Abstract
S100B is highly over-expressed in many cancers, including malignant melanoma. In such cancers, S100B binds wild-type p53 in a calcium-dependent manner, sequestering it, and promoting its degradation, resulting in the loss of p53-dependent tumor suppression activities. Therefore, S100B inhibitors may be able to restore wild-type p53 levels in certain cancers and provide a useful therapeutic strategy. In this regard, an automated and sensitive fluorescence polarization competition assay (FPCA) was developed and optimized to screen rapidly for lead compounds that bind Ca2+-loaded S100B and inhibit S100B target complex formation. A screen of 2000 compounds led to the identification of 26 putative S100B low molecular weight inhibitors. The binding of these small molecules to S100B was confirmed by nuclear magnetic resonance spectroscopy, and additional structural information was provided by x-ray crystal structures of several compounds in complexes with S100B. Notably, many of the identified inhibitors function by chemically modifying Cys84 in protein. These results validate the use of high-throughput FPCA to facilitate the identification of compounds that inhibit S100B. These lead compounds will be the subject of future optimization studies with the ultimate goal of developing a drug with therapeutic activity for the treatment of malignant melanoma and/or other cancers with elevated S100B.
Keywords: nuclear magnetic resonance, fluorescence polarization, melanoma, chlorpromazine, thimerosal, sanguinarine
Introduction
S100B, a small (21 kDa), dimeric, Ca2+- and Zn2+-binding protein, is elevated in several types of cancer including reactive gliomas, renal cell tumors, malignant mature T-cells, and malignant melanoma, where its expression correlates inversely with patient survival.1–2 While aberrant expression of S100B has been used as a cancer marker in melanoma for over 20 years, more recent discoveries indicate that S100B, and other S100 proteins (S100A1, S100A2, S100A4, S100A6), contribute to tumorigenesis by interacting with and downregulating the tumor suppressor protein, p53, and its homologs p63 and p73.3–14 Specifically, a decrease in transcription activation, as well as protein levels of ectopically expressed p53 and p53-regulated gene product, occurred in H1299 cells upon coexpression with S100B.5 The C8146A malignant melanoma cells express high levels of S100B that, when knocked down by the introduction of siRNAS100B, result in a corresponding increase in wild-type p53 protein levels and its associated gene products.6 Cancer cells with elevated levels of S100B escape p53-dependent tumor suppression pathways even when the wild-type p53 genotype is intact because the S100B-p53 interaction downregulates p53 at the protein level. In vitro, S100B was found to bind p53 directly, prevent its temperature-dependent oligomerization, and to inhibit protein kinase C (PKC)-dependent phosphorylation of the tumor suppressor.4 Therefore, a drug development program that disrupts the S100B-p53 interaction is underway as a means to restore functional p53 in cancers with elevated S100B, such as occurs in malignant melanoma that typically expresses wild-type p53.5–6
While many drugs target receptors, transporters, ion channels, and enzymes that have very discrete ligand binding sites, the development of small compounds that block the S100B-p53 interaction requires inhibition of larger protein-protein interfaces (PPIs) that were once thought to be difficult, if not impossible, to inhibit with small molecules. It is now recognized that the interfaces of most PPIs have smaller amino acid domain(s), termed “hot spots”, that contribute more energetically to the PPIs than the surrounding protein surface.15 Successful strategies to target small molecule PPI inhibitors were developed and, as a result, several PPIs have been successfully blocked, including those involving bcl-2, p53, ERK, tubulin, and several SH2 domains, to name but a few.16–17 As with these proteins, a “hot spot” on S100B was recognized in structural studies with several different S100B-peptide complexes and small compounds.18,19
Taking advantage of existing atomic resolution structural information for S100B and its Ca2+-dependent target protein interactions (see Figure 1), a high-throughput screening (HTS) method was designed and implemented to identify rapidly and reliably compounds that bind the p53 binding site on S100B and inhibit their complex formation. A peptide derived from the C-terminus of p53 residues 367–388 (p53367–388) was found to bind S100B in a Ca2+-dependent manner, inhibiting the PKC-dependent phosphorylation of the peptide in a manner identical to what is seen for full length p53.20–21 Therefore, the three-dimensional structure of this peptide bound to Ca2+-S100B was determined using nuclear magnetic resonance (NMR) spectroscopic methods, revealing that a hydrophobic cleft exposed upon Ca2+ binding to S100B is necessary for binding p53.20,22,23 Several other peptides and small molecules have been shown to interact with this hydrophobic pocket on S100B, in particular a small 12-residue peptide, TRTK-12, that was sufficient for inhibiting complex formation between S100B and full-length p53.18,19 The NMR solution and x-ray crystal structure TRTK-12 bound to S100B confirms that it binds in the same hydrophobic pocket on S100B as does p53 (see Figure 1B).24,25 With this in mind, an in vitro fluorescence polarization competition assay (FPCA) using an N-terminal 5-carboxytetramethylrhodamine (TAMRA)-labeled version of the TRTK-12 peptide (TAMRA-TRTK) was developed to screen for inhibitors that target this discrete hydrophobic pocket on Ca2+-loaded S100B.
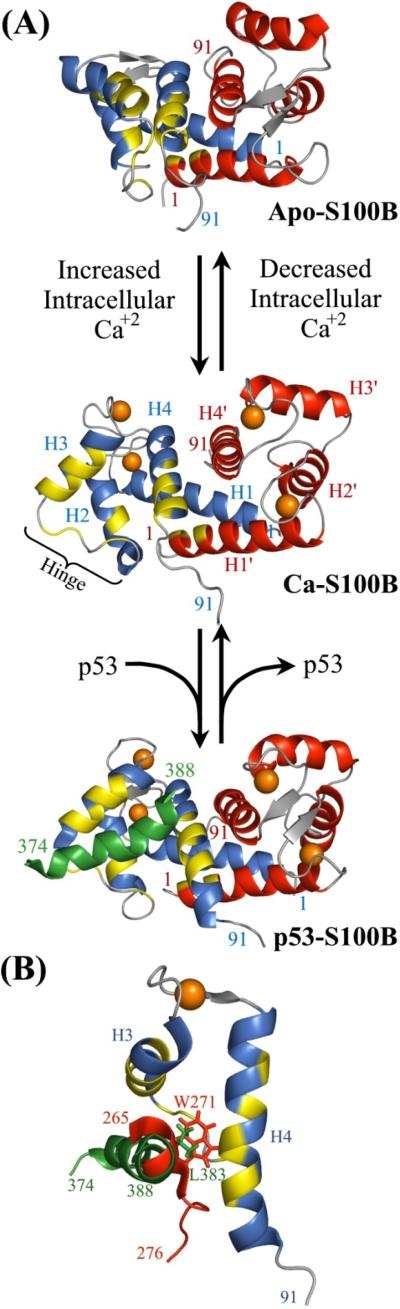
Figure 1.
The calcium-dependent interaction of S100B with p53. (A) The solution structures of, from top to bottom, apo-S100B (PDB 1SYM), Ca-S100B (PDB 1QLK), and p53-Ca-S100B (PDB 1DT7) reveal a large conformational change in S100B upon binding Ca2+, exposing a hydrophobic cleft that is capable of binding the tumor suppressor p53.22,28,58 One monomer of the dimeric S100B is shown in blue and the other in red with yellow residues, highlighting amino acids known to interact with the p53367–388 peptide. The structured region of the p53367–388 peptide formed by residues 374–388, shown in green, binds to the hydrophobic cleft between helix 3 (H3) and 4 (H4) of the Ca2+-bound protein, thus connecting Ca2+-signaling pathways with p53. There are actually two identical p53 binding site on S100B but only one is shown occupied. (B) A close-up of helix 3 (H3) and 4 (H4) have been aligned with the same region in the TRTK-12 bound Ca-S100B structure (PDB 1MWN) show that the p53 peptide (green) and the TRTK-12 peptide (red) bind the same site but with slightly different orientations.24 In addition, the large hydrophobic Trp in TRTK-12 buries itself deeper in the core of S100B than the smaller, but homologous, Leu in the p53367–388 peptide.18,24
Abbreviation: NMR, nculear magentic resonance.
Fluorescent polarization assays have been increasingly used in HTS due to their generally good sensitivity, rapid response, homogeneous format (no separation steps needed), and simple instrumentation, as reviewed by Huang and Aulabaugh.26 The principle of fluorescence polarization is well suited to studying the interaction of molecules with significantly different molecular weights, such as the S100B at 10.7 kDa and the TRTK-12 peptide at 1.5 kDa used here. The smaller probe is labeled with a fluorophore and then exposed to polarized light, leading to excitation of only those molecules in the correct orientation. The fluorescence emission is measured parallel and perpendicular to the excitation source, allowing the degree of polarization to be determined. The larger, slower, tumbling molecules will retain a higher degree of polarization, such as when the peptide probe is bound to the larger S100B, while the smaller faster-moving unbound probe will have a lower polarization. In our FPCA assay, the probe will be displaced, or competed off, by small molecules decreasing the fluorescence polarization (Figure 2).
Figure 2.
Schematic illustrating the FPCA. The FPCA uses the change in polarization of a TAMRA-labeled peptide derived from residues 265–276 of the actin capping protein CapZ (TAMRA-TRTK) that binds to S100B in the same region as p53 peptide (see Figure 1). In the presence of calcium, S100B binds TAMRA-TRTK causing the peptide to rotate slower, and the polarization values to increase. The addition of compounds that bind the same region displace the peptide, allowing it to rotate freely and decreasing the polarization value. A HTS version of this assay was used to screen for putative inhibitors of S100B, and it is also used to determine the binding affinity (KD) of the compound to S100B (see Table 2). The S100B dimer is shown in blue, the TAMRA-TRTK in red, and the Ca2+ ions are shown as orange spheres.
Abbreviations: FPCA, fluorescence polarization competition assay; high-throughput screening.
The high-throughput FPCA developed was used to screen the 2000-compound Spectrum Collection comprised of known drugs and biologically active compounds, and revealed several compounds that inhibit the TRTK-S100B interaction. NMR spectroscopic methods confirm that the compounds bind S100B, and x-ray crystal structures have been determined for three of the compounds from the library. Structures of chlorpromazine (SC0067), thimerosal (SC0322), and sanguinarine (SC0844) bound to S100B indicate that all three molecules have the same hydrophobic cleft of Ca2+-loaded S100B. Two of the structures reveal that the compounds covalently modify Cys84 in S100B, and further studies confirm that many of the inhibitors found in this screen may function in a similar mannner.
Material and methods
Materials
All chemical reagents were American Chemical Society grade or higher unless otherwise indicated. All buffers were passed through Chelex-100 (Bio-Rad, Hercules, CA) to remove trace metals. The D2O, D6-DMSO, 15NH4Cl, and 13C-labeled glucose were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). The 2000-compound Spectrum Collection of known drugs and natural products were purchased from MicroSource Discovery Systems Inc. (Gaylordsville, CT) and suspended in 100% DMSO or D6-DMSO for NMR experiments. All other chemicals and small compounds screened were purchased from LKT (St. Paul, MN), Sigma-Aldrich (St. Louis, MO), Calbiochem (San Diego, CA), or Chembridge (San Diego, CA).
