Abstract
RNA activities can be regulated by modulating the relative energies of all conformations in a folding landscape; however, it is often unknown precisely how peripheral elements perturb the overall landscape in the absence of discrete alternative folds (inactive ensemble). This work explores the effects of sequence and secondary structure in governing kinase ribozyme activity. Kin.46 catalyzes thiophosphoryl transfer from ATPγS onto the 5′ hydroxyl of polynucleotide substrates, and is regulated 10,000-fold by annealing an effector oligonucleotide to form activator helix P4. Transfer kinetics for an extensive series of ribozyme variants identified several dispensable internal single-stranded segments, in addition to a potential pseudoknot at the active site between segments J1/4 and J3/2 that is partially supported by compensatory rescue. Standard allosteric mechanisms were ruled out, such as formation of discrete repressive structures or docking P4 into the rest of the ribozyme via backbone 2′ hydroxyls. Instead, P4 serves both to complete an important structural element (100-fold contribution to the reaction relative to a P4-deleted variant) and to mitigate nonspecific, inhibitory effects of the single-stranded tail (an additional 100-fold contribution to the apparent rate constant, kobs). Thermodynamic activation parameters ΔH‡ and ΔS‡, calculated from the temperature dependence of kobs, varied with tail length and sequence. Inhibitory effects of the unpaired tail are largely enthalpic for short tails and are both enthalpic and entropic for longer tails. These results refine the structural view of this kinase ribozyme and highlight the importance of nonspecific ensemble effects in conformational regulation by peripheral elements.
Keywords: ribozyme kinetics, SELEX, RNA folding/structure, peripheral nucleotide sequences, Eyring analysis, phosphoryl transfer
INTRODUCTION
Conformational regulation of RNA activities is essential for extant RNA biology, as well as for engineering artificial RNA-based components and devices for synthetic biology. Conformational dynamics in natural RNAs have been recognized since at least the late 1970s in the form of transcriptional attenuation based on kinetic control of RNA folding (Oxender et al. 1979). Numerous riboswitches undergo metabolite-induced changes in secondary and tertiary structures to regulate transcription or translation (Edwards et al. 2007; Henkin 2008; Montange and Batey 2008; Wang and Breaker 2008). RNA conformational switching is also critical in the catalytic cycle of spliceosome-mediated pre-mRNA splicing and during ribosome assembly, where specialized helicases assist in these processes (for reviews, see Le Hir and Andersen 2008; Tarn and Chang 2009). Aptamer modules adjacent to the hammerhead ribozyme catalytic core stabilize the active structures in otherwise disordered ribozymes upon binding of their respective ligands, and they can shift the base-pairing register of two strands to favor productive (or repressed) pairing interactions in the active site (Tang and Breaker 1997; Koizumi et al. 1999; Soukup and Breaker 1999a; Win and Smolke 2008; Beisel and Smolke 2009; Win et al. 2009). Detection of specific RNA strands has been achieved by engineering large-scale rearrangement of base-pairing patterns that relieve competing inhibitory structures (Burke et al. 2002; Saksmerprome and Burke 2003, 2004; Cong and Nilsen-Hamilton 2005), or that complete required secondary structural elements (Vaish et al. 2003) in natural ribozymes. Similarly, artificial ribozymes have been selected or engineered to be activated or repressed by analytes ranging from proteins (Robertson and Ellington 2000; Robertson and Scott 2007) and oligonucleotides (Lorsch and Szostak 1994) to small organic molecules (Roth and Breaker 1998) (for reviews, see Silverman 2003; Win and Smolke 2007; Fastrez 2009). For some of these regulated RNAs, the regulation is classical allostery—transmission to the active site of analyte binding information from a distant effector site—while for others, the effector analyte is itself integral to the catalytic structure or mechanism (McCarthy et al. 2005).
Some systems of conformational regulation can be understood in terms of simple, two-state models in which the repressed state is a single, discrete structure that directly competes with the productive conformation. However, the inactive state can also reflect an ensemble of conformations that include fully unfolded molecules, partially folded “molten globules,” and nearly native states that require only local reorientation to assume the catalytically competent conformation. Recent computational advances for dealing with such ensembles include statistical mechanics approaches that enumerate and count the self-avoiding chain conformations to account for the excluded volume effect and other structural details (Cao and Chen 2006, 2009; Chen 2008; Cao et al. 2010; Liu and Chen 2010).
The chemical repertoire of natural RNA enzymes (ribozymes) is largely limited to nucleophilic displacement at substituted phosphates (such as phosphodiester exchange and hydrolysis) and at carbonyl centers (peptide bond formation within ribosomes). Artificial ribozymes derived from in vitro selection catalyze these and many additional chemical transformations. These capabilities have provided new tools for synthetic biology, propelled efforts to develop ribozyme-based therapies, and supported RNA world hypotheses that RNA preceded proteins as the dominant biological catalyst (for reviews, see Burke 2004; Chen et al. 2007; Cochrane and Strobel 2008). The recent discovery of metabolite-linked and cofactor-linked RNAs in cell extracts (Chen et al. 2009; Kowtoniuk et al. 2009) further raises the possibility that ribozymes may have important roles in metabolic transformations of small molecules in modern biology. Phosphoryl transfer is of special interest because of its ubiquitous role in cellular biology, and phosphoryl transfer ribozymes (and deoxyribozymes) have been isolated and described by us (Rhee and Burke 2004; Saran et al. 2005, 2006; Biondi et al. 2010) and by others (Lorsch and Szostak 1994, 1995; Li and Breaker 1999; Wang et al. 2002; Achenbach et al. 2005; Curtis and Bartel 2005; Chiuman and Li 2006; McManus and Li 2007, 2008). Nevertheless, there is still relatively little known about the mechanisms by which nucleic acids catalyze phosphoryl transfer or the structures that they use to do so.
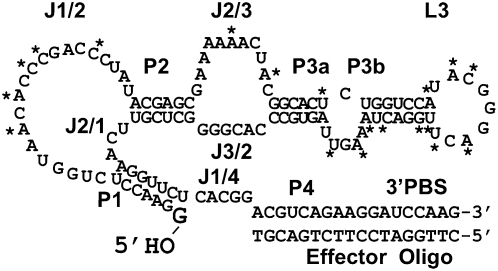
The present work dissects the structural and thermodynamic basis of catalytic activity and conformational regulation within the Kin.46 self-phosphorylating ribozyme. This RNA catalyzes transfer of the gamma-(thio)phosphoryl group of ATP (or ATPγS) onto the 5′ end of an RNA substrate. This acceptor substrate can either be a short, diffusible strand (intermolecular catalysis, or reaction in trans) or it can be covalently attached to the ribozyme (intramolecular catalysis, or reaction in cis). In either configuration, the guanosine 5′ hydroxyl group that acts as phosphoryl acceptor is held in the active site through base pairing with a 7-nucleotide internal guide sequence to form stem P1 (Fig. 1). During the initial in vitro selection of the Kin.46 ribozyme from a random-sequence library, an 18 nt DNA strand complementary to the ribozyme 3′ primer binding site (PBS) was present for affinity chromatography capture and cDNA synthesis. Formation of helix P4 upon binding of this 18mer “effector oligo” (EO) was later found to be required for optimal ribozyme activity, increasing kobs by almost 10,000-fold (Lorsch and Szostak 1994). We previously exploited P4-driven activation to explore the architecture of Kin.46, using topologically rearranged variants (Cho and Burke 2006); however, the underlying molecular basis for this activation has not been explored. It could, for example, represent completion of the core catalytic structure, classic allostery through binding at a site that is distinct from the core, or a combination of these effects.
