Abstract
Proteolysis by the ubiquitin-proteasome pathway (UPP) is now widely recognized as a molecular mechanism controlling myriad normal functions in the nervous system. Also, this pathway is intimately linked to many diseases and disorders of the brain. Among the diseases connected to the UPP are neurodegenerative disorders such as Alzheimer’s, Parkinson’s and Huntington’s diseases. Perturbation in the UPP is also believed to play a causative role in mental disorders such as Angelman syndrome. The pathology of neurodegenerative diseases is characterized by abnormal deposition of insoluble protein aggregates or inclusion bodies within neurons. The ubiquitinated protein aggregates are believed to result from dysfunction of the UPP or from structural changes in the protein substrates which prevent their recognition and degradation by the UPP. An early effect of abnormal UPP in diseases of the nervous system is likely to be impairment of synaptic function. Here we discuss the UPP and its physiological roles in the nervous system and how alterations in the UPP relate to development of nervous system diseases.
1. Introduction
The link between ubiquitin-proteasome-mediated proteolysis and diseases of the nervous system has come full circle. Originally ubiquitin immunoreactivity was observed by pathologists in diseased brain tissue. No causal link between the presence of ubiquitin and the diseased states was apparent at the time. When the role for ubiquitin in protein degradation was first discovered, it was thought that it targeted only abnormal proteins for degradation. During the 1990s a physiological role for the ubiquitin-proteasome pathway (UPP) was first established in progression of cell cycle. This was soon followed by the discoveries that connected ubiquitin-proteasome-mediated proteolysis to synaptic plasticity. Many studies have established the role of the ubiquitin pathway in various other normal functions of the brain such as development of synaptic connections. With the advancement of our understanding of normal role of the UPP in many parts of the body including the brain, causative links between protein degradation and abnormalities of the brain are now beginning to be elucidated.
In the nervous system, the UPP has been connected to several diseases such as Alzheimer’s [1], Parkinson’s [2] and Huntington’s [3] diseases. A common feature among several chronic neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), motor neuron disease, and prion encephalopathies is abnormal deposition of insoluble protein aggregates or inclusion bodies within neurons. Nuclear insoluble inclusions are also observed in disorders associated with polyglutamine repeats, such as Huntington’s disease and spinocerebellar ataxia. Some studies indicate that impairment in the UPP may be linked to disorders such as schizophrenia [4].
A hallmark of neurodegenerative disorders such as AD and PD is the presence of ubiquitin immunoreactivity in the neuronal inclusions such as Lewy bodies and neurofibriallary tangles (NFTs). Ubiquitin immunoreactivity in the intracellular inclusions results from accumulation of ubiquitinated proteins. The accumulation of ubiquitin-protein conjugates in neuropathological lesions was detected first in NFT [5] and later in inclusions observed in a whole range of neurodegenerative diseases [6]. Although different parts of the brain are affected in different neurodegenerative disorders, the presence of ubiquitinated proteins in all the neurodegenerative disorders suggests the possibility that these diseases are associated with an inability of the neuron to degrade specific proteins or accumulated protein aggregates. Impairment of the UPP may contribute to the neurodegenerative process as this proteolytic pathway is known to degrade short-lived regulatory proteins and misfolded or abnormal proteins. The ubiquitinated protein aggregates are likely to result from malfunctioning of the UPP or changes in the protein substrates themselves which renders them resistant to degradation. In addition to a possible role in neurodegeneration, the UPP is known to play role in synaptic function and synaptic plasticity [7,8]. Therefore, malfunctioning of this pathway might also lead to brain disorders without neurodegeneration such as Angelman syndrome which is characterized by mental retardation. In order to comprehend the role of the UPP in abnormalities of the nervous system, it would be helpful to understand its role in the normal nervous system. Therefore, we first describe the UPP and its physiological role in the nervous system, and then discuss the impairment of this pathway in diseases of the nervous system.
2. The ubiquitin-proteasome pathway
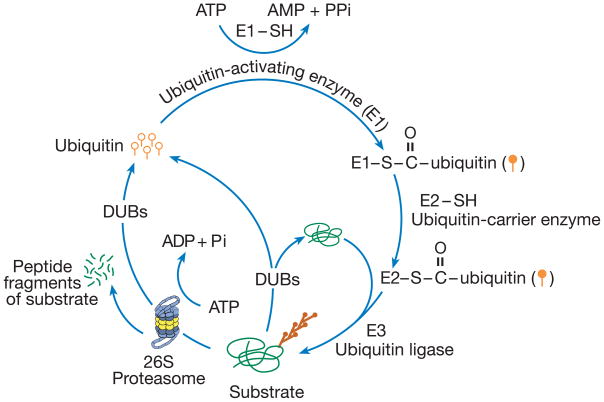
The main function of the UPP is to degrade proteins. Proteolysis by the UPP is highly regulated and specific: only proteins that are marked by attachment of ubiquitin are degraded and only at the right time. The tagging of a protein substrate with ubiquitin is regulated by the action of three enzymes: E1, E2, and E3 (So named because of the order of their elution on anion-exchange chromatography [9]). First, ubiquitin is activated by E1. Then, the activated ubiquitin is transferred to one of a family of enzymes termed E2s, which associate with E3 ubiquitin ligases. The E3s ligate the ubiquitin to a lysine (Lys) residue in the protein substrate. A second ubiquitin is covalently attached to the first ubiquitin and thus a polyubiquitin chain is formed. Ubiquitin attachment to other ubiquitins could occur through any of the seven lysine residues (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48 and Lys-63) in the ubiquitin molecules. For marking the substrate for ubiquitin-proteasome-mediated degradation, additional ubiquitins are attached to Lys-11 or Lys-48 in the first ubiquitin. Lys-63-linked polyubiquitin chains are known to have non-proteolytic function such as marking substrates for endocytosis or a role in intracellular signaling such as NFκ-B activation [10]. The functions of polyubiquitin chains with Lys-6, Lys-27, Lys-29 and Lys-33 linkages have not been elucidated [11,12]. The polyubiquitinated substrate with Lys-11 or Lys-48 linkage is recognized by the proteasome and is degraded to small peptides and amino acids [13,14] (Fig. 1).
Fig. 1.
The ubiquitin-proteasome pathway. In this proteolytic pathway, ubiquitin (single ubiquitin molecule is represented by open circles with straight tails) is selectively and covalently liked to the substrate. The enzymatic process of attaching ubiquitin to substrates is called ubiquitination or ubiquitin conjugation and depends on the action of three different classes of enzymes E1, E2 and E3. First ubiquitin is activated by E1 to form a ubiquitin-AMP intermediate. Activated ubiquitin (light orange color-filled circles with straight tails) is passed on to an E2 (ubiquitin carrier enzyme). Some E3s (ubiquitin ligases) such as HECT ligases accept ubiquitin as a thiol intermediate and then ligate it to the substrate. The RING ligases bring the E2 and substrate together and facilitate the transfer of ubiquitin from the E2 to the substrate. To the ubiquitin attached to substrate another ubiquitin is attached and thus through successive linkages of ubiquitin a polyubiquitin chain forms (conjugated ubiquitin is represented by dark orange color-filled circles and a branched pattern of tails). Polyubiquitinated substrates are degraded by a proteolytic complex called the 26S proteasome in an ATP-dependent reaction. Ubiquitin is not degraded but the polyubiquitin chain is disassembled and ubiquitin is recycled by deubiquitinating enzymes (DUBs). Before being committed to be degraded by the proteasome, ubiquitination is reversible. DUBs can disassemble the polyubiquitin chain if a substrate is ubiquitinated erroneously and prevent the degradation of the substrate.
Before a substrate is recognized by the proteasome and is fated to be degraded, ubiquitination can be reversed by deubiquitinating enzymes (DUBs). The polyubiquitin chains on the substrates degraded by the proteasome are not degraded but disassembled by DUBs that are part of or associated with the proteasome [15] (Fig. 1).
2. 1. Ubiquitin conjugation
The series of enzymatic reactions in the process of attaching a ubiquitin molecule to a substrate is called ubiquitin conjugation or ubiquitination. First reaction in this series is activation of ubiquitin by the enzyme E1. The activation of ubiquitin is ATP dependent. ATP is hydrolyzed to generate AMP, and a high energy thiol-ester ubiquitin-AMP intermediate is formed. The activated ubiquitin is then transferred to an E2, which is also called a ubiquitin carrier. The ubiquitin molecule is then transferred to an E3 which ligates ubiquitin to an ε amino group of a lysine residue in the substrate protein. A second ubiquitin molecule is then attached to an internal lysine reside in the first ubiquitin and thus a polyubiquitin chain grows[13].
2.2. Ubiquitin-conjugating Enzymes: E1, E2, and E3
Among the three classes of ubiquitin-conjugating enzymes, E1 is the least physiologically regulated. E2s are more selective and are believed to interact with specific E3s. Substrate specificity of the ubiquitin conjugation reaction is largely determined by E3s.
Originally, E2 enzymes were believed only to carry the activated ubiquitin and transfer it onto the E3s. Recent studies, however, suggest that at least some E2s can directly conjugate ubiquitin to substrates and are now commonly referred to as ubiquitin conjugating enzymes. E2s are structurally and functionally diverse. Simple eukaryotes like yeast (Saccharomyces cerevisiae) have 13 genes potentially encoding E2s. The number of E2s in mammals is estimated to be in the range of 25–30. The diversity of E2 generates some degree of specificity in the ubiquitin-conjugating reaction. E2s transfer ubiquitin to the substrates directly or in case of some ligases through generation of E3~ubiquitin thioester intermediates. The substrate-specificity of ubiquitin ligation is essentially determined by E3s. Sometimes an E3 might prefer a particular E2 by binding to a specific non-catalytic amino or carboxyl terminal extension in an E2. In such cases only specific E2–E3 pairs can ligate ubiquitin to substrates. Since the diversity of E3s is even greater than that of E2s, the combination of E2s and E3s potentially can generate a high degree of specificity [16].
There are two major classes of E3s: HECT (homologous to E6-AP C-terminus) domain E3s and RING (really interesting new gene) finger E3s.
2.2.1. HECT domain E3s
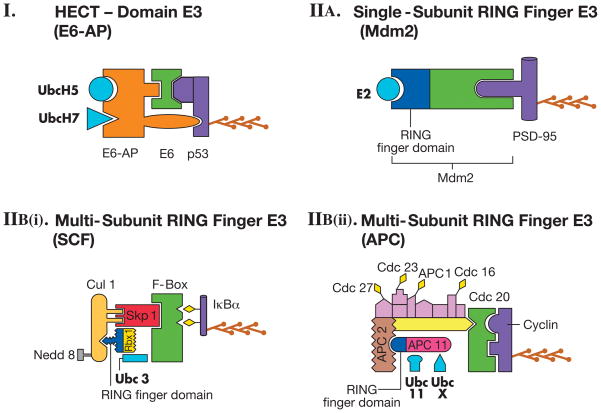
The typical example in this class of E3 is the ubiquitin ligase called E6-AP, which ligates ubiquitin to the tumor suppressor protein p53 in human papilloma virus (HPV) infected cells [17]. E6 protein, which is encoded by oncogenic strains of HPV, associates with a cellular protein called E6-AP (E6-associated protein). The C-terminal region of E6-AP contains the catalytic domain of the ubiquitin ligase. E6-AP ligase can function with either of the E2s called UbcH5 and UbcH7 (Fig. 2). Later studies found that a family of ubiquitin ligases with homology to the catalytic domain of E6-AP exists. These ubiquitin ligases are called HECT domain E3s. In addition to the HECT domain, there is another domain in many E3s called the WW domain. The WW domain-containing E3s also tend to have a C2 domain. The presence of the C2 domain is highly relevant to nervous system function because the C2 domain responds to elevation of intracellular Ca2+ and helps in translocation to the plasma membrane. Therefore, presence of this domain in neuronal HECT E3s might be critical in ligating ubiquitin to neurotransmitter receptors or proteins associated with them [7,13].