Preparation of recombinant S100B
The pET11b expression vector (Novagen Inc., Madison, WI) containing either rat S100B gene, a double-mutant (C68/84S) rat S100B gene, or the bovine S100B gene (for crystallization) was used for the production of the S100B in HMS174 (DE3) cells (Novagen Inc., Madison, WI).27 Mammalian S100B is highly conserved, rat is 95.6% homologous to bovine, with rat M7, E62, S78, and V80 being V7, S62, A78, and I80, respectively, in bovine S100B. Unlabeled and 15N-labeled S100B were prepared and purified (> 99%) under reducing conditions using procedures similar to those described previously,27,28 except that dithiothreitol (DTT) was used as a reducing agent instead of β-mercaptoethanol. The concentrations of S100B stock solutions were determined using the Bio-Rad Protein Assay (Bio-Rad Inc., Hercules, CA) using wild-type S100B of known concentration as the standard. The concentration of this S100B standard was determined by quantitative amino acid analysis (BioSynthesis Inc., Lewisville, TX). The S100B was stored at a concentration of ~ 10 mM in < 2 mM TES or HEPES, at pH 7.2, with 0.5 mM DTT at –20°C until use.
Peptides
All peptides were synthesized using solid-state peptide synthesis and their purity was determined to be > 95% by high pressure liquid chromatography and mass spectrometry (Biosynthesis Inc., Lewisville, TX). The p53 peptides were derived from the terminal 27 residues of human p53, residues 367–393 (SHLKSKKGQSTSRHKKLMFKTEGPDSD), either with the N-terminus acetylated (p53367–393), fluorescein isothiocyanate (FITC)-labeled (FITC-p53367–393), or TAMRA-labeled (TAMRA-p53367–393). The TRTK-12 peptides are derived from the CapZ protein residues 265–276 (TRTKIDWNKILS), the C-termini are amidated, and have either the N-terminus acetylated (TRTK) or TAMRA-labeled (TAMRA-TRTK). The Hdm4 peptides derived from the human homolog of Mdm4 residues 25–43 (QINQVRPKLPLLKILHAAGAQ) have the C-termini amidated, and have either the N-terminus acetylated (Mdm425–43) or TAMRA-labeled (TAMRA-Mdm425–43). Each peptide was soluble and stored in H2O at pH 7.2. The concentration of the stock solutions of unlabeled peptides were determined by quantitative amino acid analysis (Biosynthesis Inc., Lewisville, TX) and/or determined using the extinction coefficient for the methyl ester of N-acetyl tryptophan, ε280 = 5600 cm-1M-1, as previously described.20 The concentration of FITC peptides was determined at pH 8.0 using the extinction coefficient for amide-linked FITC, ε494 = 68,000 cm-1M-1, and the TAMRA peptides by using the extinction coefficient for TAMRA, ε547 = 65,000 cm-1M-1.29
Fluorescent polarization competition assay
Fluorescence polarization experiments were performed in Corning 96-well, flat bottom, black assay plates (Corning, NY) in a final volume of 200 μL. The final assay buffer used in the screens contained 2.5 μM rat S100B (or 5 μM C68/84S mutant rat S100B), 50 nM TAMRA-TRTK, 50 mM TES or HEPES, pH 7.2, 15 mM NaCl, 100 mM KCl, 10 mM CaCl2, 0.10% Triton X-100, and 5% DMSO. Automation of the liquid handling and compound addition was performed using either an eight-span pipetting head-equipped Biomek NXP, or a 96-well pipetting head-equipped Biomek FX laboratory automation workstation (Beckman-Coulter). Increasing amounts of S100B were added to determine the affinity of S100B for the peptide in these buffer conditions. The high throughput FPCA was performed at four different concentrations of compound, ie, 62.5, 31.3, 15.6, and 7.8 μM that, with the “no S100B” control and the “no inhibitor” control, could be used to estimate the IC50. More robust titrations of the “hits” from the high throughput FPCA were performed to determine more accurate IC50 values for the compounds under the same conditions. All solutions used in the FPCA were incubated for a minimum of 15 minutes at 37°C, at which time the polarization was read at 37°C from the top of the well with a BMG POLARstar fluorescent plate reader (BMG Labtech, Durham, NC) using a 544 ± 5 nm excitation and 590 ± 15 nm emission filter.
The quality and suitability of the high-throughput FPCA were evaluated using the Z factor developed by Zhang et al.30 The Z factor = 1 – (3SDb + 3SDf)/(μb-μf), where μb and μf are the mean polarization (mP) values of the bound and free probe, respectively, and SDb and SDf are the standard deviations of those values for bound and free probe, respectively. The Z factor can be any value ≤ 1, with a value of 1 being an ideal assay, ≥ 0.5 but < 1.0 being an excellent assay, and a value < 0.5 being unacceptable for our application.
The binding data were fit using a single-site binding model with Origin software (OriginLab Corp., Northampton, MA), with one peptide bound per symmetrical S100B subunit. For the fluorescence polarization competition titrations, an equation derived by Nikolovska-Coleska et al was used to calculate the KD from IC50 titrations as follows: KD = [I]50/([L]50/TAMRA-TRTKKD + [P]0/TAMRA-TRTKKD + 1) where [I]50 is the concentration of the unlabeled compound at 50% inhibition, [L]50 is the concentration of the free TAMRA-TRTK at 50% inhibition, [P]0 is the concentration of the free protein at 0% inhibition, and TAMRA-TRTKKD is the dissociation constant of the S100B-TAMRA-TRTK complex (TAMRA-TRTKKD = 1.19 ± 0.65 μM; see Figure 3).31
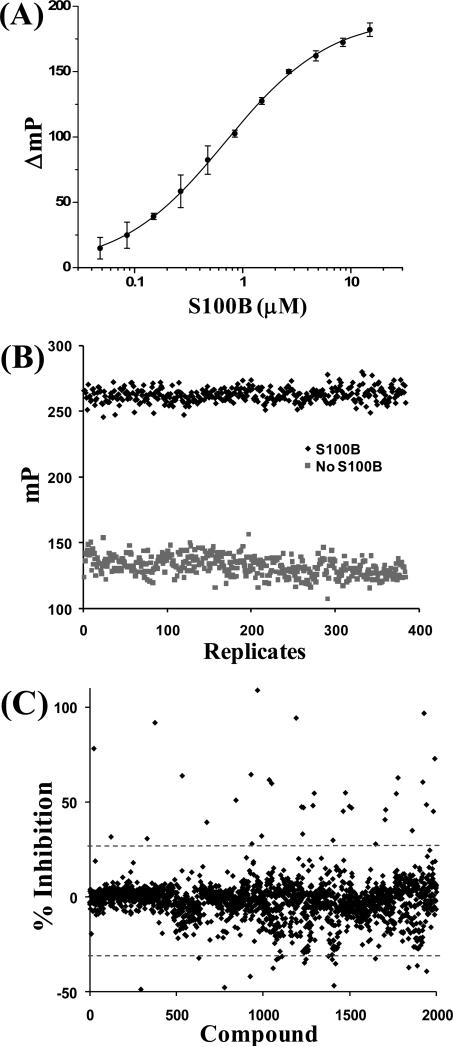
Figure 3.
Data from the FPCA assay. (A) The binding of S100B to TAMRA-TRTK was measured in the FPCA buffer conditions giving a KD = 1.19 ± 0.65 μM. (B) The Z factor of 0.71 within the acceptable range of 0.5 to 1.0 was determined for the FPCA assay with 2.5 μM S100B and 50 nM TAMRA-TRTK (see Materials and methods section for equation).30 (C) The HTS FPCA was run on four different concentrations of compounds; shown is the resulting percentage inhibition from the 62.5 μM concentration for each compound. Any compounds inhibiting greater than two standard deviations from the mean, shown with the dashed line, were tested by NMR to confirm that they bound S100B for each compound concentration, resulting in 26 inhibitors (see Table 2). There were some compounds that appeared to increase the binding, two standard deviations below the average; however, none of these compounds bound S100B, with the false signal caused by FI in the FPCA.
Abbreviations: FPCA, fluorescence polarization competition assay; NMR, nuclear magnetic resonance; FI, fluorescence interference; HTS, high-throughput screening.
Fluorescence polarization binding assay
The same buffer conditions as for the FPCA were used when determining the affinity of the different peptides for rat S100B or the C68/84S mutant rat S100B, except that an increasing concentration of S100B protein was added while monitoring the change in polarization at 544 ± 10 nm excitation and 590 ± 10 nm emission filter for TAMRA-labeled peptides and 485 ± 10 nm excitation and 520 ± 10 nm emission filter for the FITC-labeled peptide. All measurements were performed on a Varian Carey Eclipse fluorescence spectrophotometer with a control temperature cell at 37°C in a 3 × 3 mm cuvette. The binding data were fitted using a single-site binding model with Origin software (OriginLab Corp., Northampton, MA), with one peptide bound per symmetrical S100B subunit.
Nuclear magnetic resonance spectroscopy
NMR spectra were collected at 37°C with either a Bruker DMX600 NMR spectrometer (600.13 MHz for protons) or a Bruker AVANCE 800 NMR spectrometer (800.27 MHz for protons) equipped with pulsed-field gradients, four frequency channels, and triple resonance, z-axis gradient cryogenic probes. A one-second relaxation delay was used, and quadrature detection in the indirect dimensions was obtained with states-TPPI phase cycling; initial delays in the indirect dimensions were set to give zero- and first-order phase corrections of 90° and –180°, respectively.32,33 Data were processed using the processing program nmrPipe on Linux workstations.34 All proton chemical shifts are reported with respect to the H2O or HDO signal, taken to be 4.658 ppm relative to external TSP (0.0 ppm) at 37°C. The 15N chemical shifts were indirectly referenced using the zero-point frequency at 37°C of 0.10132905 for 15N-1H, as previously described.35–37
Uniformly 15N-labeled S100B was used to collect two-dimensional heteronuclear single quantum coherence (HSQC) spectra of S100B-compound complexes. Specifically, the 1H,15N-fast HSQC experiment was collected for each point in titrations of Ca2+-loaded S100B with various compounds to monitor changes in the backbone 15N and 1H resonances of S100B.38 Typical NMR samples contained 0.05–0.50 mM rat S100B (or C68/84S rat S100B), 0–5.0 mM compound, 10 mM TES or 10 mM HEPES, pH 7.2, 15 mM NaCl, 10 mM CaCl2, 0.34 mM NaN3, 5–10% D2O, and 0–5% D6-DMSO.
