SUMMARY
Pten deficiency depletes hematopoietic stem cells (HSCs) but expands leukemia-initiating cells and the mTOR inhibitor, rapamycin, blocks these effects. Understanding the opposite effects of mTOR activation on HSCs versus leukemia-initiating cells could improve anti-leukemia therapies. We found that the depletion of Pten-deficient HSCs was not caused by oxidative stress and could not be blocked by N-acetyl-cysteine. Instead, Pten deletion induced, and rapamycin attenuated, the expression of p16Ink4a and p53 in HSCs, and p19Arf and p53 in other hematopoietic cells. p53 suppressed leukemogenesis and promoted HSC depletion after Pten deletion. p16Ink4a also promoted HSC depletion but had a limited role suppressing leukemogenesis. p19Arf strongly suppressed leukemogenesis but did not deplete HSCs. Secondary mutations attenuated this tumor suppressor response in some leukemias that arose after Pten deletion. mTOR activation therefore depletes HSCs by a tumor suppressor response that is attenuated by secondary mutations in leukemogenic clones.
Keywords: Pten, hematopoietic stem cell, leukemia-initiating cell, mTOR, Ink4a/Arf, p53
INTRODUCTION
Phosphatidylinositol 3-kinase (PI-3kinase) pathway signaling promotes cell growth, proliferation, and survival by several mechanisms (Wullschleger et al., 2006; Yuan and Cantley, 2008) (see Fig. S1 for a schematic). Receptor tyrosine kinases and other signaling pathways activate PI-3kinase, which generates phosphatidylinositol-3,4,5-trisphosphate (PIP3) (Yuan and Cantley, 2008). PIP3 activates Akt, which promotes cell growth, proliferation, and survival by phosphorylating diverse substrates (Manning and Cantley, 2007), including the Tuberous Sclerosis Complex (TSC) (Inoki et al., 2002). Phosphorylation by Akt negatively regulates TSC, leading to the activation of the mammalian target of rapamycin (mTOR) kinase (Inoki et al., 2002). mTOR functions in two distinct complexes, mTORC1, which is directly inhibited by rapamycin, and mTORC2, which can be indirectly inhibited by rapamycin (Guertin and Sabatini, 2007; Sarbassov et al., 2006). mTORC1 promotes cell growth and proliferation by activating S6 kinase and inactivating 4EBP1, promoting protein synthesis (Inoki et al., 2002). mTORC2 regulates Akt activation (Guertin and Sabatini, 2007). PI-3kinase signaling is attenuated by Pten, which dephosphorylates PIP3 (Maehama and Dixon, 1998), reducing the activation of Akt, mTORC1, and S6 kinase. As a result, Pten deficiency increases the growth, proliferation, and survival of many cells (Sun et al., 1999) and Pten is commonly deleted in diverse cancers (Di Cristofano and Pandolfi, 2000).
PI-3kinase pathway signaling has divergent effects on stem cells. Conditional deletion of Pten from embryonic stem cells and neural stem cells increases cell cycle entry and self-renewal (Gregorian et al., 2009; Groszer et al., 2006; Groszer et al., 2001; Sun et al., 1999). In contrast, Pten deletion from adult HSCs increases cell cycle entry but this leads to rapid HSC depletion (Yilmaz et al., 2006; Zhang et al., 2006). We showed that this depletion was mediated by mTOR activation as rapamycin blocked the depletion of Pten-deficient HSCs (Yilmaz et al., 2006). Subsequent studies of Tsc1-deficient HSCs confirmed that increased PI-3kinase pathway signaling can drive HSCs into cycle but that this leads to mTOR-mediated HSC depletion (Chen et al., 2008; Gan et al., 2008). Pml deletion also increased HSC cycling and led to mTOR-mediated HSC depletion (Ito et al., 2008). mTOR is thus a critical modulator of stem cell maintenance, raising the question of how mTOR activation leads to stem cell depletion.
While Pten deletion leads to the depletion of normal HSCs, this leads to the generation and expansion of leukemia-initiating cells (Yilmaz et al., 2006). This makes it possible to deplete leukemia-initiating cells while rescuing normal HSC function using rapamycin in mice that are deficient for Pten or Pml (Ito et al., 2008; Yilmaz et al., 2006). A sophisticated understanding of PI-3kinase pathway signaling can therefore lead to therapies that eliminate cancer-initiating cells with limited toxicity to normal stem cells.
One mechanism by which Pten deletion and PI-3kinase pathway activation could deplete stem cells involves a tumor suppressor response. Sustained oncogenic signals can induce tumor suppressors that cause cellular senescence (Lin et al., 1998; Serrano et al., 1997). Conditional inactivation of Pten in prostate leads to the induction of p53-mediated senescence, impeding the development of prostate cancer (Chen et al., 2005). These studies raise the question of whether a similar tumor suppressor response occurs after Pten deletion in stem cells, and whether this suppresses leukemogenesis or depletes HSCs.
Another mechanism by which Pten deletion could deplete HSCs involves the inactivation of FoxO family transcription factors. When localized to the nucleus, FoxO transcription factors promote the expression of enzymes that eliminate reactive oxygen species (ROS). However, activated Akt phosphorylates FoxO proteins, leading to their retention in the cytoplasm (Biggs et al., 1999; Brunet et al., 1999) and increasing ROS levels. HSCs are particularly sensitive to the toxic effects of ROS (Ito et al., 2004; Ito et al., 2006). Deletion of FoxO1/3/4, or FoxO3a alone, from adult HSCs leads to increased ROS levels and HSC depletion that can be at least partially rescued by the antioxidant N-Acetyl-cysteine (NAC) (Miyamoto et al., 2007; Tothova et al., 2007; Yalcin et al., 2008). The depletion of Tsc1-deficient HSCs is also partly rescued by NAC treatment (Chen et al., 2008). On the other hand, constitutively active Akt did not increase ROS levels in HSCs and the depletion of these HSCs could not be rescued by NAC treatment (Kharas et al., 2010). These results raise the question of whether Pten deficiency and FoxO deficiency deplete HSCs by similar or different mechanisms.
