Abstract
Genistein, a natural isoflavone found in soybean products, has been reported to down-regulate telomerase activity and that this prevents cancer and contributes to the apoptosis of cancer cells. However, the precise molecular mechanism by which genistein represses telomerase is not clear. Here, we show that genistein inhibits the transcription of hTERT (human telomerase reverse transcriptase), the catalytic subunit of the human telomerase enzyme, in breast MCF10AT benign cells and MCF-7 cancer cells in a time- and dose-dependent manner. Three major DNA methyltransferases (DNMT1, 3a and 3b) were also decreased in genistein-treated breast cancer cells suggesting that genistein may repress hTERT by impacting epigenetic pathways. Sequential depletion of the hTERT promoter revealed that the hTERT core promoter region is responsible for the genistein-induced repression of hTERT transcription. Using a newly developed technique of chromatin immunoprecipitation (ChIP)-related bifulfite sequencing analysis, we found an increased binding of E2F-1 to the hTERT promoter is due to the site-specific hypomethylation of the E2F-1 recognition site. In addition, we found that genistein can remodel chromatin structures of the hTERT promoter by increasing trimethyl-H3K9 but decreasing dimethyl-H3K4 in the hTERT promoter. The repression of hTERT was enhanced by combination with genistein and the DNMT inhibitor, 5-aza-2′-deoxycytidine (5-aza-dCyd). These findings collectively show that genistein is working, at least in part, through epigenetic mechanisms of telomerase inhibition in breast benign and cancer cells and may facilitate approaches to breast cancer prevention and treatment using an epigenetic modulator combined with genistein.
Keywords: genistein, hTERT, telomerase, epigenetic, E2F-1, breast cancer
Breast cancer accounts for ~30% of all cancers diagnosed in the United States and leads to the second cause of cancer death in women.1 Risk factors proposed for breast cancer include individual genetic background and lifestyle elements, such as diet.2 Epidemiological studies show that Asian immigrants who consume soybean products as their traditional diet in the United States have a much lower susceptibility to breast cancer, suggesting that the development of breast cancer is partly attributable to environmental differences (especially diet) rather than genetic differences.3,4
Genistein, the most abundant isoflavone found in soybean, is a potential chemopreventive element against various types of cancers, including breast cancer.5 Possible mechanisms for the anticancer property of genistein include: prevention of DNA mutation, reduction in cancer cell proliferation, inhibition of angiogenesis, and induction of differentiation.6–9 The effects of genistein on various cancer cell lines have been extensively investigated. Studies have reported that genistein represents mixed estrogen agonist and antagonist properties that can regulate signal transduction by inhibiting protein tyrosine kinases,10 cellular oxidative stress11 and angiogenesis.6 It has been reported that genistein can inhibit cancer cell growth and induce apoptotic cell death accompanied by cell cycle arrest at G2-M phase.12 The precise molecular mechanisms responsible for these activities, however, are not clearly understood. One potential mechanism that has recently received considerable attention is that genistein may regulate gene transcription by modulating epigenetic events such as DNA methylation and/or histone acetylation.13,14 This hypothesis is supported by studies showing that dietary genistein causes epigenetic changes in mouse prostate.15,16 Genistein can also up-regulate mRNA expression of the BRCA1 tumor suppressor gene during mammary tumorigenesis, which is frequently inactivated by epigenetic events in breast cancer.17
Recently, extensive interest has been focused on the aberrant regulation of telomerase activity in tumorigenesis. Telomeres comprise the nucleoprotein complexes that cap the ends of eukaryotic chromosomes and are maintained by a specialized reverse transcriptase, telomerase.18 Human telomerase is composed of an RNA subunit known as hTER (human telomerase RNA) and a protein subunit termed hTERT (human telomerase reverse transcriptase), which is the key determinant of the enzymatic activity of human telomerase.19 Activated expression of hTERT is present in stem cells, cancer-derived cell lines, and spontaneously immortalized cells and is detectable in up to 90% of malignant tumors but is usually not detectable in normal somatic cells.20,21 Recently identified loci at the hTERT gene region have been proved to associate with lung cancer incidence indicating that aberrant genetic or epigenetic regulation of hTERT could trigger cancer development.22 Multiple mechanisms that regulate hTERT gene transcription have been elucidated recently which include a variety of cellular and viral oncogene mechanisms.23 Furthermore, epigenetic pathways such as DNA methylation and chromatin modification also play an important role in modulating hTERT transcription.24 Hypermethylation of CpG islands in gene promoters is generally associated with gene silencing.25 Paradoxically, the hTERT promoter is highly methylated in most tumor cell types, rendering hTERT active.26,27 The tumor-specific differences in methylation status of the hTERT promoter and telomerase activity may be due to the selection of cells with site-specific changes in transcriptional factor binding characteristics. Furthermore, histone acetylation/methylation has also been implicated in modulating hTERT transcription in normal and malignant human cells.28,29 However, key aspects of chromatin modifications and DNA methylation patterns, and their association with gene expression have not been thoroughly studied for hTERT, especially with regard to the effects of dietary compounds on hTERT gene regulation that may have significant potential in cancer prevention and treatment.
Previous studies have indicated that genistein can repress telomerase activity in human prostate cancer cells through transcriptional control of hTERT.30 Our studies were aimed to address the mechanisms of hTERT transcription by genistein both in breast benign-derived cells and breast cancer cells. In the present studies, we analyzed the epigenetic and genetic mechanisms of hTERT transcriptional inhibition both in breast precancerous MCF10AT cells and cancer MCF-7 cells treated with genistein. We found that genistein leads to site-specific hypomethylation at the E2F-1 recognition site in the hTERT promoter. Moreover, genistein increased the inactive chromatin marker trimethyl-H3K9 and decreased the active marker dimethyl-H3K4 to the hTERT promoter. Our findings not only reveal the mechanisms of genistein on breast cancer suppression, but more importantly, also provide new insights into the regulatory mechanisms of hTERT.