Fig. 2.
Classes/Examples of Ubiquitin Ligases (E3s). (I) HECT-Domain E3. E6-AP ubiquitin ligase in combination with E6 protein and one of the two E2s (UbcH5 or UbcH7) ligates ubiquitin to the p53 tumor suppressor protein. (IIA) Single-subunit RING Finger E3. Mdm2 ligates ubiquitin to PSD 95 with the help of an E2 enzyme. (IIBi) Multi-subunit RING finger E3. SCF ligases contain the substrate recognition site on an F-box protein. Skp1 is an adaptor that joins the F box protein to Cul1. Ring finger domain is on Rbx 1. The E2 is Ubc3. Cul 1 is modified by Nedd 8, a ubiquitin- like protein leading to an increase in the activity of the ligase complex. The substrate is phosphorylated (diamonds) IκBα. (IIBii) Multi-subunit RING finger E3. APC is a more complex example of multi-subunit RING finger E3s and has a subunit composition distinct from that of SCF. Cdc20 protein in APC has the substrate (Cyclin) recognition site. The RING finger domain is on APC11. The E2s Ubc 11 or UbcX can function with the APC ligase. In addition, several adaptor proteins, some labeled (Cdc27, Cdc23, APC 1, Cdc16) and some unlabeled, interact with Cdc20 and APC11. Diamonds on the adaptor subunits indicate phosphorylation. Polyubiquitin chain is shown on the substrates in each panel. In all panels, E2s are light blue, RING finger domains are dark blue and the substrates are purple in color.
2.2. 2. RING finger E3s
These E3s are called RING finger E3s because they contain a RING finger domain, which consists of seven cysteine residues and one histidine residue forming a single folded domain binding two zinc ions. During the past few years, numerous ubiquitin ligases were found to contain the RING finger. Unlike the HECT ligases, the RING finger ligases do not form a thioester intermediate with ubiquitin. Rather, these E3s bind both the substrate and the E2 and mediate the transfer of ubiquitin from the E2 to the substrate. It is now generally believed that the RING finger motif plays a critical role in ubiquitin ligation to substrates or to RING finger proteins themselves. The RING finger category of E3s can be subdivided into single-subunit RING finger E3s and multi-subunit RING finger E3s [14](Fig. 2).
2.2.2.1. Single-subunit RING finger E3s
Single-subunit RING finger E3s contain the RING finger domain and the substrate recognition site in the same protein. One of the well-characterized single-subunit RING finger E3s is Mdm2, which ubiquitinates a postsynaptic density protein PSD-95 in neurons [18]. Another widely studied ligase in this category is parkin which was originally discovered through its association with juvenile recessive parkinsonsism [19].
2.2.2.2. Multi-subunit RING finger E3s
There are two major examples of Multi-subunit RING finger E3s SKP1–cullin–F-box protein (SCF) complex and Anaphase-promoting complex (APC) (Fig. 2).
SCF complex contains at least four proteins: Skp1, Cul1, Roc1/Rbx1/Hrt1, and an F-box protein. The SCF-type ligases have another invariant protein called cullin. The theme appears to be that the cullins provide a scaffold on which the ligase complex is built. Cullins interact with linker proteins such as Skp1 to recruit substrate-interacting proteins such as the F-box proteins. In the center of the SCF complex is the RING finger domain-containing protein Rbx1. The ligases like SCFs are often referred to as cullin-RING ligases [20]. There are at least five different cullins in mammals. There are several F-box proteins as well. A well-characterized substrate of the SCF complex is IkBα [21].
APC also has a subunit with a RING finger domain (APC11) but this ubiquitin ligase is distinct from the SCF ligase in overall subunit combination. For example, instead of one adaptor found in SCF ligases (such as Skp1), APC has multiple subunits that serve as adaptors. Also, unlike SCF ligases, substrate phosphorylation is not an important determinant for specific substrate recognition by the APC ligase. Rather, substrate specificity of APC ligases appears to be modulated by incorporation of ‘specificity factors’ into the ligase complex. APC acts together with an E2 Ubc11 or Ubc X. One of the most studied substrates of APC is mitotic cyclin [22,23].
2.3. The Proteasome
The term proteasome is used to describe two kinds of multi-subunit proteolytic complexes, the 26S and 20S, based on their sedimentation coefficient. The 26S proteasome degrades ubiquitinated protein substrates. The 26S proteasome consists of a 20S catalytic core and two 19S regulatory particles (RP) attached to either end of the cylindrical 20S core.
2.3.1. The catalytic 20S core
In eukaryotes, the 20S core is made up of two outer rings with seven α subunits (α1 to α7) in each ring and two inner rings consisting of seven β subunits (β1 to β7). The catalytic activity of the proteasome is provided by three of the seven β subunits (β1, β2 & β5). The catalytic sites in these β subunits are located at their N-termini which are situated inside the catalytic chamber with an opening 13Ǻ in diameter [24]. Thus only an unfolded substrate can pass through this aperture. The catalytic core of the proteasome is a threonine protease [25]. The 20S proteasome can exist not only as a core of 26S, but also as a separate population that cannot degrade ubiquitinated proteins. However, the 20S proteasome by itself has chymotrypsin-like, trypsin-like, and postglutamyl peptidase activities which cleave after hydrophobic, basic, and acidic residues, respectively. The peptide-hydrolyzing activity of the 20S proteasome can itself be modulated by an 11S regulatory complex [14].
2.3.2. 19S RC
The 19S RC recognizes the polyubiquitinated substrate, and channels the substrate into the catalytic 20S core of the proteasome. It also has the capacity to regulate the activity of the catalytic core and determine the nature of the degradation process [26]. Usually one 19S RC is attached to either end of the catalytic core. Some of the 19S RC subunits are believed to channel the substrate into the catalytic chamber for degradation by unfolding the substrate. The unfolding activity is believed to be provided by the ATPases that are present in the base of the 19S RP which contains six ATPase subunits Rpt1-Rpt6 (Regulatory particle ATPase 1-6) and four non-ATPase subunits Rpn1, Rpn2, Rpn10 & Rpn13 (Regulatory particle non-ATPases 1, 2, 10 & 13). The other part of the 19S RP is called the ‘lid’ which comprises only non-ATPase subunits (Rpn3, Rpn5, Rpn6-9, Rpn11, Rpn12, & Rpn15) [14,25]. Among the Rpn subunits, Rpn11 (also called Poh1) and Rpn13 (also called Uch37) are DUBs that are integral part of the 19S RP that assist in deubiquitination of the substrate as it is unfolded and threaded into the catalytic chamber of the 20S core. Another DUB called Usp14 (also known as Ubp6) reversibly associates with the Rpn1 and stimulates substrate degradation through deubiquitination [27,28].
2.4. Deubiquitinating Enzymes
Ubiquitination reaction is reversible before the ubiquitinated protein is committed to degradation by the proteasome. Ubiquitin is removed from substrates by enzymes called DUBs. Based on protein sequence and the molecular size, initially DUBs were classified into two general classes: (1) low molecular weight (20–30K) ubiquitin C-terminal hydrolases (UCHs), and, (2) high molecular weight (around 100 K) ubiquitin-specific proteases (UBPs; also called USPs) [29]. Among the DUBs, UBPs belong to a large family containing diverse genes whereas the UCH family has fewer genes. For example, in yeast S. cerevisiae, there are 17 UBPs and 1 UCH. In the human genome, there are 63 genes encoding UBPs and 4 genes that code for UCHs. UCHs and UBPs subserve different functions in the eukaryotic cell. Although the current name for these enzymes, that is, DUBs, emphasizes the removal of ubiquitin from substrates, some DUBs, especially UCHs, function to process linearly linked ubiquitin precursors and generate monoubiquitin. DUBs are important for generating free ubiquitin at various steps of the ubiquitin–proteasome pathway. Ubiquitin is encoded by the tandemly linked polyubiquitin genes UbB and UbC [30,31]. In addition to the polyubiquitin gene, ubiquitin is also encoded by fusion of a ubiquitin-coding region to one of the two ribosomal subunits called L40 and S27. These gene products are thought to be processed by UCHs. Cleavage of isopeptide bond in the ubiquitin chains linked through Lys-48 (and presumably the ubiquitin chains linked through Lys-11) of ubiquitin serves two purposes. One is to recycle ubiquitin after it has been used for marking a substrate for degradation. Another function is to ‘edit’ the errors made by the ubiquitin-conjugating enzymes and reverse the ubiquitination reaction so that the substrate is no longer degraded. Also, editing function of DUBs probably serves to reverse the monoubiquitin attachment that marks membrane proteins for endocytosis [7,29].
In the post-genome era bioinformatics studies discovered three additional classes of DUBs: the OTU family (genes homologous to ovarian tumor gene), the Josephin/MJD family (genes containing the Josephin domain and the gene homologous to ataxin-3 which is implicated in Machado-Joseph Disease [MJD]), and the JAMM family (the genes containing the JAMM [JAB1/MPN/Mov34 metalloenzyme] domain) [15]. The roles of these classes of DUBs in the nervous system are not well understood.
3. Physiological roles of the UPP in the normal nervous system
3.1. The UPP and synaptic plasticity
Until the early 1990s no physiological or pathological role for the UPP in the nervous system was found. The first discovery of ubiquitin-proteasome-mediated degradation of a physiologically relevant substrate in the nervous system was that of R subunits of cAMP-dependent protein kinase (PKA) [32]. Since then several substrates of the UPP in the nervous system have been identified [4,7,8].
3.1.1. Degradation of the R subunits of PKA
Evidence for a role for the UPP in synaptic plasticity came from the investigation on persistent activation of PKA. Work on the biochemical mechanism of long-term facilitation [33] in Aplysia indicated that PKA was persistently activated in the absence of elevated cAMP. How is PKA activated in the absence of sustained increase in cAMP? It was found that the R subunits of PKA were decreased without any change in the catalytic (C) subunit during induction of long-term facilitation. Since there was no change in mRNA for either the R subunit or the C subunit, it was concluded that R subunits were diminished perhaps through proteolysis. What is the mechanism of R subunit degradation? Hegde et al [32] found through a series of biochemical experiments that R subunits were substrates for ubiquitination and proteasome-mediated degradation.
Moreover, a UCH (Ap-uch) that interacts with the proteasome was found to be induced by 5-HT, the neurotransmitter that induces long-term facilitation. Ap-uch was found to be critical for induction of long-term facilitation [34]. The role of UCH in long-term synaptic plasticity and long-term memory have been corroborated by experiments on long-term potentiation and hippocampus-dependent learning in mice [35].
