Abstract
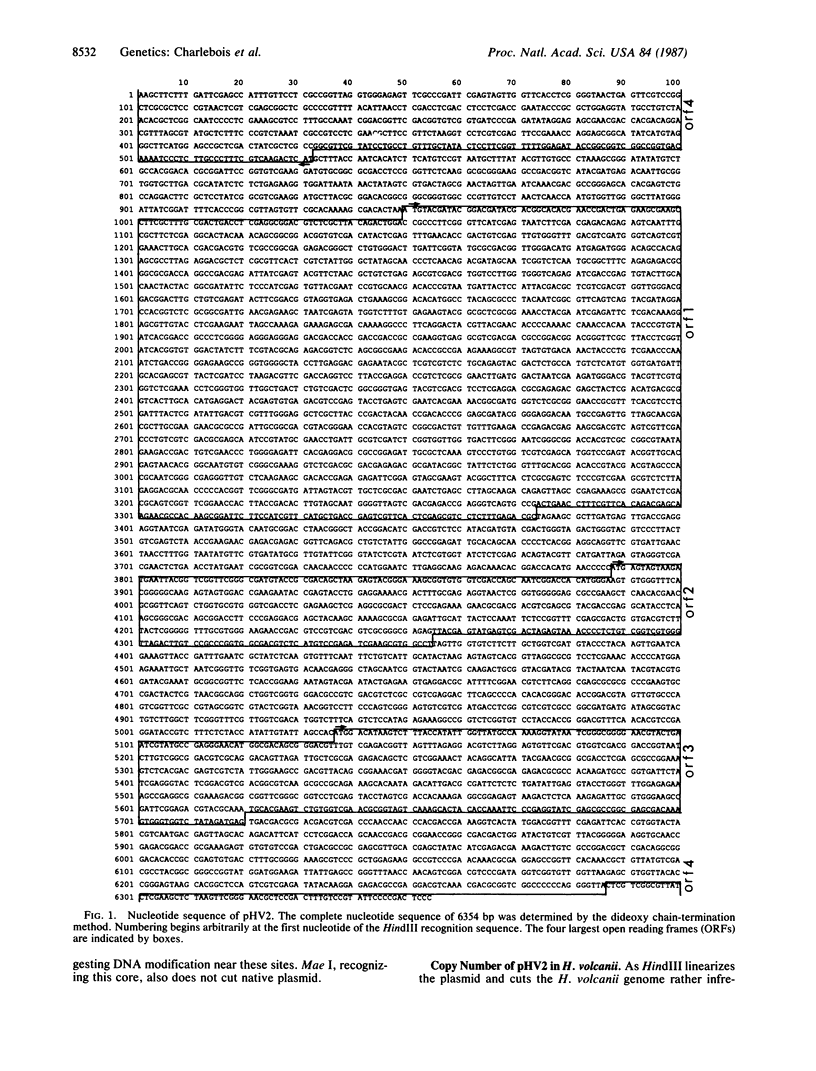
We determined the complete nucleotide sequence of the 6354-base-pair plasmid pHV2 of the archaebacterium Halobacterium volcanii. This plasmid is present in approximately six copies per chromosome. We have generated a strain, H. volcanii WFD11, cured of pHV2 by treatment of liquid cultures with ethidium bromide. We describe PEG-mediated transformation of H. volcanii WFD11 with intact pHV2 and with a form of pHV2 marked by a 93-base-pair deletion generated in vitro.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayley S. T., Morton R. A. Recent developments in the molecular biology of extremely halophilic bacteria. CRC Crit Rev Microbiol. 1978;6(2):151–205. doi: 10.3109/10408417809090622. [DOI] [PubMed] [Google Scholar]
- Bertani G., Baresi L. Genetic transformation in the methanogen Methanococcus voltae PS. J Bacteriol. 1987 Jun;169(6):2730–2738. doi: 10.1128/jb.169.6.2730-2738.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Oesterhelt D. The halo-opsin gene. II. Sequence, primary structure of halorhodopsin and comparison with bacteriorhodopsin. EMBO J. 1987 Jan;6(1):265–273. doi: 10.1002/j.1460-2075.1987.tb04749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. W., Doolittle W. F. Efficient transfection of the archaebacterium Halobacterium halobium. J Bacteriol. 1987 Mar;169(3):1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C. J., McKee A. H., Doolittle W. F. Archaebacterial heat-shock proteins. EMBO J. 1984 Apr;3(4):745–749. doi: 10.1002/j.1460-2075.1984.tb01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. P. Molecular biology of archaebacteria. J Bacteriol. 1986 Nov;168(2):471–478. doi: 10.1128/jb.168.2.471-478.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. H., Yang R. C., Wu R. An improved strategy for rapid direct sequencing of both strands of long DNA molecules cloned in a plasmid. Nucleic Acids Res. 1983 Aug 25;11(16):5521–5540. doi: 10.1093/nar/11.16.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Jones W. J., Nagle D. P., Jr, Whitman W. B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987 Mar;51(1):135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Queen C. L., Wegman M. N. Computer analysis of nucleic acid regulatory sequences. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4401–4405. doi: 10.1073/pnas.74.10.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher M. F., Strominger J. L. Structural (shape-maintaining) role of the cell surface glycoprotein of Halobacterium salinarium. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2687–2691. doi: 10.1073/pnas.73.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mevarech M., Werczberger R. Genetic transfer in Halobacterium volcanii. J Bacteriol. 1985 Apr;162(1):461–462. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullakhanbhai M. F., Larsen H. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch Microbiol. 1975 Aug 28;104(3):207–214. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- Patterson N. H., Pauling C. Evidence for two restriction-modification systems in Halobacterium cutirubrum. J Bacteriol. 1985 Aug;163(2):783–784. doi: 10.1128/jb.163.2.783-784.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Characterization of plasmids in halobacteria. J Bacteriol. 1981 Jan;145(1):369–374. doi: 10.1128/jb.145.1.369-374.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Doolittle W. F. Unusual physical organization of the Halobacterium genome. Nature. 1982 Feb 4;295(5848):384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Schnabel H., Zillig W., Pfäffle M., Schnabel R., Michel H., Delius H. Halobacterium halobium phage øH. EMBO J. 1982;1(1):87–92. doi: 10.1002/j.1460-2075.1982.tb01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C. Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1596–1600. doi: 10.1073/pnas.78.3.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. A., Blakesley R. W., Doran K., Hough C. J., Wells R. D. Purification of nucleic acids by RPC-5 ANALOG chromatography: peristaltic and gravity-flow applications. Methods Enzymol. 1983;100:368–399. doi: 10.1016/0076-6879(83)00068-3. [DOI] [PubMed] [Google Scholar]
- Toeckenius W., Kunau W. H. Further characterization of particulate fractions from lysed cell envelopes of Halobacterium halobium and isolation of gas vacuole membranes. J Cell Biol. 1968 Aug;38(2):337–357. doi: 10.1083/jcb.38.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontano A., Macchiato M. F. Probability of coding of a DNA sequence: an algorithm to predict translated reading frames from their thermodynamic characteristics. Nucleic Acids Res. 1986 Jan 10;14(1):127–135. doi: 10.1093/nar/14.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Fox G. E. Archaebacteria. J Mol Evol. 1978 Aug 2;11(3):245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]