FIGURE 1.
Secondary structural features of ribozyme Kin.46, labeled as described in the text. The 5′ hydroxyl that is phosphorylated by the ribozyme is shown in large text. Asterisks indicate positions where polymorphisms were observed in the original selection (Lorsch and Szostak 1994). All other positions were invariant.
To determine the mode of conformational activation of the Kin.46 ribozyme, the present work uses activity assays and thermodynamic analysis of Kin.46 variants. We first defined the sequence requirements for thiophosphoryl transfer and refined the secondary structure model of the Kin.46 ribozyme. We demonstrate that most of the single-stranded joining regions can be discarded without loss of activity or regulation, but that two of the joining regions base pair to form a required helix within or near the catalytic active site. Sequence and nucleotide composition of activator helix P4 have little effect on EO-mediated activation or on substrate binding energy (KmATPγS). Rather than rearranging strand complementarity to form alternate helices that compete with the active structure, we find that annealing of the EO serves two separate functions. First, formation of the P4 activator helix completes an important, but nonessential structural element that contributes ∼100-fold to Kin.46 catalytic activity relative to a helix-deleted variant. Second, sequestering the unpaired tail mitigates enthalpic and (in most cases) entropic penalties associated with the unpaired 3′ tail to stimulate kobs by another ∼100-fold. Thus, the influence of the 3′ tail on Kin.46 activity appears to be mediated through multiple underlying mechanisms that redistribute the conformational ensemble.
RESULTS
Four to seven unpaired nucleotides are required within J1/4
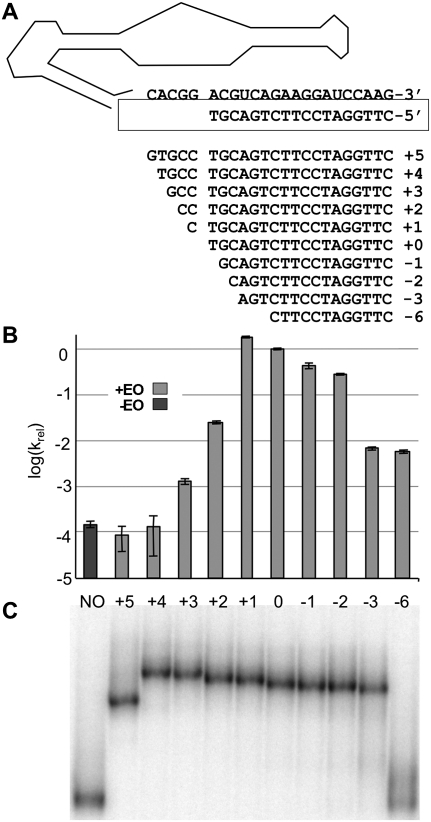
The effector oligonucleotide (EO) binds to the 3′ tail of ribozyme Kin.46 to form helix P4, which is separated from active site helix P1 by a 5 nt joining segment denoted J1/4 (“Joining segment between helices P1 and P4”). As a first step toward defining the sequence requirements for activity and activation of Kin.46, we systematically varied the number of nucleotides exposed in J1/4. The original 18 nt EO (EO0) was lengthened on its 3′ end to form new effectors denoted EO+1 to EO+5 that range in size from 19 to 23 nt. The EO was also shortened from its 3′ end (EO-1 to EO-6) to sizes of 17 to 12 nt (Fig. 2A). Product formation was then monitored, and kobs values calculated, for Kin.46 annealed to each EO (Fig. 2B). The parental ribozyme yielded a kobs value of 0.06 ± 0.01 min−1 in the presence of EO0. Omitting the effector oligonucleotide reduced kobs nearly four orders of magnitude (Fig. 2B, dark gray bar), similar to previous observations for related Kin.46 variants (Lorsch and Szostak 1994). Four of the 10 effector oligos tested (EO+1, EO0, EO-1, and EO-2) stimulated Kin.46 activity to similar degrees, with the greatest stimulation observed for EO+1 (approximately twofold faster than Kin.46 annealed to EO0). Longer EOs that pair with more of the original J1/4 region rapidly lost the ability to activate Kin.46. Similarly, activation dropped by almost two orders of magnitude as the EO was shortened to expose nucleotides from the original P4 helix. Optimal activity therefore requires that at least four nucleotides of J1/4 be unpaired, and that this unpaired segment be no longer than seven nucleotides.
FIGURE 2.
Effect on Kin.46 ribozyme activity of extending or shortening the effector oligonucleotide. For Figures 2, 3, 4, and 6, light gray bars indicate data obtained in the presence of effector oligonucleotides, and dark gray bars indicate data obtained when EO was omitted from the reaction. (A) Sequences for EO+5 through EO-6 are shown 3′-to-5′ below the sequence of Kin.46 J1/4 and PBS strands (structural elements are as detailed in Fig. 1). (B) Log(kobs) values are plotted relative to the values obtained in the presence of EO0 or without EO (sample “NO”). (C) Comparison of electrophoretic mobilities of Kin.46 with EOs of varying lengths in a 10% nondenaturing polyacrylamide gel. Labels above the lanes align with bar graphs in B. Identical patterns of relative migration were also seen for all 10 samples run on 8% and 6% native polyacrylamide gels (data not shown).
P4 formation does not induce overall conformational change
To determine whether differential stimulation observed for the various EOs was due to differences in the impact of their binding on overall ribozyme conformation, each assembled complex was subjected to nondenaturing PAGE analysis. Nine of the 10 EOs stably bind Kin.46, as evidenced by the appearance of single bands of reduced mobility in the presence of these EOs (Fig. 2C). For ribozymes bound to effector oligos EO+4 to EO-3, electrophoretic mobilities increased as the EOs were shortened, as expected from the progressively reduced masses of the annealed complexes; however, there is no sharp transition in mobility that correlates with a change in activity. Curiously, Kin.46 mobility was greater when it was annealed to EO+5 than when it was annealed to EO+4, even though the EO+5 complex is one nucleotide larger. As this change in conformation did not correspond to any change in activity, it was not further explored. The dodecamer EO-6 forms a complex with Kin.46 that is only moderately stable. The degree of binding by EO-6 in solution is sufficient to activate thiophosphoryl transfer 30-fold (Fig. 2B, cf. columns “NO” and “-6”), but it dissociates during electrophoresis to produce a smear just above the free RNA. Gel-shift analysis therefore does not reveal any large-scale conformational difference between the more activated and the less activated ribozymes. Independently, the nuclease sensitivity of Kin.46 was monitored in the presence and absence of ribonucleases S1 (cleaves single-stranded RNA), T1 (cleaves single-stranded RNA after guanosines), and V1 (cleaves double-stranded RNA). Cleavage patterns were consistent with the secondary structure shown in Figure 1, with especially prominent S1 cleavages in J1/2 and L3 (data not shown). Moreover, the only segment of the ribozyme for which the cleavage pattern changed upon addition of EO was the PBS itself upon formation of helix P4. These results are consistent with the lack of change in electrophoretic gel mobilities for different length EOs (Fig. 2C), and rule out gross changes in base-pairing patterns between the activated and nonactivated states.