Material and methods
Cell culture and cell treatment
The MCF-7 cell line was obtained from American Type Culture Collection and grown in DMEM medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Mediatech). MCF10AT human fibrocystic ER(+) breast cells31 were kindly provided by Dr. Andra R. Frost, which were derived from benign MCF10A cells transformed with T24 Ha-ras. MCF10AT cells were cultured in DMEM/F-12 medium (Mediatech) supplemented with 10 μg/ml of human insulin (Sigma, St. Louis, MO), 20 ng/ml of epidermal growth factor (Sigma), 100 ng/ml of cholera toxin (Sigma), 0.5 μg/ml of hydrocortisone (Sigma), 10% horse serum (Mediatech) and 1% penicillin/streptomycin (Mediatech). Cells were maintained in a humidified environment of 5% CO2 and 95% air at 37°C. After 24 hr seeding, attached MCF-7 and MCF10AT cells were treated with 0, 10, 25, 50 and 100 μM genistein (Sigma) for 1, 2 and 3 days, respectively. The medium with genistein was replaced every 24 hr for the duration of the experiment. Control cells received equal amounts of DMSO (Sigma) in the medium. For the combination study, MCF-7 cells were treated with 50 μM genistein or 2 μM 5-aza-dCyd (Sigma) alone or together for 3 days. For cell growth analyses, cells were trypsinized and resuspended in growth medium. Trypan Blue (0.4%) was added to the cell suspension, and both live and dead cells were counted using a hemacytometer. Cells were then used for apoptosis analysis with the Vybrant Apoptosis Assay kit #2 (Invitrogen, Carlsbad, CA). After fixation with the annexin-binding buffer, cells were stained both with Alexa Fluor Annexin V and propidium iodide (PI) according to the manufacturer's instructions. Flow cytometry was performed on a Becton Dickinson FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The fluorescence intensity of the viable cells was analyzed using CellQuest software.
Reporter gene assay of hTERT promoter activity
The PGL2-enhancer luciferase vectors containing 3 hTERT promoter constructs, P-330, P-1009, P-3996, were kindly provided by Dr. Silvia Bacchetti.32 Before transfection, MCF-7 cells were grown in 24-well culture plates with or without various concentrations of genistein treatment for 3 days. MCF-7 cells were transiently transfected with 3 hTERT promoter-luciferase constructs along with PGL2-enhancer vector (Promega, Madison, WI) as a basic control for 24 hr, respectively. Luciferase activity was measured in cell lysates by a microplate luminometer using the Dual Luciferase assay kit (Promega) according to the manufacturer's protocol. Luciferase activity was normalized by Renilla luciferase activity through co-transfection with the pRL-SV40 plasmid (Promega). Each experiment was repeated in triplicate.
Quantitative real-time PCR
Both MCF-7 and MCF10AT cells were cultured and treated as described above. Total RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Genes of interest were amplified using 5 μg of total RNA reverse transcribed to cDNA using the Superscript II kit (Invitrogen) with oligo-dT primer. In the real-time PCR step, PCR reactions were performed in triplicate with 1 μg cDNA per reaction and primers specific for hTERT (Hs00162669_ml) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs99999905_ml) provided by Inventoried Gene Assay Products (Applied Biosystems, Foster City, CA) using the Platinum Quantitative PCR Supermix-UDG (Invitrogen) in a Roche LC480 thermocycler. Thermal cycling was initiated at 94°C for 4 min followed by 35 cycles of PCR (94°C, 15 sec; 60°C, 30 sec). GAPDH was used as an endogenous control, and vehicle control was used as a calibrator. The relative changes of gene expression were calculated using the following formula: fold change in gene expression, 2−ΔΔCt = 2−{ΔCt (genistein-treated samples)−ΔCt (untreated control samples)}, where ΔCt = Ct (hTERT)−Ct (GAPDH) and Ct represents threshold cycle number.
Western blot analysis
For regular Western blot analysis, protein extracts were prepared by TRAPeze 1 × CHAPS cell lysis buffer (Chemicon International, Temecula, CA) from MCF-7 cells treated with or without genistein according to the manufacturer's protocol. Proteins (50 μg) were electrophoresed on a 10% SDS-polyacrylamide gel and transferred to nitrocellulose membranes. Membranes were probed with antibodies to hTERT (Epitomics, Burlingame, CA), DNMTs (Abcam, Cambridge, MA), c-MYC (N-262; Santa Cruz Biotechnology, Santa Cruz, CA) and E2F-1 (H-137, Santa Cruz Biotechnology), respectively, then each membrane was stripped and reprobed with GAPDH antibody (V-18, Santa Cruz Biotechnology) as loading control. Molecular weight markers were run on each gel to confirm the molecular size of the immunoreactive proteins. Immunoreactive bands were visualized using the enhanced chemiluminescence detection system (Santa Cruz Biotechnology) following the protocol of the manufacturer.
Telomeric repeat amplification protocol assay
MCF-7 and MCF10AT cells with or without genistein treatments were harvested on days 0, 1, 2 and 3 for telomerase activity analysis. The telomeric repeat amplification protocol (TRAP) assay was performed with the TRAPEZE Telomerase Detection kit (Chemicon International) as reported previously.33 All samples were incubated at 94°C for 10 min to serve as a heat inactivation control. After incubating at 30°C for 30 min, the samples were amplified by PCR (94°C for 30 sec, 59°C for 30 sec, and 72°C for 1 min) for a total of 36 cycles. The reaction was finished at 55°C for a 25 min extension step. The PCR products were electrophoresed on a 10% non-denaturing polyacrylamide gel and stained with SYBR Green (Molecular Probes, Eugene, OR).
Chromatin immunoprecipitation assay
MCF-7 and MCF10AT cells were treated with 100 μM genistein and an equal volume of DMSO as the untreated control for 3 days. Approximately 2 × 106 cells were cross-linked, with a 1% final concentration of formaldehyde (37%, Fisher Chemicals, Fairlawn, NJ) for 10 min at 37°C. ChIP assays were performed with the EZ Chromatin Immunoprecipitation (EZ ChIP™) assay kit according to the manufacturer's protocol (Upstate Biotechnology, Charlottesville, VA) as described previously.34 ChIP-purified DNA was amplified by standard PCR using primers specific for the hTERT promoter containing the 236 bp fragment: sense, 5′-CAGGACCGCGCTTCCCACG-3′ and anti-sense, 5′-GGCTTCCCACGTGCGCAGC-3′. PCR amplification was performed using the 2 × PCR Master Mix (Promega) and the reaction was initiated at 94°C for 4 min followed by 30 cycles of PCR (94°C, 30 sec; 56°C, 30 sec; 72°C, 1 min), and extended at 72°C for 5 min. After amplification, PCR products were separated on 1.5% agarose gels and visualized by ethidium bromide fluorescence using Kodak 1D 3.6.1 image software (Eastman Kodak Company, Rochester, NY). Quantitative data were analyzed using the Sequence Detection System software version 2.1 (PE Applied Bio-systems, Foster City, CA).
ChIP-bisulfite sequencing analysis
To assess the methylation status changes in the hTERT promoter of the ChIP products, sodium bisulfite methylation sequencing was performed. Approximately 50 μl of ChIP product was treated with bisulfite following the manufacture's protocol (Human Genetic Signatures, Macquarie Park, Australia). Bisulfite-modified DNA was analyzed using two primer sets spanning a region from −2ε288 to −231 of the hTERT promoter, which contained the distal E-box and E2F-1 recognition site. PCR amplifications were performed with primers BF 5′-GGTTTTTAGTGGATT-3′, BR 5′-AAAACCAAAACTTCCCAC-3′. PCR products were purified using a gel extraction kit (Qiagen) and were directly sequenced with primer BF on an automated DNA sequencer. Each sample was sequenced 3 times to determine the site-specific methylation changes in the amplified regions.
