Abstract
Mitochondrial genome sequences are important markers for phylogenetics but taxon sampling remains sporadic because of the great effort and cost required to acquire full-length sequences. Here, we demonstrate a simple, cost-effective way to sequence the full complement of protein coding mitochondrial genes from pooled samples using the 454/Roche platform. Multiplexing was achieved without the need for expensive indexing tags (‘barcodes’). The method was trialled with a set of long-range polymerase chain reaction (PCR) fragments from 30 species of Coleoptera (beetles) sequenced in a 1/16th sector of a sequencing plate. Long contigs were produced from the pooled sequences with sequencing depths ranging from ∼10 to 100× per contig. Species identity of individual contigs was established via three ‘bait’ sequences matching disparate parts of the mitochondrial genome obtained by conventional PCR and Sanger sequencing. This proved that assembly of contigs from the sequencing pool was correct. Our study produced sequences for 21 nearly complete and seven partial sets of protein coding mitochondrial genes. Combined with existing sequences for 25 taxa, an improved estimate of basal relationships in Coleoptera was obtained. The procedure could be employed routinely for mitochondrial genome sequencing at the species level, to provide improved species ‘barcodes’ that currently use the cox1 gene only.
INTRODUCTION
Next-generation sequencing (NGS) technologies allow considerably greater numbers of nucleotides to be characterized, from any given DNA sample, when compared with conventional approaches (1,2). However, in light of the number of base pairs usually needed to establish phylogenetic relationships in molecular systematics, which only requires a limited number of markers, the output from a single sequencing reaction greatly exceeds the requirements for a single taxon. In contrast, phylogenomic studies thrive on the depth and breadth of gene sequencing, but glean their data from genomic and transcriptomic approaches currently at the expense of sufficiently dense taxon sampling for most taxonomic groups (3). The application of NGS techniques in phylogenetics has the potential for reducing stochastic errors, by adding more genes (4), but has yet to provide the means of reducing systematic errors, by adding significantly more taxa for sequencing single or multiple markers routinely. Therefore, an important remaining challenge for effectively exploiting NGS in molecular systematics is to develop protocols for simultaneous analysis of multiple individuals which can be sequenced for specified portions of the genomes (5). This may be achieved in pooled samples, but presents the problem that the resulting mixed sequence information has to be related back to specific individuals in the pool of samples.
Various approaches currently exist to increase the number of samples that are analyzed in a single sequencing run. This includes the subdivision of the sequencing device, e.g. the physical separation of samples in the Roche/454 technology into up to 16 lanes (6). In addition, the samples can be labelled by ligating short indexing tag (‘barcode’) sequences prior to the sequencing run that assign each sequence read to a particular sample present in the sequenced pool (7,8). These techniques can greatly increase the number of taxa separable in a sequencing run, but they are costly because of the need to produce individual libraries for each sample prior to the pyrosequencing step, i.e. they scale linearly with the number of taxa. This greatly limits the use of ‘barcodes’ for multiplexing, except in cases of amplicon sequencing of short polymerase chain reaction (PCR) fragments (9) where the barcode tag can be fused to the primer sequence (10), some of which are commercially available as ‘multiplex identifiers’ (MIDs) for both the 454/Roche and Illumina platforms. The latter approach of amplicon sequencing is attractive, e.g. for the sequencing of mtDNA including the cox1 sequence that is frequently used as a taxonomic ‘barcode’ (11) in species identification, but it is currently limited to amplicons of <800 bp which is generally considered insufficient for phylogenetics.
A strong phylogenetic signal is usually obtained when mtDNA sequences from multiple protein-coding genes are combined. Whole mitochondrial genome sequences have been shown to resolve deep-level relationships in many groups, including insects (12–15), and their power is expected to increase with greater density of taxon sampling as widely shown for single mtDNA markers (16). Therefore, the acquisition of large numbers of whole-mt genome sequences is desirable, but sequencing still requires considerable effort. A key step is the long-range PCR amplification of large portions of the mitochondrial genomes, followed by either conventional shotgun sequencing or pyrosequencing of individual genomes. The latter has been shown to generate sequences of high quality (17), but remains costly even when sequencing plates are subdivided, while the minimum numbers of sequences obtained per genome greatly exceeds the depth of coverage needed.
Here, we trial a sequencing protocol for mixed samples of mt genomes, which could reduce the number of sequences per taxon to a level of coverage that provides a better balance of cost and data quality. Pooling samples in a single sequencing run bears the obvious risk that reads from different taxa cannot be separated by the assembler software, or that individual taxa are mis-assembled from the pool of sequence reads to create chimerical or mosaic contigs. In addition, the approach requires that each contig can be associated with a particular individual present in the pool which may be performed through similarity searches against known partial sequences or through combinatorics (18,19) in multi-sample designs. None of these steps have been explored carefully for the sequencing of whole mitochondrial genomes.
We use the Coleoptera (beetles) to trial this approach. Currently (April 2010), mitochondrial genomes for 25 species are available at GenBank, and various attempts have been made recently to establish their phylogenetic relationships (20–29). While these studies were not fully conclusive, and leave out the vast majority of the 168 currently recognized families and several of the 18 superfamilies (30), these analyses also demonstrate that mt genomes are highly appropriate to solve the elusive basal relationships in Coleoptera. These existing sequences were used for the design of universal primers for long-range PCR, followed by multiplex sequencing. A sample of 30 species representing a wide range of families of Coleoptera was included in this study and demonstrates a high success rate of obtaining complete mt genomes even when sequenced in a small (1/16th) sector of a Roche/454 sequencer.