Systematic deletion of the ribozyme 3′ PBS
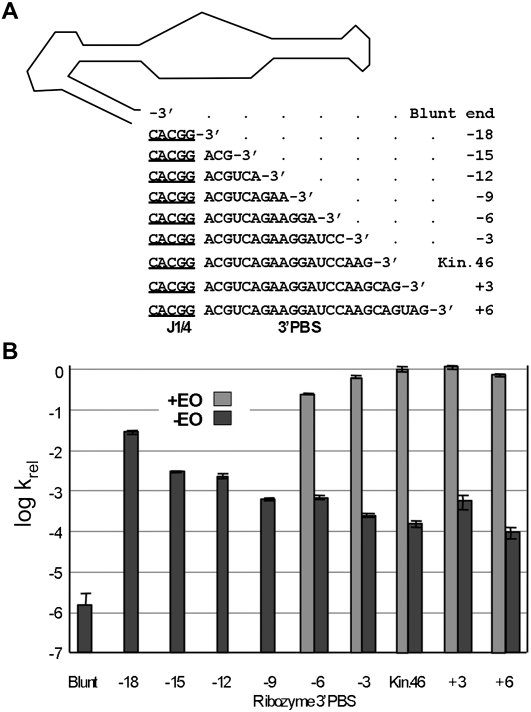
To determine whether the length of the 3′ single-stranded tail contributes to regulation, Kin.46 variants were generated that either lengthened or shortened the PBS strand of P4, and the activities of each variant were measured in the presence and absence of the corresponding EOs (Fig. 3). EOs were not used for ribozymes with 3′ PBS tails shorter than 12 nt because of instability of EO-6 in the native gel shift assays above. Removing both the PBS and J1/4 yielded a blunt-ended ribozyme (BE) that reacted nearly a million-fold more slowly than activated Kin.46 and almost two orders of magnitude more slowly than the unactivated Kin.46, clearly demonstrating the importance of J1/4. Restoring only the five nucleotides of J1/4 (ribozyme “-18” in Fig. 3) increased the apparent rate constant of the unactivated ribozyme to within two orders of magnitude of that of the parental activated ribozyme. Restoring the 3′ tail in trinucleotide increments produced a steady decline in the kobs for unactivated ribozymes, with the exception of a slight rise at +3. Thus, the ribozyme is most active with a fully-formed P4 stem, and it is least active when J1/4 is removed (106-fold diminished kobs in “blunt”) or when long, single-stranded tails are present (104-fold diminished in “-3” to “+6”), and it shows intermediate activity when both strands of the P4 stem are completely removed (102-fold diminished in “-18”).
FIGURE 3.
Effect of 3′ tail length on activity and allostery. (A) “Blunt end” indicates a ribozyme terminating at the 3′ end of P1. Numbering indicates the lengths of the 3′ tail relative to the original 3′PBS. (B) Activities for “+EO” were only measured for ribozymes “-6” through “+6.”
P4 composition and sequence
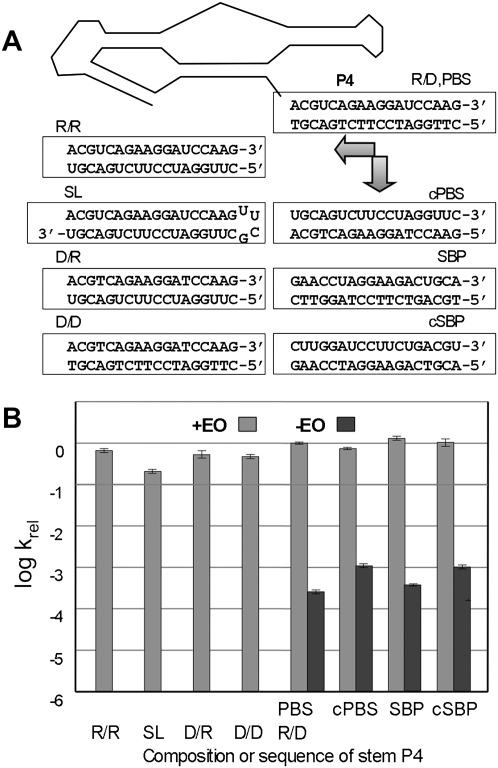
Ribose 2′ hydroxyls can participate in RNA structure through stabilization of tertiary and noncanonical secondary structures, as in the docking of stem P1 into the catalytic core of the self-splicing Group I intron (Pyle and Cech 1991). To determine whether 2′ hydroxyls are involved in conformational activation or repression of Kin.46, the composition and sequence of P4 were varied by replacing the 3′ terminal 18 ribonucleotides of the ribozyme with an oligodeoxy 3′ PBS tail (PBSD), and by using both RNA (EOR) and DNA (EOD) versions of EO0. Reaction kinetics were then measured for all four DNA/RNA combinations (Fig. 4). Activation with the original DNA effector (designated PBSR/EOD) demonstrated the highest activity, followed closely by activation with an RNA version of the effector to generate an all-RNA P4 helix (PBSR/EOR). An additional ∼4-fold reduction in kobs is observed for an all-RNA stem P4 generated as an intramolecular stem–loop structure (ribozyme “SL”) with the sequence UUCG (Fig. 4) or GAAA (data not shown). The two complexes containing a 3′ DNA tail (PBSD/EOD and PBSD/EOR) each showed approximately half the activity of the ribozyme carrying the original PBSR/EOD helix. These differences are minor compared to the effects of including or omitting the respective EO. As such, these results establish that the 2′ hydroxyls of P4 are not required for Kin.46 regulation, and that helix formation alone is sufficient for normal or near-normal activity. Further, since DNA helices are generally B-form, RNA helices are A-form, and DNA/RNA hybrid duplexes contain characteristics of both (Gyi et al. 1996), gross helical structure of P4 also appears to have only limited importance for activity, again indicating the absence or minimal participation of tertiary interactions with the P4 backbone.
FIGURE 4.
Effect of P4 helix composition on ribozyme activity. (A) P4 variants are shown below a schematic of Kin.46. “SL” is a stem–loop construct in which the ribozyme 3′ end folds back on itself to form the activating helix. Essentially identical data were obtained for an SL construct in which the UUCG loop was replaced with a slightly less stable GAAA tetraloop (Supplemental Table S1; data not shown). (B) Relative apparent initial rate constants for P4 variants shown in A. Apparent binding affinity for donor substrates is not adversely affected by altering the sequence of the 3′ tail. Km values for all four tail variants are within experimental error of each other, both in the presence or absence of EO (0.5–1.6 ± 0.4 μM) (Supplemental Table S1), and all are well below the substrate concentration used in the Eyring analysis (10 μM).
To eliminate potential specific requirements of P4 sequence in regulating Kin.46, the 3′ PBS sequence was changed to its direct complement (ribozyme “cPBS”), to its reverse sequence (“SBP”), and to its reverse complement (“cSBP”). In the presence of their corresponding EOs (Fig. 4), light gray bars), kobs values for all three mutant constructs were within a factor of two of the value for the native Kin.46. Moreover, kobs values dropped at least 1000-fold for all four ribozymes when EO was omitted from the reaction (Fig. 4, dark gray bars). Thus, sequence-specific contacts between either the single-stranded 3′ PBS or P4 helix with the rest of the ribozyme are not required either for inhibition or for activation, and the only requirement for optimal activity is a correctly positioned P4 helix.