Subsequently Chain et al [36] showed that at sensory-motor neuron synapses, injection of lactacystin, a specific proteasome inhibitor blocked induction of long-term facilitation. Since R subunit inhibit the activity of C subunits of PKA, the results were interpreted to suggest that the UPP operates to remove inhibitory constraints on formation of long-term memory. This has been corroborated by work carried out on the rat hippocampus. Lopez-Salon et al [37] demonstrated that bilateral infusion of lactacystin to the CA1 region of the rat hippocampus caused total retrograde amnesia for a one-trial avoidance learning. They also showed that total ubiquitination increases in the hippocampus 4 h after the training. These results are consistent with the idea that a decrease in some critical inhibitory proteins during long-term memory formation is mediated by the UPP. Proteolysis may play a role in other aspects of memory such as those steps in memory recall and reconsolidation. Using infusion of β-lactone into the CA1 region of the hippocampus Lee et al [38]showed that ubiquitin-proteasome-mediated degradation plays a role in destabilization of retrieved memory as well as extinction of fear memory. Several other studies have provided evidence for the role of the UPP in synaptic plasticity as well (reviewed in [8,39]).
3.1.2. The UPP and transcription: Possible roles in synaptic plasticity
Apart from the roles of the UPP in synaptic plasticity discussed above (see section 3.1.1), the UPP is likely to have a role in controlling transcription that underlies long-term synaptic plasticity. The dependence of long-term synaptic plasticity on gene transcription and new protein synthesis has been intensely studied over the past several years. What has been lacking, however, is the elucidation of the regulation of transcription that occurs in neurons that are modified by learning. For example what molecular processes determine the threshold for transcription? What determines the narrow time window of gene expression observed in many long-term memory paradigms? In many instances, the initial stimuli the neuron receives such as binding of a neurotransmitter to its receptor are the same for short-term and long-term synaptic plasticity. And yet long-term synaptic plasticity requires induction of gene expression. Long-term synaptic plasticity-producing protocols somehow must initiate a transcription cascade. Thus far the molecular processes that control threshold for transcription have not been elucidated. It is possible that the UPP operates to regulate the threshold for gene induction. Minimally the UPP could play a role in regulating transcription at two steps. One is the relief of transcription repression and the other is control of transcriptional activation. For example, it has been well established that CREB-mediated gene expression is required for long-term memory formation in Aplysia, Drosophila and mice [40–42]. It is also known that through alternative splicing, CREB gene generates activators and repressors [43]. The repressors are normally present at a higher stoichiometry in the cell than the activators [44,45]. For CREB-mediated gene expression to go forward, the repression has to be relieved. In Aplysia neurons, we have found that the repressor CREB1b is degraded by the UPP whereas the activator CREB1a is stable compared to the repressor [46]. The regulation of CREB1b degradation is likely to determine the threshold for induction of CREB-mediated gene expression and hence a threshold for long-term facilitation. With respect to removal of activator by proteolysis, previous work in Aplysia has shown that C/EBP, a transcription factor induced during long-term facilitation is degraded by the UPP [47]. The degradation of C/EBP determines a time window during which transcription must take place to induce long-term facilitation. The newly discovered role of the proteasome in transcription might be relevant to synaptic plasticity as well. We have found that proteasome inhibitors block induction of BDNF mRNA during hippocampal late phase long-term potentiation (LTP), a mammalian model of long-term synaptic plasticity [48].
There is a growing body of literature on the role of non-proteolytic roles of the proteasome in transcription [49,50]. In addition, ubiquitin-like proteins such as SUMO (small ubiquitin-related modifier) play a critical role in transcription [51,52]. With respect to nervous system function, studies on these other modes of transcriptional regulation have not been studied extensively and therefore are not reviewed here.
3.2. The UPP and developmental synaptic plasticity
Ubiquitin-proteasome-mediated proteolysis also plays a role in establishing and fine-tuning of synaptic connections during development of the nervous system. Synapse formation begins with axon growth towards the dendritic arbors or the cell body of their targets along a correct path. Establishment of mature pattern of synaptic connectivity requires retention of a subset of synapses that are originally formed. Synapse elimination likely requires pruning of axons and dendrites [53]. Consistent with this idea, in cerebellar granule cells axonal growth is inhibited by the multi-subunit ligase Cdh1-APC, which operates in the nucleus [54]. In Drosophila, a ubiquitin conjugating enzyme called UbcD1 helps prune dendrites. Also, in Drosophila, axon pruning is inhibited by mutations in the ubiquitin activating enzyme E1 or some of the proteasome subunits. Synaptic number in insect neuromuscular junction is controlled by a ubiquitin ligase highwire and a DUB called fat facets and synaptic size is determined by a subunit of APC (APC2).
4. Roles of the UPP in diseases of the nervous system
The components of the UPP have been linked to several diseases of the nervous system (Table 1). In some instances, mutations in specific genes have been linked to the etiology of the disease while in other cases impairment of UPP might be a late event in pathogenesis [55]. Although the perturbations in ubiquitin-proteasome-mediated proteolysis lead to pleiotropic effects on neurons including cell death or degeneration, one of the early effects is believed to be synaptic malfunction [56]. Many recent reports from animal studies demonstrate a role for ubiquitin-proteasome-mediated degradation of numerous substrate proteins in synaptic plasticity thus linking UPP to the normal synaptic function [7,57,58]. Although the UPP is linked to several neurodegenerative diseases including AD, PD and HD, impairment in synaptic plasticity and its connection to the UPP is better understood in AD than in other neurodegenerative disease.
Table 1.
The ubiquitin-proteasome pathway and diseases of the nervous system
| Disease | Component linked to the disease | Reference |
|---|---|---|
| Alzhemier’s disease | Ubiquitin (abnormal form) | [199] |
| Inhibition of proteasome | [113] | |
| Amyotrophic lateral sclerosis | Agrregated superoxide dismutase | [125] |
| Angelman syndrome | Mutation in E6-AP (UBE3A) ubiquitin ligase | [141,142] |
| Ataxia | USP14 (DUB) | [153] |
| Gracile axonal dystrophy | Deletion in the UCH-L1 gene | [200] |
| Huntington’s disease | Abnormal nuclear inclusions of ubiquitinated proteins | [201] |
| Parkinson’s disease | Parkin (E3, ubiquitin ligase) UCH-L1 α-synuclein (substrate) |
[19,171,202] |
| Schizophrenia | Decreased expression of UCHL1 and UBP14 | [203,204] |
| Wallerian degeneration | Possible microtubule fragmentation through the ubiquitin-proteasome pathway | [196] |
4.1. Alzheimer’s disease (AD)
AD is a neurodegenerative disorder of the central nervous system (CNS) clinically characterized by progressive loss of memory and other cognitive skills resulting in severe dementia. The condition often begins with mild memory lapses, and then gradually advances to dementia, a progressive deterioration of memory, language and most mental functions. Early during development of AD, neurological examination is normal except for cognitive dysfunction like progressive worsening of memory and other cognitive functions [59–62]. The intellectual decline is accompanied by the progressive extracellular and intracellular accumulation of insoluble fibrous material in the brain in the form of senile plaques and neurofibrillary tangles (NFTs) [63].
AD is the most prevalent neurodegenerative disorder and the most common cause of dementia [64]. Familial AD is a rare autosomal dominant disease with early onset, caused by mutations in the amyloid precursor protein (APP) and presenilins (PSEN1 & PSEN2) genes, both linked to amyloid β peptide (Aβ) metabolism [65]. Aβ is produced from APP by sequential cleavage by β-secretase (β-site APP cleaving enzyme 1; BACE1) and γ-secretase (presenilin complex), and released into the extracellular spaces [66]. Sporadic AD is common heterogeneous disease and caused by a complex interaction of genetic and environmental risk factors [65]. The pathological signs of AD include loss of medium and large pyramidal neurons, the presence of plaques and NFTs, composed of deposits of amyloid filaments and hyperphosphorylated tau respectively, surrounded by altered neurite processes and glia together with a degeneration of the neurons and loss of synapses [67,68]. Tau is a structural protein that is normally associated with microtubules. In abnormal condition like in AD, tau undergoes an abnormal posttranslational modification characterized by hyperphosphorylation. In addition, levels of tau could increase because of translational up-regulation mediated by increased S6 kinase as suggested by studies on postmortem AD brain and SH-SY5Y neuroblastoma cells [69]. Although multiple genetic disturbances are believed to cause AD, a major cause of the disease is buildup of the toxic Aβ peptide [70].
The formation of the neurofibrillary lesions is believed to produce the symptoms of the disease, which result most probably from the degeneration of nerve cells in cerebral cortex and hippocampal formation following synaptic loss. Defective synaptic function and memory loss is seen in early stages of AD and animal models of AD. Cognitive deficits are correlated with the loss of synapses [56]. During the early phase of AD there is 25–35% decrease in the density of synapses, and as the disease progresses loss of synapses is more strongly correlated with the disease than the plaques and tangles [71]. Animal models of AD mimic cognitive impairment seen in human AD patients [72,73].
Recent studies also suggest that AD begins with subtle alterations in synaptic efficacy prior to neuronal degeneration, and diffusible oligomeric assemblies of the Aβ [56] cause this neuronal dysfunction. Although there is eventual loss of synapses in both AD and animal models of AD, deficits in spatial memory and inhibition of LTP (an electrophysiological measure of synaptic strength) precede morphological alterations in the animal models, suggesting earlier biochemical changes in the disease [56,74,75]. Aβ treatment of cultured hippocampal neurons leads to the inactivation of PKA and persistence of its regulatory subunit PKAII alpha suggests that Aβ acts directly on the signaling pathways consisting of cAMP/PKA/cAMP-responsive element binding protein (CREB) required for development of late phase LTP [76]. Therefore, agents that enhance the PKA pathway have potential for the treatment of AD [76]. In transgenic mice expressing mutant amyloid precursor protein (APP), learning and memory is impaired at 9–10 months of age although no tangles are seen in the brains of these mice [77]. In mice carrying various mutations of APP, both in vitro and in vivo LTP is impaired much before detectable Aβ deposits are observed [78]. Also, cerebral microinjection of oligomers of Aβ peptide inhibits in vivo LTP in rats [79]. Moreover, Aβ dimmers isolated directly from human AD brain inhibit LTP and impair memory [80].
4.1.1. The UPP and AD
Several studies implicate neuronal UPP in the pathogenesis of AD. Abnormal deposition of highly insoluble protein aggregates or inclusion bodies within nerve cells is commonly observed in nervous tissue in association with several chronic neurodegenerative diseases including AD [81]. These inclusions bodies show ubiquitin immunoreactivity in human AD brains along with immunoreactivity to neurofilaments, tau, NEDD8 (Neural Precursor Cell Expressed, Developmentally Downregulated 8), PSEN1 and proteasome subunits [1,82–87]. Recent studies show that Lys-63-linked ubiquitin chains were found in intraneuronal inclusions in the AD brain [88]. The accumulation of ubiquitin-protein conjugates in neuropathological lesions was detected first in NFT isolated from human brain [5,89] and these NFTs best correlate with the degree of dementia [90]. It has been suggested that the UPP might also play a role in mediating the effects of inflammation in the brain and in linking it to formation of NFTs [91]. Aggregated proteins in cell line experiments tend to inhibit the UPP [55,92]. Initiation of aggregation of ubiquitinated proteins might come about by impairment of proteasome function as has been suggested by experiments using a mutation of a proteasome subunit [93].
4.1.2. The UPP and synaptic dysfunction early in AD
Memory impairment in AD is likely to result from synaptic dysfunction such as a defect in ability to change synaptic strength or synaptic plasticity. The findings from basic research on the critical function of Uchl1 (sometimes referred to as Uch-L1) in long-term synaptic plasticity [34] in invertebrates suggested the possibility of using Uchl1 for therapeutic purposes. It was found that treatment with exogenous Uchl1 can reverse the synaptic dysfunction in hippocampal slices from APP/PSEN1 mice (an animal model of AD) or synaptic plasticity impairment in hippocampal slices from normal mice caused by treatment with oligomeric Aβ [35]. Application of exogenous Uchl1 also reversed loss of spine density of hippocampal pyramidal neurons in APP/PSEN1 mice [94]. The essential role of Uchl1 in synaptic plasticity [34] and the application of Uchl1 to treat synaptic dysfunction in AD [35] suggests that perturbations in UPP components may be an important causative factor in AD.