MATERIAL AND METHODS
DNA extraction, PCR amplification and 454 pyrosequencing
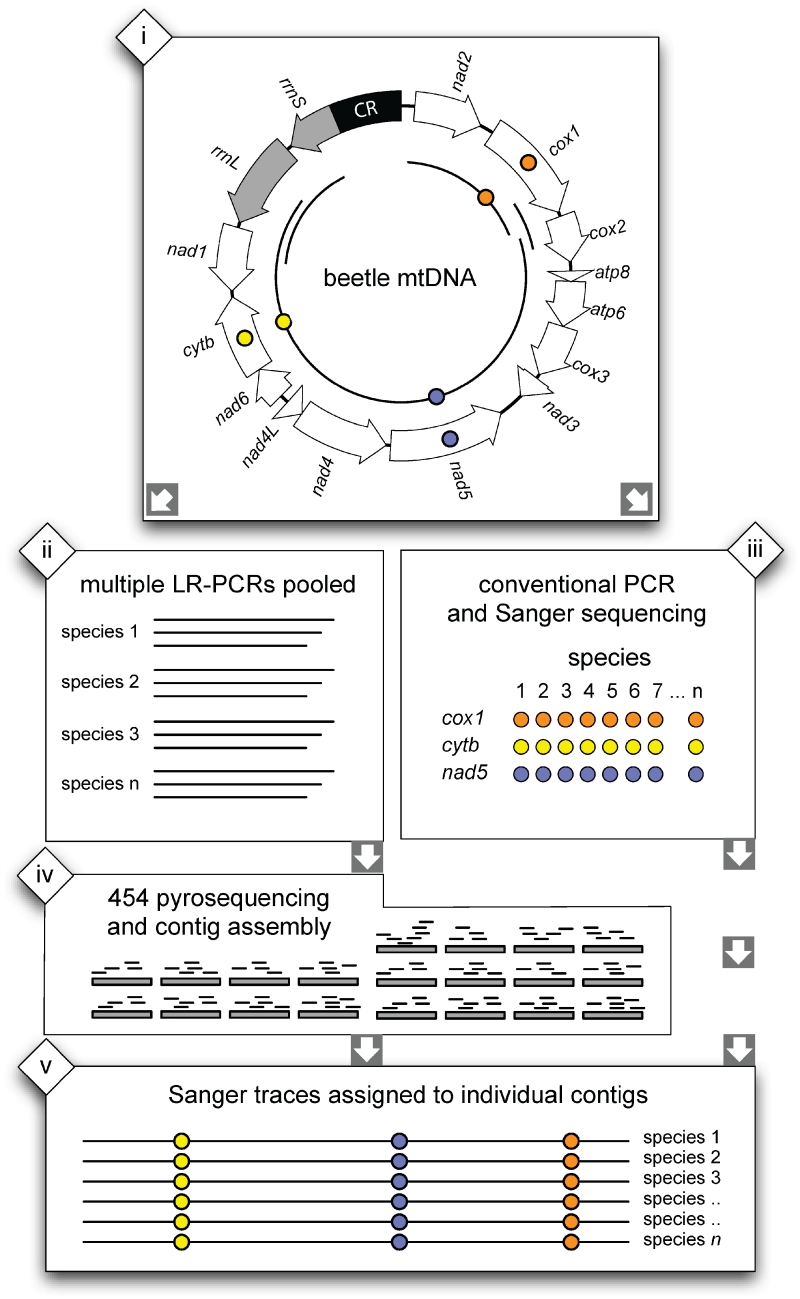
Individual beetles freshly preserved in 100% Ethanol were subjected to DNA extraction from whole or partial specimens (depending on body size) with the Qiagen DNeasy Blood and Tissue kit using spin columns. Universal PCR primers were designed based on conserved regions of available mt genome sequences of Coleoptera. All newly designed primers and PCR reaction conditions are given in Supplementary Table S1. These universal primers had a very high amplification success across all groups of Coleoptera and greatly improved PCR on existing primers, including the Pat and Jerry primers that have been widely used in beetle phylogenetics (31). A single fragment of >10 kb (3′cox1 to rrnL) amplified the bulk of the mitochondrial genome using long-range PCR amplification (Supplementary Table S2). Three further fragments were generated for amplification of regions encoding: trnM-nad2-5′cox1; cox1-cox2; and nad1-rrnS (Figure 1).
Figure 1.
Cartoon illustrating the sequencing strategy. Long-range PCR is performed, together with PCR of shorter fragments to cover all protein-coding regions (i). PCR products are pooled (ii) for 454 sequencing and contig production (iv), while in parallel conventional PCR and sequencing is performed to obtain three bait sequences (iii). In the final step the bait sequences are used to identify the respective contigs in the pool (v).
Prior to 454 pyrosequencing, amplification products obtained for individual taxa were pooled into three groups of 10 based on similar amounts of DNA (estimated from intensity of bands on an agarose gel) for each individual and four PCR fragments. This resulted in a total of 10 pools which were each purified using the QIAquick PCR Purification Kit (Qiagen). After elution the concentration of each sample was measured with a ND-1000 spectrometer (Nanodrop technologies) and equimolar amounts combined for a final pool. The mixed sample was used for library construction and pyrosequencing on two 1/16th lanes of a 454 GS FLX Titanium PicoTiterPlate following standard procedures (University of Cambridge, Department of Biochemistry).
Bioinformatics
Sff-files were pre-processed (e.g. adaptors and low quality regions were identified and trimmed), and primer sequences were masked using the program cross_match (-minmatch 10; -minscore 15) (P. Green, University of Washington; www.phrap.org). The resulting Fasta files and accompanying quality data were read into the program MIRA [Mimicking Intelligent Read Assembly; (32)] for contig generation and separation of mixed samples into individual mitogenomes. MIRA used the denovo and ‘accurate’ settings, and assumed non-uniform read distribution (-AS:urd=no). Post-MIRA joining of contigs (secondary assembly) was performed using the Phrap software package (www.phrap.org) with stringent settings to avoid mis-assembly (-forcelevel 0).
To test the validity of the assembly method from pooled samples, we used MetaSim (33) on the 25 available beetle mitochondrial genomes to simulate a Roche 454 sequence run on these taxa. A set of 30 000 reads (corresponding to the output from a single 1/16th lane of the 454 sequencer) was generated from the mixed multi-taxon data set using default settings of the simulator a mean fragment length of 284 bp (±30 bp). These fragments were subsequently assembled using the MIRA software package and the resulting assembly products were compared to the GenBank entries from which they were obtained initially. All 25 genomes were recovered without the formation of chimerae, and in the correct gene order in all cases. Differences between the original and simulated mt genome sequences affected less than 0.1% of base pairs. In addition, small indels resulted in length variation, in particular in the d-loop region where in two cases small repeats were absent in our assembly (Supplementary Table S3). Minor discrepancies also result from the way the data are simulated for sequences at both ends of the (linearized) mt genome. The average length of the resulting contigs was slightly shorter than the original data (15 679 bp instead of 15 959 bp). The simulated data also include two species belonging to the same genus Rhagophthalmus with a patristic distance of P = 0.109 for the whole fragment or P = 0.086 if a highly variable portion of ∼1000 bp is removed. Their correct assembly shows that mixed sequence pools may include closely related species.
Our scheme for analyzing pooled samples further involved conventional Sanger sequencing (ABI BigDye technology) of three short-sequence fragments for each of the 30 taxa included in the pool. These were used to validate the correct assembly of contigs and to assign the individual contigs to the taxa present in the pool (Figure 1). We obtained portions of the nad5, cox1 and cob genes whose locations are dispersed over the mt genome by PCR amplification with newly designed primers (Supplementary Table S1). These sequences were used as ‘baits’ in queries against the mitogenome contigs using the BLAST algorithm (blastn). Only contigs to which at least one Sanger sequence could be assigned with certainty were retained for further analysis. In a few cases overlapping contigs assigned to a single species were obtained and assembled by hand. Next, the contigs obtained from the combined datasets were put in the same orientation using a custom Perl script, aligned with MAFFT 4.0 (34), and annotated using the DOGMA (35) webserver. Subsequently, contigs were aligned to the gene sequences of the 25 Coleoptera mitogenomes available on GenBank. Protein coding genes for each species were excised from the alignments and inspected for frameshifts and stop codons generating conceptual translations with TransAlign (36). Each gene was individually aligned on the protein level with ClustalW and the resulting alignments were used to create nucleotide alignments in triplets matching the amino acids. Inconsistencies were edited after inspection of the raw assembly data in Eagleview (37) or Tablet (38). A small number of unresolved base calls were trimmed or masked. The newly generated sequences were submitted to GenBank (Acc. No. HQ232800-HQ232827 and HM486073).