Base-pairing interactions near the catalytic core
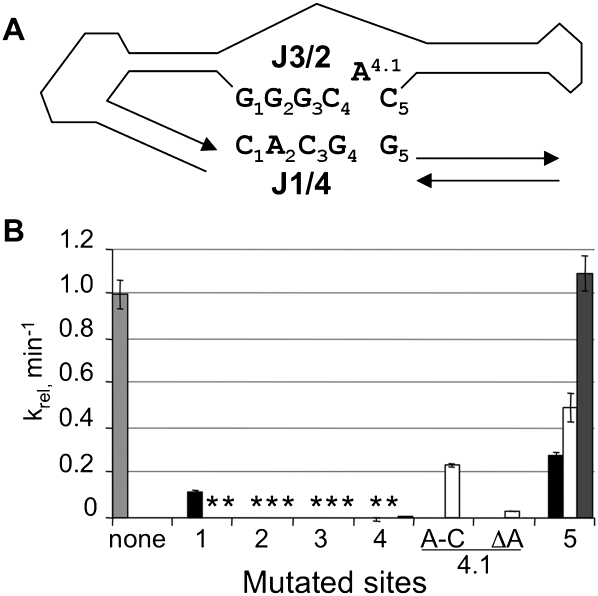
Complementarity between portions of J1/4 and joining region J3/2 raises the possibility of a pseudoknot at the catalytic active site. Potential long-range base-pairing interactions include several Watson-Crick pairs, one G•A pair, and one unpaired adenosine (Fig. 5A). Such interactions could explain the inhibitory effects of 3′-extended EOs that pair with J1/4. To assess the significance of these potential interactions, each nucleotide in J1/4 and J3/2 was mutated singly or pair-wise to its Watson-Crick complement, and catalytic activities were measured in the presence of EO0 (Fig. 5B). Single mutations at positions 2, 3, and 4 in J1/4 abolished activity, with no measurable product formation after 40 min (>1000-fold reduced activity). Similarly, mutations in J3/2 at five of the six positions inactivated the ribozyme. Mutating the unpaired adenosine at position 4.1 resulted in moderate (A-to-C substitution) or severe (deletion) reduction in activity. When compensatory point mutations were introduced simultaneously at position 5 in both J1/4 and J3/2, catalytic activity was fully rescued, consistent with formation of a productive Watson-Crick pair between J1/4 and J3/2 at this position. In contrast, catalytic activity was not rescued when compensatory mutations were introduced simultaneously in J1/4 and J3/2 for any of the positions 1 through 4. If base-pairing interactions exist at any of these sites, proper assembly of the active site may require specific structural features that cannot be readily provided by other Watson-Crick pairs. The intolerance of mutations in J1/4 and J3/2, the absolute sequence conservation at these positions within the Kin.46 family of class I kinases (Fig. 1), and the dramatically reduced activation by effector oligos that base pair with J1/4 (Fig. 2) all demonstrate that these specific nucleotides are required for optimal activity.
FIGURE 5.
Potential tertiary interactions in Kin.46. (A) Base numbering of nucleotides in J3/2 and J1/4. The rest of the ribozyme is shown schematically. (B) Activities of mutated ribozymes carrying mutations at the positions indicated, normalized to the activity of the original Kin.46, in the presence of EO0. For each mutant, the indicated position was changed to its Watson-Crick complement (A changed to U; G changed to C, etc.). The light gray bar, representing the original Kin.46 ribozyme, indicates no changes; the black bars, mutations within J1/4 only; white bars, mutations within J3/2 only; the dark gray bar, compensatory mutations within both strands, as detailed in the text. A-C is an A-to-C mutation at position 4.1; ΔA is a deletion of this same nucleotide. Where no bars are visible (*), measured activity was >1000-fold reduced relative to activated Kin.46.
Unpaired nucleotides in P3, L3, and J1/2 are dispensable for catalysis and activation
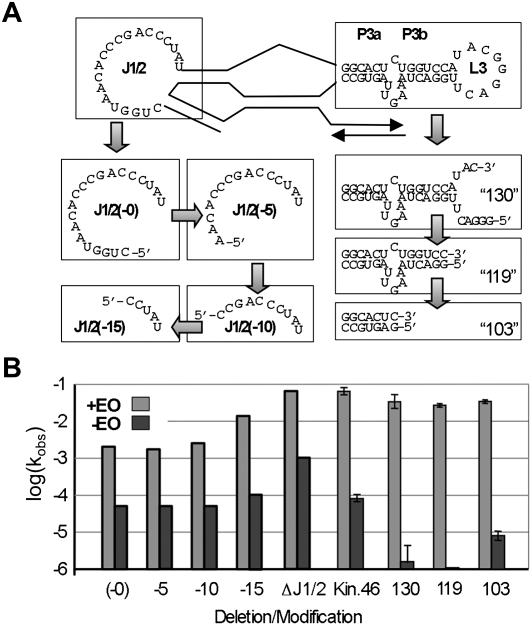
If inhibitory structures are formed as a result of specific interactions between PBS and any of the unpaired nucleotides, then internally deleted constructs should be expected to show a decreased dependence on the presence of EO. To facilitate removal of internal unpaired nucleotides that could participate in forming alternative secondary structures during EO-mediated activation, Kin.46 variants were assembled by annealing separate 5′ and 3′ fragments, yielding active ribozymes with discontinuities and deletions of various sizes in J1/2, L3, and/or P3. Strands that begin with 5′C or 5′A were generated by a “tandem ribozyme” design in which the highly efficient, self-cleaving hammerhead ribozyme, “RzB” (Saksmerprome et al. 2004), was appended onto the 5′ termini of each transcript, and the mature cleavage products that accumulated during transcription were purified (Supplemental Fig. S1). Initial mutations targeted joining segment J1/2. Previous work showed that Kin.46 can act in trans upon a 7 nt exogenous substrate when the connection between P1 and J1/2 was severed (Lorsch and Szostak 1995). That species, referred to here as “J1/2(-0),” catalyzed thiophosphoryl transfer with a kobs value that was ∼5% that the parental Kin.46 construct (Fig. 6). Activity in the presence of EO increased as J1/2 nucleotides were removed from the 5′ end of the fragmented ribozyme. When the entire J1/2 segment was deleted (“ΔJ1/2”), activity was indistinguishable from that of Kin.46 annealed to EO0, establishing that none of J1/2 is required for catalysis. Furthermore, all of these ribozymes were repressed ∼50- to 200-fold in the absence of EO0. Similarly, ribozymes carrying disruptions and/or deletions within L3 had a minimal effect on the observed rate constant in the presence of EO while retaining strong repression when EO is omitted (Fig. 6). Thus, the repressed state cannot be ascribed to specific interactions between the ribozyme 3′ tail and single-stranded nucleotides in either J1/2 or L3.
FIGURE 6.
Effect of internal deletions on split kinase ribozyme activity and allosteric activation. (A) The J1/2 loop was disconnected from the 7 nt substrate strand using hammerhead-kinase tandem ribozyme transcripts to remove 0, 5, 10, or 15 nucleotides, as indicated, or all 20 nucleotides (ΔJ1/2, structure not shown explicitly). The L3 loop was similarly interrupted and deletions were made as indicated. Numbers in the corners of each box indicate the sum of the lengths of the 5′ and 3′ segments of the annealed ribozyme. (B) Activities and allosteric activation of “split” ribozyme constructs compared to wild-type. The light gray bars indicate observed rates in the presence of EO; dark gray bars indicate rates in the absence of EO.
