Abstract
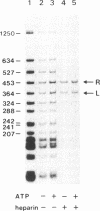
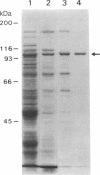
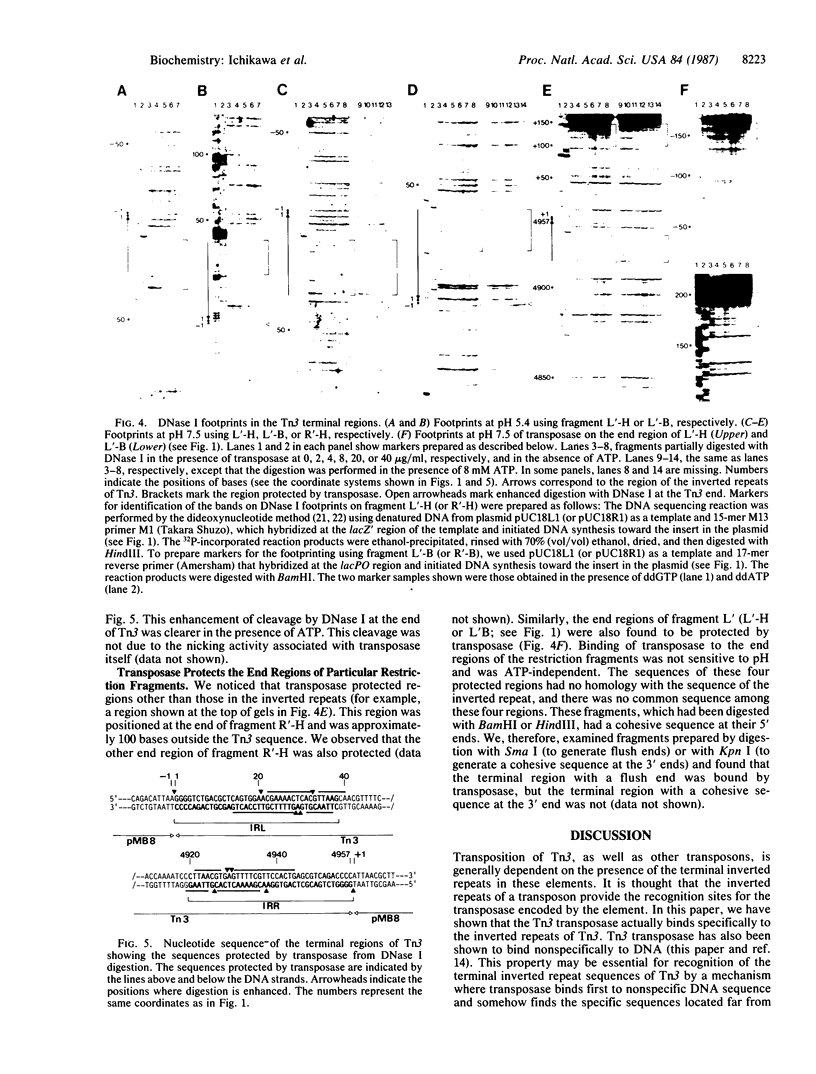
Tn3 transposase, which is required for transposition of Tn3, has been purified by a low-ionic-strength-precipitation method. Using a nitrocellulose filter binding assay, we have shown that transposase binds to any restriction fragment. However, binding of the transposase to specific fragments containing the terminal inverted repeat sequences of Tn3 can be demonstrated by treatment of transposase-DNA complexes with heparin, which effectively removes the transposase bound to the other nonspecific fragments at pH 5-6. DNase I "footprinting" analysis showed that the transposase protects an inner 25-base-pair region of the 38-base-pair terminal inverted repeat sequence of Tn3. This protection is not dependent on pH. Interestingly, binding of the transposase to the inverted repeat sequences facilitates DNase I to nick at the end of the Tn3 sequence. It was also observed that the transposase protects the end regions of restriction fragments with a cohesive sequence at their 5' end or with a flush end from DNase I cleavage. The specific and nonspecific binding of transposase to DNA is ATP-independent.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chou J., Casadaban M. J., Lemaux P. G., Cohen S. N. Identification and characterization of a self-regulated repressor of translocation of the Tn3 element. Proc Natl Acad Sci U S A. 1979 Aug;76(8):4020–4024. doi: 10.1073/pnas.76.8.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Lemaux P. G., Casadaban M. J., Cohen S. N. Transposition protein of Tn3: identification and characterisation of an essential repressor-controlled gene product. Nature. 1979 Dec 20;282(5741):801–806. doi: 10.1038/282801a0. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Cloning of the A gene of bacteriophage Mu and purification of its product, the Mu transposase. J Biol Chem. 1985 Feb 10;260(3):1832–1835. [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Fennewald M. A., Gerrard S. P., Chou J., Casadaban M. J., Cozzarelli N. R. Purification of the Tn3 transposase and analysis of its binding to DNA. J Biol Chem. 1981 May 25;256(10):4687–4690. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Heffron F., Falkow S. Identification of the protein encoded by the transposable element Tn3 which is required for its transposition. Nature. 1979 Dec 20;282(5741):797–801. doi: 10.1038/282797a0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. Transposition of Tn3 and related transposons. Cell. 1983 Jan;32(1):3–5. doi: 10.1016/0092-8674(83)90490-7. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Huang C. J., Heffron F., Twu J. S., Schloemer R. H., Lee C. H. Analysis of Tn3 sequences required for transposition and immunity. Gene. 1986;41(1):23–31. doi: 10.1016/0378-1119(86)90263-5. [DOI] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Morita C., Heffron F. Transposon Tn3 encodes a site-specific recombination system: identification of essential sequences, genes, and actual site of recombination. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4041–4045. doi: 10.1073/pnas.78.7.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCormick M., Wishart W., Ohtsubo H., Heffron F., Ohtsubo E. Plasmid cointegrates and their resolution mediated by transposon Tn3 mutants. Gene. 1981 Nov;15(2-3):103–118. doi: 10.1016/0378-1119(81)90120-7. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Rosenbloom M., Schrempf H., Goebel W., Rosen J. Site specific recombination involved in the generation of small plasmids. Mol Gen Genet. 1978 Feb 16;159(2):131–141. doi: 10.1007/BF00270886. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Grindley N. D. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981 Sep;25(3):721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Resolution of cointegrates between transposons gamma delta and Tn3 defines the recombination site. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3428–3432. doi: 10.1073/pnas.78.6.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart W. L., Broach J. R., Ohtsubo E. ATP-dependent specific binding of Tn3 transposase to Tn3 inverted repeats. Nature. 1985 Apr 11;314(6011):556–558. doi: 10.1038/314556a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]