Group epitope mapping via saturation transfer difference (STD) NMR was completed for the chlorpromazine-S100B complex in a manner similar to that described previously.39 Specifically, during the two-second presaturation pulse, the on-resonance irradiation of the protein was performed at a chemical shift of -0.4 ppm and the off-resonance irradiation was applied at 30 ppm, where no protein signals were present. As a control, the STD experiments were collected in the absence of a T1ρ filter, and as expected, the one-dimensional spectrum of holo-S100B was fully restored. The final sample contained 50 μM S100B, 1.0 mM SC0067, 10 mM CaCl2, 5% D6-DMSO, 99.98% D2O, 15 mM NaCl, 0.34 mM NaN3, and 10mM Tris D11, at pH 7.2. The STD data were collected at 25°C to achieve more efficient saturation of the protein. Proton assignments of chlorpromazine were confirmed in the above conditions in the absence and presence of S100B using two-dimensional total correlation spectroscopy experiments.40
Protein crystallization
Diffraction quality crystals for the chlorpromazine-Ca2+-S100B complex were obtained via sitting drop vapor diffusion at 22°C by mixing 2 μL of S100B protein (40 mg/mL S100B, 7.5 mM CaCl2, 4.0 mM SC0067, 20 mM cacodylate buffer at pH 7.2) with a 2 μL of reservoir solution (200 mM CaCl2, 0.1 M Bis-Tris buffer at pH 7.0, and 42% MPD) and equilibrating for 30–60 days. After crystals formed, they were cryoprotected in a harvest solution (200 mM CaCl2, 4.0 mM chlorpromazine, 0.1 M Bis-Tris buffer at pH 7.0, 45% MPD, and 5% glycerol) for 30–60 seconds and then flash-cooled in liquid nitrogen. The SC0332-Ca2+-S100B (2–3 day crystal formation) and SC0844-Ca2+-S100B (15–30 day crystal formation) complexes were crystallized in a similar manner using complex-specific protein buffers (SC0332-Ca2+-S100B, 40 mg/mL S100B, 15 mM CaCl2, 4.0 mM SC0332, and 20 mM cacodylate buffer at pH 7.2; SC0844-Ca2+-S100B, 40 mg/mL S100B, 7.5 mM CaCl2, 4.0 mM SC0844, 20 mM Tris buffer, at pH 7.2, reservoir solutions, ie, SC0332-Ca2+-S100B, 50 mM MgCl2, 0.1 M HEPES buffer at pH 7.4, and 34% PEGMME550; SC0844-Ca2+-S100B, 7.5 mM CaCl2, 0.2 M Li2SO4, 62.5 mM cacodylate buffer at pH 8.5, and 38% PEG3350) and harvest solutions (SC0332-Ca2+-S100B, 7.5 mM CaCl2, 50 mM MgCl2, 4.0 mM SC0332, 0.1 M HEPES buffer at pH 7.4, 34% PEGMME550, and 5% glycerol; SC0844-Ca2+-S100B, 7.5 mM CaCl2, 0.1 M Li2SO4, 0.5 mM SC0844, 27.5 mM Tris buffer at pH 8.5, 39% PEG3350, and 5% glycerol). Space groups and unit cell parameters are given in Table 1. SC0067-Ca2+-S100B, SC0332-Ca2+-S100B, and SC0844-Ca2+-S100B had four, one, and two S100B subunits in the asymmetric unit, respectively.
Table 1.
Diffraction and refinement statistics.
| SC0067 Ca2+-S100B | SC0322 Ca2+-S100B | SC0844 Ca2+-S100B | |
|---|---|---|---|
| Diffraction statistics | |||
| Space group | P21 | C2221 | P3221 |
| Cell dimensions a,b,c (Å) | 28.6, 59.1, 104.0 | 35.2, 88.8, 59.0 | 46.3, 46.3, 172.4 |
| Cell angles α, β, γ (deg) | 90, 92.5, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 103.7–2.04 (2.09–2.04) | 44.41–1.79 (1.84–1.79) | 57.45–1.85 (1.89–1.85) |
| Unique reflections (n) | 20393 (1190) | 7229 (188) | 18153 (1202) |
| Completeness (%) | 96.86 (79.95) | 84.36 (30.40) | 99.27 (93.01) |
| Rsyma | 0.075 (0.428) | 0.057 (0.308) | 0.055 (0.519) |
| Average I/σ | 14.14 (2.94) | 29.09 (2.31) | 37.48 (2.80) |
| Multiplicity | 4.0 (4.0) | 5.9 (3.4) | 9.6 (6.4) |
| Refinement statistics | |||
| Rcrysb (%) | 23.7 (19.7) | 19.8 (23.6) | 19.5 (24.1) |
| Rfreeb (%) | 29.1 (29.3) | 24.2 (28.8) | 21.8 (24.1) |
| Protein atoms | 2887 | 704 | 1481 |
| Water molecules | 82 | 47 | 127 |
| Nonhydrogen atoms | 3014 | 779 | 1658 |
| Mean B-values (Å2) | |||
| Overall | 44.54 | 48.52 | 33.90 |
| Protein atoms | 43.48 | 48.48 | 33.61 |
| Water molecules | 47.03 | 63.46 | 37.48 |
| Ca2+ ions | 39.69 | 43.58 | 31.29 |
| small molecules | 113.50 | 56.37, 91.30 | 33.62 |
| Root-mean-squared difference | |||
| Bond length (Å) | 0.017 | 0.012 | 0.012 |
| Bond angles (Å) | 1.557 | 1.427 | 2.166 |
| Ramachandran plot (%)c | |||
| Most favored | 94.9 | 91.5 | 95.2 |
| Additionally allowed | 5.1 | 6.1 | 4.8 |
| Generously allowed | 0.0 | 2.4 | 0.0 |
| PDB identification | 3LKO | 3LK1 | 3LLE |
Numbers in parentheses represent the last outer shell.
Rsym = ΣhΣi(|Ii(h)| - |{l(h)}|)/ΣhΣiIj(h), where Ii(h) = observed intensity, and {l(h)} = mean intensity obtained from multiple measurements.
Rcrys and Rfree = Σ||Fo| - |Fc||/Σ||Fo|, where |Fo| = observed structure factor amplitude and |Fc| = calculated structure factor amplitude for the working and test sets, respectively.
For SC0833-Ca2+-S100B the calculations had 160 residues in the most favored region and eight residues in additionally allowed regions.
X-ray data collection, model building, and refinement
X-ray data for SC0067-Ca2+-S100B were collected at 100 K using an in-house x-ray generator (MSC Micromax 7, Rigaku, TX) and a Raxis-4++ image plate detector (Rigaku, TX). X-ray data for crystals SC0332-Ca2+-S100B and SC0844-Ca2+-S100B were collected remotely at Stanford Synchrotron Radiation Laboratory (beamlines 9-1 and 12-2, respectively, Menlo Park, CA) using an ADSC Quantum-315 CCD detector (Area Detector Systems Corp., Poway, CA) and a MAR325 CCD detector (Marresearch, Germany), respectively. The reflection intensities were integrated and scaled with the HKL2000 suite of computer programs.41 The crystals of SC0067-Ca2+-S100B, SC0332-Ca2+-S100B, and SC0844-Ca2+-S100B diffracted to 2.04 Å, 1.79 Å, and 1.85 Å resolution, respectively. Preliminary phases were obtained via molecular replacement techniques using the structure of Ca2+-bound S100B (PDB file: 1MHO) as a search model and the computer program phaser from the CCP4 program suite.42,43 Refinement and model building was completed using REFMAC5 and COOT.44,45 The locations of the SC inhibitors and several water molecules were determined by visual inspection of electron density maps calculated with 2mFo-DFc and mFo-DFc coefficients using COOT45. It was evident from visual inspection of the 2mFo-DFc and mFo-DFc electron density maps that SC0067 was bound to only two monomers of the four in the asymmetric unit and the occupancy of both chlorpromazine molecules was 1.0. The mercury for the SC0332 molecule was covalently linked to the Cys84 with the mercury at an occupancy of 0.25. The other half of SC0332 found in the hydrophobic pocket was set to an occupancy of 1, except for the sulfur which was set to an occupancy of 0.25. SC0844 occupancy was set to 1.0 and was covalently attached to the Cys84 sulfur similar to the mercury of SC0332 (see Table 1). The stereochemistry was checked with the programs WHATCHECK and PROCHEK.46,47 The quaternary structure and accessible surface areas were analyzed using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/cgibin/piserver). The coordinates for SC0067-Ca2+-S100B, SC0332-Ca2+-S100B, and SC0844-Ca2+-S100B x-ray structure were deposited in the Protein Databank and assigned the following accession numbers: 3LK0 for SC0067-Ca2+-S100B, 3LK1 for SC0332-Ca2+-S100B, and 3LLE for SC0844-Ca2+-S100B.48,49 Figures were generated with the program PyMol (http://www.pymol.org).
Results and discussion
Fluorescence polarization competition assay development
Fluorescence polarization assays have been used for HTS applications due to their high sensitivity, rapid readout, homogenous format, and relatively simple instrumentation. However, such assays require optimization to develop a rapid and sensitive method for efficient and reliable screening of large compound libraries. Some of these issues are not necessarily exclusive to polarization assays, or even fluorescent-based assays, but had to be addressed when developing the assay for discovering S100B inhibitors.
Fluorescence polarization competition assay buffer
The buffer originally used was based on the conditions used for the NMR structure determination of peptide-bound S100B (10 mM Tris, pH 6.5, 15 mM NaCl, 10 mM CaCl2) but was modified to represent intracellular conditions better with the addition of 100 mM KCl.22,24 The pH and buffer was switched to 50 mM HEPES, pH 7.2, in place of Tris, pH 6.5, again to mimic better intracellular pH and to provide a higher buffering capacity as necessary to prevent compounds from changing pH. This is important during an HTS scenario in which the pH is not trivial to adjust after the addition of each compound. It was also necessary to add 5% DMSO and 0.1% Triton X-100 to provide the optimal conditions for compound solubility.
Typically, compound libraries used for HTS screens are prepared in 100% DMSO due to its ability to dissolve a wide range of compounds. The FPCA assay will be used to screen relatively high concentrations of compound, so even weakly binding compounds including those that bind with low micromolar affinities could be identified; under such conditions, the final concentration of DMSO would also increase. Therefore, it was necessary to determine a concentration of DMSO that appreciably solubilized the compounds in aqueous solutions, but had a minimal affect on peptide binding.50 With this in mind, the affinity of S100B for target peptide was tested under varying concentrations of DMSO. In the final FPCA, 5% DMSO was used because it only decreased the affinity for the peptide minimally (< 2.0- fold) and allowed a high enough concentration of compound to be screened, consistent with previous studies showing that the use of 5% DMSO or greater increases the solubility of some of the compounds in aqueous solutions.50
The inclusion of Triton X-100 in the FPC assay was done to reduce false positives caused by compounds that themselves form aggregates and, in turn, sequester proteins non-specifically. Compounds such as these, termed “promiscuous binders”, need to be minimized, as previously described.51,52 The FPCA was tested on the same library of compounds in the absence and presence of either 0.05% or 0.10% Triton X-100 in order to identify “promiscuous binders”. While both 0.10% and 0.05% Triton X-100 effectively reduced such false positives in the FPCA, it was decided that 0.10% Triton X-100 would be used in the final assay as a cautious measure. As an additional benefit, the addition of Triton X-100, like the small amount of DMSO, also increased the solubility of many of the compounds.