Some other studies have suggested a role for Uchl1 in AD as well. For example, the genetic defect in gracile axonal dystrophy (gad) in mice has been shown to be caused by an in-frame deletion including exons 7 and 8 of Uchl1, encoding the ubiquitin carboxyl-terminal hydrolase isozyme Uchl1. The gad mice show ataxia (lack of coordination in muscular movements) characterized by dragging of the hind legs. In the sensory and motor neurons of gad mice accumulation of Aβ and ubiquitin-positive deposition occurs in a manner reminiscent of AD pathology [95,96]. The studies by Osaka et al [97] demonstrated that Uchl1 ensures ubiquitin stability within neurons. Also, UCHL1 protein level is downregulated in idiopathic Parkinson’s disease (PD) as well as AD brains [98]. Full-length human UCHL1 appears to be a major target of oxidative damage in AD and PD brains, and the enzyme is extensively modified by carbonyl formation, methionine oxidation, and cysteine oxidation [99]. Also, evidence from studies on Chinese Han population indicates the genetic association of serine to tyrosine mutation (S18Y polymorphism) in the UCHL1 gene and sporadic AD. This study concluded that Y allele and YY genotype of the S18Y in the UCHL1 gene may have protective effect against sporadic AD in female subjects [100].
Studies on transgenic mice expressing an aberrant ubiquitin called UBB+1 (ubiquitin with 20 extra amino acid residues at its C-terminus, which is implicated in AD) show memory deficits by about 9 months of age even though these mice do not show overt neurological deficits [101]. Since UBB+1 impairs proteasome function (see section 4.1.3), memory deficits in UBB+1 transgenic mice might be the result of synaptic dysfunction preceding the appearance of memory impairment.
4.1.3. The UPP and proteolytic defects in AD
The ubiquitinated protein aggregates found in AD brains are likely to result from a malfunction or overload of the ubiquitin-proteasome dependent protein degradation pathway or from structural changes in the protein substrates, halting their degradation. Presence of ubiquitinated proteins in all the AD cases [102–104] leads us to speculate that AD is associated with an inability of neurons to degrade specific proteins or accumulated protein aggregates.
How does a defect in UPP in AD come about? An unusual ubiquitin-conjugating enzyme, E2-25K/Hip-2, was found to be an intermediary of Aβ-mediated toxicity and E2-25K/Hip-2 enzymatic activity was required for inhibition of the proteasome in an animal model containing APP mutations [105]. Ubiquitin mutant, UBB+1 (a potent inhibitor of proteasome which is observed in AD brains) also functionally interacted with E2-25K/Hip-2 mediated neurotoxicity [105]. Additional studies found that E2-25K/Hip-2 is likely to stabilize Caspase-12 via inhibition of the proteasome (through accumulation of polyubiquitin conjugates and ubiquitinated UBB+1) and thus mediate toxicity caused by Aβ [106]. It has been found that Aβ can bind and inhibit the proteasome and thus block degradation of ubiquitin-conjugated proteins [107]. In in vitro experiments, Aβ peptide has been shown to inhibit the proteasome [108]. Intraneuronally accumulated Aβ peptide in APP/PSEN1 mutant neuronal cell culture is also shown to be able to inhibit the proteasome and deubiquitinating enzymes [109]. Ubiquitin immunoreactivity detected in axonal spheroids and dendritic compartments are associated to the elevated intraneuronal Aβ peptide and axonopathy seen in transgenic mouse model of mutant APP/PSEN1 [110,111]. Also, it is thought that other causative factors in AD such as paired helical filaments (PHF) of tau protein impair proteasome function [92]. In the brains of AD patients, proteasome function has been shown to be reduced mostly in the areas critical for long-term memory formation such as hippocampus, parahippocampal gyrus superior and middle temporal gyri and inferior parietal lobule but not in other areas such as the occipital lobe [112]. Another study showed that PHF of tau in brains of AD patients, as well as in vitro assembly of PHF using human recombinant tau protein both inhibited proteasome activity [113].
Taking the studies on early cognitive impairment in AD patients and animal models of AD together with the investigations on impairment of the UPP in AD, one could argue that the early synaptic defects result from impairment of proteolysis. There is considerable evidence for a role of UPP in synaptic plasticity [8,39]. Although a secondary role for UPP in AD pathology cannot be ruled out, given the role of UPP in synaptic plasticity and in maintaining the integrity of the synapse, perturbations in these functions might first lead to synaptic dysfunction and eventually loss of synapses.
Presence of ubiquitin and its association with tau in NFT’s and senile plaques in AD is common factor [81]. Accumulated ubiquitin is also present in Lewy bodies characteristic to some forms of the disease [85]. In all these cases, the role of tau and other putative target proteins in the pathogenesis is not understood. Also, it is still not clear whether aberration in proteolysis plays a causative role or only a secondary role [114]. Conflicting reports also exist on the E3 that ubiquitinates tau. Two reports claim that ubiquitin ligase, carboxyl terminus of Hsp 70-interacting protein (CHIP) targets tau [115,116] whereas another study argues that ubiquitin ligase TRAF6 is the ligase that conjugates ubiquitin to tau [117]. The discrepancy in the results perhaps could be resolved if the phosphorylation state of tau and the type of ubiquitin linkage that occurs on tau with each ligase are determined. CHIP has been shown to target hyperphosphorylated tau and TRAF6 ligases causes polyubiquitination via Lys-63-linked ubiquitin. Thus, the two ligases might act on the same substrate under different conditions.
A more direct relationship between the ubiquitin system and pathogenesis of AD was established with the discovery of UBB+1 which results from a frameshift mutation in the ubiquitin transcript that elongates the C-terminus of ubiquitin. The UBB+1 protein has been observed in the brains of AD patients [118]. UBB+1 is an efficient substrate for polyubiquitination, however, causing formation of polyubiquitin chains that cannot be disassembled by deubiquitinating enzymes. Polyubiquitin chains with UBB+1 potently inhibit the degradation of a polyubiquitinated substrate by the 26S proteasome [119] and acts as proteasome inhibitor in cells in a dose-dependent manner [120]. This report also suggests that the inhibitory activity of UBB+1 may be an important determinant of neurotoxicity and contribute to an environment that favors the accumulation of misfolded proteins [121,122]. In UBB+1 transgenic mice protein changes in the brain are remarkably similar to that in the human AD brain [101]. In addition, other genes implicated in AD such as presenilins (PSENs) 1 and 2 which are critical for processing APP are linked to UPP. Both PSEN1 and PSEN2 are targets for the UPP [123]. It is not known whether aberration in the degradation of PSEN1 and PSEN2 plays any role in the pathogenesis of AD.
4.2. Amyotrophic Lateral Sclerosis (ALS)
ALS is a disease of the neurons (motor neurons) that control muscle movement. Degeneration of neurons causes muscle atrophy eventually impairing the movement of people afflicted with the disease. Mutations of Copper-zinc superoxide dismutase1 (SOD1) have been associated with some cases of autosomal dominant familial ALS and some cases of sporadic ALS. Several studies have linked UPP to turnover of SOD1 but the observations have not yielded a clear picture. Some studies reported that mutant SOD1 proteins are degraded more rapidly than wild type SOD1 by the UPP [124,125]. Consistent with this idea, dorfin and NEDL1 ubiquitin ligases were shown to ubiquitinate mutant but not wild type SOD1 [126,127]. Di Noto L et al [128], however, reported that metal free SOD1 is degraded by 20S proteasome in vitro without requiring ubiquitination. In their study, monomeric forms of both wild type and mutant SOD1 were susceptible to degradation by the proteasome [128]. Investigations by others reported comparable degradation of both wild type and mutant SOD1 proteins by macroautophagy as well as by the proteasome [129].
Overexpression of the putative SOD1 E3 ligase dorfin can inhibit cell death induced by the mutant SOD1 protein [126]. Gene expression profiling of spinal cords from sporadic ALS patients indicated that genes associated with the UPP (dorfin and ubiquitin-like protein 5), oxidative toxicity, transcription, neuronal differentiation and inflammation might function in the pathogenesis of sporadic ALS [130]. It is possible that the expression of dorfin ligase is increased in an attempt to enhance clearance of the mutant SOD1. Recent studies also reported that heat-shock protein 70 (Hsp-70) or heat-shock cognate Hsc70, and CHIP play a role in proteasomal degradation of mutant SOD1 [131]. In addition, Urushitani et al. [132] found that oxidative damage increases the degree of ubiquitination of mutant SOD1 protein and proteasome activity decreases following one week of expression of a mutant SOD1 gene in cultured cells. Toxicity of mutant SOD1 protein aggregates is controversial, however. Lee et al. [133] found that aggregates of mutant SOD1 did not cause cell death. Also, others found that motor neurons from wild type animals and motor neurons from transgenic mice with mutant SOD1 were equally viable [134]. Some studies, however, have demonstrated that proteasome inhibition results in increased cell death in human cells expressing mutant SOD1 protein [135,136]. Recent work has suggested a connection between SOD mutation and impairment of proteasome. In transgenic mice with SOD gene mutation G93A, expression of proteasome subunits is decreased and the UPP function is impaired in spinal motor neurons [137]. Another molecule TAR-DNA-binding protein-43 (TDP-43), which has been linked to ALS, is a substrate for ubiquitination. It has been suggested that impairment of the UPP might play a role in neurodegeneration caused by TDP-43 dysfunction [138,139]. The role of protein aggregates in familial ALS remains unclear, however. As with other neurodegenerative diseases, the exact connection between SOD1 protein aggregates, the UPP, and disease progression remains to be elucidated.
4.3. Angelman Syndrome (AS)
AS is a neurological disorder with symptoms such as mental retardation, unusually happy demeanor, susceptibility to epileptic seizures, and abnormal gait [140]. Occurrence of AS is estimated to be 1 in 15,000 births. In about 65–75% of AS patients, maternal deletions at chromosome 15q11-q13 are found. Other types of genetic abnormalities such as uniparental disomy and imprinting mutations are also observed in AS, each of which accounts for about 3–5% of the cases. It has been found that the defects occur in a gene called UBE3A [141,142]. Point mutations in UBE3A are found in about 4–6% of the AS cases [143]. UBE3A is a maternally imprinted gene with brain specific expression from the maternal allele [144,145]. UBE3A gene encodes a ubiquitin ligase which had been previously identified as E6-AP (E6-Associated Protein) ubiquitin ligase. E6-AP is the cellular protein that associates with a human papilloma virus protein called E6. E6-AP in association with E6 degrades the tumor suppressor protein p53 [146]. Apart from p53, E6-AP is known to attach ubiquitin to at least three other substrates, RAD23, a human homolog of a yeast DNA repair protein [147]; multi copy maintenance protein 7 (MCM7) which is thought to function in chromosome replication [148]; and E6-AP itself [149].