Statistical analyses
Data from Real-time PCR and luciferase assays were derived from at least 3 independent experiments. For quantification of ChIP products, Kodak 1D 3.6.1 image software was used. The protein levels were quantified by optical densitometry using ImageJ Software version 1.36b (http://rsb.info.nih.gov/ij/). Statistical significance between treatment and control groups was evaluated using Student's t-test, and values were presented as mean ± SE. p < 0.05 was considered significant.
Results
Transcription suppression of hTERT by genistein treatment occurred in a time- and dose-dependent manner
Several studies have shown previously that genistein has anti-proliferative effects on breast cancer cells, however, few investigations have examined hTERT transcriptional regulation by genistein. We initiated our study to first determine the optimal dose of genistein that will induce hTERT repression to the breast MCF10AT benign fibrocystic cells and MCF-7 cancer cells. As shown in Figure 1a, we observed a dose-dependent cell growth inhibition with genistein treatment both in MCF10AT and MCF-7 cells. In addition, the cell growth was completely inhibited at 50 μM and 100 μM of genistein after 3 days treatment without induction of cell death in MCF10AT and MCF-7 cells, respectively. In MCF10AT cells, 100 μM of genistein treatment caused a slight but non-significant increase in cell growth inhibition compared with 50 μM, indicating that the concentration of 50 μM has a more efficient effect in MCF10AT cells for studying the mechanisms of hTERT expression regulation. We have also performed the apoptosis assay (Supporting information) and found that genistein could not induce apoptosis under the aforementioned concentrations both in MCF10AT and MCF-7 cells. These results suggest that applying genistein under the aforementioned concentrations is capable of completely inhibiting cell growth without inducing apoptosis within the 3 days examined.
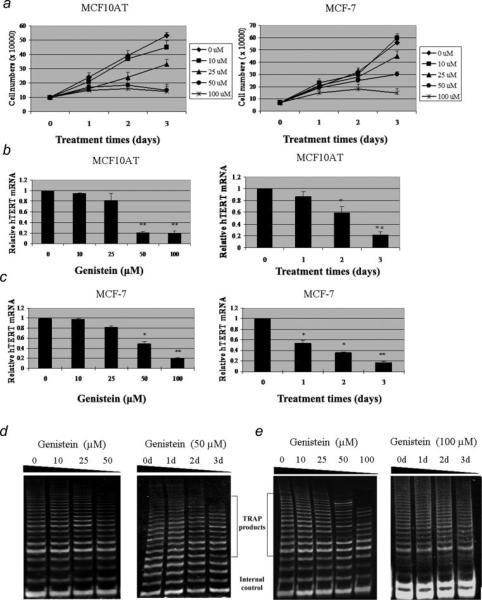
Figure 1.
Dose- and time-dependent inhibition of hTERT expression and telomerase activity in MCF10AT and MCF-7 cells treated with genistein. a, Genistein-induced cell growth inhibition in breast tumor cells without induction of cell death. MCF10AT (left) and MCF-7 (right) cells were plated in triplicate wells and exposed to various concentrations of genistein (0, 10, 25, 50 and 100 μM) for 3 days. Viable cells were counted by Trypan Blue staining using a hemacytometer. b and c, Dose- (left) and time-dependent (right) alterations of hTERT mRNA expression in MCF10AT and MCF-7 cells, respectively. Left, 24 hr after plating, cells were exposed to various concentrations of genistein. Right, the MCF10AT and MCF-7 cells were treated with genistein at a concentration of 50 μM and 100 μM for 3 days, respectively. Cell pellets were collected and subjected to real-time PCR and TRAP assay analysis. Relative quantification was performed by quantitative real-time PCR. Data are in triplicate from 3 independent experiments and were normalized to GAPDH and calibrated to levels in untreated samples. Bars, SE; *p < 0.05; **p < 0.001, significantly different from control. d and e, Dose- (left) and time-dependent (right) effect of genistein on telomerase activity in MCF10AT and MCF-7 cells. The internal control was evaluated for equal loading. Representative photograph from an experiment was repeated 3 times.
We also observed that genistein decreased hTERT transcription both in MCF10AT and MCF-7 cells in a dose- (left) and time-dependent (right) manner, as shown in the Figures 1b and 1c. In MCF10AT cells, similar significant hTERT suppression effects were observed both at 50 μM and 100 μM (Fig. 1b), which is consistent with our previous studies of cell growth inhibition. In addition, we found that the dose of 100 μM(p < 0.001) has a more effective impact on hTERT inhibition in MCF-7 cells (Fig. 1c). Furthermore, we observed overwhelming hTERT suppression that occurred on the third day of treatment both in MCF10AT and MCF-7 cells at the genistein concentration of 50 μM and 100 μM, respectively. These results indicate that genstein-induced growth inhibition of breast cancer cell is not through the apoptosis pathway, but rather, involved in regulating gene expression such as that of the hTERT gene, which is consistent with the previous studies.30
It is well known that high activity of telomerase is present in the majority of cancer cells, which allows the cells to survive and proliferate. The genistein-induced repression of hTERT, the key enzymatic component of telomerase, prompted us to investigate the alteration of telomerase activity with genistein treatment in benign and malignant breast cancer cells. Hence, we conducted our subsequent experiments to investigate whether genistein elicits any effect on telomerase activity.
Genistein repressed telomerase activity in MCF-7 cells
To address whether genistein could repress telomerase activity in breast cancer cells, we performed TRAP assays on MCF10AT and MCF-7 cells at the aforementioned days and various genistein concentrations. As illustrated in Figures 1d and 1e, the presence of telomerase activity was shown by the 6-bp ladder banding pattern that started at 61 bp. A 56-bp internal control band to monitor the PCR efficiency was visible in every sample. We observed that genistein repressed telomerase activity both in MCF10AT and MCF-7 cells dose- (left) and time-dependently (right). Up to 50% reduction in telomerase activity was observed on the third day of genistein treatment compared with untreated control cells, which revealed a delay in repression of telomerase activity relative to its mRNA product that is due to a ~24 hr half-life of the telomerase enzyme.35 These findings are consistent with our previous results showing that genistein induced hTERT suppression, which strongly suggest that repression of telomerase activity both in benign and cancer-derived cells by genistein is due to down-regulation of hTERT expression.