Selection of fluorescently-labeled peptide probes
Several peptides have been found to bind S100B with different affinities, including one derived from p53, CapZ (TRTK-12), Hdm2, and Hdm4.18 These peptides were also synthesized with a specific N-terminal FITC or TAMRA label. To screen for inhibitors of the S100B-p53 interaction, it seemed logical to use a peptide based on the well-characterized C-terminal peptide of p53, and initially the FPCA was performed with a peptide comprising residues 367–393 of human p53 with a single FITC conjugated to its amino-terminus (FITC-p53367–393). This peptide was shown to bind to Ca2+-S100B by fluorescence polarization, and could be competed off with the unlabeled p53, TRTK-12, and Hdm4 peptides, showing that the four peptides bind in the same location on S100B. However, the KD of FITC-p53367–393 was found to have a significantly higher affinity than the unlabeled version, indicative of the FITC moiety contributing to the binding interaction. In addition, the fluorescence lifetime of the FITC varies with pH, so any slight change in pH could result in a false-positive reading. Lastly, it is also documented that FITC is sensitive to the solutions redox state, which is another source of false-positive readings, particularly with numerous compounds in the Spectrum Collection library.53 Thus, for compounds such as these, a time-dependence and sensitivity to reducing agents was noted, again due to the physical characteristics of FITC responding to the solutions redox state and not as the result of binding to S100B. To avoid these problems with the use of FITC, the N-terminal TAMRA-labeled peptides were used here.
A TAMRA-labeled version of the p53 peptide (TAMRA- p53367–393), TRTK-12 (TAMRA-TRTK), and Hdm4 (TAMRA-Hdm4) were synthesized, and their affinities for S100B determined under assay conditions. The TAMRA- p53367–393, TAMRA-TRTK, and TAMRA-Hdm4 were found to have KD values of 25.39 ± 2.97 μM, 1.19 ± 0.65 μM (see Figure 3A), and 3.08 ± 0.39 nM, respectively. The particularly high affinity of the Hdm4 peptide was surprising, and the peptide can be used in secondary screens for the highest binding inhibitors discovered. However, this peptide would not be useful for screening large libraries because the probe concentration would have to be < 1 nM, which is too low to make accurate polarization readings for many commercially available plate readers. On the other hand, the relatively low affinity binding of the TAMRA-p53367–393 peptide would require high concentrations of S100B that would be prohibitive to complete a screen of a large compound library. However, the affinity of the TAMRA-TRTK peptide for S100B was ideally suited to accommodate sufficient screening ability without the need for extremely large quantities of protein. For these reasons, it was chosen for use in the FPCA assay. The dynamic range for measuring polarization for this peptide in the free and bound state was also favorable, with the free TAMRA-TRTK peptide giving a polarization value of 134.2 ± 5.1 mP that increased to 308.2 ± 7.4 mP with saturating S100B (> 99.5% occupancy). For the most sensitive polarization competition assay, the fraction of [ligand bound]/[total ligand] should be between 0.5 and 0.8, with the total ligand (TAMRA-TRTK) being ≤ 2 × KD;54 therefore, 2.5 μM S100B was used with 50 nM TAMRA-TRTK for a fractional occupancy of 0.67, within the range. When the FPCA assay was run under these conditions, a Z factor of 0.71 is obtained, which is within the acceptable range of 0.5–1.0 (see Material and Methods section for calculation).30
High-throughput screen using FPCA
As illustrated (see Figure 2), the TAMRA-TRTK FPCA was used to screen the 2000-compound Spectrum Collection library, and 26 compounds were identified as possible S100B inhibitors. The assay was done in a high-throughput mode using four different concentrations of compound, ie, 62.5, 31.3, 15.6, and 7.8 μM. Under these conditions, compounds that inhibited the binding by at least twice the standard deviation from the mean values of all the tested compounds were considered to be top candidate inhibitors (see Figure 3C). Using these selection criteria, there were 39 (2.0%), 39 (2.0%), 37 (1.9%), and 32 (1.6%) compounds from the 2000 total that were selected for secondary screens for the 62.5, 31.3, 15.6, and 7.8 μM concentrations, respectively. Several other compounds were initially identified because they appeared to increase the binding to the peptide. However, upon closer inspection, these compounds were causing fluorescence interference (FI) with the assay. The FI could be due to the compound having overlapping fluorescence, quenching the fluorophore, and/or changing the fluorescence lifetime of the probe. Once these compounds were eliminated from further consideration, a total of 49 compounds were selected for secondary screening using NMR, with 26 compounds meeting the selection criteria. In the end, it was determined that all 26 compounds shown to bind S100B using the NMR secondary screening method would have been selected using either the 31.3 or 62.5 μM concentrations alone. Nonetheless, the use of four concentrations is still in use because it also allows for an estimation of the IC50, and also allows for easy identification of FI in the FPCA that might not be seen using a single concentration. However, for very large screens, to save material, the assay can be reliably completed using only 31.3 or 62.5 μM.
Secondary screening of compounds by NMR
The monitoring of NMR chemical shift perturbations for 1H and 15N resonances in S100B upon the addition of compound was used as a secondary screen of the top 49 compounds identified from the FPCA. These NMR data were important to eliminate false positives as well as to provide residue-specific information for mapping the compound binding site on Ca2-bound S100B. For such measurements, recombinant S100B protein was produced with 15N ammonium chloride as its only nitrogen source, so backbone 15N amide and 1HN protons could be readily detected in the presence and absence of the small molecule inhibitor using the 1H-15N edited HSQC experiment. However, collecting HSQC data such as these is not as rapid as with the FPCA, such that it is most useful as a secondary screen when only a limited number of samples need to be examined. For example, the automatic sample changers commercially available for NMR spectrometers typically hold 60 to 250 samples, with each sample requiring two 10-minute periods of data collection. Here, the samples were all prepared using 10-fold excess compound versus S100B (100 μM; 10 mM Ca2+). These data were compared with a S100B spectrum in the absence of compound, and only compounds that caused changes in the chemical shift values and/or caused loss of peaks due to chemical exchange broadening were considered to be S100B inhibitors. In addition to these data providing confirmation compound binding, the NMR chemical shift perturbations also provide residue-specific data that are useful for defining the compound binding site on the protein in solution, particularly given that the high resolution structure of Ca2+-S100B is known (see Figure 6A).
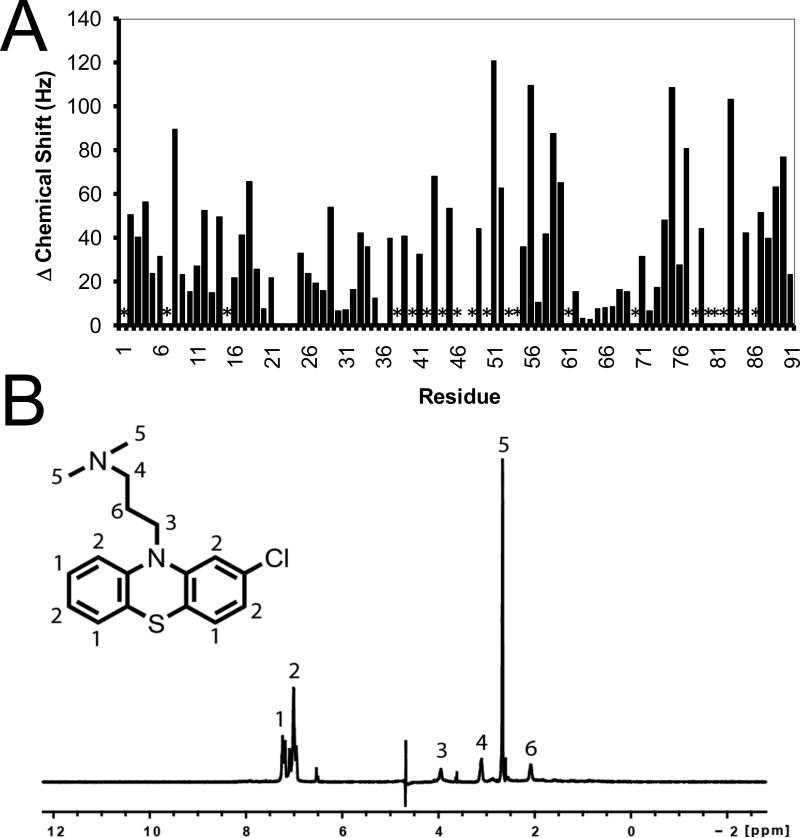
Figure 6.
NMR data for the SC0067-S100B complex. (A) The binding of chlorpromazine (SC0067) to Ca2+-S100B was monitored by HSQC NMR titrations and the combined perturbation in the chemical shifts of the backbone amide and proton were graphed. The asterisk indicates the peak was lost due to chemical exchange broadening. (B) The binding of chlorpromazine (SC0067) to Ca2+-S100B, as monitored by saturation transfer difference, NMR spectroscopy showing the strongest peaks for the atoms closest to S100B.
Abbreviations: HSQC, heteronuclear single quantum coherence; NMR, nuclear magnetic resonance;
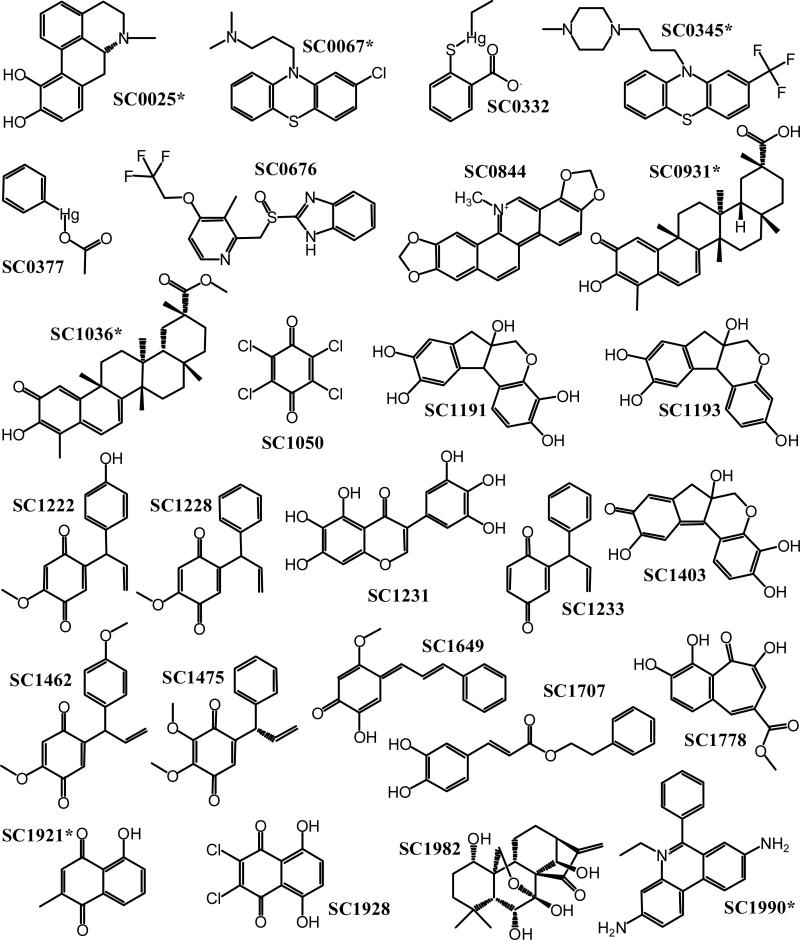
The NMR screening confirmed that 26 of the 49 compounds identified in the FPCA assay were indeed binding Ca2+-bound S100B. Many of the “false-positive” hits originally identified in the FPCA were subsequently found to possess intrinsic fluorescence that directly interfered with fluorescence polarization (ie, FI). However, nine compounds initially selected in the FPCA, but later shown to have FI in the FPCA, did in fact still bind S100B as determined by NMR. Therefore, it is not recommended to have a prescreen to eliminate compounds that potentially have FI in the FPCA. For example, two compounds, SC0931 and SC1036, are structurally very similar. In a quantitative titration using the FPCA, SC0931 was found to bind to Ca2+-S100B with the highest affinity (KD = 0.04 ± 0.01 μM) of all the compounds that were discovered here (0.04 μM < KD < 1500 μM); whereas the affinity of SC1036 could not be determined quantitatively in the FPCA due to FI. Thus, this compound showed some binding in the initial screen and was found to bind S100B by NMR. If a prescreen that eliminated the testing of compounds with FI, then SC1036 would not have been included in the NMR screen and hence not discovered as a potential lead. In addition, it should also be noted that the intrinsic fluorescence of a compound, while interfering with quantitative interpretation of the FPCA screen, is useful for monitoring binding via direct fluorescence changes (ie, as was the case with SC0844, see Table 2 and Figure 10). For the other nine compounds that had FI but were determined to bind S100B via NMR, only very rough estimates of their binding affinities could be obtained at this time, and they remain compounds of interest for further investigation. In order to provide further structural details of how the compounds identified interact with S100B, several of the compound-S100B complexes were crystallized and their structures determined at atomic resolution.