Phylogenetic analyses
Alignments on the nucleotide and amino-acid level from individual genes were concatenated into a combined data set for phylogenetic analyses. The choice of tree building methods took into account the rate heterogeneity and compositional bias in particular in 3rd codon positions, broadly following methods described in Pons et al. (2010). That study showed the best partitioning strategy for concatenated mitochondrial protein coding genes in beetles is by codon positions (1st: 2nd: 3rd). Hence concatenated sequences were either split into three codon partitions, or into two partitions whereby 3rd positions were removed and 1st positions were RY coded. The latter reduces compositional biases and homoplastic substitutions (39).
Tree searches with Bayesian methods were conducted with the parallel version of MrBayes 3.1.2 (40) performing two independent searches of four chains. Each of these used the default priors starting with random trees and running three heated and one cold Markov chains for 2 million generations sampled at intervals of 1000 generations. The convergence of parameters was assessed with Tracer 1.4 (41) and the convergence of posterior clade probabilities with AWTY (42). After discarding burn-in samples, trees from the two independent runs were combined in a single majority consensus topology using the sumt command in MrBayes and the frequencies of the nodes in a majority rule tree were taken as a posteriori probabilities (40). The same partition strategies were used to analyze the data under the maximum likelihood criterion in RAxML 7.2.6 (43). The best evolutionary model for each partition was selected by jModelTest (44) under the Bayesian Information Criterion. The results suggested an independent GTR+I+G model and nucleotide composition for each partition except for the RY recoded 1st codon sites for which a F81+I+G model was implemented.
Bayesian analyses were also conducted at both the nucleotide and the amino-acid level using the CAT model in PhyloBayes (45). This model estimates the distribution of site-specific effects underlying each data set by combining infinite K categories of site-specific rates (46) and site-specific profiles over the 4 nucleotide frequencies or the 20 amino acid frequencies (47). The global rates of nucleotide variation were inferred from the data (CAT–GTR settings). The convergence of split frequencies was assessed with the ‘bpcomp’ command and effective sample size for all parameters with the ‘tracecomp’ command. The first 1000 samples were discarded as burn-in and then sampled every 10 generations. Independent runs were considered to have converged when the maximum split frequency was <0.1 and effective sample size was >100 (45).
RESULTS
Sequence analysis
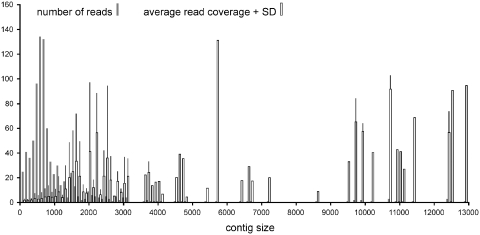
The long-range and short-range PCR products of 30 species of Coleoptera (Table 1) were pooled in roughly equimolar concentrations for construction of a single library, to be run on two lanes (1/16th) of the 454 sequencer. In total 71 647 pyrosequencing reads were generated (33 848 for lane 1; 37 799 for lane 2), with an average length of 280 bp (SD: 168) for lane 1 and 374 bp (SD: 167) for lane 2. Three separate sequence assemblies were carried out using MIRA for lane 1, lane 2 and the combined data set. They resulted in 452, 509 and 829 contigs, respectively. However, most of these contigs were shorter than 1000 bp and showed a low average read coverage (Figure 2). Average depth of coverage was higher for longer contigs, with a maximum of 155 for a contig of 1920 bp in the combined analysis. The largest contigs covering the bulk of the mt genome sequences (below) had a coverage in the range of 10–100× or more (Figure 2).
Table 1.
Specimens used in this study
| BMNH | Family | Species | Country | State/more for place | Place | Collection date |
|---|---|---|---|---|---|---|
| 840125 | Hydraenidae | Hydraena sp. | South Africa | Western Cape | Bergrivier | 11/11/09 |
| 840179 | Meloidae | Meloidae gen. sp. | South Africa | Western Cape | Kirstenbosch | 14/11/09 |
| 840193 | Hydrochidae | Hydrochus sp. | South Africa | Western Cape | Bergrivier | 11/11/09 |
| 840194 | Carabidae | Trechini gen. sp. | South Africa | Western Cape | Bergrivier | 11/11/09 |
| 840198 | Phalacridae | Phalacridae gen. sp. | South Africa | Western Cape | 10 km S. Gordon's Bay | 6/11/09 |
| 840202 | Cerambycidae | Clostomerus claviger | South Africa | Western Cape | Bontebok N.P. | 9/11/09 |
| 840203 | Tenebrionidae | Eutrapela ruficollis | South Africa | Western Cape | Bontebok N.P. | 9/11/09 |
| 840206 | Scraptiidae | Anaspis sp. | South Africa | Western Cape | De Hoop N.R. | 31/10/09 |
| 840207 | Megalopodidae | Zeugophora sp. | South Africa | Western Cape | De Hoop N.R. | 31/10/09 |
| 840208 | Dermestidae | Anthrenus sp. | South Africa | Western Cape | De Hoop N.R. | 31/10/09 |
| 840215 | Chrysomelidae | Peltoptera acromnalis | South Africa | Western Cape | Hoogekraalpas | 7/11/09 |
| 840216 | Elmidae | Elmidae gen. sp. | South Africa | Western Cape | Hoogekraalpas | 7/11/09 |
| 840449 | Heteroceridae | Heterocerus fenestratus | Slovakia | Stúrovo | Kamenica n. H. | 30/4/04 |
| 840452 | Eulichadidae | Eulichas sp. | Philippines | Mindanao | Mt. Apo. | 22/01/07 |
| 840454 | Eucnemidae | Melasis buprestoides | Czech Republic | Olomouc Distr. | Velký Týnec | 9/5/03 |
| 840457 | Lycidae | Merolycus dentipes | South Africa | Eastern Cape | Silaka Nature Reserve | 11/12/07 |
| 840459 | Drilidae | Drilus flavescens | Malta | Weid Babu | – | 4/4/04 |
| 840462 | Lampyridae | Drilaster sp. | Japan | Osaka Pref. | Iwakiyama | 16/6/02 |
| 840465 | Cantharidae | Cantharis pellucida | Czech Republic | Olomouc Distr. | Velký Týnec | 8/5/01 |
| 840466 | Nosodendridae | Nosodendron sp. | Indonesia | Sumatera Jambi Prov. | Gn. Kerinci | 22/01/05 |
| 840469 | Anobiidae | Ptinus rufipes | Czech Republic | Olomouc Distr. | Naklo | Mar-05 |
| 840470 | Lymexylonidae | Hyloecetus dermestoides | Czech Republic | Zlin Distr. | Valašské Klobouky | 14/5/03 |
| 840476 | Cerylonidae | Cerylon histeroides | Slovakia | Stúrovo | Hegyfarok | 30/4/04 |
| 840477 | Byturidae | Byturus ochraceus | Slovakia | Stúrovo | Hegyfarok | 30/4/04 |
| 840479 | Erotylidae | Tritoma bipustulata | Czech Republic | Moravia | Břeclav | 10/8/04 |
| 840483 | Boridae | Boros schneideri | Slovakia | Badin | – | Nov-09 |
| 840485 | Mycetohagidae | Mycetophagus quadripustulatus | Slovakia | Biescady Nat. Park | Nova Sedlica | 6/07/05 |
| 840487 | Anthicidae | Omonadus floralis | Czech Republic | Olomouc Distr. | Moravičany | 7/6/05 |
| 840491 | Oedemeridae | Oedemera virescens | Czech Republic | Olomouc Distr. | Mladeč | 30/4/05 |
| 840493 | Pyrochroidae | Ischalia sp. | N. Laos | Oudom Xai | 2002 |
Figure 2.