Mental retardation in human AS patients suggests synaptic malfunction. In support of this idea, mice with deficiency of Ube3a maternal allele exhibit impairment of LTP and contextual learning. Previous studies showed an indirect effect of Ube3a on a molecule critical for synaptic plasticity, namely, Calcium/Calmodulin-dependent protein kinase II (CaMKII) has been observed, however. In the hippocampus of AS mouse, an increase in inhibitory autophosphorylation (on Thr305 & Thr306) of CaMKII occurs leading to reduction in kinase activity and dissociation of CaMKII from postsynaptic density [150]. Also, reduction in inhibitory autophosphorylation of CaMKII in AS mice rescues the neurological deficits in AS model mice [151]. So far no explanation as to how mutation in Ube3a causes alteration in CaMKII has been obtained. Another molecule critical for synaptic plasticity Arc has been shown to be a substrate for Ube3a and it has been suggested that Ube3a regulates excitatory synapse development by targeting Arc [152].
4.4. Ataxia
There is a direct link between mutation in a DUB called ubiquitin-specific protease 14 (Usp14) and ataxia. Mice with homozygous recessive mutations Usp14 develop ataxia and severe tremors by 2–3 weeks of age, followed by hindlimb paralysis and death by 6–10 weeks of age [153]. The mutation severely reduces the expression of Usp14 to about 5–10% of the levels found in wild type mice. The function of Usp14 is believed to be recycling of ubiquitin through disassembly of polyubiquitin chains to the monomeric form of ubiquitin. In support of this idea, levels of monomeric ubiquitin are reduced in the brains of Usp14 mutant mice [154]. In contrast to other neurodegenerative disorders such as Parkinson’s disease and SCA1 in humans and gad in mice, neither ubiquitin-positive protein aggregates nor neuronal cell loss is detectable in the CNS of these mice. Instead, they have defects in synaptic transmission in both the central and peripheral nervous systems. Neuron-specific expression of Usp14 rescues the growth defects and lethality [155]. The expression of transgene also rescues motor defects to some extent although some defects in motor coordination remain. Incomplete rescue of motor defects has been attributed to lack of expression of the Usp14 transgene in cerebellar Purkinje cells [155]. Additional evidence gathered by Chen et al [156]showed that Usp14 is critical for maintaining synaptic ubiquitin levels at neuromuscular junctions.
4.5. Huntington’s disease (HD)
HD is caused by mutations in a gene called Huntingtin. The disease is caused by abnormal expansion of CAG repeats which encode long stretches of glutamine (polyglutamine). In addition to HD there are about several other known polyglutamine diseases such as spinocerebellar ataxia, and, spinal and bulbar muscular atrophy [81].
HD can also be viewed as a disease of the synapse. In HD patients cognitive deficits appear much before the clinical symptoms of the disease [157–161]. This phenomenon is reproduced in mouse models of HD as well. For example, in transgenic mice carrying exon 1 of human HD mutation, hippocampal plasticity is significantly altered and deficits in spatial learning are observed. Deficits in synaptic plasticity occur before the overt neurological phenotype is observed [162]. Introduction of HD-like CAG repeats into murine huntingtin causes behavioral abnormalities as well [163]. Another study reported abnormalities in synaptic vesicle fusion machinery in HD mutant mice [164]. Even ectopic expression of an N-terminal fragment with 150 glutamine residues in Aplysia neurons impaired long-term facilitation without affecting basal synaptic transmission or short-term facilitation [165].
Is the UPP impaired in HD? Altered proteasome function was observed when mutant huntingtin was expressed in Neuro2A cells under the control of an inducible promoter [166]. A convincing demonstration was obtained using expression of huntingtin with a stretch of 103 glutamines in human embryonic kidney (HEK) 293 cells. Expression of mutant huntingtin with 103 glutamine in HEK 293 cells causes aggregate formation, accumulation of ubiquitinated proteins and cell cycle arrest whereas expression of shorter stretch of polyglutamine (25 glutamines) had markedly less effect on all these parameters [167]. Compelling evidence for impairment of the UPP came from a thorough study that used mass spectrometry techniques specifically to identify polyubiquitin chains that are formed through Lys-48 linkage of ubiquitin. This study showed accumulation of polyubiquitin chains in HD. This study revealed that Lys-48 linked ubiquitin chains accumulate in brains of R6/2 transgenic mice (widely used mouse model of HD), a knock-in mouse model (Q150/Q150) of HD, and in brains of HD patients [168].
How do polyglutamine-containing proteins inhibit the proteasome? One explanation is that polyglutamine containing proteins act as direct inhibitors of the proteasome [81]. An alternative explanation is that the UPP is overwhelmed by the load of aggregated or misfolded proteins. Since the nuclear inclusions in HD show ubiquitin immunoreactivity suggest that perhaps ubiquitin conjugation to aggregated protein is taking place and the proteasome is unable to efficiently degrade the ubiquitinated proteins. In support of this idea, proteins inclusions from isolated neurons and brains of conditional HD transgenic mice can be revered if the transgene is turned off. The disappearance of inclusion is proteasome dependent because the proteasome inhibitor lactacystin inhibits the reversal process [169]. Also, when green fluorescent protein containing polyglutamine is expressed in SH-SY5Y cells, basal proteasome activity is not impaired but the ability of the proteasome to respond to stress such as heat shock is dramatically impaired [170].
Based on the results from various studies described above, it is clear that the failure of the UPP ultimately contributes to cell death or degeneration in polyglutamine diseases. This process is probably progressive. Initial formation of aggregates with proteins with long polyglutamine stretches perhaps impairs proteasome activity which in turn leads to accumulation of more protein aggregates and thus misfolded proteins could build up in the cell. Initially, however, impairment of the proteasome is likely to have effect on synaptic properties of the neuron because as discussed elsewhere in this review, proteasome activity in various subcellular compartments of the neuron is essential for normal synaptic function and plasticity.
4.6. Parkinson’s disease
Protein products of four of the genes associated with Parkinson’s disease, namely those of α-synuclein, UCH-L1, DJ-1 and parkin [19,171–174] are linked to the UPP. α-synuclein is part of the Lewy bodies, the intracellular inclusions seen in the brains of Parkinson’s disease patients. Lewy bodies contain high amounts of ubiquitinated proteins including ubiquitinated α-synuclein. UCH-L1 is an enzyme of the ubiquitin pathway. DJ-1 is a substrate for conjugation to SUMO-1, a protein related to ubiquitin. SUMOylation of DJ-1 appears to be essential for its function [175]. The amino acid sequence of parkin protein contains a UbL domain at its N-terminus. Also, parkin has been shown to be a ubiquitin ligase. Three of the genes linked to Parkinson’s disease have direct synaptic connection. Parkin is present in postsynaptic densities. The Aplysia homologue of UCH-L1 is present in presynaptic terminals (Hegde et al., unpublished observations). Synuclein is associated with synaptic vesicles [176]. There is also evidence indicating that PD might be affect synaptic function adversely. In parkin deficient mice, synaptic excitability was reduced in striatal spiny neurons [177]. Also, parkin, which is a ubiquitin ligase [178] has been shown to ubiquitinate synaptotagmin XI [179].
Studies investigating possible connections of parkin to neurodegeneration have found that parkin is a key regulator of the aggresome-autophagy pathway [180–182]. Parkin targets several misfolded proteins to the aggresome-autophagy pathway through Lys-63 linked polyubiquitination. Parkin also promotes ubiquitination and degradation of a polyglutamine-expanded ataxin-3 and reduces its cellular toxicity [183,184]. Ataxin-3 which is implicated in Machado-Joseph Disease (MJD)/spinocerebellar ataxia-3 (SCA3), is a DUB that preferentially cleaves Lys-63 linkages [185]. The DUB activity of ataxin-3 is enhanced by its ubiquitination [186]. Both parkin and ataxin-3 participate in the aggresome formation to remove the misfolded proteins via autophagy [187]. It will be interesting to know the possible role of ubiquitination and deubiquitination activity of these proteins in regulating their own turnover and in the early synaptic failure seen in PD or MJD/SCA3. These studies could also potentially provide an explanation for parkinsonian symptom seen in MJD patients [188].
A link between proteasome dysfunction and neurodegeneration in PD has been provided by an elegant mouse model using targeted conditional depletion of 26S proteasome through inactivation of a 19S proteasome subunit Psmc1 (also called Rpt2/S4). In these mice, 26S proteasome depletion led to neurodegeneration and Lewy-like inclusions. In the brain of these mice intraneuronal inclusions contained ubiquitin and α-synuclein and the inclusions displayed striking similarities to Lewy bodies on the brains of human PD patients [189]. A connection between the proteasome and PD was also provided by a genetic study of German PD patients which found variations in intron 5 of a gene encoding the proteasome subunit S6 ATPase more frequently in early-onset PD patients relative to late-onset PD patients [190].
4.7. Neurodegeneration through autophagy: The ubiquitin connection
Impairment in proteolysis as a possible cause of neurodegeneration seen in many diseases of the nervous system has been previously suggested [3,81]. Several studies during the past few years point to a possible new connection between the UPP and neurodegeneration through autophagy. While the UPP mediates non-lysosomal protein degradation in the cell, autophagy is the process through which part of the cytoplasm is delivered to the lysosomes to be degraded. Basal autophagy prevents toxic effects of misfolded or abnormal proteins sequestering them inside autophagosomes (the double-membrane vesicle that engulfs portions of the cytoplasm or organelles) that deliver them to the lysosome for degradation [3]. Some of the proteins critical for autophagy are part of a conjugation system similar to that of ubiquitin. For example, Atg12 is activated by Atg7, a E1-like molecule, and transferred to Atg10, an E2-like enzyme, and conjugated to Atg5 through an isopeptide bond. Atg5 appears to be required for vesicle formation and completion which is required for autophagy. Mice lacking Atg7 specifically in the CNS showed neurodegeneration and showed motor deficits [191]. Atg5 null mutation in mice also causes deficits in motor function [192]. In these mice accumulation of ubiquitin immunoreactive cytoplasmic inclusion bodies in neurons is observed. Although the UPP and autophagy were thought to work in parallel, recent investigations suggest a functional link between the two. For example, proteasome inhibition activates autophagy and suppression of autophagy causes polyubiquitinated protein aggregates [193]. Studies on a Drosophila model of the neurodegenerative disease spinobulbar muscular atrophy have provided a mechanistic link between the UPP and autophagy. Flies containing a mutation in a proteasome subunit showed neurodegeneration which was rescued by overexpression of histone deacetylase 6 (HDAC6). HDAC6-mediated rescue of neurodegeneration occurred only if autophagy in these flies was intact [194]. Since HDAC6 is known to bind misfolded, polyubiquitinated proteins, it might shuttle these proteins to autophagosomes. Another polyubiquitin binding protein called p62/SQSTM1 might also connect polyubiquitinated proteins to autophagy [195].
4.8. Wallerian Degeneration
Wallerian degeneration is observed at the distal portion of a transected axon following injury. Previous studies have shown that application of proteasome inhibitors slows Wallerian degeneration [196]. Other studies have shown that there are differences between naturally occurring developmental axon degeneration and injury-induced axon degeneration. For example, the WldS protein that protects against Wallerian degeneration has no effect on developmental axon pruning whereas it protects against injury-induced degeneration of the same axons at the same developmental stage. WldS protein is a product of a chimeric gene that is found in laboratory (C57BL) mice which encodes ubiquitination factor E4B (also known as UFD2/E4) fused to nicotinamide mononucleotide adenyltransferase (Nmnat). UFD2/E4 functions in polyubiquitination and Nmnat is an enzyme that facilitates NAD synthesis. In contrast to the function of WldS, the UPP appears to be required for both developmental axon pruning and injury-induced axon degeneration [197].