We found that Pten deficiency and FoxO1/3/4 deficiency lead to HSC depletion by different mechanisms. We were unable to detect FoxO3a inactivation or a significant increase in ROS levels in HSCs after Pten deletion. We were not able to rescue the depletion of Pten-deficient HSCs by treating with NAC. Instead, Pten deletion induced the expression of p16Ink4a and p53 in HSCs, and p19Arf and p53 in other hematopoietic cells. Analysis of compound mutant mice revealed that deficiency for p19Arf, or p16Ink4a/p19Arf, or p53 (but not p16Ink4a) accelerated leukemogenesis after Pten deletion. Moreover, deficiency for p16Ink4a, or p16Ink4a/p19Arf, or p53 (but not p19Arf) autonomously prolonged the reconstituting capacity of HSCs after Pten deletion. Our results demonstrate there are multiple mechanisms by which increased PI-3kinase pathway signaling can deplete HSCs, including an mTOR-mediated tumor suppressor response that occurs in HSCs after Pten deletion and that is attenuated by secondary mutations in leukemogenic clones.
RESULTS
Akt activation after Pten deletion activates mTORC1 but does not inactivate FoxO3a
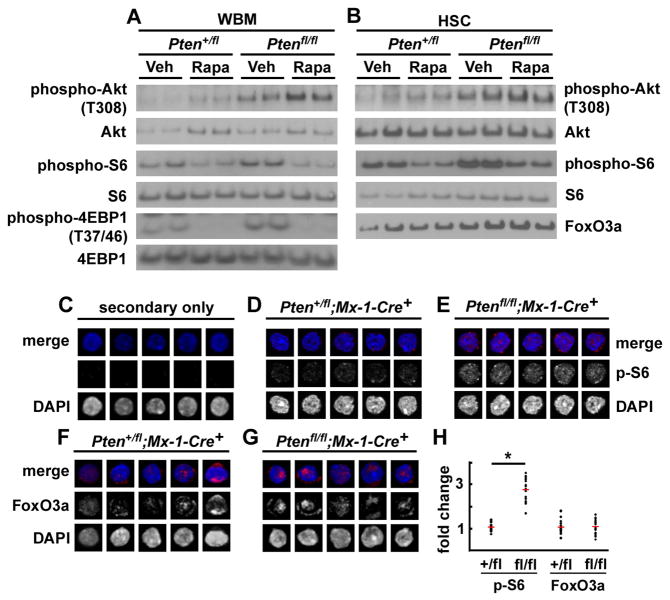
We conditionally deleted Pten from HSCs and other hematopoietic cells in young adult Ptenfl/fl;Mx-1-Cre+ mice and Pten+/fl;Mx-1-Cre+ littermate controls by administering seven doses of polyinosine-polycytidine (pIpC) over 14 days (Yilmaz et al., 2006). Pten-deleted mice and littermate controls were then treated for a week with daily injections of rapamycin or vehicle. Rapamycin treatment and Pten deletion both increased Akt activation in bone marrow cells and in HSCs based on Western blots performed in c-kit+Flk-2−Lin−Sca-1+CD48− cells, a population highly enriched for HSCs (Christensen and Weissman, 2001; Kiel et al., 2005) (Fig. 1A, B). The increase in Akt phosphorylation after rapamycin treatment is consistent with the attenuation of a negative feedback loop from S6 kinase to IRS-1 as a result of reduced mTORC1 activation (Harrington et al., 2004). Rapamycin does not, therefore, rescue the reconstituting activity of Pten-deficient HSCs by normalizing Akt activation.
Figure 1. Pten deletion activated Akt and mTORC1 signaling in HSCs but FoxO3a was not inactivated.
(A, B) Pten deletion increased phospho-Akt (T308), phospho-S6, and phospho-4EBP1 (T37/46) levels in whole bone marrow cells (A) as well as in c-kit+Flk-2−Lin−Sca-1+CD48− HSCs (B) as expected. Rapamycin treatment tended to further increase phospho-Akt levels, but decreased phospho-S6, and phospho-4EBP1 (T37/46) levels, as expected. Quantification demonstrated that Pten deletion increased phospho-Akt levels by 2.6-fold and phospho-S6 levels by 1.5-fold by in HSCs. Rapamycin treatment further increased phospho-Akt levels by 1.7-fold in HSCs and decreased phospho-S6 levels by 40% in HSCs. Total protein levels of FoxO3a did not decrease with Pten deletion and were unaffected by rapamycin treatment (B). Each lane contained protein extracted from 40,000 sorted cells. (C–H) Staining of sorted CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs with secondary antibody alone (C), or primary and secondary antibody against phospho-S6 (D–E) or FoxO3a (F–G). Phospho-S6 staining was significantly elevated in Ptenfl/flMx-1-Cre+ HSCs as compared to Pten+/flMx-1-Cre+ control HSCs, as expected (D–E, H; *, p<0.0001 by Student’s t-test), but the level and subcellular localization of FoxO3a staining did not differ between Ptenfl/flMx-1-Cre+ and control HSCs (F–H). We analyzed 10–30 HSCs from 1–2 mice/genotype in each of 3 independent experiments. In a similar assay, culture of HSCs in medium containing SCF and TPO did lead to decreased total FoxO3a levels and cytoplasmic localization (Fig. S3). See also Figures S1, S2, S3.
Pten deletion did not appear to deplete HSCs by negatively regulating FoxO expression. By western blot neither Pten deletion nor rapamycin treatment appeared to significantly affect total FoxO3a levels in c-kit+Flk-2−Lin−Sca-1+CD48− HSCs (Fig. 1B), or phospho-FoxO3a levels, total FoxO1 levels, or phospho-H2AX levels in c-kit+Lin−Sca-1+ cells (Fig. S2). In control experiments, when CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs (Kiel et al., 2005) were sorted from control and Pten-deleted mice and stimulated with SCF and TPO in culture for 24 hours they showed an overall decrease in FoxO3a staining as well as decreased nuclear staining and increased cytoplasmic staining (Fig. S3). In contrast, when HSCs were stained immediately after isolation from mice, we detected no change in FoxO3a expression or subcellular localization after Pten deletion (Fig. 1F–H): FoxO3a continued to localize to the nucleus.
mTORC1 activity was clearly elevated after Pten deletion based on increased phospho-S6 levels in freshly isolated bone marrow cells and HSCs (Fig. 1A, 1B, 1D, 1E, 1H). This increase in phospho-S6 levels was attenuated by rapamycin treatment (Fig. 1A, 1B). These changes were further confirmed by increased phospho-4EBP1 levels in bone marrow cells after Pten deletion (Fig. 1A). This increase in phospho-4EBP1 levels was also attenuated by rapamycin treatment (Fig. 1A). Thus, Pten deletion increased mTORC1 and S6 kinase activation in a manner that was reversed by rapamycin, suggesting that the changes that lead to HSC depletion after Pten deletion are mediated by this branch of the PI-3kinase pathway.