Number of reads and average sequence coverage of contigs in various size classes. The light bars represent the mean number of sequence reads covering each nucleotide. The dark bars show the number of reads in a given size class. Note the very large number of short contigs, while there are very few long contigs in a given size class mostly representing a single species.
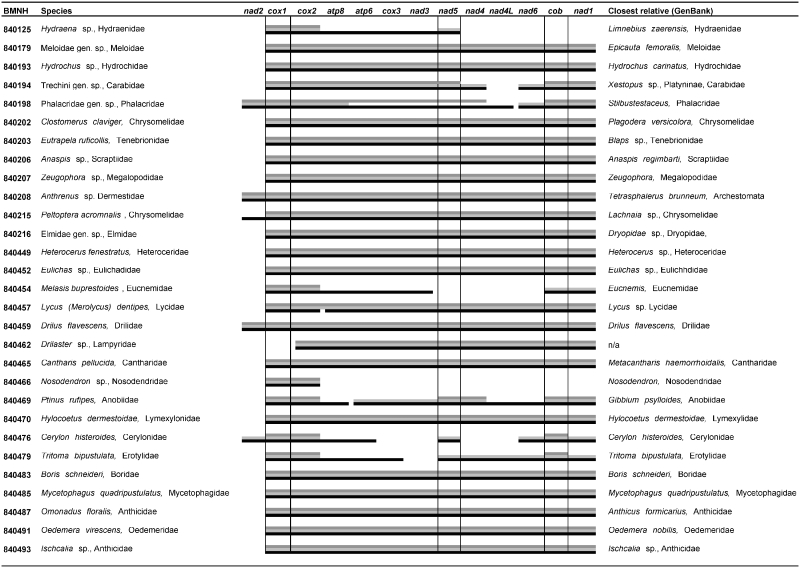
The MIRA contigs were parsed through an additional round of assembly using Phrap to combine the initial contigs into larger segments. These secondary contigs were assigned a species identity using cox1, nad5 and cob bait sequences obtained independently for the same specimens with conventional Sanger sequencing (Figure 1). The average length of contigs retained at this step was 7031 bp (SD: 4661), 7573 bp (SD: 4650) and 7366 bp (SD: 4562) for the three assemblies from lane 1, lane 2 and both lanes combined. Contigs covering the full-length cox1 to nad1 fragment were retrieved for 20 species that had matches to all three bait sequences in at least one of the three assemblies. In addition, 6 and 10 contigs with two and one bait matches were obtained, resulting in nearly complete mitogenomes for three additional species (see Figure 3 for details). Where multiple baits could be assigned to a particular contig, these consistently were of the same species, as hoped, showing that reads from mixed samples were correctly assembled into individual mitogenomes. However, there were two exceptions; in lane 2, a contig of 12 131 bp not only matched three baits belonging to Boridae (BMNH840483), but also contained the cox1 bait of Erotylidae (BMNH840479). Visual inspection of the assembly data showed MIRA attached a 1483-bp fragment containing cox1 and cox2 to the rrnL side of the contig. This erroneous connection appeared to be caused by two chimerical amplification products containing parts of the physically distant rrnL and cox2 genes. A second mis-assembly was observed in the combined data set where a fragment allocated to species BMNH840487 was joined with an unassigned contig containing three genes (cox3-atp6-cox2) by Phrap, forming a fragment of 14 604 bp. This mis-assembly was not detected by incongruous bait sequences but in the annotation of gene orders using DOGMA. In both cases, mis-assembly was easily resolved, and showed the gene order of contigs to be conserved and identical to that reported for other Coleoptera.
Figure 3.
Extent of contigs produced by lane 1 (dark grey bars, top), lane 2 (light grey bars, middle) and both lanes combined (black bars, bottom). Also given is the species identification and the closest relative in phylogenetic analysis against the 3′ half of cox1 of ∼5000 species of Coleoptera.
However, DOGMA analysis also revealed one contig with deviant gene order, in a specimen BMNH840477 (Byturus ochraceus). Closer investigation of this contig of 6381 bp showed that, in addition to a deviant gene order, this contig also contained a non-mitochondrial gene fragment (Wolbachia baseplate assembly protein J: BAP) with a gene order: cob-nad1-BAP-partial rrnL-nad5-nad4. This incorrect contig most likely arose from PCR artefacts, as inspection of the assembly data (ACE file) suggests the errors not to be caused in the sequence assembly process. This was supported by partial Sanger sequencing (data not shown) and by the fact that most PCR fragments generated for this species deviated from the expected length when visualised on agarose gels. The contig and its assigned species were excluded from further analyses.
Although the majority of mitogenomes could be assembled using data generated on a single 1/16th lane, several partially assembled fragments were joined only after increasing the number of reads by combining the data from a second lane (Figure 3). The same holds for the nad2 fragment which was assembled successfully in only 5 out of 30 cases. Inspection of the raw data set revealed that nad2 fragments were massively underrepresented. Using nad2 sequences from 25 Coleoptera mitogenomes as query, only ∼350 reads on average were found to be present in the combined datasets with the BLAST algorithm (e-value <0.005), which is only ∼0.5% of the reads generated. Failures of complete assembly are therefore likely the result of concentration effects, probably due to differences in the proportion of various PCR fragments used in construction of the pooled library.
Species assignment of the mixed 454 contigs was also attempted against existing sequence data. Sequences of the 3′ end of cox1 were retrieved for >5000 species of Coleoptera available at GenBank. An aligned matrix was generated, together with the corresponding sequences from the contigs, and a maximum parsimony tree was generated using TNT 1.1 (48). Based on this tree, most 454 contigs were correctly assigned to a sister taxon at the species, genus or subfamily level (Figure 3). The database composition is uneven in regard to major lineages of Coleoptera, and poorly overlapping with the specimens used for mitogenome sequencing which were partly from a poorly represented local fauna (South Africa). Yet, in no case was the phylogenetic position of a sequence closer to the closest relatives of a different species in the data set, i.e. given the taxonomic spread of species analyzed here, the phylogenetic placement produced the correct species assignment for all contigs.