5. The UPP and nervous system disease: Challenges and major outstanding questions
In spite of the fact that extensive research has been done on the role of the UPP in non-neuronal systems, our understanding regarding the role of the UPP in physiological and pathological functions in the nervous system is in its early stages and many questions remain unanswered and various challenges lie ahead. In terms of identifying the substrates and enzymes of the UPP that play a role in normal neuronal function and in abnormalities of the nervous system there is much more work to be done compared to what has been accomplished so far. Moreover, our knowledge regarding the already identified substrates and the enzymes of the UPP is incomplete. For example, APC appears to have nuclear as well as synaptic roles but it is not clear how this ligase is locally regulated in different neuronal compartments. Spatial and temporal regulation of the proteasome function in neurons needs to be elucidated as well.
With respect to the diseases and disorders of the nervous system, we need to understand how abnormal proteins and protein aggregates cause synaptic dysfunction and eventual synaptic loss and how the UPP fits into this picture. The overlap of symptoms in certain neurodegenerative diseases can be better understood if we can elucidate how the molecules implicated in the diseases interact with each other. For example, understanding interactions between parkin and ataxin-3 might shed light on parkinsonian symptoms seen in MJD patients. Another major challenge is to develop animal models that faithfully mimic the human disease. If we are able to comprehend the exact role of the UPP in etiology of a given neurodegenerative disease, then we might be able to create animal models of the disease which can be used for testing possible drug targets including the components of the UPP.
6. The elements of the UPP as possible therapeutic targets
Could we envisage targeting enzymes of the UPP or the proteasome as potential therapeutic targets to treat nervous system disease? In principle, it should be possible to use the UPP components as “druggable” targets if a clear causative link between a UPP component and a disease can be established. Among the enzymes that conjugate ubiquitin to substrates, E3s are the best potential therapeutic targets because they mainly determine substrate specificity. Because the substrate binding region imparts specificity to E3s, allosteric modification of this region (or modification of the active site) by small molecules so that specific E3s will have either increased affinity or decreased affinity towards specific substrates is one way of controlling the ubiquitinated substrate accumulation [2]. Selective engineering of the ubiquitin proteasome pathway components to degrade specific accumulated ubiquitinated proteins or protein aggregate and their delivery to specific affected region may also provide an alternative approach to small allosteric molecules. If defective function or deficiency of a UPP component is linked to a nervous system disease such as ataxia gene therapy with administration of specific components of the UPP might be a possibility.
A potential new area for the discovery of drug targets for treating neurodegenerative diseases is the proteasome. Although many inhibitors of the proteasome are available, no effective drugs exist that can stimulate the proteasome. Since abnormal protein aggregation and inhibition of proteasome is a common feature of AD and other neurodegenerative diseases, enhancement of proteasome activity by small molecules might be an efficient way to remove the aggregates that accumulate in the brain [2]. Removal of protein aggregation by the proteasome can be achieved by employing the following strategies: (1) By increasing the activity of the proteasome through increase in assembly of 19S and 20S complex of proteasome; (2) By stimulating the recognition of ubiquitinated proteins in protein aggregate; (3) By overexpressing or modulating the chaperone activity of either chaperonins or ATPase subunits of 19S to unfold the aggregated proteins; and (4) By stimulating the catalytic activity of the 20S core of the proteasome through small molecules [2]. In support of this idea, a study showed that Resveratrol, a naturally occurring polyphenol found in the grapes and red wine is known to lower the Aβ in cell lines by promoting the proteasome-dependent degradation of Aβ suggesting possible use of this compound for therapeutic intervention in AD [198]. A thorough understanding of the role of the UPP in the normal nervous system and elucidation of precise pathological role of ubiquitin-mediated proteolysis in disease states would be essential for future development of therapeutic approaches.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Upadhya SC, Hegde AN. Ubiquitin-proteasome pathway components as therapeutic targets for CNS maladies. Curr Pharm Des. 2005;11:3807–3828. doi: 10.2174/138161205774580651. [DOI] [PubMed] [Google Scholar]
- 3.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 4.Upadhya SC, Hegde AN. Role of the ubiquitin proteasome system in Alzheimer’s disease. BMC Biochem. 2007;8(Suppl 1):S12. doi: 10.1186/1471-2091-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori H, Kondo J, Ihara Y. Ubiquitin is a component of paired helical filaments in Alzheimer’s disease. Science. 1987;235:1641–1644. doi: 10.1126/science.3029875. [DOI] [PubMed] [Google Scholar]
- 6.Lowe J, Blanchard A, Morrell K, Lennox G, Reynolds L, Billett M, Landon M, Mayer RJ. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson’s disease, Pick’s disease, and Alzheimer’s disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol. 1988;155:9–15. doi: 10.1002/path.1711550105. [DOI] [PubMed] [Google Scholar]
- 7.Hegde AN. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog Neurobiol. 2004;73:311–357. doi: 10.1016/j.pneurobio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 9.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 10.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 11.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 12.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 14.Hegde AN. Ubiquitin-Dependent Protein Degradation. In: Mander L, Lui H-W, editors. Comprehensive Natural Products II Chemistry and Biology. Vol. 5. Elsevier; Oxford: 2010. pp. 699–752. [Google Scholar]
- 15.Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391:163–169. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 17.Beer-Romero P, Glass S, Rolfe M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene. 1997;14:595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- 18.Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 20.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K, Kawakami T, Tateishi K, Yashiroda H, Chiba T. Control of IkappaBalpha proteolysis by the ubiquitin-proteasome pathway. Biochimie. 2001;83:351–356. doi: 10.1016/s0300-9084(01)01237-8. [DOI] [PubMed] [Google Scholar]
- 22.Hershko A. Mechanisms and regulation of the degradation of cyclin B. Philos Trans R Soc Lond B Biol Sci. 1999;354:1571–1575. doi: 10.1098/rstb.1999.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang G, Yu H, Kirschner MW. Control of mitotic transitions by the anaphase-promoting complex. Philos Trans R Soc Lond B Biol Sci. 1999;354:1583–1590. doi: 10.1098/rstb.1999.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y. Toward an atomic model of the 26S proteasome. Curr Opin Struct Biol. 2009;19:203–208. doi: 10.1016/j.sbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 26.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 28.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 30.Baker RT, Board PG. The human ubiquitin gene family: structure of a gene and pseudogenes from the Ub B subfamily. Nucleic Acids Res. 1987;15:443–463. doi: 10.1093/nar/15.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker RT, Board PG. The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res. 1991;19:1035–1040. doi: 10.1093/nar/19.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegde AN, Goldberg AL, Schwartz JH. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: a molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci U S A. 1993;90:7436–7440. doi: 10.1073/pnas.90.16.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenberg SM, Castellucci VF, Bayley H, Schwartz JH. A molecular mechanism for long-term sensitization in Aplysia. Nature. 1987;329:62–65. doi: 10.1038/329062a0. [DOI] [PubMed] [Google Scholar]
- 34.Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–126. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 35.Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O. Ubiquitin Hydrolase Uch-L1 Rescues beta-Amyloid-Induced Decreases in Synaptic Function and Contextual Memory. Cell. 2006;126:775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 36.Chain DG, Casadio A, Schacher S, Hegde AN, Valbrun M, Yamamoto N, Goldberg AL, Bartsch D, Kandel ER, Schwartz JH. Mechanisms for generating the autonomous cAMP-dependent protein kinase required for long-term facilitation in Aplysia. Neuron. 1999;22:147–156. doi: 10.1016/s0896-6273(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza, Izquierdo I, Pasquini JM, Medina JH. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001;14:1820–1826. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, Choi SL, Lee SH, Kim H, Kaang BK. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science. 2008;319:1253–1256. doi: 10.1126/science.1150541. [DOI] [PubMed] [Google Scholar]
- 39.Hegde AN. The ubiquitin-proteasome pathway and synaptic plasticity. Learn Mem. 2010;17:314–327. doi: 10.1101/lm.1504010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 41.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 42.Yin JC, Wallach JS, Del VM, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 43.Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- 44.Habener JF, Miller CP, Vallejo M. cAMP-dependent regulation of gene transcription by cAMP response element-binding protein and cAMP response element modulator. Vitam Horm. 1995;51:1–57. doi: 10.1016/s0083-6729(08)61037-7. [DOI] [PubMed] [Google Scholar]
- 45.Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 46.Upadhya SC, Smith TK, Hegde AN. Ubiquitin-proteasome-mediated CREB repressor degradation during induction of long-term facilitation. J Neurochem. 2004;91:210–219. doi: 10.1111/j.1471-4159.2004.02707.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto N, Hegde AN, Chain DG, Schwartz JH. Activation and degradation of the transcription factor C/EBP during long-term facilitation in Aplysia. J Neurochem. 1999;73:2415–2423. doi: 10.1046/j.1471-4159.1999.0732415.x. [DOI] [PubMed] [Google Scholar]
- 48.Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem. 2008;15:335–347. doi: 10.1101/lm.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 50.Hegde AN, Upadhya SC. Proteasome and transcription: a destroyer goes into construction. Bioessays. 2006;28:235–239. doi: 10.1002/bies.20379. [DOI] [PubMed] [Google Scholar]
- 51.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 52.Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–1392. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen-Cory S. The developing synapse: construction and modulation of synaptic structures and circuits. Science. 2002;298:770–776. doi: 10.1126/science.1075510. [DOI] [PubMed] [Google Scholar]
- 54.Stegmuller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 55.Layfield R, Cavey JR, Lowe J. Role of ubiquitin-mediated proteolysis in the pathogenesis of neurodegenerative disorders. Ageing Res Rev. 2003;2:343–356. doi: 10.1016/s1568-1637(03)00025-4. [DOI] [PubMed] [Google Scholar]
- 56.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 57.Bingol B, Schuman EM. Synaptic protein degradation by the ubiquitin proteasome system. Curr Opin Neurobiol. 2005;15:536–541. doi: 10.1016/j.conb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 58.Patrick GN. Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Curr Opin Neurobiol. 2006;16:90–94. doi: 10.1016/j.conb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 59.Bartus RT, Dean RL, III, Beer B, Lippa AS. The cholinergic hypothesis of geriatric memory dysfunction. Science. 1982;217:408–414. doi: 10.1126/science.7046051. [DOI] [PubMed] [Google Scholar]