The hTERT core promoter is responsible for genistein-induced repression of hTERT expression
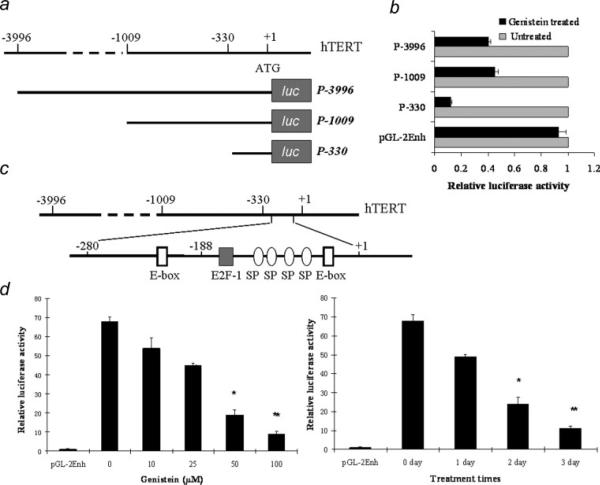
To explore the molecular mechanism of genistein-induced repression of telomerase activity, we examined hTERT promoter activity using the luciferase reporter system in breast cancer cells. Luciferase reporter plasmids contain either the full-length (3.9 kb) hTERT 5′ regulatory region (pGL2-3996) or deleted constructs of hTERT promoter elements including pGL2-1009 and pGL2-330 (Fig. 2a). As indicated in Figure 2b, the inhibition of pGL2-330, pGL2-1009 and pGL2-3396 transfected cells induced hTERT promoter activity between the genistein-treated and untreated MCF-7 cells at 12.6-, 7.9- and 8.6-fold compared with the basic vector. Further investigations on the hTERT promoter covered in the pGL2-330 construct revealed several transcription factor recognition sites in this region including E2F-1 and E-box (c-MYC) binding sties (Fig. 2c), which indicate that this particular region in the hTERT promoter may be associated with hTERT transcription regulation by genistein. We therefore used the pGL2-330 construct to examine the effect of genistein on hTERT promoter activity in MCF-7 cells. We observed that genistein treatment significantly decreased hTERT transcription activity in a dose- and time-dependent manner in MCF-7 cells as shown in Figure 2d, which is highly consistent with our former results of genistein-induced repression both of hTERT expression and telomerase activities. These results collectively suggest that the minimal hTERT promoter contains the core element that is responsible for genistein-dependent repression of hTERT transcription activity.
Figure 2.
Genistein represses hTERT transcription in breast cancer cells. a, Deletion constructs of the hTERT promoter. The A of the translation start codon (ATG) is designated as +1, and the name of each deletion construct was assigned according to the length of sequences upstream of the hTERT ATG. b, MCF-7 cells were transiently transfected with each of these luciferase plasmids together with Renilla luciferase (pRL-SV40) plasmids as the internal controls for 24 hr in normal growth media and then treated with or without 100 μM genistein for another 48 hr, after which luciferase activities assays were performed. Relative luciferase activity was normalized by Renilla luciferase activity. Luciferase activity in genistein-untreated samples was normalized to 1.0. C, The location of E2F-1, E-box and Sp1 binding sites in the hTERT proximal promoter. d, Dose- and time-dependent inhibition of hTERT promoter activity in response to genistein. MCF-7 cells were transiently transfected with P-330 with various concentrations (left) or 100 μM (right) of genistein for 72 hr. Relative luciferase activity was determined as mentioned above. Luciferase activity in control samples transfected with basic plasmid (pGL2-enh) was normalized to 1.0. Values are the means of 3 independent experiments. Bars, SE; *p < 0.05; **p < 0.001, significantly different from control.
Genistein down-regulated the expression of DNA methyltransferases
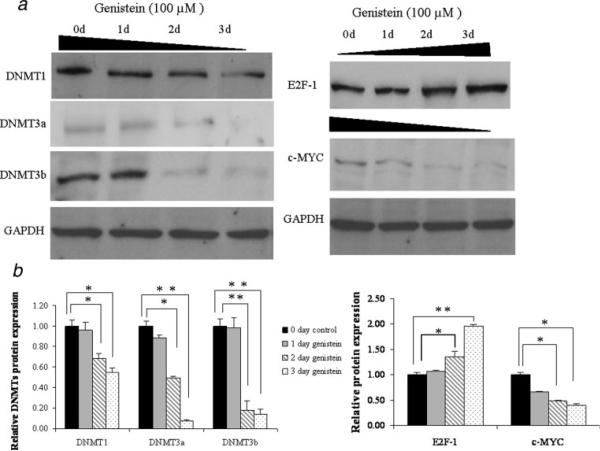
Our previous studies have shown EGCG, the major polyphenol from green tea, inhibits the activity of DNMT1 and this can lead to hypomethylation of the hTERT promoter.34 To determine whether genistein as a key polyphenolic compound in soybean has similar activities, we explored the effects of genistein on the expression of the DNMTs. Cellular proteins were extracted from MCF10AT and MCF-7 cells after 3 days of treatment with 50 μM and 100 μM genistein, respectively. Western-blot analysis revealed that genistein treatment significantly decreased the expression of DNMT1, DNMT3a and DNMT3b, the 3 main DNMTs, at 2 and 3 days of treatment with time-dependent manner in MCF-7 cells (Figs. 3a and 3b, left). In MCF10AT cells, a similar trend in DNMT1 expression was observed, however, no signals of DNMT3a and DNMT3b were found in western-blot analysis (data not shown). These results indicate that genistein may play an important role in epigenetic gene regulation via modulating DNMTs activities.
Figure 3.
Expressions of DNMTs and the main transcription factors of hTERT, E2F-1 and c-MYC, in response to genistein. a, the protein levels of DNMTs (left) and transcription factors, E2F-1 and c-MYC (right) were determined by western-blot analysis. MCF-7 cells proteins were extracted after exposure to 100 μM genistein for 3 days. Protein lysates (50 μg) were resolved on 12% SDS-PAGE, transferred onto nitro-cellulose membrane, and probed with antibody to DNMT1, DNMT3a, DNMT3b, E2F-1 and c-MYC. Membranes were reprobed with anti-GAPDH antibody to ensure for equal loading. Representative photograph from an experiment was repeated 3 times. b, Densitometry values for relative protein expressions of DNMTs (left) and transcription factors (right) were quantified and normalized to GAPDH internal controls. Bars, SE; *p < 0.05; **p < 0.001, significantly different from zero day control.