Phylogenetic analysis
The aligned data matrix comprises 12 out of 13 protein-coding mitochondrial genes; the nad2 and the 5′ end of cox1 were available only for five taxa and therefore omitted. We included 28 species, leaving out Byturus ochraceus because of apparent in vitro rearrangements and Nosodendron sp. whose sequence was insufficiently complete. In addition, all 25 full-length mitogenomes available on GenBank were included in the alignment for a matrix of 53 taxa and 9477 nucleotide sites, equal to 3159 amino acid positions. We used unweighted parsimony as baseline for the analysis, against which model-based approaches can be compared. In all nucleotide-based analyses we applied independent models to each codon position, rather than gene-by-gene partitioning (29). In addition, we performed analyses after removing 3rd codon sites and RY coding of 1st positions, to obtain data sets of reduced levels of saturation and lower nucleotide composition bias.
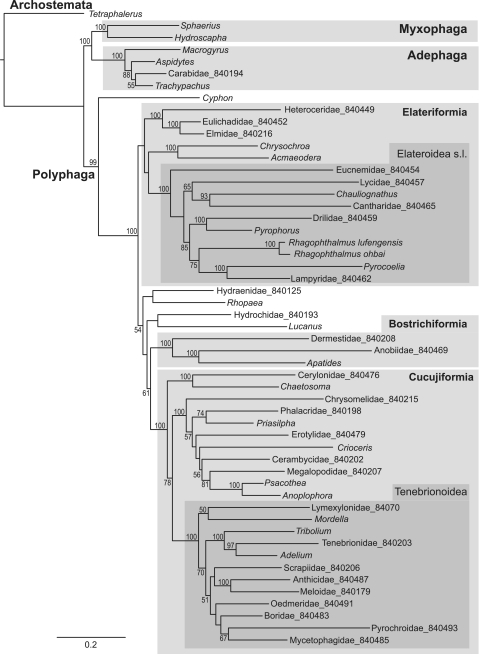
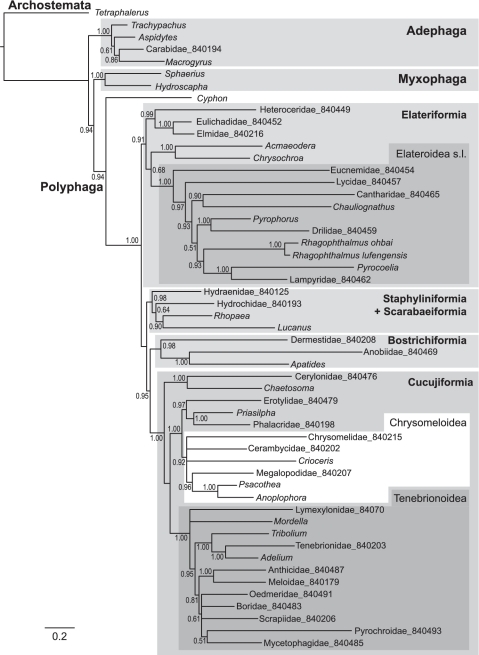
The various coding schemes produced a remarkable improvement in line with the predictions that existing biases in sequence variation, in particular in 3rd codon positions, confound the tree searches. Equal weighting of all positions under parsimony resulted in a tree that was the least consistent with current understanding of Coleoptera relationships (Supplementary Figure S1). Specifically, the divergent sequence of Tetraphalerus (Archostemata) grouped incorrectly within a derived lineage of Polyphaga, as observed previously (29,49), while several of the expected higher taxa are paraphyletic. Applying model-based tree building methods, either under likelihood in RAxML (Supplementary Figure S2, Figure 4) or Bayesian methods in MrBayes (Supplementary Figures S3 and S4), resulted in improved recovery of key groups, in particular after removal of 3rd positions and RY coding of 1st positions (Figure 4). Only the latter coding scheme recovers the correct position of Archostemata distant from Polyphaga. Finally, the CAT model (implemented in Phylobayes) allows the number of rate categories to vary freely, each of which can vary under an independent GTR model. This model resulted in further improvements both when applied at the nucleotide level (Supplementary Figure S4) and amino acid level (Figure 5). We consider the resulting tree the most accurate reflection of coleopteran relationships, as it recovers the greatest number of higher-level groups established by morphology and recent molecular data.
Figure 4.
ML tree from RaxML, with 1st position RY coded and 3rd positions removed. Major taxa are marked in shades of grey.
Figure 5.
Bayesian analysis on amino acid variation performed with the Phylobase software.
Under this final coding scheme, the relationships of the four suborders conform to the traditional view of Myxophaga + Polyphaga (50), which was also supported by EST-based ribosomal protein sequences (51). Recent studies that were mainly based on 18S rRNA generally support Myxophaga + Archostemata (31), while studies of mitogenomes so far mostly support Myxophaga + Adephaga (29,49). The latter relationship is also observed in virtually all analyses conducted here based on nucleotide data (except in Phylobase which puts groups Archostemata + Myxophaga; Supplementary Figure S5). Within Polyphaga, we obtained Cyphon (Eucinetoidea) as sister to all other Polyphaga, confirming existing studies (31), and the monophyly of the Series Elateriformia and Cucujiformia, represented by 15 and 23 taxa, respectively. Internal relationships within Elateriformia were recovered including the monophyly of Byrrhoidea and Elateroidea s.l. (52). The Elateriformia are the sister to the other four Series of Polyphaga, with a close relationship of Staphyliniformia and Scarabaeiformia (=Haplogastra), and the Bostrichiformia as sister to Cucujiformia, the very large radiation that includes about half of the modern Coleoptera. Within Cucujiformia, we find the monophyly of Tenebrionoidea (including a single representative of the small superfamily Lymexyloidea) but the polyphyly of Cucujoidea, as expected from recent studies (31). The Chrysomeloidea (leaf beetles and longhorns) was also recovered with Phylobase analysis on amino acid sequences, but not in any of the other procedures.
We also assessed the contribution of individual genes to the phylogenetic signal of the full data set. This was performed in a parsimony framework using partitioned Bremer support (PBS) (53). The total PBS of each gene was summed and normalized for the number of base pairs and informative sites (Table 2). PBS was overall strongly positive across loci, indicating that various genes contribute to the tree in a similar way. This confirms indirectly that the genome assemblies are accurate, as chimerical sequences would result in phylogenetic conflict in some portions of the tree evident in loci that are physically adjacent. However, the PBS was strongly negative at several nodes for various genes. In particular, the cox1 and nad4L genes showed a negative PBS over the entire tree (subtracting 155.1 and 64.9 steps, given a total PBS of 953 steps). The negative signal was high also if normalized for the number of (informative) nucleotide sites in a gene and the total number of steps contributed by the gene to the combined analysis tree (Table 2). The cox1 locus was identified here directly by the bait sequences, i.e. the incongruence of this gene with the remainder of the mitochondrial genome cannot be due to the incorrect incorporation of this locus into the contig. Hence, the conflict suggests the greater effect of confounding biases in this gene compared to most others in the mitochondrial genome, as already established from detailed studies of mitochondrial rate variation in Coleoptera (29).
Table 2.