- 60.Graeber MB, Kosel S, Egensperger R, Banati RB, Muller U, Bise K, Hoff P, Moller HJ, Fujisawa K, Mehraein P. Rediscovery of the case described by Alois Alzheimer in 1911: historical, histological and molecular genetic analysis. Neurogenetics. 1997;1:73–80. doi: 10.1007/s100480050011. [DOI] [PubMed] [Google Scholar]
- 61.Moller HJ, Graeber MB. The case described by Alois Alzheimer in 1911. Historical and conceptual perspectives based on the clinical record and neurohistological sections. Eur Arch Psychiatry Clin Neurosci. 1998;248:111–122. doi: 10.1007/s004060050027. [DOI] [PubMed] [Google Scholar]
- 62.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perry G, Rizzuto N, utilio-Gambetti L, Gambetti P. Paired helical filaments from Alzheimer disease patients contain cytoskeletal components. Proc Natl Acad Sci U S A. 1985;82:3916–3920. doi: 10.1073/pnas.82.11.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selkoe DJ. The molecular pathology of Alzheimer’s disease. Neuron. 1991;6:487–498. doi: 10.1016/0896-6273(91)90052-2. [DOI] [PubMed] [Google Scholar]
- 65.Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 66.Iwata N, Higuchi M, Saido TC. Metabolism of amyloid-beta peptide and Alzheimer’s disease. Pharmacology & Therapeutics. 2005;108:129–148. doi: 10.1016/j.pharmthera.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 67.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 69.An WL, Cowburn RF, Li L, Braak H, Alafuzoff I, Iqbal K, Iqbal IG, Winblad B, Pei JJ. Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer’s disease. Am J Pathol. 2003;163:591–607. doi: 10.1016/S0002-9440(10)63687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 71.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 72.Hsia AY, Masliah E, McConlogue L, Yu GQ, Tatsuno G, Hu K, Kholodenko D, Malenka RC, Nicoll RA, Mucke L. Plaque-independent disruption of neural circuits in Alzheimer’s disease mouse models. Proc Natl Acad Sci U S A. 1999;96:3228–3233. doi: 10.1073/pnas.96.6.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larson J, Lynch G, Games D, Seubert P. Alterations in synaptic transmission and long-term potentiation in hippocampal slices from young and aged PDAPP mice. Brain Research. 1999;840:23–35. doi: 10.1016/s0006-8993(99)01698-4. [DOI] [PubMed] [Google Scholar]
- 75.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Van Den Haute C, Checler F, Godaux E, Cordell B, Van Leuven F. Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. Journal of Biological Chemistry. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 76.Vitolo OV, Sant’Angelo A, Costanzo V, Battaglia F, Arancio O, Shelanski M. Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc Natl Acad Sci U S A. 2002;99:13217–13221. doi: 10.1073/pnas.172504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 78.Chapman PF, White GL, Jones MW, Cooper-Blacketer D, Marshall VJ, Irizarry M, Younkin L, Good MA, Bliss TVP, Hyman BT, Younkin SG, Hsiao KK. Impaired synaptic plasticity and learning in aged amyloid precursor protein transgenic mice. Nature Neuroscience. 1999;2:271–276. doi: 10.1038/6374. [DOI] [PubMed] [Google Scholar]
- 79.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 80.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 82.Mayer RJ, Tipler C, Arnold J, Laszlo L, Al-Khedhairy A, Lowe J, Landon M. Endosome-lysosomes, ubiquitin and neurodegeneration. Adv Exp Med Biol. 1996;389:261–9. 261–269. doi: 10.1007/978-1-4613-0335-0_33. [DOI] [PubMed] [Google Scholar]
- 83.Busciglio J, Hartmann H, Lorenzo A, Wong C, Baumann K, Sommer B, Staufenbiel M, Yankner BA. Neuronal localization of presenilin-1 and association with amyloid plaques and neurofibrillary tangles in Alzheimer’s disease. J Neurosci. 1997;17:5101–5107. doi: 10.1523/JNEUROSCI.17-13-05101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chui DH, Shirotani K, Tanahashi H, Akiyama H, Ozawa K, Kunishita T, Takahashi K, Makifuchi T, Tabira T. Both N-terminal and C-terminal fragments of presenilin 1 colocalize with neurofibrillary tangles in neurons and dystrophic neurites of senile plaques in Alzheimer’s disease. J Neurosci Res. 1998;53:99–106. doi: 10.1002/(SICI)1097-4547(19980701)53:1<99::AID-JNR10>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 85.Lowe J. Establishing a pathological diagnosis in degenerative dementias. Brain Pathol. 1998;8:403–406. doi: 10.1111/j.1750-3639.1998.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Checler F, da Costa CA, Ancolio K, Chevallier N, Lopez-Perez E, Marambaud P. Role of the proteasome in Alzheimer’s disease. Biochim Biophys Acta. 2000;1502:133–138. doi: 10.1016/s0925-4439(00)00039-9. [DOI] [PubMed] [Google Scholar]
- 87.Dil KA, Kito K, Abe Y, Shin RW, Kamitani T, Ueda N. NEDD8 protein is involved in ubiquitinated inclusion bodies. J Pathol. 2003;199:259–266. doi: 10.1002/path.1283. [DOI] [PubMed] [Google Scholar]
- 88.Paine S, Bedford L, Thorpe JR, Mayer RJ, Cavey JR, Bajaj N, Sheppard PW, Lowe J, Layfield R. Immunoreactivity to Lys63-linked polyubiquitin is a feature of neurodegeneration. Neurosci Lett. 2009;460:205–208. doi: 10.1016/j.neulet.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 89.Perry G, Friedman R, Shaw G, Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc Natl Acad Sci U S A. 1987;84:3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 91.Arnaud L, Robakis NK, Figueiredo-Pereira ME. It may take inflammation, phosphorylation and ubiquitination to ‘tangle’ in Alzheimer’s disease. Neurodegener Dis. 2006;3:313–319. doi: 10.1159/000095638. [DOI] [PubMed] [Google Scholar]
- 92.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Z, Arnaud L, Rockwell P, Figueiredo-Pereira ME. A single amino acid substitution in a proteasome subunit triggers aggregation of ubiquitinated proteins in stressed neuronal cells. J Neurochem. 2004;90:19–28. doi: 10.1111/j.1471-4159.2004.02456.x. [DOI] [PubMed] [Google Scholar]
- 94.Smith DL, Pozueta J, Gong B, Arancio O, Shelanski M. Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proc Natl Acad Sci U S A. 2009;106:16877–16882. doi: 10.1073/pnas.0908706106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ichihara N, Wu J, Chui DH, Yamazaki K, Wakabayashi T, Kikuchi T. Axonal Degeneration Promotes Abnormal Accumulation of Amyloid Beta-Protein in Ascending Gracile Tract of Gracile Axonal Dystrophy (Gad) Mouse. Brain Research. 1995;695:173–178. doi: 10.1016/0006-8993(95)00729-a. [DOI] [PubMed] [Google Scholar]
- 96.Wu J, Ichihara N, Chui DH, Yamazaki K, Kikuchi T. Ubiquitin immunoreactivity in the central nervous system of gracile axonal dystrophy (GAD) mouse. No To Shinkei. 1995;47:881–885. [PubMed] [Google Scholar]
- 97.Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, Harada T, Hara Y, Kimura I, Chiba S, Namikawa K, Kiyama H, Noda M, Aoki S, Wada K. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 98.Barrachina M, Castano E, Dalfo E, Maes T, Buesa C, Ferrer I. Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiology of Disease. 2006;22:265–273. doi: 10.1016/j.nbd.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 99.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 100.Xue S, Jia J. Genetic association between Ubiquitin Carboxy-terminal Hydrolase-L1 gene S18Y polymorphism and sporadic Alzheimer’s disease in a Chinese Han population. Brain Res. 2006;1087:28–32. doi: 10.1016/j.brainres.2006.02.121. [DOI] [PubMed] [Google Scholar]
- 101.Fischer DF, Rvan D, Pvan T, Hobo B, Verhage MC, van der Schors RC, Li KW, Jvan M, Hol EM, van Leeuwen FW. Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol Aging. 2009;30:847–863. doi: 10.1016/j.neurobiolaging.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Wang LN, Zhu MW, Gui QP, Li XH. An analysis of the causes of dementia in 383 elderly autopsied cases. Zhonghua Nei Ke Za Zhi. 2003;42:789–792. [PubMed] [Google Scholar]
- 103.van Leeuwen FW, Pvan T, Sonnemans MA, Hobo B, Mann DM, Cvan B, Kumar-Singh S, Cras P, Leuba G, Savioz A, Maat-Schieman ML, Yamaguchi H, Kros JM, Kamphorst W, Hol EM, de Vos RA, Fischer DF. Frameshift proteins in autosomal dominant forms of Alzheimer disease and other tauopathies. Neurology. 2006;66:S86–S92. doi: 10.1212/01.wnl.0000193882.46003.6d. [DOI] [PubMed] [Google Scholar]
- 104.Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, Coslett HB, Chatterjee A, Hurtig HI, Karlawish JH, Rosen HJ, Van D, Lee VVM, Miller BL, Trojanowski JQ, Grossman M. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59:952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song S, Kim SY, Hong YM, Jo DG, Lee JY, Shim SM, Chung CW, Seo SJ, Yoo YJ, Koh JY, Lee MC, Yates AJ, Ichijo H, Jung YK. Essential role of E2-25K/Hip-2 in mediating amyloid-beta neurotoxicity. Mol Cell. 2003;12:553–563. doi: 10.1016/j.molcel.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 106.Song S, Lee H, Kam TI, Tai ML, Lee JY, Noh JY, Shim SM, Seo SJ, Kong YY, Nakagawa T, Chung CW, Choi DY, Oubrahim H, Jung YK. E2-25K/Hip-2 regulates caspase-12 in ER stress-mediated Abeta neurotoxicity. J Cell Biol. 2008;182:675–684. doi: 10.1083/jcb.200711066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shringarpure R, Grune T, Sitte N, Davies KJ. 4-Hydroxynonenal-modified amyloid-beta peptide inhibits the proteasome: possible importance in Alzheimer’s disease. Cell Mol Life Sci. 2000;57:1802–1809. doi: 10.1007/PL00000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gregori L, Hainfeld JF, Simon MN, Goldgaber D. Binding of amyloid beta protein to the 20 S proteasome. J Biol Chem. 1997;272:58–62. doi: 10.1074/jbc.272.1.58. [DOI] [PubMed] [Google Scholar]
- 109.Almeida CG, Takahashi RH, Gouras GK. Beta-amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J Neurosci. 2006;26:4277–4288. doi: 10.1523/JNEUROSCI.5078-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wirths O, Weis J, Kayed R, Saido TC, Bayer TA. Age-dependent axonal degeneration in an Alzheimer mouse model. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 111.Wirths O, Weis J, Szczygielski J, Multhaup G, Bayer TA. Axonopathy in an APP/PS1 transgenic mouse model of Alzheimer’s disease. Acta Neuropathol (Berl) 2006;111:312–319. doi: 10.1007/s00401-006-0041-4. [DOI] [PubMed] [Google Scholar]
- 112.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 113.Keck S, Nitsch R, Grune T, Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer’s disease. J Neurochem. 2003;85:115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 114.Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10(Suppl):S2–9. S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 115.Dickey CA, Yue M, Lin WL, Dickson DW, Dunmore JH, Lee WC, Zehr C, West G, Cao S, Clark AM, Caldwell GA, Caldwell KA, Eckman C, Patterson C, Hutton M, Petrucelli L. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J Neurosci. 2006;26:6985–6996. doi: 10.1523/JNEUROSCI.0746-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sahara N, Murayama M, Mizoroki T, Urushitani M, Imai Y, Takahashi R, Murata S, Tanaka K, Takashima A. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J Neurochem. 2005;94:1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- 117.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 118.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, Grosveld FG, Peter J, Burbach H, Hol EM. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 119.Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R. Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2000;97:9902–9906. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pvan T, de Vrij FM, Schuurman KG, Dantuma NP, Fischer DF, van Leeuwen FW, Hol EM. Dose-dependent inhibition of proteasome activity by a mutant ubiquitin associated with neurodegenerative disease. J Cell Sci. 2007;120:1615–1623. doi: 10.1242/jcs.03438. [DOI] [PubMed] [Google Scholar]
- 121.Rde P, Fischer DF, Maat-Schieman ML, Hobo B, de Vos RA, Brunt ER, Hol EM, Roos RA, van Leeuwen FW. Accumulation of aberrant ubiquitin induces aggregate formation and cell death in polyglutamine diseases. Hum Mol Genet. 2004;13:1803–1813. doi: 10.1093/hmg/ddh188. [DOI] [PubMed] [Google Scholar]
