.1042/BST0341067. [DOI] [PubMed] [Google Scholar]
- 31.Rappas M, Bose D, Zhang X. Bacterial enhancer-binding proteins: unlocking sigma54-dependent gene transcription. Curr. Opin. Struct. Biol. 2007;17:110–116. doi: 10.1016/j.sbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Studholme DJ, Dixon R. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Hoover TR. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 2001;4:138–144. doi: 10.1016/s1369-5274(00)00179-x. [DOI] [PubMed] [Google Scholar]
- 34.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 35.Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 36.Hoch JA, Silhavy TJ. Two-component Signal Transduction. Washington, DC: ASM Press; 1995. [Google Scholar]
- 37.Caccavo F, Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. Nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 1994;60:3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 39.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 40.Ueki T, Inouye S. Transcriptional activation of a heat-shock gene, lonD, of Myxococcus xanthus by a two-component histidine-aspartate phosphorelay system. J. Biol. Chem. 2002;277:6170–6177. doi: 10.1074/jbc.M110155200. [DOI] [PubMed] [Google Scholar]
- 41.Ueki T, Inouye S. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J. Biol. Chem. 2005;280:32279–32284. doi: 10.1074/jbc.M507191200. [DOI] [PubMed] [Google Scholar]
- 42.Ueki T, Lovley DR. Genome-wide gene regulation of biosynthesis and energy generation by a novel transcriptional repressor in Geobacter species. Nucleic Acids Res. 2010;38:810–821. doi: 10.1093/nar/gkp1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehio M, Knorre A, Lanz C, Dehio C. Construction of versatile high-level expression vectors for Bartonella henselae and the use of green fluorescent protein as a new expression marker. Gene. 1998;215:223–229. doi: 10.1016/s0378-1119(98)00319-9. [DOI] [PubMed] [Google Scholar]
- 44.Kovach ME, Phillips RW, Elzer PH, Roop RMII, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 45.Barrios H, Valderrama B, Morett E. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studholme DJ, Buck M, Nixon T. Identification of potential sigmaN-dependent promoters in bacterial genomes. Microbiology. 2000;146:3021–3023. doi: 10.1099/00221287-146-12-3021. [DOI] [PubMed] [Google Scholar]
- 47.Leang C, Krushkal J, Ueki T, Puljic M, Sun J, Juárez K, Núñez C, Reguera G, DiDonato R, Postier B, et al. Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens. BMC Genomics. 2009;10:331. doi: 10.1186/1471-2164-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inouye M, Dutta R. Histidine Kinases in Signal Transduction. New York: Academic Press; 2003. [Google Scholar]
- 49.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lonergan DJ, Jenter HL, Coates JD, Phillips EJP, Schmidt TM, Lovley DR. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovley DR, Phillips EJ, Lonergan DJ, Widma PK. Fe(III) and S0 reduction by Pelobacter carbinolicus. Appl. Environ. Microbiol. 1995;61:2132–2138. doi: 10.1128/aem.61.6.2132-2138.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 54.Farnham PJ, Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981;9:563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Studholme DJ, Pau RN. A DNA element recognized by the molybdenum-responsive transcription factor ModE is conserved in Proteobacteria, green sulphur bacteria and Archaea. BMC Microbiol. 2003;3:24. doi: 10.1186/1471-2180-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodionov DA, Dubchak I, Arkin A, Alm E, Gelfand MS. Reconstruction of regulatory and metabolic pathways in metal-reducing δ-proteobacteria. Genome Biol. 2004;5:R90. doi: 10.1186/gb-2004-5-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol. Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reittzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 59.Bender RA. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol. Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 60.Reitzer LJ, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc. Natl Acad. Sci. USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoover TR, Santero E, Porter S, Kustu S. The integration host factor stimulates interaction of RNA polymerase with NifA, the transcriptional activator for nitrogen fixation operons. Cell. 1990;5:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 62.Chen P, Reitzer LJ. Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli: stimulation by phosphorylation and the binding of ATP. J. Bacteriol. 1995;177:2490–2496. doi: 10.1128/jb.177.9.2490-2496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minchin SD, Austin S, Dixon RA. The role of activator binding sites in transcriptional control of the divergently transcribed nifF and nifLA promoters from Klebsiella pneumoniae. Mol. Microbiol. 1988;2:433–442. doi: 10.1111/j.1365-2958.1988.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 64.Egger LA, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 65.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 66.Drew R, Lowe N. Positive control of Pseudomonas aeruginosa amidase synthesis is mediated by a transcription anti-termination mechanism. J. Gen. Microbiol. 1989;135:817–823. doi: 10.1099/00221287-135-4-817. [DOI] [PubMed] [Google Scholar]
- 67.Wilson SA, Wachira SJ, Drew RE, Jones D, Pearl LH. Antitermination of amidase expression in Pseudomonas aeruginosa is controlled by a novel cytoplasmic amide-binding protein. EMBO J. 1993;12:3637–3642. doi: 10.1002/j.1460-2075.1993.tb06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Hara BP, Norman RA, Wan PTC, Roe SM, Barrett TE, Drew RE, Pearl LH. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 1999;18:5175–5186. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chai W, Stewart V. NasR, a novel RNA-binding protein, mediates nitrateresponsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader in vitro. J. Mol. Biol. 1998;283:339–351. doi: 10.1006/jmbi.1998.2105. [DOI] [PubMed] [Google Scholar]
- 70.Chai W, Stewart V. RNA Sequence Requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J. Mol. Biol. 1999;292:203–216. doi: 10.1006/jmbi.1999.3084. [DOI] [PubMed] [Google Scholar]
- 71.Shu CJ, Ulrich LE, Zhulin IB. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 2003;28:121–124. doi: 10.1016/S0968-0004(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 72.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl Acad. Sci. USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCleary WR, Stock JB, Ninfa AJ. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elsen S, Swem LR, Swem DL, Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 2004;68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 2007;65:1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mouser PJ, Holmes DE, Perpetua LA, Didonato R, Postier B, Liu A, Lovley DR. Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. ISME J. 2009;3:454–465. doi: 10.1038/ismej.2008.126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.