Genistein down-regulated the expression of c-MYC, but up-regulated E2F-1
To identify the direct mechanism of genistein on hTERT transcription regulation, we performed western-blotting to examine E2F-1 and c-MYC expression with genistein treatment. We started with E2F-1 and c-MYC because they are the most important repressor and activator of hTERT, respectively, whose binding depends on the methylation status of the hTERT promoter.36,37 As shown in Figures 3a and 3b right, the protein level of E2F-1 was increased significantly at 2 and 3 days of treatment, whereas c-MYC was decreased prominently in response to genistein during the same time interval both in MCF10AT (data not shown) and MCF-7 cells. These results indicate that expression alterations of key transcription factors caused by genistein may be a potential factor in hTERT transcription regulation. It may not be the only reason, however, since the methylation-sensitive behavior of E2F-1 and c-MYC also affects the binding of these transcription factors to the hTERT promoter depending on methylation status of the recognition sites. This is experimentally relevant in light of our results showing the inducible changes of DNMTs expression in response to genistein. We therefore performed ChIP assay and bisulfite sequencing to analyze the binding alterations of transcription factors and methylation status of the hTERT promoter in the following studies.
Genistein treatment altered the binding of E2F-1 and c-MYC to the hTERT promoter
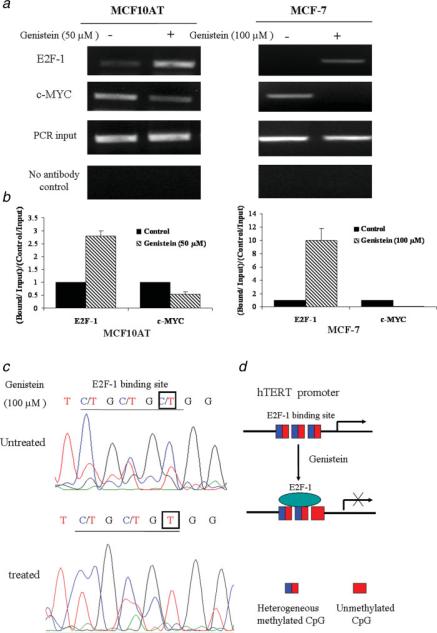
Since we had shown that the expressions of hTERT as well as DNMTs were markedly decreased in response to genistein, we therefore sought to explore the impact that this could have on the binding of transcription factors to the hTERT promoter. Using chromatin immunoprecipitation (ChIP) techniques, we observed that the binding of E2F-1 to the hTERT promoter was increased by 2.8- and 10-fold, whereas binding of c-MYC was decreased to 0.54- and 0.08-fold compared with the untreated samples in the MCF10AT and MCF-7 cells, respectively (Figs. 4a and 4b). The results suggest that the binding alterations of these transcription factors likely contributed to the suppression or down-regulation of hTERT due to genistein treatment.
Figure 4.
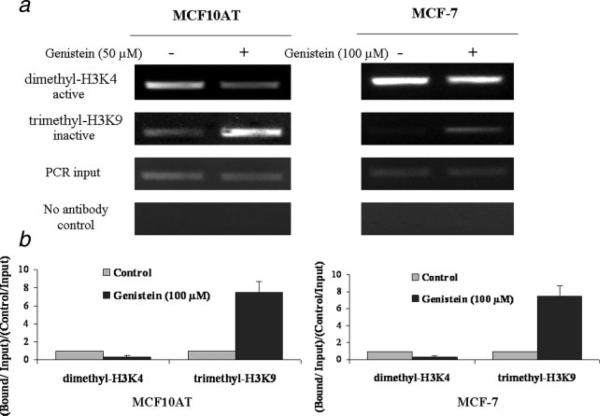
Genistein alters the recruitment of transcriptional factors to the hTERT promoter. a, Chromatin DNA from MCF10AT and MCF-7 cells was immunoprecipitated with antibodies against E2F-1 and c-MYC together with no antibody controls. MCF10AT (left) and MCF-7 cells (right) were treated with 50 μM and 100 μM of genistein, respectively, for 3 days and analyzed by ChIP assay together with untreated control cells. The purified DNA was amplified by PCR with the use of hTERT promoter primers. Inputs came from the total DNA and served as the same ChIP PCR conditions. Representative photograph from an experiment was repeated in triplicate. b, ChIP data were calculated from the corresponding DNA fragments amplified by PCR; columns, mean; bars, SD. Enrichment was calculated as the ratio between the net intensity of each bound sample divided by the input and the untreated control sample divided by the input (bound/input)/(control/input). c, DNA methylation patterns of E2F-1 binding site in the hTERT promoter in response to genistein. ChIP-Bisulfite sequencing shows that genistein-treated DNA is associated with hypomethylated CpG dinucleotides of the E2F-1 binding site, whereas untreated DNA is associated with heterogenerous CpG. d, Schematic presentation of E2F-1 preferentially binding to the unmethylated E2F-1 binding site in the hTERT promoter in response to genistein. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The binding of E2F-1 to the hTERT promoter was increased due to hypomethylation of the E2F-1 recognition site in response to genistein
Since genistein was more effective on the transcription factor binding to the hTERT promoter in MCF-7 cells than MCF10AT cells according to the ChIP results, we therefore used MCF-7 cells to perform the bisulfite sequencing analysis. To elucidate the effects of methylation on transcription factor binding to the hTERT promoter, we developed a novel ChIP-bisulfite sequencing approach by using ChIP purified DNA as a template for bisulfite treatment. By using this novel technique, we analyzed the methylation status of the hTERT promoter in the region −298 to −31 which contains many CpG dinucleotides and overlapping transcription factor binding sties. We found that the CpG dinucleotides of the E2F-1 binding site close to the start point switched to hypomethylation in response to genistein (Figs. 4c and 4d). However, there was no methylation change in both the proximal and distal c-MYC binding sites (E-box) in the hTERT promoter (data not shown). Because E2F-1 cannot bind to hypermethylated recognition sites,38 these results suggest that genistein-induced hypomethylation of the E2F-1 recognition site allows for the E2F-1 binding to the hTERT promoter which in turn led to the repression of hTERT expression.
Genistein caused chromatin remodeling at the hTERT promoter
In addition to DNA methylation as the primary mechanism of epigenetic control, histone acetylation and methylation also play important roles in hTERT gene regulation. Hyperacetylated histones and histone H3 methylated at lysine 4 are common markers of active chromatin, whereas, hypoacetylated histones and methylation of histone H3 at lysine 9 and 27 are generally associated with inactive genes. Therefore, to explore whether these chromatin makers affect hTERT gene expression in response to genistein, we performed ChIP assays in MCF10AT and MCF-7 cells with and without genistein treatment. We analyzed the hTERT promoter by using antibodies for both transcriptionally active (acetyl-H3K9 and dimethyl-H3K4) and inactive (trimethyl-H3K9) markers of chromatin. The results indicated that genistein increased trimethyl-H3K9, the inactive chromatin marker, to the hTERT promoter by 2.1- and 7.5-fold, whereas it decreased dimethyl-H3K4, the active chromatin marker, to 0.4- and 0.28-fold compared with the DMSO controls in the MCF10AT and MCF-7 cells, respectively (Fig. 5). However, no obvious change was found in acetyl-H3 between the genistein-treated and -untreated MCF10AT and MCF-7 cells (data not shown). Collectively, these results suggest that genistein may influence hTERT transcription regulation through chromatin remodeling primarily through alterations of methylation in specific histone residues within the hTERT promoter.