Pten deletion increases ROS levels in thymocytes but not in HSCs or bone marrow cells
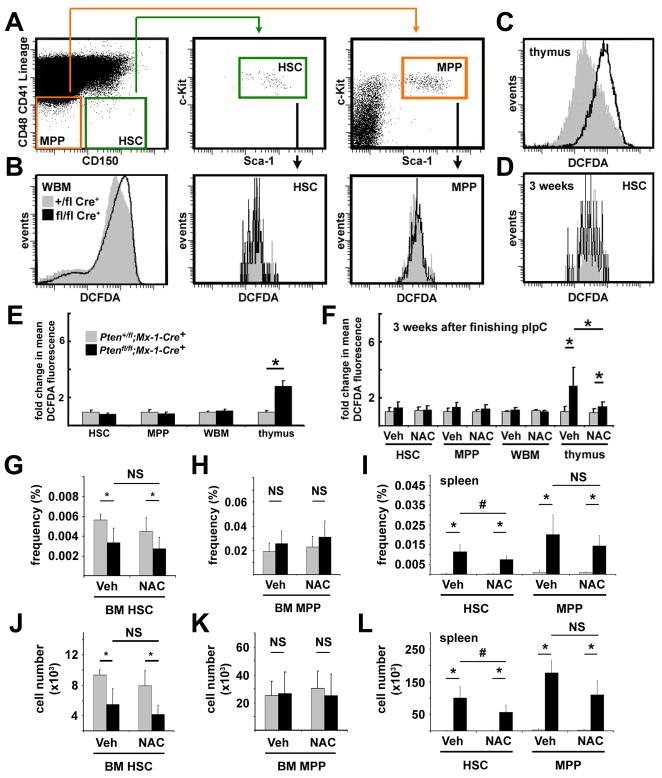
We assessed the intracellular levels of ROS in CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs, CD150−CD48−CD41−Lin−c-kit+Sca-1+ transiently reconstituting multipotent progenitors (MPPs) (Kiel et al., 2008), bone marrow cells, and thymocytes from Pten-deleted mice and littermate controls one week after finishing pIpC treatment. After Pten deletion, we detected a significant increase in ROS levels in thymocytes (Fig. 2C, 2E; 2.8-fold, p<0.05) but not in bone marrow cells, HSCs, or MPPs (Fig. 2B, E). Three weeks after finishing pIpC treatment we also did not detect an increase in ROS levels within unfractionated bone marrow cells, HSCs, or MPPs (Fig. 2D, F). Only mice showing no signs of neoplasms were used in these experiments.
Figure 2. Pten deletion significantly increased ROS levels in thymocytes but not in HSCs, MPPs, or whole bone marrow cells and NAC treatment failed to rescue the depletion of HSCs.
(A) Gating scheme used to assess intracellular ROS levels in CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs and CD150−CD48−CD41−Lin−c-kit+Sca-1+ MPPs. 7 (B) or 21 (D) days after finishing pIpC treatment (7 doses of pIpC over 14 days), DCFDA staining of whole bone marrow cells, HSCs, and MPPs did not significantly differ between Ptenfl/flMx-1-Cre+ and Pten+/flMx-1-Cre+ control mice. (C) In contrast, thymocytes from Ptenfl/flMx-1-Cre+ mice did exhibit significantly greater DCFDA staining than thymocytes from Pten+/flMx-1-Cre+ controls. (E) Mean DCFDA fluorescence levels showed no evidence of increased ROS levels in HSCs, MPPs, or bone marrow cells, but a significant (*, p<0.05 by Student’s t-test) increase in ROS levels within thymocytes. Similar experiments performed 21 days after finishing pIpC treatment yielded similar results (F). Daily subcutaneous injections of NAC after pIpC treatment did not significantly affect DCFDA staining of HSCs, MPPs, or bone marrow cells, but did significantly reduce DCFDA staining of thymocytes (F). The frequency (G) and absolute number (J) of CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs in the bone marrow declined significantly after Pten deletion but were not affected by NAC. The frequency (H) and absolute number (K) of CD150−CD48−CD41−Lin−c-kit+Sca-1+ MPPs in the bone marrow were not affected by Pten deletion or NAC. The frequency (I) and absolute number (L) of HSCs and MPPs in the spleen significantly increased after Pten deletion. The increase in HSCs was slightly but significantly (#, p<0.05) attenuated by NAC treatment but the increase in MPPs was not significantly affected (I, L). Data (mean±standard deviation) are from 4 independent experiments with 1–2 mice/genotype/treatment. See also Figure S4.
The increase in ROS levels in the thymus after Pten deletion was attenuated by daily subcutaneous injection of the anti-oxidant NAC, but NAC did not affect ROS levels in bone marrow cells, HSCs, or MPPs (Fig. 2F). It is unclear why Pten deletion increased ROS levels in thymocytes but not in HSCs or WBM cells, but ROS regulation is known to differ among hematopoietic cells (Ito et al., 2004; Ito et al., 2006; Tothova et al., 2007). We were also unable to detect significant effects of Pten deletion on the expression of FoxO target genes (Tothova et al., 2007) or antioxidant response genes in bone marrow cells, HSCs, or MPPs (Fig. S4C–H).