Analysis of PBS for each protein coding gene and the combined mitogenome
| Gene | Total PBS | total sites | Inform. sites | Number steps | PBS/sites | PBS/inform. sites | PBS/steps |
|---|---|---|---|---|---|---|---|
| atp6 | 33.6 | 678 | 435 | 5339 | 0.0495 | 0.0772 | 0.0062 |
| atp8 | −7.5 | 174 | 126 | 1857 | −0.0431 | −0.0595 | −0.0040 |
| cob | 63.1 | 1137 | 696 | 8620 | 0.0554 | 0.0906 | 0.0073 |
| cox1 | −155.1 | 822 | 442 | 5437 | −0.1886 | −0.3509 | −0.0285 |
| cox2 | 83.9 | 690 | 451 | 5362 | 0.1215 | 0.1860 | 0.0156 |
| cox3 | 119.9 | 786 | 480 | 5923 | 0.1525 | 0.2497 | 0.0202 |
| nad1 | 212 | 957 | 626 | 7267 | 0.2215 | 0.3386 | 0.0291 |
| nad3 | 150 | 351 | 247 | 3263 | 0.4273 | 0.6072 | 0.0459 |
| nad4 | 150.6 | 1347 | 947 | 11 254 | 0.1118 | 0.1590 | 0.0133 |
| nad4l | −64.9 | 294 | 218 | 2376 | −0.2207 | −0.2977 | −0.0273 |
| nad5 | 240.7 | 1716 | 1168 | 14 066 | 0.1402 | 0.2060 | 0.0171 |
| nad6 | 126.7 | 525 | 428 | 5610 | 0.2413 | 0.2960 | 0.0225 |
| total | 953 | 9477 | 6264 | 76 374 | 0.1005 | 0.1521 | 0.0124 |
DISCUSSION
This study demonstrates the relative ease with which the full complement of mitochondrial protein coding genes can be obtained, and the power of this data type to resolve basal relationships within the Coleoptera, the largest group of insects. The method is universally applicable to any group of organisms wherever long-range PCR fragments are obtained for multiple taxa. Our method is different from existing approaches in several ways. First, unlike most existing studies of mt genomes [but see (54)], we used universal, mildly degenerate primers for all taxa. Long-range PCR is often thought to be most successful with long (∼30 nt), species-specific (non-degenerate) primers. However, our primers are not only much shorter but also contain degenerate positions that work for long-range PCR on a wide range of beetle species, thus greatly reducing the complexity of the amplification process. Second, we show that sequencing is possible in mixtures from which unique contigs can be separated bioinformatically. The fact that up to 30 taxa could be sequenced in 1/16 of a pyrosequencing plate (i.e. 480 taxa in a full plate) will greatly extend the application of NGS technology in molecular systematics and taxonomy. Third, it was possible to associate individual contigs unequivocally to particular species in the pool through the use of short bait sequences which also established the correct assembly of contigs by associating distant portions of the mt genome to the same sample in the pool. This obviates the need for independent construction of libraries using different ‘barcode’ tags for indexing the pooled individuals (5,55).
The quality of the contig sequences obtained was excellent, given a target sequencing depth of ∼50× (calculated assuming 13 kb per mt genome × 30 species = 390 kb of unique sequence, and an expected 20 Mb per sector of a 454 plate). Most sequences perfectly maintained the reading frame for all protein-coding regions, although the well established problem of long oligo-T nucleotide repeats required manual editing in several cases. The resulting contigs were obtained reliably at this level of sequence coverage, even without the doubling of the number of sequence reads in the combined analysis of both sectors on the 454 sequencer, which had little effect on the recovery of full-length contigs for the majority of species. However, where partial contigs were obtained from a single sector, this was generally improved by combining both sets. As the relevant contigs had low coverage, but increased coverage by doubling the number of sequence reads, this was likely due to differences in concentration of PCR fragments used in the library construction. Although every effort was made to obtain equimolar quantities of the various fragments based on gel intensity and measures of optical density, long-range PCR reactions may be prone to generate minor products that were carried over to the mixed libraries and confound the concentrations of the target DNA. In fact, our use of universal primers, while simplifying the generation of a data set with broad taxonomic coverage, may increase the occurrence of such minor products and hence the proportion of unassignable, comparatively short contigs (Figure 2). A further inconvenience was the uneven success of the long-range PCR across the diversity of taxa which required the use of slightly different primer combinations (from the universal primer set in Supplementary Table S1) for various target taxa and the use of conventional PCR to obtain missing regions on either end of the large fragment (Figure 1). In particular, the recovery the nad2 fragment was very limited, possibly because of incorrect estimates of the DNA concentrations. Finally, we did not make an effort to obtain the control region, as it is not usually informative for deep-level phylogenetics and may complicate the assembly of contigs because this region often is characterized by sequence repeats and extreme AT bias.
Despite some missing portions in several target taxa, the contigs themselves were likely to be assembled correctly. First, in a simulation experiment starting from fully sequenced mt genomes on GenBank (Supplementary Table 1) we established that these genomes are correctly reassembled by the MIRA software without rearrangements or formation of inter-taxon chimerae, even if closely related species are included in the data set. The assembly was problematic in a few cases only for the repetitive d-loop region which is not sequenced here. The newly sequenced data behave very similar as the simulated sequences, except that fragments were not present in stoichiometric amounts and minor PCR fragments were present as contaminants. This apparently created additional problems of assembly and resulted in shortened contigs, but larger contigs could be built by a secondary assembly using Phrap. When this step was included, many additional full-length contigs were obtained, without any evidence of mis-assembly. The conservative settings for the assembler are a desirable feature, to avoid spurious contigs, but in some cases the resulting products may require manual intervention to decide for or against contig building.
In addition to the simulation study, support for the correct assembly of the newly obtained sequences was based on the following facts. First, the correct gene order to match those in known mitogenomes of Coleoptera, except in a single case (Byturus ochraceus, Byturidae, BMNH480477) which apparently resulted from in vitro rearrangements in the PCR. Second, the use of three bait sequences confirmed the same species identity of various portions of the contigs except for one case, and the gene order analysis revealed a second exception, but these discrepancies were easily resolved by visual inspection of the MIRA-assembled contigs. Third, the phylogenetic signal of the sequences was strong and largely uniform amongst various genes as assessed with the PBS, which is not to be expected if contigs were chimeric products assembled from different taxa. The greatest discrepancy with the combined-analysis tree was for the cox1 gene which was one of the bait sequences whose species associations are not in question. In addition, wherever negative PBS values were encountered, these are not concentrated around adjacent loci which would be expected if chimeric contigs had been produced.
The success of sequencing the full set of mitochondrial protein coding regions has several practical implications. The procedure greatly simplifies the amplification of most of the mitochondrial genome, and overcomes the problem of sequencing across some of the most variable regions for which no universal primers exist. Primer walking and classical Sanger sequencing technology in a data set of the magnitude produced here would require hundreds of species-specific primers (49). In addition, our finding that contigs can be obtained reliably from mixtures of PCR products also simplifies the process and greatly reduces the cost. A major concern was that chimeric contigs might be produced in the assembly, but this was found to occur only in exceptional cases and was easy to spot and eliminate. Tagging of PCR products using paired ends might be used to confirm the correct assembly of contigs, but we here establish the assembly is sufficiently secure that such step is not necessary. Yet, paired end sequencing will give greater confidence in the data by providing additional verification of contigs, and may allow more closely related taxa to be multiplexed.