Figure 5.
Genistein modulates the histone status of hTERT in breast cancer cells. α, Genistein treated and untreated MCF10AT and MCF-7 cells were analyzed by ChIP assay using the chromatin markers, dimethyl-H3K4 (active), trimethyl-H3K9 (inactive) and no antibody controls. The PCR primers and conditions were used as described above. Representative photograph from an experiment was repeated in triplicate. b, Histone modification enrichment were calculated from the corresponding DNA fragments amplified by PCR as described previously; columns, mean; bars, SD.
The effects of combination of genistein and 5-aza-dCyd on the suppression of hTERT
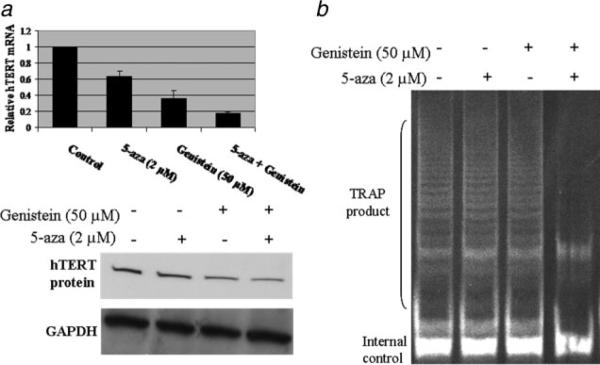
In our previous studies, we found that genistein could down-regulate the expression levels of DNMTs (Fig. 3a). We next explored the combined treatments of genistein with an important DNMTs inhibitor, 5-aza-dCyd, to analyze hTERT expression and telomerase activities. MCF-7 cells were treated with 50 μM genistein for 3 days alone, followed by a treatment with 2 μM 5-aza-dCyd for two additional days as combination samples. As indicated in Figure 6, both genistein and 5-aza-dCyd caused a reduction in hTERT expression and telomerase activity, however, genistein alone induced a more pronounced hTERT inhibition than 5-aza-dCyd alone. More importantly, the effect of combination treatment with genistein and 5-aza-dCyd was higher with 82% hTERT inhibition than that of each agent alone, suggesting an additive action on hTERTM gene regulation.
Figure 6.
Effects of a combination of genistein and 5-aza-dCyd (5-aza) on the repression of hTERT and telomerase activity. a, Genistein enhances the repression of hTERT expression combined with 5-aza. MCF-7 cells were treated with or without either 50 μM genistein or 2 μM5-aza alone or together for 3 days and cultured for an additional day in fresh medium. Quantitative real-time PCR (upper) and western-blot (bottom) were performed. b, Genistein in combination with 5-aza enhances the repression of telomerase activity in an additive manner. TRAP assays were performed after the treatment described above. Representative photograph from an experiment was repeated in triplicate.
Discussion
Various molecular mechanisms for the inhibitory effects of the soybean product, genistein, on many types of cancers have been proposed. In the present study, we focused on the molecular mechanism of genistein-induced repression of telomerase activity both in breast precancerous cells and breast cancer cells. Our results clearly show that genistein can repress hTERT expression via epigenetic mechanisms that involved both DNA hypomethylation and chromatin remodeling in the hTERT promoter. These mechanisms in turn repress telomerase activity both in breast precancerous cells and breast cancer cells, although the effects were more pronounced in breast cancer cells. Therefore, our results indicate that genistein, a multifunctional dietary component, can not only prevent breast cancer development, but also more importantly could be applied as a potential anti-cancer drug administrated in future breast cancer therapy.
An increasing number of findings have defined the hTERT gene as an important target of cancer therapy.39 In our studies with genistein, a dietary component involved in breast cancer prevention, we observed a dose- and time-dependent decrease of hTERT mRNA and protein, as well as telomerase activity in MCF10AT and MCF-7 cells. These studies are consistent with a recent report showing the suppression of hTERT transcription by genistein in prostate cancer cells30 and provide a key mechanism for this effect.
Aberrant regulation of the hTERT gene always results in over-expression of hTERT product and leads to telomerase activation which in turn triggers cancer development including lung, renal and breast tumors.20–22 Multiple mechanisms involved in hTERT gene transcription regulation have been proposed, which include cellular and viral oncogenic factors, as well as the epigenetic pathways.40 Furthermore, the presence of dense CpG sites and abundant transcription factor binding sites in the hTERT core promoter allow the accessibility of epigenetic modulators and/or transcription factors to regulate hTERT expression.23,41 However, unlike most human gene promoters in which CpG island hypermethylation leads to gene silencing, the hTERT promoter is highly methylated in most cancer cell lines, rendering hTERT transcriptionally active.27 In our present studies, we found that genistein can regulate hTERT transcription not only by modulating the expression of essential transcription factors, such as E2F-1 and c-MYC, but also by altering the methylation status that leads to hTERT gene silencing. Further studies clearly demonstrated that genistein treatment decreased the capacity of DNMTs to maintain methylation, leading to site-specific demethylation of the proximal E2F-1 binding site in the hTERT promoter, thus allowing the methylation-sensitive repressor to bind and block transcription as we have shown in Figure 4. However, not all the CpGs of the hTERT promoter are affected, including the E-box, the c-MYC binding site, which suggests that the decreased recruitment of c-MYC to the hTERT promoter may be due to the direct effect of genistein on c-MYC expression. Another explanation may be that the DNMTs preferentially methylate the CpGs contained in the E2F-1 sites and that disruption of DNMTs activity initially causes a loss of methylation in that area. Consistent with our previous studies on the green tea polyphenol, EGCG, showing that EGCG could impact epigenetic regulation by inducing hypomethylation of the E2F-1 binding site in the hTERT promoter,34 our current studies indicate that genistein may also modulate hTERT gene regulation and telomerase activity through a similar mechanism.