NAC does not rescue major hematopoietic defects after Pten deletion
Consistent with the normal ROS levels in HSCs and most other hematopoietic cells, daily treatment with NAC for 3 weeks after pIpC treatment did not rescue the changes in spleen, thymus, or bone marrow cellularity after Pten deletion (Fig. S4A, B). NAC treatment also did not rescue the depletion of bone marrow HSCs (Fig. 2G, J). MPP frequency and number in the bone marrow and spleen were not significantly affected by Pten deletion or NAC treatment (Fig. 2H, I, K, L). The frequency and absolute number of HSCs and MPPs increased significantly in the spleen three weeks after pIpC treatment (Fig. 2I, L), consistent with the extramedullary hematopoiesis that arises after Pten deletion (Yilmaz et al., 2006). NAC attenuated the increase in splenic HSCs, though HSC frequency and number still increased significantly in the spleen (Fig. 2I, L). NAC also had little effect on other changes in hematopoiesis after Pten deletion (Fig. S5A–C). The inability of NAC to rescue hematopoietic defects after Pten deletion contrasts with the ability of rapamycin to do so (Yilmaz et al., 2006).
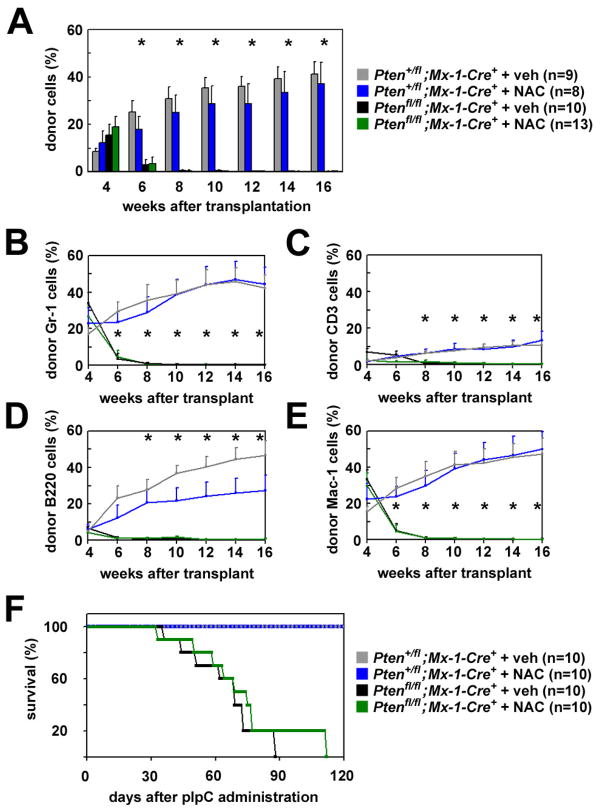
To test whether NAC rescues the function of Pten-deleted HSCs, we treated Ptenfl/fl;Mx-1-Cre+ and Pten+/fl;Mx-1-Cre+ mice with 7 doses of pIpC over 14 days, then transplanted 10 donor CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs along with a radioprotective dose of recipient bone marrow cells into irradiated recipients. Half of the recipients received daily subcutaneous injections of NAC, while the other half received daily saline injections, starting on the day of transplantation. In three independent experiments, all recipients of control HSCs, whether treated with NAC or vehicle, were long-term multilineage reconstituted by donor cells (Fig. 3A–E). All of the recipients of Pten-deleted HSCs showed transient multilineage reconstitution, irrespective of whether they were treated with NAC (Fig. 3A–E). The inability of NAC to rescue Pten deficient HSC frequency or function suggests oxidative stress is unlikely to be the primary determinant of HSC depletion after Pten deletion.
Figure 3. NAC treatment did not restore the reconstituting capacity of HSCs or block leukemogenesis after Pten-deletion.
(A–E) After pIpC treatment, 10 donor CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs were transplanted into lethally irradiated recipients along with 300,000 recipient bone marrow cells, and recipients were maintained on daily injections of NAC or vehicle beginning the day after transplantation. Control (Pten+/flMx-1-Cre+) HSCs gave high levels of long-term multilineage reconstitution by donor Gr-1+ myeloid (B), CD3+ T (C), B220+ B (D), and Mac-1+ myeloid cells (E) in all recipients, irrespective of NAC treatment. Pten-deleted (Ptenfl/flMx-1-Cre+) HSCs gave transient multilineage reconstitution in all recipients, and significantly (*, p<0.05 by Student’s t-test) lower levels of donor reconstitution in all lineages (BE), irrespective of NAC treatment. NAC treatment did not significantly affect reconstitution levels from either Pten-deleted or control HSCs. Data represent mean±SEM from 3 independent experiments. (F) In 2 independent experiments, 1×106 unexcised donor cells from Ptenfl/flMx-1-Cre+ or Pten+/flMx-1-Cre+ mice were transplanted into irradiated recipient mice. Six weeks later Pten was deleted by pIpC treatment, then recipients were given daily injections of NAC or vehicle. NAC treatment did not prolong the survival of mice or delay the onset of leukemia after Pten deletion. None of the recipients of control cells developed neoplasms but all mice with Pten-deficient cells had MPD and/or T-ALL when they died, irrespective of NAC treatment. See also Figure S5.
To assess whether NAC treatment reduces leukemogenesis after Pten deletion, we transplanted 1×106 bone marrow cells from untreated Ptenfl/fl;Mx-1-Cre+ mice or Pten+/fl;Mx-1-Cre+ controls along with 500,000 recipient bone marrow cells into irradiated recipients. Six weeks later, we treated the recipients with seven doses of pIpC over 14 days followed by daily NAC or saline injection. In two independent experiments, recipients of control cells all survived for 120 days and showed no signs of leukemogenesis (n=20, Fig. 3F). In contrast, all 20 recipients of Pten-deleted bone marrow died with myeloproliferative disease (MPD) and/or T-cell acute lymphoblastic leukemia (T-ALL) 33 to 112 days after pIpC treatment, irrespective of whether they were treated with NAC (Fig. 3F; Fig.. S5D–I). NAC treatment did not significantly affect the lifespan of mice after Pten deletion (Fig. 3F) in contrast to rapamycin, which prevents leukemogenesis and extends the lifespan of Pten-deleted mice (Yilmaz et al., 2006).