Entropic and enthalpic contributions to transition state activation energies upon annealing effector oligos
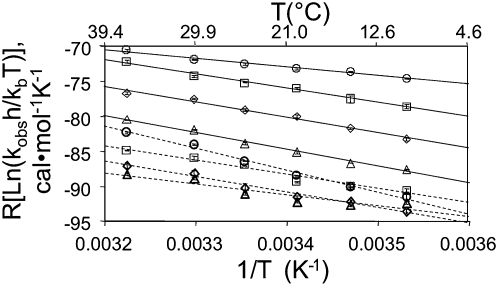
The mechanism by which a given mutation contributes to activity or to activation should be reflected in the enthalpy and/or entropy changes required to achieve the transition state for the rate-limiting step(s) in each reaction. Therefore, kobs values were calculated for ribozyme in the presence or absence of EO at various temperatures from 10°C–37°C. Plotting the log of the apparent rate constant against the inverse temperature (Fig. 7) allowed calculation of the entropy (ΔS‡) and enthalpy (ΔH‡) of activation for each condition using Eyring analysis (Equation 1, below), which extends the Arrhenius temperature dependence of reaction rates to include the transition state (Wynne-Jones and Eyring 1935), with ΔH‡ given by the slope of the plot and ΔS‡ by the extrapolated y-intercept. Eyring plots remained linear throughout the temperature range for most mutants analyzed, as expected if the rate-limiting step remains unchanged over this temperature range. For the parental Kin.46 ribozyme in the presence of EO0, we calculated ΔH‡ to be 12.3 ± 0.9 kcal/mol and ΔS‡ to be –31 ± 3 entropy units (eu). These values are similar to those obtained previously for a trans-acting version of this ribozyme (11 kcal/mol and –33 eu, respectively) (Lorsch and Szostak 1995), and they can be used to calculate a ΔG‡ value of 21 ± 1 kcal/mol at 20°C. In the absence of effector oligo, the three corresponding values shift to 17 ± 3 kcal/mol, –34 ± 10 eu, and 27 ± 4 kcal/mol, respectively. Annealing the effector oligo thus favors both the enthalpic and entropic components of ΔG‡, with a larger contribution from the change in ΔH‡ than from the change in TΔS‡. The similar calculated ΔΔG‡ values for ribozymes with altered 3′ tails (cPBS, SBP, and cSBP) (Supplemental Fig. S2; Supplemental Table S1) reflects their similar degrees of activation upon annealing to their respective EOs (Fig. 4). Interestingly, energetically equivalent stabilization of transition states is achieved for these 3′ tail variants by nonequivalent contributions from the activation enthalpies and entropies.
FIGURE 7.
Eyring plots of autokinase activity by Kin.46 mutants with alterations in 3′ tail length. Circles with solid lines, represent original Kin.46 in the presence of EO0. All other reactions are in the absence of EO. Squares with solid lines indicate ribozyme “-18” (18 nucleotides removed from the 3′ end of the original Kin.46); diamonds with solid lines indicate “-15”; triangles with solid lines indicate “-12”; circles with dashed lines indicate “-9”; squares with dashed lines indicate “-6”; diamonds with dashed lines indicate “-3”; triangles with dashed lines represent original Kin.46 without EO. Error bars reflect standard deviations of three or more measurements at each temperature.
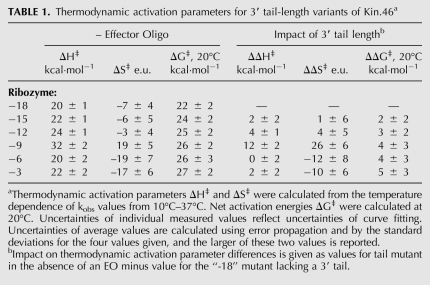
Enthalpic destabilization by short 3′ tails
The 3′ tail-length variants had substantial and systematic impact on overall kinetic behavior, especially for the ribozymes with the shortest 3′ tails (Fig. 3); they therefore represent an ideal platform for further assessing the underlying mechanisms by which P4 and unpaired 3′ tails contribute to ribozyme activity. Using ΔH‡ and ΔS‡ values from Eyring analysis, the calculated ΔG‡ values increased with tail length, as expected from the detrimental impact of increasing tail length on initial velocity. Where the trend in reaction rate contants was most striking (ribozymes –18, –15, –12, and –9), the increase in ΔG‡ values is due to a steady, corresponding increase in ΔH‡ values, which is only partially offset by a modest decrease in the entropic cost of the reaction (Table 1). For ribozymes with still longer 3′ tails (–6, –3, and –0/PBS), the apparent enthalpic cost of accessing the transition state abruptly decreases, along with a concomitant increase in the entropic penalty. This entropy/enthalpy compensation accounts for the fact that changes in tail length have little impact on the reaction at 20°C for unactivated ribozymes with long tails.
TABLE 1.
Thermodynamic activation parameters for 3′ tail-length variants of Kin.46a
DISCUSSION
The present work systematically dissects the sequences and secondary structures required for catalytic activity and oligonucleotide-mediated regulation of the Kin.46 ribozyme. We find that joining region J1/4 cannot be removed (Fig. 3, “blunt”), and that activity is lost if J1/4 is mutated or if too little or too much of it is paired with the effector oligo (Fig. 2). These observations are consistent with potential pairing between J1/4 and J3/2, which is supported by compensatory mutational rescue at position 5 (Fig. 5). The other positions in strands J1/4 and J3/2 are highly sensitive to mutation, even when potential base-pairing is retained, suggesting that they are integral to the active site. In contrast, the single-stranded nucleotides within J1/2, L3, and the bulges within P3 are all nonessential for catalysis, consistent with the fact that 17 of 19 observed polymorphisms in the originally published sequence set are located in these structural elements (asterisks in Fig. 1).
A simple mechanism for achieving oligonucleotide-induced regulation would be for the ribozyme to assume a specific, repressed secondary or tertiary structure in the absence of effector, and then to switch to a different, active secondary structure upon binding to the EO. This is essentially the mechanism utilized by many riboswitches to effect gene regulation, and by hammerhead ribozymes engineered to be responsive to oligonucleotides (Burke et al. 2002; Saksmerprome and Burke 2003, 2004) and some small molecule effectors (Soukup and Breaker 1999a; Win and Smolke 2008). However, all of the mutations we analyzed that were capable of binding an effector oligo were strongly stimulated by binding EO, irrespective of the sequence or composition of the 3′ tail, and irrespective of removing the single-stranded J1/2, L3, and P3 bulge nucleotides (Figs. 4 and 5). Native gel electrophoresis revealed no gross conformational rearrangement correlating with loss of activity (Fig. 2C), and EO binding did not alter nuclease sensitivity, other than at the EO-binding site in the 3′ tail (not shown). Thus, the inactive state of the unactivated ribozyme is unlikely to represent a specific structure, and instead likely reflects an ensemble of near-native folds that position the substrates nonproductively.
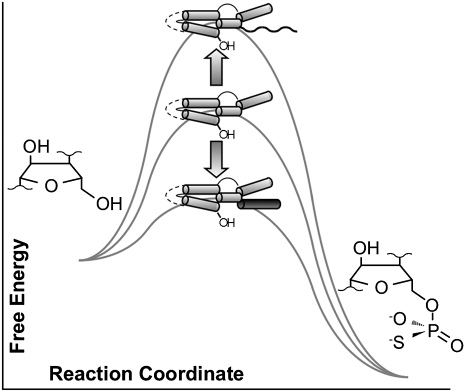
Rather than rearranging strand complementarity to form alternate helices that compete with the active structure, these data identify two roles for the P4 activator helix. In discussing these roles, it is convenient to consider the P4-deleted form of the ribozyme (ΔP4) as a reference state (Fig. 8). First, P4 acts as a peripheral element that sequesters the unpaired tail and mitigates enthalpic and (in most cases) entropic penalties associated with the unpaired 3′ tail, stimulating the observed rate constant by another ∼100-fold (kobs(ΔP4)/kobs(Kin.46 -EO) ≈ 100). Thus, the unpaired 3′ tail plays a direct, nonspecific inhibitory role that is overcome by annealing with EO. Second, the proximal portion of helix P4 helps to stabilize the active site. Addition of the P4 activator helix to the ΔP4 ribozyme completes a structural element that contributes ∼100-fold to catalysis (kobs(Kin.46 +EO)/kobs(ΔP4) ≈ 100).