Epigenetic modification may affect the accessibility of hTERT by a specific transcription factor. Alternatively, excess amounts of a particular transcription factor in a specific cell type or aberrant recruitment of that transcription factor to the hTERT promoter may interfere with the epigenetic stability of the hTERT promoter that may affect telomerase activity. The interactions between genetic and epigenetic factors will form a permissive or inhibitive condition for hTERT transcription depending on the specific cellular context. To elucidate these interactions, a new technique, ChIP-Bisulfite Sequencing Analysis (ChIP-BSA) was introduced in our studies for the first time, which is believed to resolve the technical problem by combining ChIP bound with the transcription factor and bisulfite sequencing analysis of the complex methylation pattern of hTERT. Our current findings presented here strongly suggest that genistein may play an important role in the epigenetic control of hTERT through hypomethylation at specific recognition sites for certain transcription factors within the hTERT promoter, indicating a cross-talk between genetic and epigenetic regulation in controlling hTERT transcription in response to genistein.
Epigenetic processes involving chromatin modification, such as histone acetylation, methylation and phosphorylation, are believed to play important roles in controlling hTERT gene transcription.28,29 Recent studies have proved that genistein can also influence the epigenetic regulation in prostate cancer cells.14 Chromatin analysis of the hTERT promoter in breast cancer cells provides another explanation for the complicated hTERT gene regulation in response to genistein. In terms of the chromatin histone methylation markers, we found that the inactive marker, trimethyl-H3K9, was enriched; whereas the active marker dimethyl-H3K4 was depleted throughout the hTERT promoter in response to genistein. However, there is no difference in the acetylated active marker of chromatin, H3, between the genistein-treated and -untreated breast cancer cells in the hTERT promoter region. This is consistent with a weak inhibitory effect of genistein on histone deacetylase (HDAC) activity as suggested by Fang et al.13 On the other hand, our results suggest that, at least for hTERT, genistein may be more effective in influencing the chromatin histone methylation pattern in determining hTERT expression.
Genistein, the soybean component, has been shown to inhibit tumorigenesis in different organs and cells. In our present studies, mechanisms of genistein-induced hTERT repression involving in DNA methylation regulation were proposed. Therefore, the inhibitory effects of genistein on DNA methyltransferases may have intrinsic synergistic effects on hTERT combined with other DNA methyltransferase inhibitors, such as 5-aza-dCyd.42 Indeed, our results (Fig. 6) show that genistein, at 50 μM, enhanced the activity of lower concentrations of 5-aza-dCyd (2 μM) to suppress hTERT and telomerase activity, which indicates that the additive effects between genistein and other epigenetic modulators could facilitate the availability of breast cancer prevention and therapy.
Geinistein bioavailability and concentrations that are used in this study are ongoing important issues and consuming dietary soybean could be potentially favorable in breast cancer prevention and therapy. First, our results in Figure 1a and supporting information show that the genistein concentration in this study can lead to cell growth arrest without induction of cell death, indicating that genstein could exert its anti-cancer effects through gene regulation such as we have shown for hTERT rather than inducing cell apoptosis. Secondly, genistein could be selectively concentrated in breast tissue with high distributions of estrogen receptors (ERα and ERβ) in this tissue because of the higher binding affinity of genistein for these receptors.43–45 Therefore the consumption of modest amounts of isolated isoflavone can exert potentially important biological effects such as epigenetic changes in human breast tissue. Thirdly, in vivo studies have consistently shown that effective genistein levels were lower than those of in vitro models46 indicating that genistein could act as prodrug-like molecules that undergo structural changes that favor potency against telomerase through impacting epigenetic regulation. On the basis of these observations, we therefore believe that dietary genistein could be applied in breast cancer prevention and therapy and its mechanism of action is likely through targeting the expression of the hTERT component of telomerase by affecting epigenetic regulation.
In summary, the findings from this study illustrate the multi-factorial mechanisms by which genistein induces repression of telomerase activity in both breast precancerous and cancer cells. Both the direct regulation of key transcription factors and epigenetic modulations are involved in genistein-induced repression of telomerase activity. More importantly, for the first time we found genistein treatment leads to site-specific hypomethylation of the hTERT promoter including E2F-1 binding sites resulting in an increase in binding of this repressor. We have also shown for the first time that genistein treatment can regulate the methylation status of histones in the hTERT promoter, further contributing to hTERT down-regulation. Genistein also shows an additive effect on hTERT repression in combination with another DNA methyltransferase inhibitor, such as 5-aza-dCyd. These findings have important implications for the application of genistein in breast cancer chemoprevention and therapy. Future efforts aimed at determining the appropriate administration of genistein and elucidating the further anti-cancer mechanisms are needed in vivo.
Supplementary Material
Acknowledgements
The authors thank Dr. Silvia Bacchetti (Department of Pathology and Molecular Medicine, McMaster University, Canada) for the luciferase constructs used in this investigation. They greatly appreciate Dr. Andra R. Frost (Department of Pathology and Cell Biology, University of Alabama at Birmingham) for providing the MCF10AT cells in this study.
Grant sponsor: National Cancer Institute; Grant number: RO1 CA129415; Grant sponsors: Susan G. Komen for the Cure, and the Glenn Foundation for Medical Research.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Zafonte BT, Hulit J, Amanatullah DT, Albanese C, Wang C, Rosen E, Reutens A, Sparano JA, Lisanti MP, Pestell RG. Cell-cycle dysregulation in breast cancer: breast cancer therapies targeting the cell cycle. Front Biosci. 2000;5:D938–D961. doi: 10.2741/zafonte. [DOI] [PubMed] [Google Scholar]
- 2.Diet, nutrition, and cancer. Executive summary of the report of the committee on diet, nutrition, and cancer. Assembly of life sciences, national research council. Cancer Res. 1983;43:3018–23. [PubMed] [Google Scholar]
- 3.Lee HP, Gourley L, Duffy SW, Estéve J, Lee J, Day NE. Dietary effects on breast-cancer risk in Singapore. Lancet. 1991;337:1197–200. doi: 10.1016/0140-6736(91)92867-2. [DOI] [PubMed] [Google Scholar]
- 4.Fang CY, Tseng M, Daly MB. Correlates of soy food consumption in women at increased risk for breast cancer. J Am Diet Assoc. 2005;105:1552–8. doi: 10.1016/j.jada.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Barnes S. Effect of genistein on in vitro and in vivo models of cancer. J Nutr. 1995;125:777S–783S. doi: 10.1093/jn/125.3_Suppl.777S. [DOI] [PubMed] [Google Scholar]