Pten deletion leads to a tumor suppressor response in HSCs
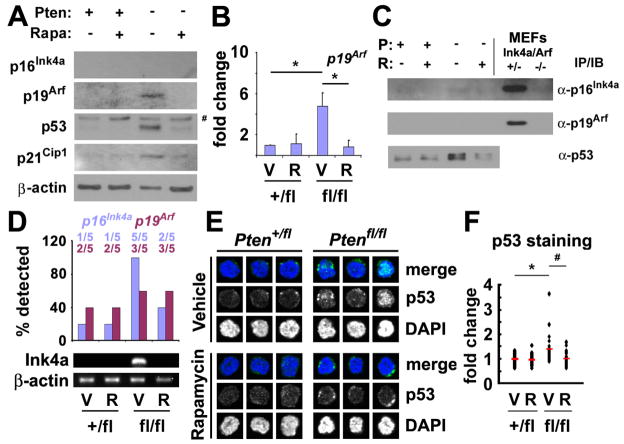
We tested whether Pten deletion induced a tumor suppressor response by assessing the expression of p16Ink4a, p19Arf, p53, and p21Cip1 in splenocytes. We did not detect p16Ink4a protein expression in splenocytes with or without Pten deletion (Fig. 4A). We did detect very low levels of p16Ink4a transcript in splenocytes, but transcript levels were not significantly affected by Pten deletion or rapamycin treatment (Fig. S6A). In contrast, we observed clear increases in p19Arf, p53, and p21Cip1 protein after Pten deletion (Fig. 4A). These increases were attenuated or eliminated by rapamycin, suggesting that the expression of these tumor suppressors was elevated as a consequence of mTOR activation (Fig. 4A). We confirmed significantly increased expression of p19Arf (Fig. 4B) and p21Cip1 (Fig. S6B) by qPCR in Pten-deficient splenocytes, and that these increases were eliminated by rapamycin treatment. We observed no effect of Pten deletion or rapamycin treatment on p53 transcript levels (Fig. S6C), consistent with the observation that p53 expression is mainly regulated post-transcriptionally (Olsson et al., 2007).
Figure 4. Pten deletion increased p19Arf, p21Cip1, and p53 expression in splenocytes, and p16Ink4a and p53 in HSCs, and rapamycin attenuated these increases.
(A) Levels of p16Ink4a, p19Arf, p21Cip1, and p53 were assessed by Western blot in unfractionated splenocytes from Ptenfl/flMx-1-Cre+ mice and Pten+/flMx-1-Cre+ controls 14 days after pIpC treatment. Mice were given daily injections of rapamycin or vehicle after pIpC treatment ended. Analysis of p53-deficient MEFs (data not shown) indicated that the upper band (#) was not specific for p53 but the lower band was. This blot is representative of 3 independent experiments. (B) We also observed increased p19Arf transcript levels by qPCR in splenocytes after Pten deletion and this effect was attenuated by rapamycin treatment (mean±SD from 3 independent experiments). (C) 4 weeks after pIpC treatment ended, 2×106 Lin−c-kit+ stem/progenitor cells were sorted from control and Pten-deleted mice and cell lysates were immunoprecipitated using antibodies against p16Ink4a, p19Arf, and p53 before Western blotting. p16Ink4a and p53 levels increased in Pten-deleted cells, but we detected no increase in p19Arf. MEFs that were deficient or heterozygous for p16Ink4a/p19Arf were used as negative and positive controls. (D) p16Ink4a transcript could always be amplified from Pten deficient CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs (5 of 5 samples) but usually not from control (1 of 5) or rapamycin-treated Pten deficient samples (2 of 5) 4 weeks after Pten deletion. p19Arf transcripts could only be amplified from about half of the samples, irrespective of Pten deletion or rapamycin treatment (data are from 5 independent experiments). (E, F) 4 weeks after pIpC treatment ended, CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs from Pten-deleted mice exhibited higher levels of immunofluorescence for p53 than control HSCs or Pten deleted HSCs treated with rapamycin. (F) The average staining intensity for p53 increased 1.4-fold (*, p<0.008 by Student’s t-test) in Pten-deleted HSCs as compared to control HSCs. Rapamycin treatment rescued this effect (#, p<0.003 by Student’s t-test; data are from 30 HSCs per group compiled from 2 independent experiments). None of the mice studied in this figure showed any signs of hematopoietic neoplasms. See also Figure S6.
We hypothesized that this tumor suppressor response inhibited leukemogenesis but promoted the depletion of HSCs after Pten deletion, and that rapamycin rescued HSC depletion by attenuating the tumor suppressor response in HSCs. Since p16Ink4a and p19Arf are expressed at extraordinarily low levels in primary somatic cells and only limited amounts of protein are available from rare HSCs, we sorted 2×106 Lin−c-kit+ hematopoietic stem/progenitor cells, immunoprecipitated p16Ink4a, p19Arf, and p53, then performed Western blots to assess the levels of p16Ink4a, p19Arf, and p53. We observed p16Ink4a and p53 expression in the Pten-deficient Lineage−c-kit+ cells but not in wild-type cells or cells treated with rapamycin, but did not detect p19Arf expression in wild-type or Pten-deficient Lin−c-kit+ cells (Fig. 4C).
p16Ink4a transcript levels also increased in CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs after Pten deletion and this increase was attenuated by rapamycin treatment (Fig. 4D). We were unable to detect a clear increase in p19Arf expression after Pten deletion (Fig. 4D). We also stained CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs with anti-p53 antibody on slides to assess the level of p53 expression on a cell-by-cell basis. Pten deficient HSCs that had not been treated with rapamycin exhibited significantly increased p53 staining (Fig. 4E, F). Together, our results suggest that p16Ink4a and p53, but not p19Arf, are induced in HSCs after Pten deletion.