FIGURE 8.
Effect of activator helix P4 and 3′ tail on transition state stabilization by Kin.46. Relative to the tail-less “-18” ribozyme (center), adding an unpaired 3′ tail increases the barrier to RNA-catalyzed thiophosphorylation (top), while adding both strands of P4 decreases the free energy barrier (bottom).
The nature of the inhibition by the single-stranded tail is a function of tail length. There is a strong inverse correlation between tail length and activity in the absence of EO (Fig. 3); ΔS‡ becomes more favorable as the tail is lengthened from “-18” to “-9,” implying that less ordering is required to take place to access the transition state. The values of ΔH‡, on the other hand, become increasingly disfavorable for the unactivated ribozymes as the tail is lengthened, and this enthalpic trend accounts for the initial decline in reactivity (Table 1). If these short-tail ribozymes are forming inhibitory interactions that need to be broken to access the transition state, the mutational analysis above establishes that such interactions must be nonspecific with respect to sequence and backbone composition. Activation parameters for the –9 variant are significantly different from the other ribozymes. Although not explored in detail, this discontinuity may arise from the differential contributions of tail length to activation parameters for long and short tails, and the intermediate position of the –9 tail between these two sets.
Optimization of ribozyme activity often includes removing unstructured peripheral nucleotide segments that decrease the functionally folded fraction. For cis-reactive Kin.46 in the absence of EO, autothiophosphorylation was improved by trimming the single-stranded 3′ PBS tail (Fig. 3, dark gray bars), and for trans-reactive ribozymes, phosphorylation of the 7 nt substrate was improved by trimming J1/2 (Fig. 6, light gray bars). Similarly, Unrau and coworkers improved a nucleotide synthase ribozyme through random internal deletions to tighten the fold (Chapple et al. 2003), and the utility of long random-sequence libraries was found to be limited by a combination of misfolding and reduced RT/PCR amplification efficiency (Coleman and Huang 2005). Superfluous flanking sequences can clearly contribute to formation of alternatively paired structures that compete with the productive fold in many cases. However, the present work shows that these floppy flanks can also lead to nonspecific perturbation that increases either the enthalpic (short and long tails) or entropic (long tails) cost of accessing the transition state.
What is the transition state that is being monitored in these analyses? Previous work with Kin.46 suggested that the rate-limiting step is formation of a metaphosphate intermediate in a largely dissociative chemical mechanism (Lorsch and Szostak 1995) (denoted “SN1”-like in older nomenclatures [Ingold 1953, 1969], or “DN+AN” in more recent nomenclature [Guthrie 1988]). This conclusion was based on the sensitivity of the reaction rate to the nonbridging sulfur atom, insensitivity of the rate to pH (ruling out nucleophile deprotonation or leaving group protonation as being rate limiting), and the equivalent binding affinities of ADP and ATPγS (ruling out ground-state perturbation). However, the pH data may be less informative than originally believed, as nucleophile activation is sometimes considered not to be a major catalytic strategy of phosphoryl transfer enzymes, based on equivalent rates of uncatalyzed phosphorylation among organic alcohols with widely ranging pKa values (Admiraal and Herschlag 1995). Furthermore, while uncatalyzed phosphoryl transfers exhibit near-zero entropies of activation (Thatcher and Kluger 1978; Benkovic and Richardson 1987), the values measured here and previously (Lorsch and Szostak 1995) are large and negative. Therefore, steps other than chemistry may be rate-limiting for these phosphotransferase ribozymes.
Conformational dynamics can also make major contributions to reaction rates. Reactions at phosphate centers are strongly dependent upon distance (<3Å) and geometry (optimal near 180°), as has been demonstrated previously in relation to intramolecular phosphodiester exchange (Soukup and Breaker 1999b). Similarly, the 2′ and 5′ OH at the scissile phosphates within minimal hammerhead ribozymes must reorient from the ground-state conformation for in-line attack (Pley et al. 1994; Scott et al. 1995; Murray et al. 1998). The slow conformational change masks the faster chemical step and produces the overall monophasic kinetics of “well-behaved” hammerhead ribozymes. Peripheral loop-loop interactions facilitate this alignment in natural hammerheads (Martick and Scott 2006). Furthermore, the relative reactivities of active site water and imidazole in wild type and His-to-Gly mutants of nucleoside diphosphate kinase has been ascribed to nucleophile positioning in the active site (Admiraal et al. 1999, 2001). For kinase ribozymes, precise positioning of the (thio)phosphoryl donor and acceptor substrates is similarly expected to be a major determinant of the observed reaction rates. We conclude, then, that the role of helix P4 in activating the Kin.46 ribozyme is to shift the energetic landscape among the ensemble of near-native conformations to increase the frequency with which the ribozyme accesses highly-reactive geometries that productively juxtapose the 5′OH acceptor and (thio)phosphoryl donor species.
MATERIALS AND METHODS
Materials
Transcription templates and other single-stranded DNA were synthesized by Integrated DNA Technologies. RNA was generated by run-off transcription using T7 RNA polymerase with [α-32P]UTP (MP Biomedicals) and eluted from denaturing 8% polyacrylamide gels. RNA strands with 5′ ends that are incompatible with efficient transcription by T7 RNA polymerase (such as 5′ AACAC…) were generated by appending the highly efficient self-cleaving hammerhead ribozyme, RzB (Saksmerprome et al. 2004) onto the 5′ end of the transcript and gel-purifying the cotranscriptional cleavage product (Supplemental Fig. S1). Transcripts containing the 5′ guanosine that acts as phosphoryl acceptor were generated by dephosphorylating gel-purified transcripts with calf intestinal phosphatase (CIP) (New England Biolabs). Control RNA containing a 5′ thiophosphate were generated enzymatically by incubating 5′-guanosine transcripts with polynucleotide kinase (PNK, New England Biolabs) and ATPγS to effect the transfer of the γ-thiophosphoryl group from ATPγS to the RNA 5′ hydroxyl. RNA-DNA hybrid ribozymes were made by annealing a DNA oligo with a long 5′ overhang that then served as template for RNA-primed DNA synthesis by Klenow exo− (New England Biolabs) (Huang and Szostak 1996). We followed Igloi (Igloi 1988) for the synthesis of ([N-acryloylamino]phenyl) mercuric chloride (APM) from (4-aminophenyl) mercuric acetate (Sigma-Aldrich) and acryloyl chloride (Fluka). Denaturing 6% APM-PAGE gels were cast with a layer (∼4 − 5 cm) containing 24 μg APM/mL polyacrylamide solution (∼60 μM APM) (Rhee and Burke 2004). After polymerization, a non-APM polyacrylamide layer was stacked on top to create an interface for the separation and quantitation of thiophosphorylated RNA. Tris(2-carboxyethyl)phosphine (TCEP) was purchased from Pierce. Additional enzymes were purchased from New England Biolabs and Promega. All other chemicals were from Sigma-Aldrich.