Table 2.
Summary of the 26 compounds confirmed to bind S100B by NMR.
| Compound | NMRa | IC50 (μM)b | KD (μM)c | Cys dependent |
|---|---|---|---|---|
| SC0025 | Yes | FI | FI | No |
| SC0067 | Yes | 2222.0 ± 1364.3 | 696.9 ± 427.9 | No |
| SC0332 | Yes | 177.7 ± 16.5 | 55.2 ± 5.1 | Yes |
| SC0345 | Yes | 1576.7 ± 274.3 | 494.3 ± 86.0 | No |
| SC0377 | Yes | 4.4 ± 2.2 | 0.78 ± 0.39 | Yes |
| SC0676 | Yes | 606.6 ± 370.7 | 189.8 ± 116.0 | Yes |
| SC0844 | Yes | 35.9 ± 16.4 | 10.7 ± 4.9 (5.8 ± 0.2d) | Yes |
| SC0931 | Yes | 2.0 ± 0.6 | 0.04 ± 0.01 | No |
| SC1036 | Yes | FI | FI | No |
| SC1050 | Yes | 7.6 ± 2.1 | 1.79 ± 0.50 | Yes |
| SC1191 | Yes | FI | FI | Yes |
| SC1193 | Yes | FI | FI | Yes |
| SC1222 | Yes | 1926.1 ± 1050.6 | 604.0 ± 329.5 | Yes |
| SC1228 | Yes | 832.0 ± 381.3 | 260.6 ± 119.4 | Yes |
| SC1231 | Yes | FI | FI | Yes |
| SC1233 | Yes | 4689.2 ± 4521.2 | 1471.4 ± 1418.7 | Yes |
| SC1403 | Yes | 1615.1 ± 906.0 | 506.4 ± 284.1 | Yes |
| SC1462 | Yes | 420.1 ± 66.8 | 131.3 ± 20.9 | Yes |
| SC1475 | Yes | 133.1 ± 65.0 | 41.2 ± 20.1 | Yes |
| SC1649 | Yes | FI | FI | Yes |
| SC1707 | Yes | FI | FI | Yes |
| SC1778 | Yes | FI | FI | Yes |
| SC1921 | Yes | 13.0 ± 3.5 | 3.5 ± 0.9 | No |
| SC1928 | Yes | 728.4 ± 698.7 | 228.1 ± 218.8 | Yes |
| SC1982 | Yes | FI | FI | Yes |
| SC1990 | Yes | 1349.2 ± 698.7 | 422.9 ± 219.0 | No |
The compounds identified in the initial screen were confirmed to bind S100B via NMR HSQC experiments.
Secondary titrations using the FPCA were done to determine IC50 of the compounds, however, fluorescence interference (FI), from overlapping fluorescence, quenching, etc..., prevented an exact IC50 from being determined for certain compounds.
When available, the IC50 from the FPCA was used to calculate the KD using the equation as described by Nikolovska-Coleska et al31.
Change in the compounds intrinsic fluorescence was used to determine the KD.
Figure 10.
The x-ray structure of the SC0844-Ca2+-S100B complex. (A) Ribbon and surface diagram of sanguinarine (SC0844)-Ca2+-S100B, illustrating the location of SC0844 (SC0844, orange; nitrogen, blue; oxygen, red; sulfur, orange) covalently bound to Cys84. (B and C) The electron density maps calculated with the 2mFo-DFc coefficients (1.0σ) for SC0833 in each monomer of the biological dimer. (D) Residues in Ca2+-S100B involved in coordinating SC0844. (Protein Databank accession number: 3LLE)
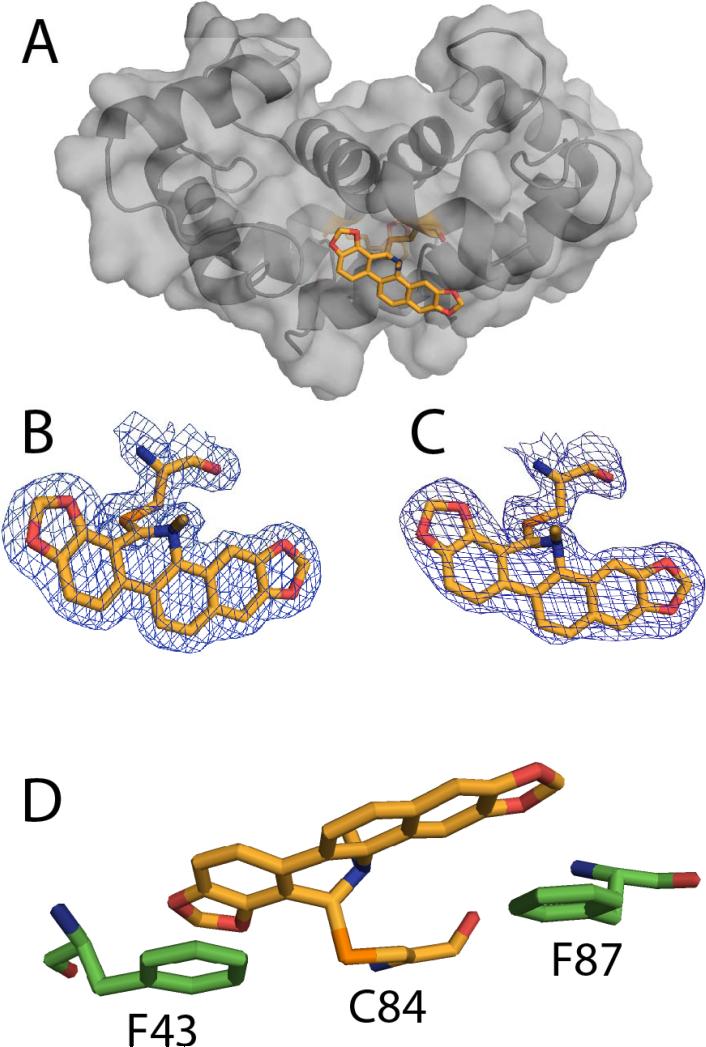
Structural studies of the chlorpromazine-Ca2+-S100B complex
The x-ray structure of chlorpromazine bound to Ca2+-S100B was solved at 2.04 Å resolution (see Figure 5 and Table 1). In the chlorpromazine-Ca2+-S100B structure, the asymmetric unit included four subunits of S100B monomers designated A–D; models A and B of the monomers formed the biologically active dimeric form of S100B. Each monomer in the Ca2+-S100B structure contained Met-0 to Glu-89, eight calcium ions, and two chlorpromazine molecules. Models A and D, each a monomer representing half of the biological dimer, had a bound chlorpromazine molecule. In the final refined models, residues of S100B in the chlorpromazine-Ca2+-S100B complex were nearly all in the most favorable region of their respective Ramachandran plots (95%) with the remaining residues falling into the allowed region (5%, see Table 1).
Figure 5.
X-ray structure of the chlorpromazine (SC0067)-Ca2+-S100B complex. (A) Surface diagram illustrating the location of the chlorpromazine molecule. (B) Ribbon diagram illustrating the asymmetric unit of SC0067-Ca2+-S100B having only two S100B monomers occupied with chlorpromazine. (C and D) Electron density maps calculated with the 2mFo-DFc coefficients (1.0σ) for each of the chlorpromazine molecules modeled. (E) Residues of S100B with 5 Å of chlorpromazine are illustrated. (Protein Databank accession number: 3LK0)
As expected, the two EF-hand calcium binding domains in each subunit were found to coordinate two Ca2+ ions, as found previously with the typical EF-hand of S100B in the Ca2+-bound “open” conformation and the dimer interface aligned as a symmetric X-type four helix bundle comprising helices 1, 1’ and 4, 4’, respectively (see Figure 1).27,55–58 The global fold for S100B in all four monomers in the chlorpromazine-Ca2+-S100B complex was similar to that reported for Ca2+-bound S100B, Zn2+, Ca2+-S100B, and several other Ca2+-loaded S100 proteins.59–62 The binding of chlorpromazine compound caused only minor structural perturbations, as evidenced by low root-mean-square distance (RMSD) values between the chlorpromazine-bound monomers in the asymmetric unit compared with unbound structures in the asymmetric unit (mainchainRMSD≈0.27 Å).42,58 Specifically, each subunit of S100B in the chlorpromazine-Ca2+-S100B complexes contained four helices (helix 1, E2-G19; helix 2, K28-L40; helix 3, E49-D61; helix 4, D69-F88) and one small antiparallel β-sheet (strand 1, K26-K28; strand 2, D69-E67).