Deficiency for p19Arf or p53, but not p16Ink4a, accelerates leukemogenesis
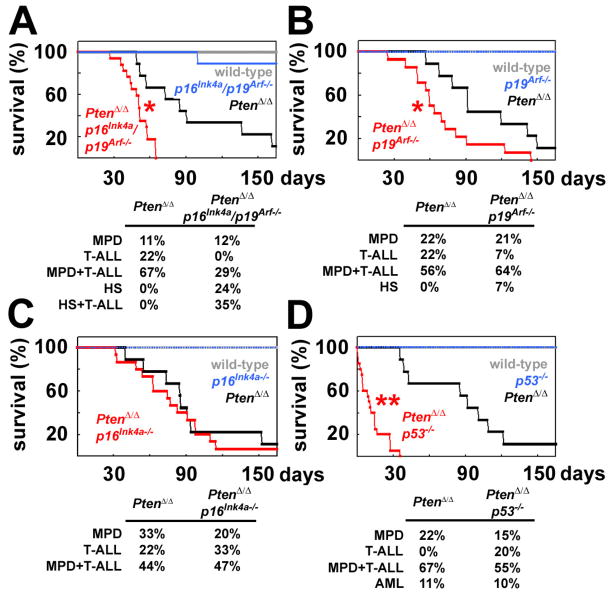
Conditional deletion of Pten from p16Ink4a/p19Arf-deficient or p53-deficient backgrounds led to leukemogenesis (T-ALL, MPD, and/or histiocytic sarcoma) and to the death of mice within 14 days of pIpC treatment, much faster than after deletion of Pten from mice with wild-type tumor suppressors (data not shown). To avoid the rapid death of compound mutant mice and to compare the rates of leukemogenesis from limited numbers of mutant cells in a wild-type environment, we transplanted 1×106 unexcised donor (CD45.2+) bone marrow cells from wild-type, p16Ink4a/p19Arf−/−, Ptenfl/fl;Mx-1-Cre+, and Ptenfl/fl;Mx-1-Cre+;p16Ink4a/p19Arf−/− mice into irradiated wild-type recipient mice along with 500,000 recipient (CD45.1+) bone marrow cells. Six weeks after transplantation, when the donor cells had stably engrafted, all recipients were treated with 7 injections of pIpC over 14 days to induce Pten-deletion. Recipients of wild-type and p16Ink4a/p19Arf−/− bone marrow cells almost all survived for the duration of the experiment (165 days) with no signs of leukemogenesis (Fig. 5A). In contrast, recipients of Ptenfl/fl;Mx-1-Cre+ cells died 49 to 162 days after ending pIpC treatment (Fig. 5A). Recipients of Ptenfl/fl;Mx-1-Cre+;p16Ink4a/p19Arf−/− cells died significantly (p<0.02) more quickly, 27 to 65 days after ending pIpC treatment (Fig. 5A). Histology confirmed that the mice had MPD and/or histiocytic sarcoma and/or T-ALL when they died. These data indicate that deficiency for p16Ink4a and p19Arf accelerates leukemogenesis after Pten deletion.
Figure 5. Deficiency for p19Arf or p53, but not p16Ink4a, accelerated leukemogenesis after Pten-deletion.
1×106 donor bone marrow cells from mice with the indicated genotypes were transplanted into irradiated recipient mice along with 500,000 recipient bone marrow cells. Six weeks after transplantation, all recipients were treated with pIpC and their survival was monitored over time (up to 165 days after pIpC treatment ended). (A) Recipients of Ptenfl/flMx-1-Cre+p16Ink4a/p19Arf−/− cells (displayed as PtenΔ/Δp16Ink4a/p19Arf−/−) exhibited significantly (*, p<0.02 by Student’s t-test) accelerated death as compared to recipients of Ptenfl/flMx-1-Cre+ cells (PtenΔ/Δ). Mice were sacrificed when moribund and their hematopoietic tissues analyzed. The neoplasms observed in each mouse at the time of sacrifice included myeloproliferative disease (MPD), T-ALL, MPD+T-ALL, histiocytic sarcoma (HS), and HS+T-ALL. (B) Recipients of Ptenfl/flMx-1-Cre+p19Arf−/− cells (PtenΔ/Δp19Arf−/−) exhibited significantly (*, p<0.02 by log-rank test) accelerated death from leukemogenesis as compared to recipients of Ptenfl/flMx-1-Cre+ cells (PtenΔ/Δ). (C) Recipients of Ptenfl/flMx-1-Cre+p16Ink4a (PtenΔ/Δp16Ink4a) cells died at a similar rate and with similar neoplasms as recipients of Ptenfl/flMx-1-Cre+ cells (PtenΔ/Δ). (D) Recipients of Ptenfl/flMx-1-Cre+p53−/− cells (PtenΔ/Δp53−/−) exhibited significantly (**, p<0.0001 by log-rank test) accelerated death from leukemogenesis as compared to recipients of Ptenfl/flMx-1-Cre+ cells (PtenΔ/Δ). Data are from 3 independent experiments with a total of 9 mice/genotype except for compound mutant mice, which had 14–20 mice/genotype.
To better understand the relative contributions of p16Ink4a and p19Arf to the suppression of leukemogenesis after Pten deletion we performed the same experiment using mice that were compound mutants for Pten and p16Ink4a (Fig. 5C) or Pten and p19Arf (Fig. 5B). p16Ink4a deficiency did not significantly affect the rate at which mice died or the spectrum of hematopoietic neoplasms that arose after pIpC treatment (Fig. 5C). In contrast, p19Arf deficiency did significantly (p<0.02) increase the rate at which mice died after pIpC treatment (Fig. 5B). These results suggest that p19Arf suppresses leukemogenesis after Pten deletion, consistent with the increase in p19Arf expression in splenocytes after Pten deletion (Fig. 4A).
To assess the role of p53 we performed the same experiment using mice that were compound mutant for Pten and p53 (Fig. 5D). Recipients of Ptenfl/fl;Mx-1-Cre+;p53−/− cells died much more quickly after pIpC treatment as compared to recipients of Ptenfl/fl;Mx-1-Cre+ cells (p<0.0001; Fig. 5D). All mice were confirmed to have MPD, AML, and/or T-ALL when they died (Fig. 5D). These results suggest that p53 suppresses leukemogenesis after Pten deletion.
p16Ink4a and p53 promote the depletion of HSCs after Pten deletion
To test whether the tumor suppressors that inhibit leukemogenesis after Pten deletion also act cell-autonomously within HSCs to promote their depletion, we transplanted 10 donor CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs from mice with each of the genetic backgrounds depicted in Figure 6 into irradiated wild-type recipient mice, along with a radioprotective dose of 300,000 recipient bone marrow cells. The HSCs were isolated from the donor mice after they had been treated with 3 injections of pIpC over 6 days.