Kinetics and native gel electrophoresis
Kin.46 RNA was purified as above, and self-thiophosphorylation was carried out in 50 mM MgCl2, 150 mM PIPES (pH adjusted to 7.0 with KOH), and 200 mM KCl (400 mM total K+ including KOH). All polynucleotides, including Kin.46 RNA (1 μM final concentration) and effector oligo (2 μM final), were heated together for 1 min at 90°C in KCl and PIPES and allowed to cool to room temperature (20°C − 22°C). For reactions assaying internal deletions within the ribozyme, final concentrations were 1 μM ribozyme 5′ half; 2 μM ribozyme 3′ half; and 4 μM effector oligo (when included). Immediately prior to reaction, MgCl2 was added and a zero time-point aliquot removed. For the remaining RNA, autothiophosphorylation was initiated by the addition of ATPγS to a final concentration of 10 mM, except where indicated, along with an equal concentration of TCEP. TCEP was included in all reactions to maintain a reducing environment, which we have found to be necessary for reproducible APM-PAGE of thiophosphorylated RNA. We previously determined the effectiveness of TCEP as a reductant for use in APM-PAGE and Kin.46 kinetics, as well as its superiority over the more commonly used dithiothreitol (DTT) (Rhee and Burke 2004). Aliquots were removed at various times and quenched on ice in an equal volume of stop buffer (100 mM EDTA, 80% formamide, 0.01% xylene cyanol and bromophenol blue). Thiophosphorylated RNA was resolved by denaturing APM-PAGE and quantified by PhosphorImager (Molecular Dynamics). Fraction RNA thiophosphorylated was normalized to the PNK-thiophosphorylated control RNA. Reactions went to nearly 100% at long times, and were monophasic throughout when fit to standard single-exponential kinetic equation. However, it was impractical to allow the slowest reactions to develop enough curvature to extract exponential rate constants. Therefore, to allow a consistent analytical method to be applied across all RNA species, apparent reaction rate constants were calculated from linear regressions of data representing <15% normalized product formation. Over this time scale, the two methods yielded very similar apparent rate constants, differing by no more than 5%–10%.
Native gel electrophoresis
For samples analyzed by native gel electrophoresis, RNA was folded as above and an aliquot was removed along with the zero time-point into an equal volume of nondenaturing loading buffer (0.25% bromophenol blue and xylene cyanol in 30% glycerol [v/v]). Nondenaturing polyacrylamide gel electrophoresis was performed at room temperature in 1 × TBE running buffer.
Eyring analysis
Kinetic assays were performed at 10, 15, 20, 25, 30, and 37°C as described above with the following changes. After the 90°C heating step, polynucleotides were allowed to equilibrate for 10 min at the appropriate temperature in either a heat block (± 0.5°C), incubator (± 0.2°C), or thermocycler (± 0.1°C). We previously determined that the pH change over this temperature range was negligible (± 0.103 pH units) (Rhee and Burke 2004). Apparent rate constants were fit to the linear form of the Eyring equation:
where R = gas constant, kobs = observed rate constant, h = Planck's constant, kB = Boltzmann's constant, T = temperature, ΔH‡ = enthalpy of activation, and ΔS‡ = entropy of activation (Wynne-Jones and Eyring 1935). The free energy of activation, ΔG‡, may then be found from the standard relation,
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
Supplemental text: materials; determination of Michaelis-Menten parameters; unpaired nucleotides in P3 and L3 are dispensable both for catalysis and for activation; alternate calculation of the effects of Kin.46 mutations on activation parameters; Eyring analysis of 3' tail sequence variants.
Supplemental Tables and Figures: Table S1, thermodynamic activation parameters for Kin.46 3' tail sequence variants; Table S2, Michaelis-Menton parameters for 3' tail variants of Kin.46; Figure S1, tandem ribozyme constructs for generating sub-optimal 5' RNA ends; Figure S2, Eyring plots of autokinase activity by Kin.46 mutants with alterations in 3' tail sequence.
ACKNOWLEDGMENTS
This work was supported by NASA Exobiology Award NAG5-12360 and by a grant from the David and Lucile Packard Foundation to D.H.B. We thank then-undergraduate students Samay Jain and Kelly Woodard for extensive technical assistance early in the project.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2302810.
REFERENCES
- Achenbach J, Jeffries G, McManus S, Billen L, Li Y 2005. Secondary-structure characterization of two proficient kinase deoxyribozymes. Biochemistry 44: 3765–3774 [DOI] [PubMed] [Google Scholar]
- Admiraal S, Herschlag D 1995. Mapping the transition state for ATP hydrolysis: implications for enzymatic catalysis. Chem Biol 2: 729–739 [DOI] [PubMed] [Google Scholar]
- Admiraal S, Schneider B, Meyer P, Janin J, Véron M, Deville-Bonne D, Herschlag D 1999. Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase. Biochemistry 38: 4701–4711 [DOI] [PubMed] [Google Scholar]
- Admiraal S, Meyer P, Schneider B, Deville-Bonne D, Janin J, Herschlag D 2001. Chemical rescue of phosphoryl transfer in a cavity mutant: A cautionary tale for site-directed mutagenesis. Biochemistry 40: 403–413 [DOI] [PubMed] [Google Scholar]
- Beisel C, Smolke C 2009. Design principles for riboswitch function. PLoS Comput Biol 5: e1000363 doi: 10.1371/journal.pcbi.1000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovic T, Richardson K 1987. Mechanism and theory in organic chemistry, 3rd ed Harper & Row, New York [Google Scholar]
- Biondi E, Nickens DG, Warren S, Saran D, Burke DH 2010. Convergent donor and acceptor substrate utilization among kinase ribozymes. Nucleic Acids Res doi: 10.1093/nar/gkq433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DH 2004. RNA-catalyzed genetics. In The genetic code and the origin of life (ed. de Pouplana L Ribas), pp. 48–74 Kluwer Academic/Plenum Publishers, Georgetown, TX [Google Scholar]
- Burke DH, Ozerova NDS, Nilsen-Hamilton M 2002. Allosteric hammerhead TRAP ribozymes. Biochemistry 41: 6588–6594 [DOI] [PubMed] [Google Scholar]
- Cao S, Chen S 2006. Predicting RNA pseudoknot folding thermodynamics. Nucleic Acids Res 34: 2634–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Chen S 2009. Predicting structures and stabilities for H-type pseudoknots with interhelix loops. RNA 15: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Giedroc D, Chen S 2010. Predicting loop-helix tertiary structural contacts in RNA pseudoknots. RNA 16: 538–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple K, Bartel D, Unrau P 2003. Combinatorial minimization and secondary structure determination of a nucleoside synthase ribozyme. RNA 9: 1208–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S 2008. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu Rev Biophys 37: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li N, Ellington A 2007. Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers 4: 633–655 [DOI] [PubMed] [Google Scholar]
- Chen Y, Kowtoniuk W, Agarwal I, Shen Y, Liu D 2009. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol 5: 879–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuman W, Li Y 2006. Evolution of high-branching deoxyribozymes from a catalytic DNA with a three-way junction. Chem Biol 13: 1061–1069 [DOI] [PubMed] [Google Scholar]
- Cho B, Burke D 2006. Topological rearrangement yields structural stabilization and interhelical distance constraints in the Kin.46 self-phosphorylating ribozyme. RNA 12: 2118–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane J, Strobel S 2008. Riboswitch effectors as protein enzyme cofactors. RNA 14: 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman T, Huang F 2005. Optimal random libraries for the isolation of catalytic RNA. RNA 2: 129–136 [DOI] [PubMed] [Google Scholar]