It was necessary to model the chlorpromazine molecule into sparse electron density in only two of the asymmetric units of S100B, with each molecule having an occupancy of 1. Modeling chlorpromazine into the electron density maps calculated with 2mFo-DFc and mFo-DFc coefficients showed that the chlorpromazine molecule was oriented in the hydrophobic pocket of Ca2+-S100B with the tricyclic moiety nearby several hydrophobic residues on the hinge (H42 and F43) and helix 4 (A83, C84, F87, and F88). The dimethylamine moiety of chlorpromazine was found to be extending toward the C-terminal end of S100B (F87 and F88). Overall, the S100B-chlorpromazine interaction was stabilized via hydrophobic interactions, with residues in loop 2 (H42, F43) and in helix 4 (A83, C84, F87, F88) contributing most to binding (see Figure 5). The x-ray structure of the chlorpromazine-Ca2+-S100B complex is fully consistent with chemical shift perturbation and saturation transfer difference data collected in solution by NMR (see Figure 6). The binding of chlorpromazine to Ca2+-S100B was measured by monitoring perturbations of backbone 1H-15N correlations in two-dimensional 1H-15N NMR experiments (see Figure 6A). In HSQC titrations with chlorpromazine the 1H-15N HSQC correlations of Ca2+-S100B shifted for several residues in helix 1 (E2, E4, V8, D12, F14, S18), loop 1 (K29), loop 2 (F43, E45, E49, and E51), helix 3 (V52, V56, T59, and L60) and helix 4 (M74, A75, V77, M79, A83, F87, E89, and H90). The strength of the STD-NMR signals for the methyl group protons from the rings of the molecule, and the methyl protons on the tertiary nitrogen interact closely with S100B (see Figure 6B). In addition, the location of the chlorpromazine overlaps the binding site found for other S100B inhibitors (see Figure 7).19,60
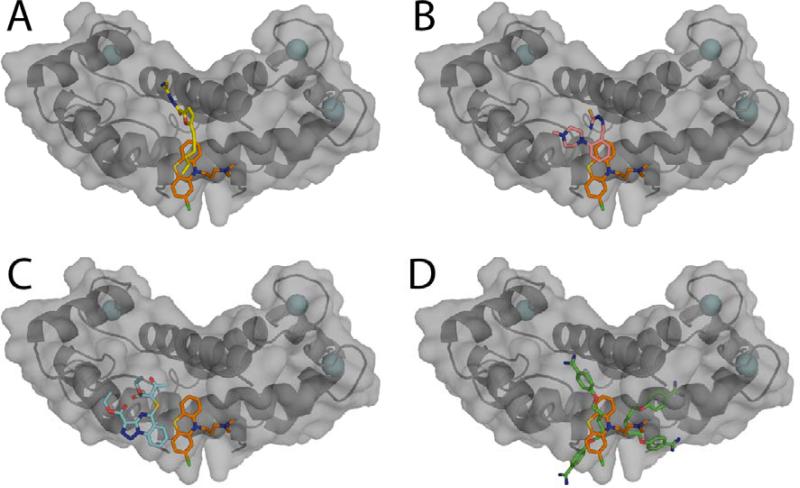
Figure 7.
Comparison of x-ray structures of small molecules bound to S100B. Surface diagrams of chlorpromazine (SC0067)-Ca2+-S100B x-ray structure is shown overlaid with other small molecules previously crystallized with Ca2+-S100B: (A) SBi132, (B) SBi279, (C) SBi523, and (D) pentamidine.19,60
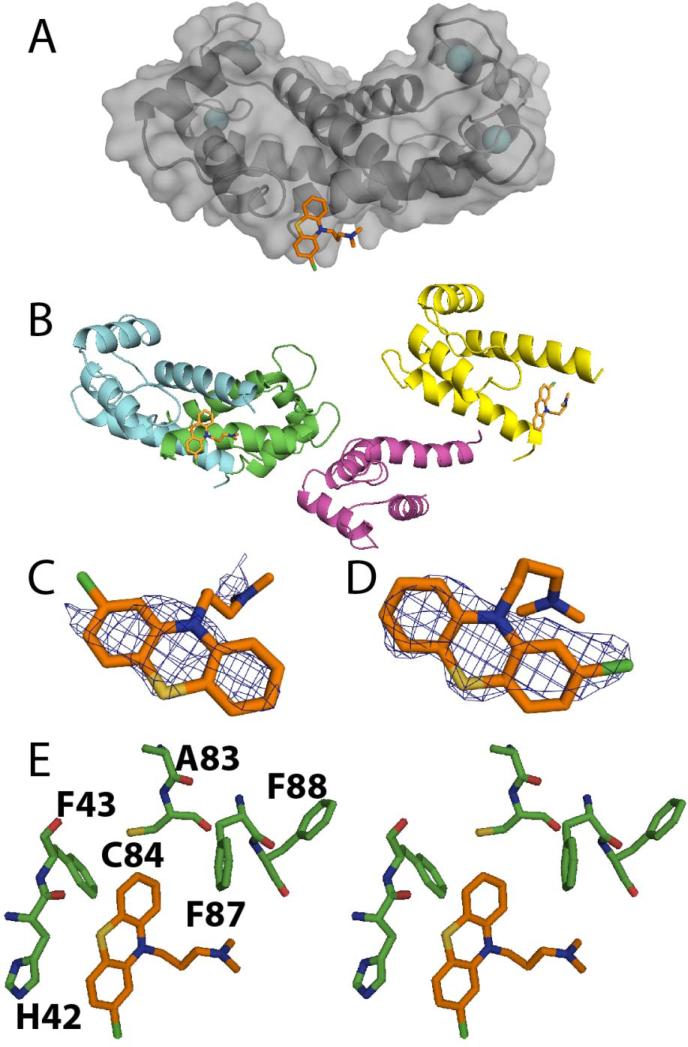
Structural studies of thimerosal-Ca2+-S100B
The x-ray structure of SC0332 bound to Ca2+-S100B was solved in the C2221 space group at 1.79 Å resolution (see Figure 8 and Table 1). The asymmetric unit for the SC0332-Ca2+-S100B structure included one monomer of S100B containing 88 residues (Met-0 to Phe-87), two calcium ions, two SC0833 molecules, and 71 water molecules. The global fold and Ca2+ ion coordination of S100B in the SC0322-Ca2+-S100B complex were very similar to that reported previously for Ca2+-S100B.42,58 Specifically, each subunit of S100B in the SC0322-Ca2+-S100B complex contained four helices (helix 1, E2-G19; helix 2, K28-L40; helix 3, E49-D61; helix 4, D69-F88) and one small antiparallel β-sheet (strand 1, K26-K28; strand 2, D69-E67). The two EF-hand Ca2+-binding domains in each subunit were found to coordinate two Ca2+ ions as found previously with the typical EF-hand of S100B in the Ca2+-bound “open” conformation and the dimer interface aligned as a symmetric X-type four helix bundle comprising helices 1, 1’ and 4, 4’, respectively.27,55–58
Figure 8.
The x-ray structure of thimerosal SC0332-Ca2+-S100B. (A) Ribbon and surface diagram of SC0332-Ca2+-S100B illustrating the location of SC0322 (SC0322, orange; nitrogen, blue; oxygen, red; sulfur, orange) with one mercury moiety covalently bound to Cys84 and the benzene in the hydrophobic pocket. (B) The electron density maps calculated with the 2mFo-DFc coefficients (1.0σ) for SC0322 in each site. (C) Residues of S100B within 4Ǻ of SC0322 bound to benzene site and depicting the distance from Cys84 that is modified by the mercury. (Protein Databank accession number: 3LK1)
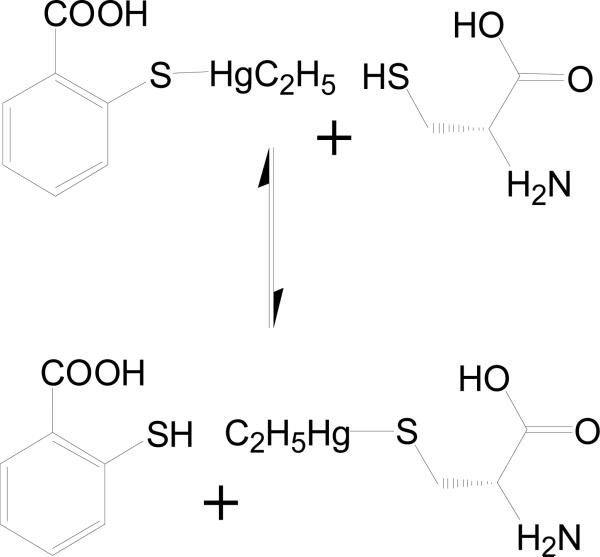
Thimerosal (SC0332) is known to covalently modify cysteine residues in proteins forming ethylmercury adducts and releasing thiosalicylic acid (see Figure 9), so it was no surprise that one of the two cysteines in S100B, Cys84, was covalently modified.63 However, visual inspection of the 2mFo-DFc and mFo-DFc electron density maps show that in addition to the ethylmercury adduct on Cys84, the thiosalicylic acid moiety remained associated with S100B near helix 3 and the N-terminus of helix 4. In addition, it should be noted that the mercury atom and thiosalicylic acid moiety had to be modeled with 0.25 occupancy. With these parameters, the electron density for the two hydrolyzed products of the SC0332 molecule were well defined with reasonable mean B-values (Å2; C21: 32.42 and 89.86; C2221: 56.37 and 91.30) when compared with the average B-factor for all protein atoms (C21: 38.92; C2221: 48.48; see Table 1). Nearly all of the residues of S100B (94.0%) in the SC0332-Ca2+-S100B complex were in the most favorable region of the Ramachandran plot with the remaining 6.0% of the residues in the allowed region (see Table 1). The thiosalicylic acid was oriented in the hydrophobic pocket adjacent to a p53 peptide-binding site as described previously (see Figure 1).64,65 The ring interacted with several hydrophobic residues on the helix 3 (V56, T59, and L60) and helix 4 (F76, M79, and I80).
Figure 9.
Mechanism of covalent modification of cysteine by thimerosal (SC0322), resulting in the formation of ethylmercury adducts while releasing thiosalicylic acid.63
Due to the interaction seen in the x-ray crystal structure, thiosalicylic acid alone was next tested for its ability to bind S100B. While it did not significantly inhibit the binding of TAMRA-TRTK in the FPCA even at 2.5 mM, it did cause significant chemical shift perturbations of the HSQC NMR spectra in the fast-exchange regime. The addition of 1 mM thiosalicylic acid to 100 μM S100B caused shifts in the end of helix 2 into the hinge region (N38, L40, S41), a flexible loop between helix 2 and 3, helix 3 (V56 and L60), and helix 4 (A75, F76, V77, S78, M79, T81, T82, A83) in amino acids consistent with the position of the thiosalicylic acid moiety in the SC0332-Ca2+-S100B x-ray crystal structure.