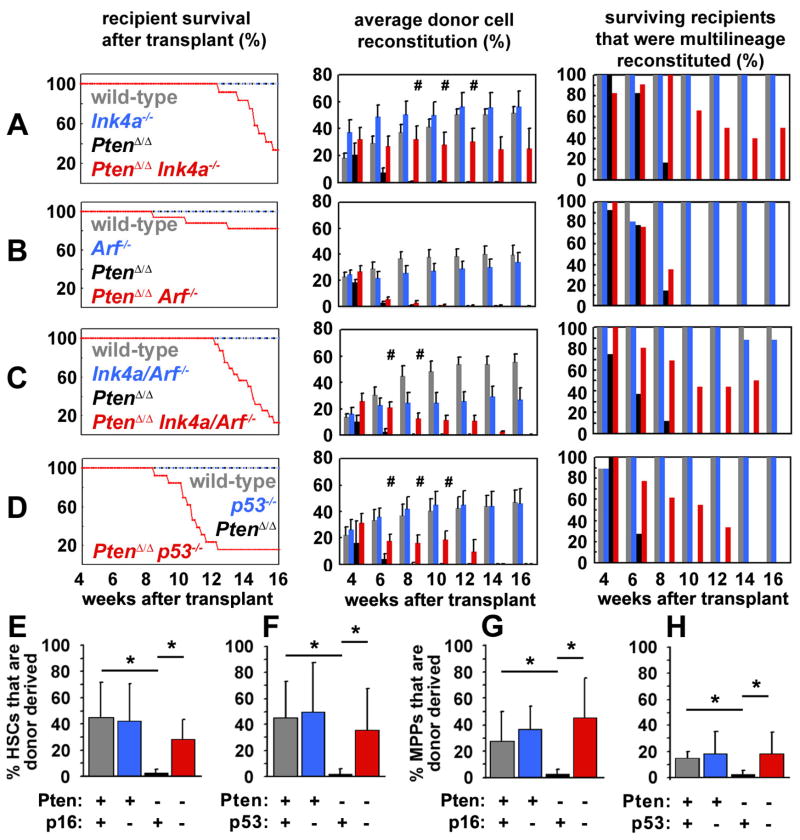
Figure 6. Deficiency for p16Ink4a or p53 prolonged the reconstituting capacity of Pten-deficient HSCs.
(A) 10 CD150+CD48−CD41−Lin−c-kit+Sca-1+ cells were sorted from mice with each of the indicated genotypes after pIpC treatment and co-injected with 300,000 recipient bone marrow cells into irradiated recipient mice. The survival, donor cell reconstitution levels (mean±SEM), and percentage of surviving recipients with multilineage reconstitution by donor cells were monitored for 16 weeks after transplantation. In all experiments, recipients of wild-type cells (A–D) and recipients of p16Ink4a-deficient cells (A), p19Arf-deficient cells (B), p16Ink4a/p19Arf-deficient cells (C) or p53-deficient cells (D) survived for the duration of the experiment and showed high levels of long-term multilineage reconstitution by donor cells. In contrast, recipients of Pten-deficient cells showed only transient multilineage reconstitution for 4 to 8 weeks in each experiment (A–D). HSCs that were compound mutant for Pten in addition to p16Ink4a (A), p16Ink4a/p19Arf (C) or p53 (D) gave significantly (#, p<0.05 by Student’s t-test) higher levels of donor cell reconstitution and multilineage reconstitution for a significantly longer period of time as compared to HSCs that were deficient only for Pten. The degree of donor cell reconstitution provided by Ptenfl/fl;Mx-1-Cre+;p16Ink4a cells (A) did not significantly differ from Ptenfl/fl;Mx-1-Cre+;p16Ink4a/p19Arf cells (C). p19Arf deficiency did not significantly affect the duration or level of reconstitution by Pten-deficient HSCs (B). Compound mutant mice that died had MPD, T-ALL, and/or histiocytic sarcoma at the time of death. All data are from 3 independent experiments with a total of 7–17 recipients per treatment. (E–H) Mice were transplanted with mutant HSCs as described above then sacrificed 8 weeks later to assess the frequency of donor CD150+CD48−CD41−Lin−c-kit+Sca-1+ HSCs and CD150−CD48−CD41−Lin−c-kit+Sca-1+ MPPs. Donor HSCs and MPPs were not detectable by this time point in the absence of Pten but depletion was rescued by either p16Ink4a deficiency (E,G) or p53 deficiency (F,H; *, p<0.05 by Student’s t-test). These data are from 3 independent experiments with a total of 3–8 recipients per treatment. Consistent with Figure 6, transplantation of higher doses of Pten-deficient CD150+CD48−CD41−Lin−c-kit+Sca-1+ cells into irradiated wild-type mice led to the development of leukemia in a higher proportion of the recipient mice (Table S1). See also Figure S7 and Table S1.
p16Ink4a deficiency prolonged the reconstituting capacity of Pten-deficient HSCs. Wild-type and p16Ink4a-deficient HSCs gave long-term multilineage reconstitution by donor cells in all recipient mice (Fig. 6A). In contrast, Pten-deleted (Ptenfl/flMx-1-Cre+) HSCs only gave transient multilineage reconstitution for 6 to 8 weeks after transplantation, consistent with our prior study (Yilmaz et al., 2006). Surprisingly, compound mutant Ptenfl/flMx-1-Cre+p16Ink4a−/− HSCs gave significantly longer multilineage reconstitution than Ptenfl/flMx-1-Cre+ HSCs (for 8 to 16 weeks, p<0.002; Fig. 6A) with significantly higher donor cell reconstitution levels than the levels of reconstitution from Ptenfl/flMx-1-Cre+ HSCs (Fig. 6A). Sixteen weeks after transplantation 4 recipients of Ptenfl/flMx-1-Cre+p16Ink4a−/− HSCs remained alive and 2 of these mice remained multilineage reconstituted by donor cells. In a separate experiment, we confirmed that CD150+CD48−CD41−Lin−c-kit+Sca-1+ donor HSCs (Fig. 6E) and CD150−CD48−CD41−Lin−c-kit+Sca-1+ donor MPPs (Fig. 6G) could be recovered 8 weeks after transplantation from mice transplanted with 10 Ptenfl/flMx-1-Cre+p16Ink4a−/− HSCs but not with 10 Ptenfl/flMx-1-Cre+ HSCs. These data indicate that p16Ink4a promotes the depletion of Pten-deficient HSCs.