Modifications of the technique could even further reduce the effort needed, in particular to avoid the time consuming production of bait sequences. For example, the PCR products could be subjected to conventional sequencing to obtain a partial sequence of both ends with which to conduct species assignment of contigs analogous to the bait sequences used here. Alternatively, short nucleotide indexing tags may be added to the long-range amplification primers which then identify a given contig as the product of a specific PCR reaction via the primers at either end; a set of PCR primer pairs with one of 30 different tags could easily be designed and one each used in each multiplexed 1/16th sector of the 454 run. As we have shown, contig assignment could also be performed without a bait sequence via existing sequences of related species available at GenBank. This assignment was unequivocal for the current data set for the cox1 gene which has high taxonomic representation in GenBank. This provides an even simpler procedure in cases where the multiplexed samples are phylogenetically distant compared to the taxonomic coverage of the database. The possibility of an approximate species assignment based on phylogenetic placement is also encouraging for amplification from environmental samples where the identity of species may be assessed against an increasing number of known mitochondrial genomes.
A final topic is the use of this procedure as a high-throughput application to be achieved with fully automated editing. Molecular systematics data are notoriously difficult to assemble and frequently require manual editing, e.g. when using EST data (56). However, with increasing sequencing power and greater sequencing depth, these problems might be reduced and a fully automated editing process could be applied. The bioinformatics steps used here are straightforward, using the MIRA and Phrap assemblers whose output is then used for Blastn searches against bait sequences and other mitochondrial databases, while alignment is conducted with standard software such as Clustal and Transalign. These programs can easily be linked up in a pipeline that invokes all steps sequentially without manual intervention. Incorrect contigs (as the small number of errors that were resolved manually; see above) could be eliminated by filtering the contigs based on maximum size or duplication of gene content. However, the arcane molecular phylogenetics that is a prerequisite for obtaining accurate trees probably is less easily automated. Here, we were able to use a strategy for phylogenetic analysis developed in a recent paper (29) and could show that the new data can be exploited best when analyzed in a similar fashion. Procedures of model choice and tree search require the use of a variety of non-standard programs and automation of this aspect may be difficult.
With the new sequence data and highly appropriate algorithms for tree building, we are now in a position where mitochondrial genomes can be assembled to resolve the deep relationships of Coleoptera. The primary objective was to establish a methodology to scale up the sequencing of mitochondrial genomes for dense taxon sampling ultimately needed to resolve relationships at the level of families and subfamilies. Taxa selected here, therefore, were from a range of taxonomic distances, to establish what is the minimum divergence for contig separation and identification. It appears none of them were too close phylogenetically to confound the assembly process. We focused on two fairly narrow portions of the tree, the Elateriformia and Tenebrionoidea. Both groups included species already sequenced in previous studies. Seven types of phylogenetic analyses were performed, to deal with the effects of rate heterogeneity and compositional bias in mitogenomes of Coleoptera (29,49). We obtained a clear progression in the recovery of established groups with increasing model complexity and removal of the most homoplastic changes (3rd positions and transitions in 1st positions). In agreement with this, the most complex CAT model, applied at the amino acid level, produced the most satisfactory trees (Figure 5), as amino acid coding largely overcomes the problem of compositional biases, combined with the freely varying number of rate classes in the model. When data sets are expanding rapidly, the development of models and their implementation in fast algorithms is as important as the development of inexpensive sequencing methodology.
CONCLUSIONS
This study demonstrates that several hundred full mitochondrial genomes can be gathered in a single sequencing run on the 454/Roche platform without the need for an expensive ligation step for index tagging (‘barcoding’). Alternative NGS platforms may be used also, but current technologies produce much shorter-sequence reads, which permits the assembly of contigs only against a reference sequence that is more closely related to the focal sequence than any others in the mixture (57). Mitochondrial genome sequences have accumulated slowly, mostly for phylogenetically distant groups, because of the difficulty to obtain the sequence information. Existing data have presented severe limitations when used to assess deep-level relationships, e.g. on the level of the Insecta or Hexapoda (13,58). However, within these groups where levels of saturation are more manageable, the phylogenetic information content of this molecule is very high, and is likely to increase further with denser taxon sampling. The possibility to sequence pools of fragments and to separate such mixtures reliably in silico, is a major step towards high-throughput sequencing of mitogenomes and other sources of long PCR amplicons. The cost of a whole-mt genome sequence using this technology was only about 10× greater than the generation of double-stranded Sanger sequences from a single PCR fragment, such as those obtained for the cox1 ‘barcode’ marker (11). Mitogenome sequences therefore in future may be obtained for each species and become the standard for taxonomic sequencing, much like the cox1 barcodes today.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Natural History Museum (DIF and SIF programmes); Natural Environment Research Council (NE/F006225/1); Leverhulme Trust (F/00696/P). Funding for open access charge: Institutional funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Peter Hammond for specimens used in this study, to Chris Barton for database searches, Nick Brown for help with mt genome annotation and to Douglas Chesters for a Perl script. Andie Hall helped to develop long-range PCR protocols. We thank the NHM sequencing facility (ABI3700), and Shilo Dickens at the Department of Biochemistry, University of Cambridge, for 454 sequencing.