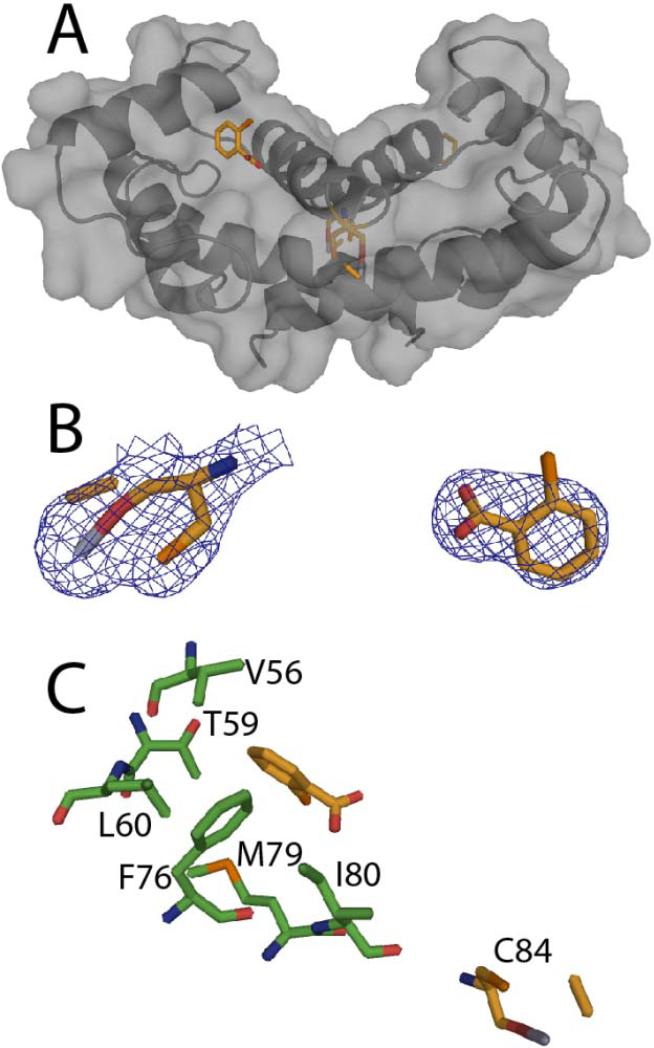
Structural studies of the sanguinarine-Ca2+-S100B complex
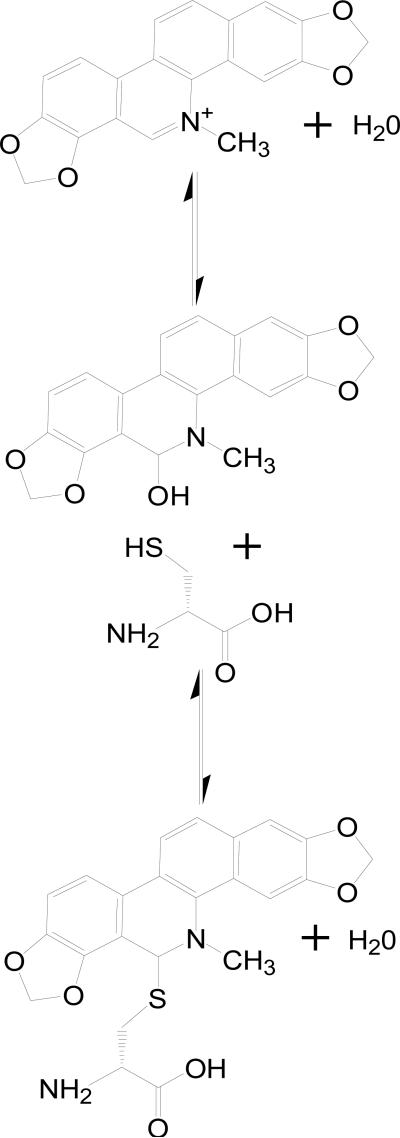
The x-ray structure of SC0844 bound to Ca2+-S100B was solved at 1.85 Å (see Figure 10 and Table 1). The asymmetric unit for the SC0844-Ca2+-S100B structure included the biologic homodimer each consisting of 90 residues per monomer (Met-0 to Glu-89), four calcium ions (two per monomer), one SC0844 molecule, and 19 water molecules (see Figure 10 and Table 1). It was evident from visual inspection of the electron density maps calculated with 2mFo-DFc and mFo-DFc coefficients that the only way to model SC0844 accurately was to link the molecule to Cys84 covalently. The covalent modification of Cys84 by SC0844 observed here was not surprising based on previous observations that cysteine residues in proteins are modified by a structurally related compound, chelerythrine (see Figure 11).66 The SC0844 molecule was found adjacent to the crystallographic two-fold axis and modeled with an occupancy of 1.0. Thus, the interactions between SC0844 and each S100B dimer were identical across the two-fold axis. Furthermore, the electron density for SC0844 was well defined (see Figure 10) with reasonable mean B-values (Å2; 33.62), which were comparable with the average B-factor for all protein atoms (33.61; see Table 1). Modeling SC0844 into the electron density maps calculated with 2mFo-DFc and mFo-DFc coefficients showed that the SC0844 molecule was oriented in the hydrophobic pocket of Ca2+-S100B covalently linked to Cys84, with the five rings lying flat across the hydrophobic pocket. Other than the covalent link with Cys84, the other two major interactions are ring stacking with Phe-43 from the flexible hinge region connecting helix 2 and helix 3, and with Phe-87 from the C-terminal region on helix 4. Surprisingly, the C-terminal end of helix 4 bent approximately 45° towards helix 1 of the other dimer. This conformational change was necessary for Phe-87 to ring stack with SC0844, and is the largest conformational change seen in S100B as a result of compound binding thus far.19,60
Figure 11.
Mechanism of covalent modification of cysteine by sanguinarine (SC0844) based on the modification of cysteines in proteins by the related compound chelerythrine.66
Nearly all of the residues in SC0844-Ca2+-S100B (95.2%) were in the most favorable region of the Ramachandran plot, with the remaining 4.8% of the residues in the allowed region (see Table 1). While the global fold and Ca2+-ion coordination of S100B in the SC0844-Ca2+-S100B complex were very similar to that reported previously for Ca2+-S100B (see Figure 11), the binding of the compound causes a significant bend in helix 4 following residue Thr-81.42,58 With the bend induced in helix 4, each subunit of S100B in the SC0844-Ca2+-S100B complex now contained five helices (helix 1, E2-G19; helix 2, K28-L40; helix 3, E49-D61; helix 4a, D69-T81; helix 4b, T82-H85) and one small antiparallel β-sheet (strand 1, K26-K28; strand 2, D69-E67). The two EF-hand calcium binding domains in each subunit were found to coordinate two Ca2+-ions as found previously with the typical EF-hand of S100B in the Ca2+-bound “open” conformation and the dimer interface aligned as a symmetric X-type four helix bundle comprising helices 1, 1’ and 4, 4’, respectively.27,55–58
Cysteine dependence of compound binding
It became clear with the completion of the x-ray crystal structures of SC0332 and SC0844 that covalent modification of Cys84 within the hydrophobic binding pocket is likely a common mechanism of the present S100B inhibitors. Therefore, the FPCA and NMR screens were repeated on the 26 compounds found to interact with S100B using a mutant of S100B, C68/84S, containing no cysteines. The C68/84S mutant S100B is properly folded, as judged by very minimal changes in its NMR HSQC spectrum, although it has a slightly decreased affinity for TAMRA-TRTK (KD is 2.42 ± 0.46 μM versus 1.19 ± 0.65 μM for the wild type). The majority of the S100B inhibitors, 19 out of 26, did not inhibit the binding of TAMRA-TRTK to the C68/84S mutant S100B, and these results were confirmed by NMR experiments showing the compounds no longer bound the mutant protein (see Table 2). While there are drugs in use that will covalently modify proteins, they are generally avoided due to their potential lack of specificity. However, other groups have taken the approach of creating libraries of drugs that bind to cysteine residues on purpose, even mutating their protein to contain a cysteine in order to get the structure of compound-protein complexes such as these. Such data, in any case, provide a useful starting point for building a specific inhibitor, as it will here for S100B.67
Conclusion
The FPCA developed here can be used in HTS to identify S100B inhibitors. Screening of the relatively small spectrum collection resulted in several inhibitors that were confirmed by NMR. It was important to show by an independent method that the compounds identified in the FPCA directly bind Ca2+-loaded S100B. For this purpose, NMR spectroscopy was chosen because chemical shift perturbations or chemical exchange broadening, as measured in 1H-15N HSQC experiments, occur for amino acid residues that directly interact with the ligand and/or as a result of more distant structural changes in the protein caused by the compound binding.68 The combination of screening methods provided 26 compounds that inhibited S100B in vitro. However, for the small collection examined here, most of the molecules that showed binding were found to modify at least one of the two cysteine residues in S100B covalently. Such molecules and the resulting three-dimensional structures determined here will be useful probes for the protein binding site that may lead to the design of better compounds, with the long-term goal of developing novel therapeutic agents for cancers such as malignant melanoma. However, compounds such as chlorpromazine (SC0067), which do not react covalently with S100B, will likely represent better leads. The fact that the binding site of chlorpromazine on Ca2+-S100B overlaps with those discovered for other S100B inhibitors (see Figure 7) suggests that this region of S100B represents a common “hot spot” for S100B inhibitors.19,60
Future studies will focus on discovering more lead compounds that inhibit S100B using the FPCA in vitro binding assay, including applying this method in combination with computer-aided drug design. The lead compounds presented here, as well as others discovered in larger screens, will then be further developed by modifying the compounds based on structure/activity relationships. It may be possible by tethering two or more lead compounds to derive a compound that binds S100B more tightly and with higher specificity.
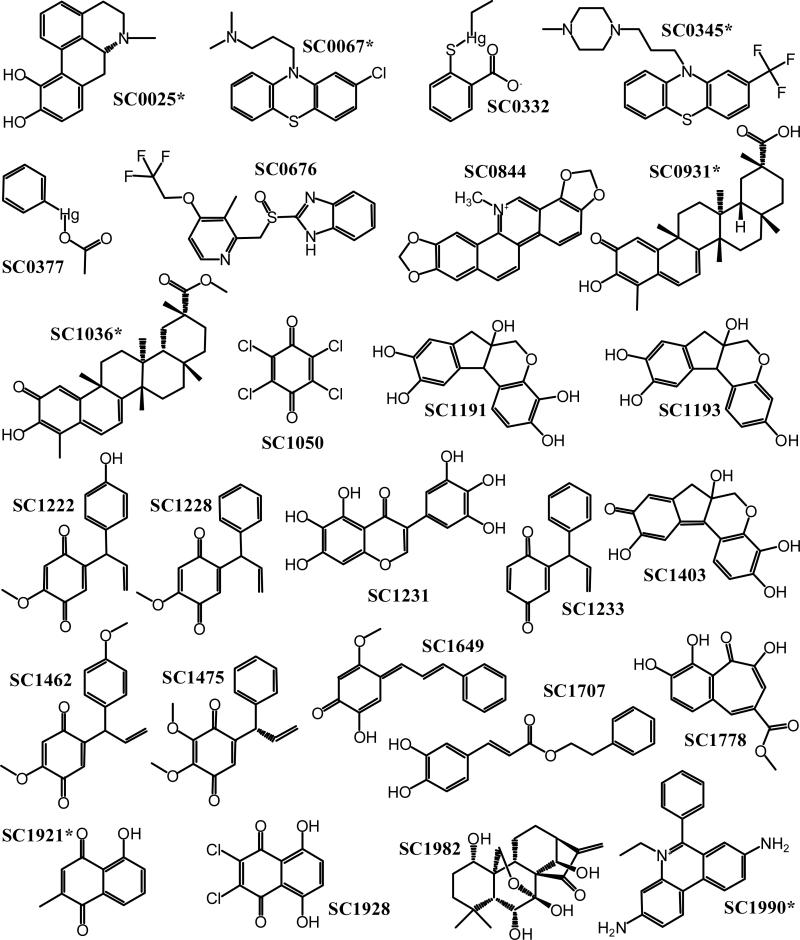
Figure 4.
Molecular structures of the S100B inhibitors. The chemical structures of the 26 compounds confirmed to bind S100B (see Table 2) are illustrated with an asterisk, indicating that those compounds bound the cysteine-free S100B mutant, C68/84S. The inhibition mechanism of the other compounds involves the modification of cysteines in S100B.
Acknowledgments
These studies were supported by grants from the National Institutes of Health (GM58888; CA107331 to DJW) and from the American Cancer Society (CDD 107745 to DJW). The NMR spectrometers used in these studies were purchased, in part, with funds from shared instrumentation grants from the NIH (S10 RR15741 to DJW).