- 122.Lindsten K, de Vrij FM, Verhoef LG, Fischer DF, van Leeuwen FW, Hol EM, Masucci MG, Dantuma NP. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J Cell Biol. 2002;157:417–427. doi: 10.1083/jcb.200111034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim TW, Pettingell WH, Hallmark OG, Moir RD, Wasco W, Tanzi RE. Endoproteolytic cleavage and proteasomal degradation of presenilin 2 in transfected cells. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 124.Hoffman EK, Wilcox HM, Scott RW, Siman R. Proteasome inhibition enhances the stability of mouse Cu/Zn superoxide dismutase with mutations linked to familial amyotrophic lateral sclerosis. J Neurol Sci. 1996;139:15–20. [PubMed] [Google Scholar]
- 125.Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Niwa J, Ishigaki S, Hishikawa N, Yamamoto M, Doyu M, Murata S, Tanaka K, Taniguchi N, Sobue G. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J Biol Chem. 2002;277:36793–36798. doi: 10.1074/jbc.M206559200. [DOI] [PubMed] [Google Scholar]
- 127.Miyazaki K, Fujita T, Ozaki T, Kato C, Kurose Y, Sakamoto M, Kato S, Goto T, Itoyama Y, Aoki M, Nakagawara A. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J Biol Chem. 2004;279:11327–11335. doi: 10.1074/jbc.M312389200. [DOI] [PubMed] [Google Scholar]
- 128.Di NL, Whitson LJ, Cao X, Hart PJ, Levine RL. Proteasomal degradation of mutant superoxide dismutases linked to amyotrophic lateral sclerosis. J Biol Chem. 2005;280:39907–39913. doi: 10.1074/jbc.M506247200. [DOI] [PubMed] [Google Scholar]
- 129.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 130.Ishigaki S, Niwa J, Ando Y, Yoshihara T, Sawada K, Doyu M, Yamamoto M, Kato K, Yotsumoto Y, Sobue G. Differentially expressed genes in sporadic amyotrophic lateral sclerosis spinal cords--screening by molecular indexing and subsequent cDNA microarray analysis. FEBS Lett. 2002;531:354–358. doi: 10.1016/s0014-5793(02)03546-9. [DOI] [PubMed] [Google Scholar]
- 131.Urushitani M, Kurisu J, Tateno M, Hatakeyama S, Nakayama K, Kato S, Takahashi R. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J Neurochem. 2004;90:231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- 132.Urushitani M, Kurisu J, Tsukita K, Takahashi R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. J Neurochem. 2002;83:1030–1042. doi: 10.1046/j.1471-4159.2002.01211.x. [DOI] [PubMed] [Google Scholar]
- 133.Lee JP, Gerin C, Bindokas VP, Miller R, Ghadge G, Roos RP. No correlation between aggregates of Cu/Zn superoxide dismutase and cell death in familial amyotrophic lateral sclerosis. J Neurochem. 2002;82:1229–1238. doi: 10.1046/j.1471-4159.2002.01056.x. [DOI] [PubMed] [Google Scholar]
- 134.Vlug AS, Jaarsma D. Long term proteasome inhibition does not preferentially afflict motor neurons in organotypical spinal cord cultures. Amyotroph Lateral Scler Other Motor Neuron Disord. 2004;5:16–21. doi: 10.1080/14660820310016804. [DOI] [PubMed] [Google Scholar]
- 135.Aquilano K, Rotilio G, Ciriolo MR. Proteasome activation and nNOS down-regulation in neuroblastoma cells expressing a Cu,Zn superoxide dismutase mutant involved in familial ALS. J Neurochem. 2003;85:1324–1335. doi: 10.1046/j.1471-4159.2003.01783.x. [DOI] [PubMed] [Google Scholar]
- 136.Hyun DH, Lee M, Halliwell B, Jenner P. Proteasomal inhibition causes the formation of protein aggregates containing a wide range of proteins, including nitrated proteins. J Neurochem. 2003;86:363–373. doi: 10.1046/j.1471-4159.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- 137.Cheroni C, Marino M, Tortarolo M, Veglianese P, De BS, Fontana E, Zuccarello LV, Maynard CJ, Dantuma NP, Bendotti C. Functional alterations of the ubiquitin-proteasome system in motor neurons of a mouse model of familial amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:82–96. doi: 10.1093/hmg/ddn319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang IF, Wu LS, Shen CK. TDP-43: an emerging new player in neurodegenerative diseases. Trends Mol Med. 2008;14:479–485. doi: 10.1016/j.molmed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 139.Buratti E, Baralle FE. The molecular links between TDP-43 dysfunction and neurodegeneration. Adv Genet. 2009;66:1–34. doi: 10.1016/S0065-2660(09)66001-6. [DOI] [PubMed] [Google Scholar]
- 140.Williams CA. Neurological aspects of the Angelman syndrome. Brain Dev. 2005;27:88–94. doi: 10.1016/j.braindev.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 141.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 142.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 143.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 144.Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 145.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 146.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- 148.Kuhne C, Banks L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem. 1998;273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- 149.Nuber U, Schwarz SE, Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–649. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- 150.Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, Sweatt JD. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila FR, Jiang YH, Elgersma Y, Weeber EJ. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–282. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- 152.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–425. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 154.Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 155.Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA, Wilson SM. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–11431. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen PC, Qin LN, Li XM, Walters BJ, Wilson JA, Mei L, Wilson SM. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci. 2009;29:10909–10919. doi: 10.1523/JNEUROSCI.2635-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mohr E, Brouwers P, Claus JJ, Mann UM, Fedio P, Chase TN. Visuospatial cognition in Huntington’s disease. Mov Disord. 1991;6:127–132. doi: 10.1002/mds.870060207. [DOI] [PubMed] [Google Scholar]
- 158.Foroud T, Siemers E, Kleindorfer D, Bill DJ, Hodes ME, Norton JA, Conneally PM, Christian JC. Cognitive scores in carriers of Huntington’s disease gene compared to noncarriers. Ann Neurol. 1995;37:657–664. doi: 10.1002/ana.410370516. [DOI] [PubMed] [Google Scholar]
- 159.Lange KW, Sahakian BJ, Quinn NP, Marsden CD, Robbins TW. Comparison of executive and visuospatial memory function in Huntington’s disease and dementia of Alzheimer type matched for degree of dementia. J Neurol Neurosurg Psychiatry. 1995;58:598–606. doi: 10.1136/jnnp.58.5.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lawrence AD, Sahakian BJ, Hodges JR, Rosser AE, Lange KW, Robbins TW. Executive and mnemonic functions in early Huntington’s disease. Brain. 1996;119(Pt 5):1633–1645. doi: 10.1093/brain/119.5.1633. [DOI] [PubMed] [Google Scholar]
- 161.Lawrence AD, Hodges JR, Rosser AE, Kershaw A, ffrench-Constant C, Rubinsztein DC, Robbins TW, Sahakian BJ. Evidence for specific cognitive deficits in preclinical Huntington’s disease. Brain. 1998;121(Pt 7):1329–1341. doi: 10.1093/brain/121.7.1329. [DOI] [PubMed] [Google Scholar]
- 162.Murphy KP, Carter RJ, Lione LA, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shelbourne PF, Killeen N, Hevner RF, Johnston HM, Tecott L, Lewandoski M, Ennis M, Ramirez L, Li Z, Iannicola C, Littman DR, Myers RM. A Huntington’s disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum Mol Genet. 1999;8:763–774. doi: 10.1093/hmg/8.5.763. [DOI] [PubMed] [Google Scholar]
- 164.Morton AJ, Faull RL, Edwardson JM. Abnormalities in the synaptic vesicle fusion machinery in Huntington’s disease. Brain Res Bull. 2001;56:111–117. doi: 10.1016/s0361-9230(01)00611-6. [DOI] [PubMed] [Google Scholar]
- 165.Lee JA, Lim CS, Lee SH, Kim H, Nukina N, Kaang BK. Aggregate formation and the impairment of long-term synaptic facilitation by ectopic expression of mutant huntingtin in Aplysia neurons. J Neurochem. 2003;85:160–169. doi: 10.1046/j.1471-4159.2003.01650.x. [DOI] [PubMed] [Google Scholar]
- 166.Jana NR, Zemskov EA, Wang G, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10:1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- 167.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 168.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 169.Martin-Aparicio E, Yamamoto A, Hernandez F, Hen R, Avila J, Lucas JJ. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington’s disease. J Neurosci. 2001;21:8772–8781. doi: 10.1523/JNEUROSCI.21-22-08772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ding Q, Lewis JJ, Strum KM, Dimayuga E, Bruce-Keller AJ, Dunn JC, Keller JN. Polyglutamine expansion, protein aggregation, proteasome activity, and neural survival. J Biol Chem. 2002;277:13935–13942. doi: 10.1074/jbc.M107706200. [DOI] [PubMed] [Google Scholar]
- 171.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 172.Polymeropoulos MH. Genetics of Parkinson’s disease. Ann N Y Acad Sci. 2000;920:28–32. doi: 10.1111/j.1749-6632.2000.tb06901.x. [DOI] [PubMed] [Google Scholar]
- 173.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 174.Cookson MR. Pathways to Parkinsonism. Neuron. 2003;37:7–10. doi: 10.1016/s0896-6273(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 175.Shinbo Y, Niki T, Taira T, Ooe H, Takahashi-Niki K, Maita C, Seino C, Iguchi-Ariga SM, Ariga H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006;13:96–108. doi: 10.1038/sj.cdd.4401704. [DOI] [PubMed] [Google Scholar]
- 176.Lotharius J, Brundin P. Pathogenesis of Parkinson’s disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- 177.Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, Gajendiran M, Roth BL, Chesselet MF, Maidment NT, Levine MS, Shen J. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 178.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 179.Huynh DP, Scoles DR, Nguyen D, Pulst SM. The autosomal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum Mol Genet. 2003;12:2587–2597. doi: 10.1093/hmg/ddg269. [DOI] [PubMed] [Google Scholar]
- 180.Lim KL, Dawson VL, Dawson TM. Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 181.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4:85–87. doi: 10.4161/auto.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chin LS, Olzmann JA, Li L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans. 2010;38:144–149. doi: 10.1042/BST0380144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 184.Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Olzmann JA, Li L, Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem. 2008;15:47–60. doi: 10.2174/092986708783330692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Buhmann C, Bussopulos A, Oechsner M. Dopaminergic response in Parkinsonian phenotype of Machado-Joseph disease. Mov Disord. 2003;18:219–221. doi: 10.1002/mds.10322. [DOI] [PubMed] [Google Scholar]
- 189.Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M, Soane T, Layfield R, Sheppard PW, Ebendal T, Usoskin D, Lowe J, Mayer RJ. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wahl C, Kautzmann S, Krebiehl G, Strauss K, Woitalla D, Muller T, Bauer P, Riess O, Kruger R. A comprehensive genetic study of the proteasomal subunit S6 ATPase in German Parkinson’s disease patients. J Neural Transm. 2008;115:1141–1148. doi: 10.1007/s00702-008-0054-3. [DOI] [PubMed] [Google Scholar]
- 191.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 192.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 193.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 195.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhai Q, Wang J, Kim A, Liu Q, Watts R, Hoopfer E, Mitchison T, Luo L, He Z. Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron. 2003;39:217–225. doi: 10.1016/s0896-6273(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 197.Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O’Leary DD, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 198.Marambaud P, Zhao H, Davies P. Resveratrol promotes clearance of Alzheimer’s disease amyloid-beta peptides. J Biol Chem. 2005;280:37377–37382. doi: 10.1074/jbc.M508246200. [DOI] [PubMed] [Google Scholar]
- 199.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, Grosveld FG, Peter J, Burbach H, Hol EM. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 200.Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 201.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 202.Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 203.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]