- 6.Fotsis T, Pepper M, Adlercreutz H, Fleischmann G, Hase T, Montesano R, Schweigerer L. Genistein, a dietary-derived inhibitor of in vitro angiogenesis. Proc Natl Acad Sci USA. 1993;90:2690–4. doi: 10.1073/pnas.90.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okura A, Arakawa H, Oka H, Yoshinari T, Monden Y. Effect of genistein on topoisomerase activity and on the growth of [Val 12]Haras-transformed NIH 3T3 cells. Biochem Biophys Res Commun. 1988;157:183–9. doi: 10.1016/s0006-291x(88)80030-5. [DOI] [PubMed] [Google Scholar]
- 8.Messina M, McCaskill-Stevens W, Lampe JM. Addressing the soy and breast cancer relationship: review, commentary, and workshop proceedings. J Natl Cancer Inst. 2006;98:1275–84. doi: 10.1093/jnci/djj356. [DOI] [PubMed] [Google Scholar]
- 9.Shon YH, Park SD, Nam KS. Effective chemopreventive activity of genistein against human breast cancer cells. J Biochem Mol Biol. 2006;39:448–51. doi: 10.5483/bmbrep.2006.39.4.448. [DOI] [PubMed] [Google Scholar]
- 10.Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem. 1987;262:5592–5. [PubMed] [Google Scholar]
- 11.Wei H, Wei L, Frenkel K, Bowen R, Barnes S. Inhibition of tumor promoter-induced hydrogen peroxide formation in vitro and in vivo by genistein. Nutr Cancer. 1993;20:1–12. doi: 10.1080/01635589309514265. [DOI] [PubMed] [Google Scholar]
- 12.Pagliacci MC, Smacchia M, Migliorati G, Grignani F, Riccardi C, Nicoletti I. Growth-inhibitory effects of the natural phyto-oestrogen genistein in MCF-7 human breast cancer cells. Eur J Cancer. 1994;30A:1675–82. doi: 10.1016/0959-8049(94)00262-4. [DOI] [PubMed] [Google Scholar]
- 13.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a. RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11:7033–41. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 14.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, Hirata H, Li LC, Zhao H, Okino ST, Place RF, Pookot D, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–44. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 15.Mentor-Marcel R, Lamartiniere CA, Eltoum IE, Greenberg NM, Elgavish A. Genistein in the diet reduces the incidence of poorly differentiated prostatic adenocarcinoma in transgenic mice (TRAMP) Cancer Res. 2001;61:6777–82. [PubMed] [Google Scholar]
- 16.Day JK, Bauer AM, DesBordes C, Zhuang Y, Kim BE, Newton LG, Nehra V, Forsee KM, MacDonald RS, Besch-Williford C, Huang TH, Lubahn DB. Genistein alters methylation patterns in mice. J Nutr. 2002;132:2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 17.Cabanes A, Wang M, Olivo S, DeAssis S, Gustafsson JA, Khan G, Hilakivi-Clarke L. Prepubertal estradiol and genistein exposures up-regulate BRCA1 mRNA and reduce mammary tumorigenesis. Carcinogenesis. 2004;25:741–8. doi: 10.1093/carcin/bgh065. [DOI] [PubMed] [Google Scholar]
- 18.Greider CM, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 19.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–19. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 20.Meyerson M, Counter CL, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–95. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 21.Kanaya T, Kyo S, Takakura M, Ito H, Namiki M, Inoue M. hTERT is a critical determinant of telomerase activity in renal-cell carcinoma. Int J Cancer. 1998;78:539–43. doi: 10.1002/(sici)1097-0215(19981123)78:5<539::aid-ijc2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24:1167–76. doi: 10.1093/carcin/bgg085. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Saldanha SN, Pate MS, Andrews LG, Tollefsbol TO. Epigenetic regulation of human telomerase reverse transcriptase promoter activity during cellular differentiation. Genes Chromosomes Cancer. 2004;41:26–37. doi: 10.1002/gcc.20058. [DOI] [PubMed] [Google Scholar]
- 25.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 26.Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–90. [PubMed] [Google Scholar]
- 27.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101:335–41. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 28.Cong YS, Bacchetti S. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J Biol Chem. 2000;275:35665–8. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]
- 29.Hou M, Wang X, Popov N, Zhang A, Zhao X, Zhou R, Zetterberg A, Björkholm M, Henriksson M, Gruber A, Xu D. The histone deacetylase inhibitor trichostatin A derepresses the telomerase reverse transcriptase (hTERT) gene in human cells. Exp Cell Res. 2002;274:25–34. doi: 10.1006/excr.2001.5462. [DOI] [PubMed] [Google Scholar]
- 30.Jagadeesh S, Kyo S, Banerjee PP. Genistein represses telomerase activity via both transcriptional and posttranslational mechanisms in human prostate cancer cells. Cancer Res. 2006;66:2107–15. doi: 10.1158/0008-5472.CAN-05-2494. [DOI] [PubMed] [Google Scholar]
- 31.Heppner GH, Wolman SR. MCF-10AT: a model for human breast cancer development. Breast J. 1999;5:122–9. doi: 10.1046/j.1524-4741.1999.00136.x. [DOI] [PubMed] [Google Scholar]
- 32.Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137–42. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Berletch JB, Green JG, Pate MS, Andrews LG, Tollefsbol TO. Telomerase inhibition by retinoids precedes cytodifferentiation of leukemia cells and may contribute to terminal differentiation. Mol Cancer Ther. 2004;3:1003–9. [PubMed] [Google Scholar]
- 34.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103:509–19. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holt SE, Aisner DL, Shay JW, Wright WE. Lack of cell cycle regulation of telomerase activity in human cells. Proc Natl Acad Sci USA. 1997;94:10687–92. doi: 10.1073/pnas.94.20.10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crowe DL, Nguyen DC, Tsang KJ, Kyo S. E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 2001;29:2789–94. doi: 10.1093/nar/29.13.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Popov N, Hou M, Wang Q, Björkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci USA. 2001;98:3826–31. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. 2000;97:6481–6. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed A, Tollefsbol TO. Telomeres, telomerase, and telomerase inhibition: clinical implications for cancer. J Am Geriatr Soc. 2003;51:116–22. doi: 10.1034/j.1601-5215.2002.51019.x. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Lai S, Andrews LG, Tollefsbol TO. Genetic and epigenetic modulation of telomerase activity in development and disease. Gene. 2004;340:1–10. doi: 10.1016/j.gene.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 42.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–95. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 43.Kuiper GG, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–70. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 44.Cassidy A, Faughnan M. Phyto-oestrogens through the life cycle. Proc Nutr Soc. 2000;59:489–96. doi: 10.1017/s0029665100000719. [DOI] [PubMed] [Google Scholar]
- 45.Gustafsson JA. Therapeutic potential of selective estrogen receptor modulators. Curr Opin Chem Biol. 1998;2:508–11. doi: 10.1016/s1367-5931(98)80127-0. [DOI] [PubMed] [Google Scholar]
- 46.Day JK, Bauer AM, DesBordes C, Zhuang Y, Kim BE, Newton LG, Nehra V, Forsee KM, MacDonald RS, Besch-Williford C, Huang TH, Lubahn DB. Genistein alters methylation patterns in mice. J Nutr. 2002;132:2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.