Abbreviations
- NMR
nuclear nagnetic resonance
- cmpd
compound
- cys
cysteine
- FI
fluorescence interference
- HSQC
heteronuclear single quantum coherence
- FPCA
fluorescence polarization competition assay
Footnotes
Disclosure
The authors report no conflict of interest in this work.
References
- 1.Weber DJ, Rustandi RR, Carrier F, Zimmer DB. Interaction of Dimeric S100B(bb) with the Tumor Suppressor Protein: A Model for Ca-dependent S100-target Protein Interactions. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. [Google Scholar]
- 2.Harpio R, Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin Biochem. 2004;37(7):512–8. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Springall DR, Gu J, Cocchia D, et al. The value of S-100 immunostaining as a diagnostic tool in human malignant melanomas. A comparative study using S-100 and neuron-specific enolase antibodies. Virchows Arch A Pathol Anat Histopathol. 1983;400(3):331–43. doi: 10.1007/BF00612194. [DOI] [PubMed] [Google Scholar]
- 4.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc Natl Acad Sci U S A. 1992;89(23):11627–31. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J, Blake M, Tang C, et al. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J Biol Chem. 2001;276(37):35037–41. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 6.Lin J, Yang Q, Yan Z, et al. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J Biol Chem. 2004;279(32):34071–77. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 7.Delphin C, Ronjat M, Deloulme JC, et al. Calcium-dependent interaction of S100B with the C-terminal domain of the tumor suppressor p53. J Biol Chem. 1999;274(15):10539–44. doi: 10.1074/jbc.274.15.10539. [DOI] [PubMed] [Google Scholar]
- 8.Hibi K, Fujitake S, Takase T, et al. Identification of S100A2 as a target of the DeltaNp63 oncogenic pathway. Clin Cancer Res. 2003;9(11):4282–5. [PubMed] [Google Scholar]
- 9.Slomnicki LP, Nawrot B, Lesniak W. S100A6 binds p53 and affects its activity. Int J Biochem Cell Biol. 2009;41(4):784–90. doi: 10.1016/j.biocel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 10.van Dieck J, Teufel DP, Jaulent AM, et al. Posttranslational modifications affect the interaction of S100 proteins with tumor suppressor p53. J Mol Biol. 2009;394(5):922–30. doi: 10.1016/j.jmb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 11.van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem. 2009;284(20):13804–11. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dieck J, Brandt T, Teufel DP, Veprintsev DB, Joerger AC, Fersht AR. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene. 2010;29(14):2024–35. doi: 10.1038/onc.2009.490. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Fernig DG, Rudland PS, Sparks A, Wilkinson MC, Barraclough R. Binding to intracellular targets of the metastasis-inducing protein, S100A4 (p9Ka). Biochem Biophys Res Commun. 2001;286(5):1212–7. doi: 10.1006/bbrc.2001.5517. [DOI] [PubMed] [Google Scholar]
- 14.Grigorian M, Andresen S, Tulchinsky E, et al. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: Functional consequences of their interaction. J Biol Chem. 2001;276(25):22699–708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- 15.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 16.Gadek TR, Nicholas JB. Small molecule antagonists of proteins. Biochem Pharmacol. 2003;65(1):1–8. doi: 10.1016/s0006-2952(02)01479-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhong S, Macias AT, MacKerell AD., Jr. Computational identification of inhibitors of protein-protein interactions. Curr Top Med Chem. 2007;7(1):63–82. doi: 10.2174/156802607779318334. [DOI] [PubMed] [Google Scholar]
- 18.Wilder PT, Lin J, Bair CL, et al. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763(11):1284–97. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 19.Charpentier TH, Wilder PT, Liriano MA, et al. Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and x-ray crystallography. Biochemistry. 2009;48(26):6202–12. doi: 10.1021/bi9005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry. 1998;37(7):1951–60. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 21.Wilder PT, Rustandi RR, Drohat AC, Weber DJ. S100B(betabeta) inhibits the protein kinase C-dependent phosphorylation of a peptide derived from p53 in a Ca2+-dependent manner. Protein Sci. 1998;7(3):794–8. doi: 10.1002/pro.5560070330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta). Nat Struct Biol. 2000;7(7):570–4. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 23.Rustandi RR, Baldisseri DM, Drohat AC, Weber DJ. Structural changes in the C-terminus of Ca2+-bound rat S100B (beta beta) upon binding to a peptide derived from the C-terminal regulatory domain of p53. Protein Sci. 1999;8(9):1743–51. doi: 10.1110/ps.8.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;324(5):1003–14. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 25.Charpentier TH, Thompson LE, Liriano MA, et al. The effects of CapZ peptide (TRTK-12) binding to S100B-Ca(2+) as examined by NMR and x-ray crystallography. J Mol Biol. 2010;396(5):1227–43. doi: 10.1016/j.jmb.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Aulabaugh A. Application of fluorescence polarization in HTS assays. Methods Mol Biol. 2009;565:127–43. doi: 10.1007/978-1-60327-258-2_6. [DOI] [PubMed] [Google Scholar]
- 27.Amburgey JC, Abildgaard F, Starich MR, Shah S, Hilt DC, Weber DJ. 1H, 13C and 15N NMR assignments and solution secondary structure of rat Apo-S100 beta. J Biomol NMR. 1995;6(2):171–9. doi: 10.1007/BF00211781. [DOI] [PubMed] [Google Scholar]
- 28.Drohat AC, Amburgey JC, Abildgaard F, Starich MR, Baldisseri D, Weber DJ. Solution structure of rat apo-S100B(beta beta) as determined by NMR spectroscopy. Biochemistry. 1996;35(36):11577–88. doi: 10.1021/bi9612226. [DOI] [PubMed] [Google Scholar]
- 29.Konecsni T, Kilar F. Monitoring of the conjugation reaction between human serum transferrin and fluorescein isothiocyanate by capillary electrophoresis. J Chromatogr A. 2004;1051(1-2):135–9. doi: 10.1016/j.chroma.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 30.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 31.Nikolovska-Coleska Z, Wang R, Fang X, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332(2):261–73. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 32.Marion D, Driscoll PC, Kay LE, et al. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1 beta. Biochemistry. 1989;28(15):6150–6. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 33.Bax A, Ikura M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the alpha-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J Biomol NMR. 1991;1(1):99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Edison AS, Abildgaard F, Westler WM, Mooberry ES, Markley JL. Practical introduction to theory and implementation of multinuclear, multidimensional nuclear magnetic resonance experiments. Methods Enzymol. 1994;239:3–79. doi: 10.1016/s0076-6879(94)39003-7. [DOI] [PubMed] [Google Scholar]
- 36.Live DH, Davis DG, Agosta WC, Cowburn D. J Am Chem Soc. 1984;(106):1939–41. [Google Scholar]
- 37.Spera S, Ikura M, Bax A. Measurement of the exchange rates of rapidly exchanging amide protons: Application to the study of calmodulin and its complex with a myosin light chain kinase fragment. J Biomol NMR. 1991;1(2):155–65. doi: 10.1007/BF01877227. [DOI] [PubMed] [Google Scholar]
- 38.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108(1):94–8. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 39.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123(25):6108–17. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 40.Bax A, Davis DG. Practical aspects of two-dimensional transverse NOE spectroscopy. Journal of Magnetic Resonance. 1985;63:207–13. [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Matsumura H, Shiba T, Inoue T, Harada S, Kai Y. A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain. Structure. 1998;6(2):233–41. doi: 10.1016/s0969-2126(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 43.Vagin AA, Isupov MN. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1451–6. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–91. [Google Scholar]
- 47.Laskowski RA, MacArthur MW, Thornton JM. Validation of protein models derived from experiment. Curr Opin Struct Biol. 1998;8(5):631–9. doi: 10.1016/s0959-440x(98)80156-5. [DOI] [PubMed] [Google Scholar]
- 48.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by x-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 10):1833–9. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 50.Danley DE. Crystallization to obtain protein-ligand complexes for structure-aided drug design. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 6):569–75. doi: 10.1107/S0907444906012601. [DOI] [PubMed] [Google Scholar]
- 51.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1(3):146–8. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 52.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45(8):1712–22. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 53.Ou B, Hampsch-Woodill M, Prior RL. Development and validation of an improved oxygen radical absorbance capacity assay using fluorescein as the fluorescent probe. J Agric Food Chem. 2001;49(10):4619–26. doi: 10.1021/jf010586o. [DOI] [PubMed] [Google Scholar]
- 54.Huang X. Fluorescence polarization competition assay: The range of resolvable inhibitor potency is limited by the affinity of the fluorescent ligand. J Biomol Screen. 2003;8(1):34–8. doi: 10.1177/1087057102239666. [DOI] [PubMed] [Google Scholar]
- 55.Baudier J, Glasser N, Gerard D. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100 alpha alpha, S100a (alpha beta), and S100b (beta beta) protein: Zn2+ regulates Ca2+ binding on S100b protein. J Biol Chem. 1986;261(18):8192–8203. [PubMed] [Google Scholar]
- 56.Kligman D, Hilt DC. The S100 protein family. Trends Biochem Sci. 1988;13(11):437–43. doi: 10.1016/0968-0004(88)90218-6. [DOI] [PubMed] [Google Scholar]
- 57.Strynadka NC, James MN. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–98. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 58.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37(9):2729–40. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 59.Wilder PT, Varney KM, Weiss MB, Gitti RK, Weber DJ. Solution structure of zinc- and calcium-bound rat S100B as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44(15):5690–5702. doi: 10.1021/bi0475830. [DOI] [PubMed] [Google Scholar]
- 60.Charpentier TH, Wilder PT, Liriano MA, et al. Divalent metal ion complexes of S100B in the absence and presence of pentamidine. J Mol Biol. 2008;382(1):56–73. doi: 10.1016/j.jmb.2008.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396(2):201–14. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber DJ, Rustandi RR, Carrier F, Zimmer DB. Interaction of Dimeric S100B(bb) with the Tumor Suppressor Protein: A Model for Ca-dependent S100-target Protein Interactions. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. [Google Scholar]
- 63.Wu X, Liang H, O'Hara KA, Yalowich JC, Hasinoff BB. Thiol-modulated mechanisms of the cytotoxicity of thimerosal and inhibition of DNA topoisomerase II alpha. Chem Res Toxicol. 2008;21(2):483–93. doi: 10.1021/tx700341n. [DOI] [PubMed] [Google Scholar]
- 64.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B. Nat Struct Biol. 2000;7(7):570–4. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 65.Markowitz J, Chen I, Gitti R, et al. Identification and characterization of small molecule inhibitors of the calcium-dependent S100B-p53 tumor suppressor interaction. J Med Chem. 2004;47:5085–93. doi: 10.1021/jm0497038. [DOI] [PubMed] [Google Scholar]
- 66.Lou H, Ookhtens M, Stolz A, Kaplowitz N. Chelerythrine stimulates GSH transport by rat Mrp2 (Abcc2) expressed in canine kidney cells. Am J Physiol Gastrointest Liver Physiol. 2003;285(6):G1335–44. doi: 10.1152/ajpgi.00271.2003. [DOI] [PubMed] [Google Scholar]
- 67.Erlanson DA, Braisted AC, Raphael DR, et al. Site-directed ligand discovery. Proc Natl Acad Sci U S A. 2000;97(17):9367–72. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41(1):1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]