- Cong X, Nilsen-Hamilton M 2005. Allosteric aptamers: Targeted reversibly attenuated probes. Biochemistry 44: 7945–7954 [DOI] [PubMed] [Google Scholar]
- Curtis E, Bartel D 2005. New catalytic structures from an existing ribozyme. Nat Struct Mol Biol 12: 994–1000 [DOI] [PubMed] [Google Scholar]
- Edwards T, Klein D, Ferré-D'Amaré A 2007. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol 17: 273–279 [DOI] [PubMed] [Google Scholar]
- Fastrez J 2009. Engineering allosteric regulation into biological catalysts. ChemBioChem 10: 2824–2835 [DOI] [PubMed] [Google Scholar]
- Guthrie R 1988. Systems for symbolic representation of reaction mechanisms (Recommendations 1988). Pure Appl Chem 61: 23–56 [Google Scholar]
- Gyi J, Conn G, Lane A, Brown T 1996. Comparison of the thermodynamic stabilities and solution conformations of DNA.RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry 35: 12538–12548 [DOI] [PubMed] [Google Scholar]
- Henkin T 2008. Riboswitch RNAs: Using RNA to sense cellular metabolism. Genes Dev 22: 3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Szostak JW 1996. A simple method for 3′-labeling of RNA. Nucleic Acids Res 24: 4360–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi G 1988. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27: 3842–3849 [DOI] [PubMed] [Google Scholar]
- Ingold C 1953. Structure and mechanism in organic chemistry, 1st ed Cornell University Press, Ithaca, NY [Google Scholar]
- Ingold C 1969. Structure and mechanism in organic chemistry, 2nd ed Cornell University Press, Ithaca, NY [Google Scholar]
- Koizumi M, Soukup GA, Kerr JN, Breaker RR 1999. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat Struct Biol 6: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kowtoniuk W, Shen Y, Heemstra J, Agarwal I, Liu D 2009. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc Natl Acad Sci 106: 7768–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Andersen G 2008. Structural insights in the exon junction complex. Curr Opin Struct Biol 18: 112–119 [DOI] [PubMed] [Google Scholar]
- Li Y, Breaker RR 1999. Phosphorylating DNA with DNA. Proc Natl Acad Sci 96: 2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen S 2010. Computing the conformational entropy for RNA folds. J Chem Phys 132: 235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Szostak JW 1994. In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature 371: 31–36 [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Szostak JW 1995. Kinetic and thermodynamic characterization of the reaction catalyzed by a polynucleotide kinase ribozyme. Biochemistry 34: 15315–15327 [DOI] [PubMed] [Google Scholar]
- Martick M, Scott W 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T, Plog M, Floy S, Jansen J, Soukup J, Soukup G 2005. Ligand requires for glmS ribozyme self-cleavage. Chem Biol 12: 1221–1226 [DOI] [PubMed] [Google Scholar]
- McManus S, Li Y 2007. Multiple occurrences of an efficient self-phosphorylating deoxyribozyme motif. Biochemistry 46: 2198–2294 [DOI] [PubMed] [Google Scholar]
- McManus S, Li Y 2008. A deoxyribozyme with a novel guanine quartet pseudoknot structures. J Mol Biol 375: 960–968 [DOI] [PubMed] [Google Scholar]
- Montange R, Batey R 2008. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37: 117–133 [DOI] [PubMed] [Google Scholar]
- Murray J, Terwey D, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott W 1998. The structural basis of hammerhead ribozyme self-cleavage. Cell 92: 665–673 [DOI] [PubMed] [Google Scholar]
- Oxender D, Zurawski G, Yanofsky C 1979. Attenuation inthe Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci 76: 5524–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley H, Flaherty K, McKay DB 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74 [DOI] [PubMed] [Google Scholar]
- Pyle AM, Cech TR 1991. Ribozyme recognition of RNA by tertiary interactions with specific ribose 2′-OH groups. Nature 350: 628–631 [DOI] [PubMed] [Google Scholar]
- Rhee S, Burke D 2004. Tris(2-carboxyethyl)phosphine stabilization of RNA: comparison with dithiothreitol for use with nucleic acid and thiophosphoryl chemistry. Anal Biochem 325: 137–143 [DOI] [PubMed] [Google Scholar]
- Robertson M, Ellington A 2000. Design and optimization of effector-activated ribozyme ligases. Nucleic Acids Res 28: 1751–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M, Scott W 2007. The structural basis of ribozyme-catalyzed RNA assembly. Science 315: 1549–1553 [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR 1998. An amino acid as a cofactor for a catalytic polynucleotide. Proc Natl Acad Sci 95: 6027–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksmerprome V, Burke DH 2003. Structural flexibility and the thermodynamics of helix exchange constrain attenuation and allosteric activation of hammerhead ribozyme TRAPs. Biochemistry 42: 13879–13886 [DOI] [PubMed] [Google Scholar]
- Saksmerprome V, Burke DH 2004. Deprotonation stimulates productive folding in allosteric TRAP hammerhead ribozymes. J Mol Biol 341: 685–694 [DOI] [PubMed] [Google Scholar]
- Saksmerprome V, Roychowdhury-Saha M, Khvorova A, Jayasena S, Burke DH 2004. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA 10: 1916–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saran D, Nickens D, Burke D 2005. A trans acting ribozyme that phosphorylates exogenous RNA. Biochemistry 44: 15007–15016 [DOI] [PubMed] [Google Scholar]
- Saran D, Held D, Burke D 2006. Multiple-turnover thio-ATP hydrolase and phospho-enzyme intermediate formation activities catalyzed by an RNA enzyme. Nucleic Acids Res 34: 3201–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W, Finch J, Klug A 1995. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002 [DOI] [PubMed] [Google Scholar]
- Silverman S 2003. Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA 9: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup G, Breaker R 1999a. Engineering precision RNA molecular switches. Proc Natl Acad Sci 96: 3584–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup G, Breaker R 1999b. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Breaker RR 1997. Rational design of allosteric ribozymes. Chem Biol 4: 453–459 [DOI] [PubMed] [Google Scholar]
- Tarn W, Chang T 2009. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol 6: 17–20 [DOI] [PubMed] [Google Scholar]
- Thatcher G, Kluger R 1978. Mechanism and catalysis of nucleophilic substitution in phosphate esters. Adv Phys Org Chem 25: 99–265 [Google Scholar]
- Vaish N, Jadhav V, Kossen K, Pasko C, Andrews L, McSwiggen J, Polisky B, Seiwert S 2003. Zeptomole detection of a viral nucleic acid using a target-activated ribozyme. RNA 9: 1058–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Breaker R 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168 [DOI] [PubMed] [Google Scholar]
- Wang W, Billen LP, Li Y 2002. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem Biol 9: 507–517 [DOI] [PubMed] [Google Scholar]
- Win M, Smolke C 2007. RNA as a versatile and powerful platform for engineering genetic regulatory tools. Biotechnol Genet Eng Rev 24: 311–346 [DOI] [PubMed] [Google Scholar]
- Win M, Smolke C 2008. Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win M, Liang J, Smolke C 2009. Frameworks for programming biological function through RNA parts and devices. Chem Biol 16: 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne-Jones W, Eyring H 1935. The absolute rate of reactions in condensed phases. J Chem Phys 3: 132 [Google Scholar]