REFERENCES
- 1.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genomics Human Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 2.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. Roy. Soc. B. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips AJ, Simon C. Simple, efficient, and non-destructive DNA extraction protocol for arthropods. Ann. Entomol. Soc. Amer. 1995;88:281–283. [Google Scholar]
- 5.Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jex AR, Hall RS, Littlewood DTJ, Gasser RB. An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Res. 2010;38:522–533. doi: 10.1093/nar/gkp883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, Fire AZ. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc.Roy.Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 13.Nardi F, Spinsanti G, Boore JL, Carapelli A, Dallai R, Frati F. Hexapod origins: monophyletic or paraphyletic? Science. 2003;299:1887–1889. doi: 10.1126/science.1078607. [DOI] [PubMed] [Google Scholar]
- 14.Cameron SL, Barker SC, Whiting MF. Mitochondrial genomics and the new insect order Mantophasmatodea. Mol. Phylogenet. Evol. 2006;38:274–279. doi: 10.1016/j.ympev.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Cameron SL, Lambkin CL, Barker SC, Whiting MF. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 2007;32:40–59. [Google Scholar]
- 16.Agnarsson I, May-Collado LJ. The phylogeny of Cetartiodactyla: the importance of dense taxon sampling, missing data, and the remarkable promise of cytochrome b to provide reliable species-level phylogenies. Mol. Phylogenet. Evol. 2008;48:964–985. doi: 10.1016/j.ympev.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 17.Jex AR, Hu M, Littlewood DTJ, Waeschenbach A, Gasser RB. Using 454 technology for long-PCR based sequencing of the complete mitochondrial genome from single Haemonchus contortus (Nematoda) BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-11. Art. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson N, Gabriel S. Combinatorics and next-generation sequencing. Nat. Biotechnol. 2009;27:826–827. doi: 10.1038/nbt0909-826. [DOI] [PubMed] [Google Scholar]
- 19.Erlich Y, Chang K, Gordon A, Ronen R, Navon O, Rooks M, Hannon GJ. DNA Sudoku-harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 2009;19:1243–1253. doi: 10.1101/gr.092957.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich M, Muqim N. Sequence and phylogenetic analysis of the complete mitochondrial genome of the flour beetle Tribolium castanaeum. Mol. Phylogenet. Evol. 2003;26:502–512. doi: 10.1016/s1055-7903(02)00335-4. [DOI] [PubMed] [Google Scholar]
- 21.Stewart JB, Beckenbach AT. Phylogenetic and genomic analysis of the complete mitochondrial DNA sequence of the spotted asparagus beetle Crioceris duodecimpunctata. Mol. Phylogenet. Evol. 2003;26:513–526. doi: 10.1016/s1055-7903(02)00421-9. [DOI] [PubMed] [Google Scholar]
- 22.Kim KG, Hong MY, Kim MJ, Im HH, Kim MI, Bae CH, Seo SJ, Lee SH, Kim I. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol. Cells. 2009;27:429–441. doi: 10.1007/s10059-009-0064-5. [DOI] [PubMed] [Google Scholar]
- 23.Bae JS, Kim I, Sohn HD, Jin BR. The mitochondrial genome of the firefly, Pyrocoelia rufa: complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol. Phylogenet. Evol. 2004;32:978–985. doi: 10.1016/j.ympev.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Amoldi FGC, Ogoh K, Ohmiya Y, Viviani VR. Mitochondrial genome sequence of the Brazilian luminescent click beetle Pyrophorus divergens (Coleoptera: Elateridae): mitochondrial genes utility to investigate the evolutionary history of Coleoptera and its bioluminescence. Gene. 2007;405:1–9. doi: 10.1016/j.gene.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Ogoh K, Ohba N, Liang XC, Ohmiya Y. Mitochondrial genomes of two luminous beetles, Rhagophthalmus lufengensis and R. ohbai (Arthropoda, Insecta, Coleoptera) Gene. 2007;392:196–205. doi: 10.1016/j.gene.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Sheffield NC, Song H, Cameron L, Whiting MF. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 2008;25:2499–2509. doi: 10.1093/molbev/msn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheffield NC, Song HJ, Cameron SL, Whiting MF. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 2009;58:381–394. doi: 10.1093/sysbio/syp037. [DOI] [PubMed] [Google Scholar]
- 28.Hong MY, Jeong HC, Kim MJ, Jeong HU, Lee SH, Kim I. Complete mitogenome sequence of the jewel beetle, Chrysochroa fulgidissima (Coleoptera: Buprestidae) Mitochondrial DNA. 2009;20:46–60. doi: 10.1080/19401730802644978. [DOI] [PubMed] [Google Scholar]
- 29.Pons J, Ribera I, Bertranpetit J, Balke M. Nucleotide substitution rates for the full set of mitochondrial protein-coding genes in Coleoptera. Mol. Phylogenet. Evol. 2010;56:796–807. doi: 10.1016/j.ympev.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence JF, Newton AF. In: Biology, Phylogeny, and Classification of Coleoptera. Pakaluk J, Slipinski SA, editors. Warzawa: Museum i Instytut Zoologii PAN; 1995. pp. 779–1066. [Google Scholar]
- 31.Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, Wild R, Hammond PM, Ahrens D, Balke M, Caterino MS, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 32.Chevreux B, Wetter T, Suhai S. Computer Science and Biology: Proceedings of the German Conference on Bioinformatic(GCB) 1999. pp. 45–56. [Google Scholar]
- 33.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim - a sequencing simulator for genomics and metagenomics. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0003373. Art. No. e3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Asimenos G, Toh H. In: Methods in Molecular Biology. Posada D, editor. Vol. 537. Totowa, NJ: Humana Press; 2009. pp. 39–64. [DOI] [PubMed] [Google Scholar]
- 35.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 36.Bininda-Emonds ORP. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang WC, Marth G. EagleView: A genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–1543. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet-next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol. Phylogenet. Evol. 2006;38:100–116. doi: 10.1016/j.ympev.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 41.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7 doi: 10.1186/1471-2148-7-214. Art. 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A, Ludwig T, Meier H. Raxml-iii. A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 44.Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 45.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 46.Huelsenbeck JP, Suchard MA. A nonparametric method for accommodating and testing across-site rate variation. Syst. Biol. 2007;56:975–987. doi: 10.1080/10635150701670569. [DOI] [PubMed] [Google Scholar]
- 47.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 48.Goloboff P, Farris S, Nixon K. TNT (Tree analysis using New Technology) Cladistics. 2004;20:84. [Google Scholar]
- 49.Song H, Sheffield NC, Cameron SL, Miller KB, Whiting MF. When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst. Entomol. 2010;35:429–448. [Google Scholar]
- 50.Crowson RA. The Natural Classification of the Families of Coleoptera. London: Nathaniel Lloyd & Co.; 1955. [Google Scholar]
- 51.Hughes J, Longhorn SJ, Papadopoulou A, Theodorides K, de Riva A, Mejia-Chang M, Foster PG, Vogler AP. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles) Mol. Biol. Evol. 2006;23:268–278. doi: 10.1093/molbev/msj041. [DOI] [PubMed] [Google Scholar]
- 52.Bocakova M, Bocak L, Hunt T, Teraväinen M, Vogler AP. Molecular phylogenetics of Elateriformia (Coleoptera): evolution of bioluminescence and neoteny. Cladistics. 2007;23:477–496. [Google Scholar]
- 53.Baker RH, DeSalle R. Multiple sources of molecular characteristics and the phylogeny of Hawaiian drosophilids. Syst. Biol. 1997;46:654–673. doi: 10.1093/sysbio/46.4.654. [DOI] [PubMed] [Google Scholar]
- 54.Roehrdanz RL, Degrugillier ME. Long sections of mitochondrial DNA amplified from fourteen orders of insects using conserved polymerase chain reaction primers. Ann. Entomol. Soc. Amer. 1998;91:771–778. [Google Scholar]
- 55.Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7 doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longhorn SJ, Pohl HW, Vogler AP. Ribosomal protein genes of holometabolan insects reject the Halteria, instead revealing a close affinity of Strepsiptera with Coleoptera. Mol. Phylogenet. Evol. 55:846–859. doi: 10.1016/j.ympev.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 57.McComish BJ, Hills SFK, Biggs PJ, Penny D. Index-free de novo assembly and deconvolution of mixed mitochondrial genomes. Genome Biol. Evol. 2010;2:410–424. doi: 10.1093/gbe/evq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cameron SL, Miller KB, D'Haese CA, Whiting MF, Barker SC. Mitochondrial genome data alone are not enough to unambiguously resolve the relationships of Entognatha, Insecta and Crustacea sensu lato (Arthropoda) Cladistics. 2004;20:534–557. doi: 10.1111/j.1096-0031.2004.00040.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.