Recipient mice that were transplanted with Ptenfl/flMx-1-Cre+p16Ink4a−/− HSCs did begin to die with MPD and/or T-ALL 12 to 16 weeks after transplantation whereas recipients of Ptenfl/flMx-1-Cre+ HSCs did not develop neoplasms (Fig. 6A). This difference partly reflects the fact that Ptenfl/flMx-1-Cre+ HSCs no longer had any donor cell chimerism by 12 weeks after transplantation and therefore were unable to develop donor cell leukemias (Fig. 6A). Therefore, p16Ink4a deficiency increased the opportunity for leukemogenesis by prolonging the reconstituting capacity of Pten-deleted HSCs. These data might also reflect a limited role for p16Ink4a in the suppression of leukemogenesis after Pten deletion.
p19Arf deficiency did not rescue the depletion of Pten-deficient HSCs. Wild-type and p19Arf-deficient HSCs gave long-term multilineage reconstitution by donor cells in all recipient mice (Fig. 6B). In contrast, Pten-deleted (Ptenfl/flMx-1-Cre+) HSCs only gave transient multilineage reconstitution for 6 to 8 weeks after transplantation. Compound mutant Ptenfl/flMx-1-Cre+p19Arf−/− HSCs also gave transient multilineage reconstitution for only 4 to 8 weeks and at levels that were not significantly different from those observed from Ptenfl/flMx-1-Cre+ HSCs (Fig. 6B). This indicates that p19Arf is not required for the depletion of HSCs after Pten deletion, consistent with our failure to detect p19Arf expression in Pten deficient HSCs (Fig. 4C, D). Only a minority of recipients of Ptenfl/flMx-1-Cre+p19Arf−/− HSCs died with leukemia (beginning 8 weeks after transplantation), reflecting the transient reconstitution by these cells (Fig. 6B).
Consistent with the partial rescue of HSC depletion by p16Ink4a deficiency, we also observed a partial rescue of HSC depletion by p16Ink4a/p19Arf deficiency. Wild-type and p16Ink4a/p19Arf-deficient HSCs gave long-term multilineage reconstitution by donor cells in almost all recipient mice (Fig. 6C). In contrast, Pten-deleted (Ptenfl/flMx-1-Cre+) HSCs only gave transient multilineage reconstitution for 4 to 8 weeks after transplantation. Compound mutant Ptenfl/flMx-1-Cre+p16Ink4a/p19Arf−/− HSCs gave significantly longer multilineage reconstitution than Ptenfl/flMx-1-Cre+ HSCs (for up to 14 weeks, p<0.01; Fig. 6C), with significantly higher donor cell reconstitution levels than from Ptenfl/flMx-1-Cre+ HSCs (Fig. 6C). These results are consistent with the data above in suggesting that p16Ink4a depletes HSCs after Pten deletion. There were no statistically significant differences in the extent to which reconstitution was rescued by p16Ink4a deficiency as compared to p16Ink4a/p19Arf deficiency, although p16Ink4a/p19Arf deletion was more leukemogenic (Fig. 6A, C).
p53 deficiency also prolonged the reconstituting capacity of Pten-deficient HSCs. Wild-type and p53-deficient HSCs gave long-term multilineage reconstitution by donor cells in all recipient mice for at least 16 weeks while Pten-deleted (Ptenfl/flMx-1-Cre+) HSCs gave transient multilineage reconstitution for 4 to 6 weeks after transplantation (Fig. 6D). Compound mutant Ptenfl/flMx-1-Cre+p53−/− HSCs gave significantly longer multilineage reconstitution than Ptenfl/flMx-1-Cre+ HSCs (for up to 12 weeks, p<0.002; Fig. 6D), with significantly higher donor cell reconstitution levels than from Ptenfl/flMx-1-Cre+ HSCs (Fig. 6D). In a separate experiment, we confirmed that donor HSCs (Fig. 6F) and MPPs (Fig. 6H) could be recovered 8 weeks after transplantation of 10 Ptenfl/flMx-1-Cre+p53−/− HSCs but not after transplantation of 10 Ptenfl/flMx-1-Cre+ HSCs. p53 therefore contributes to the depletion of Pten-deficient HSCs in addition to suppressing leukemogenesis, consistent with its increased expression after Pten deletion in splenocytes and HSCs (Fig. 4).
The precise mechanisms by which p16Ink4a and p53 promote the depletion of HSCs after Pten deletion are uncertain. We did not detect any effect of Pten deletion or rapamycin treatment on the frequency of whole bone marrow cells or HSCs undergoing cell death (Fig. S7A) or expressing senescence-associated β-galactosidase (Fig. S7C–E). Indeed, we observed an increase in the frequency of dividing HSCs after Pten deletion (rather than a decrease as would be expected if senescence were occurring), and this increase was rescued by rapamycin treatment (Fig. S7B). These results suggest that increased p16Ink4a and p53 expression in dividing HSCs is incompatible with HSC maintenance, perhaps because these tumor suppressors promote maturation out of the stem cell pool.
Pten deficient leukemias have mutations that attenuate the tumor suppressor response
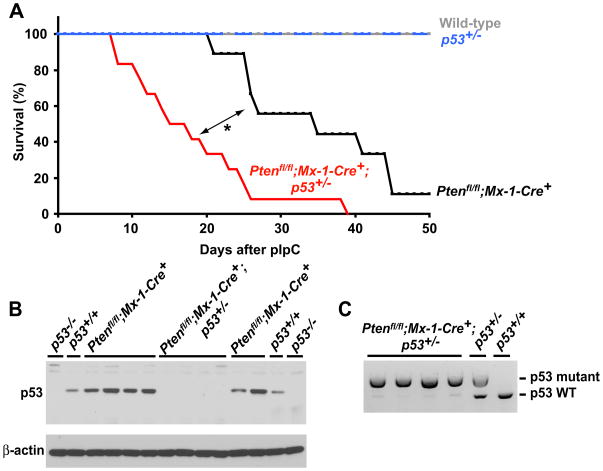
To test whether Pten deficient leukemias acquire secondary mutations that attenuate the tumor suppressor response we tested whether there was a selection pressure to inactive p53 in these leukemias. The tumor suppressors could be attenuated by many mechanisms that would silence or inactivate the tumor suppressors or other genes in the same pathways. Since it would be impossible to test all such mechanisms we circumvented this problem by studying leukemogenesis in a sensitized, p53 heterozygous background. If there is selective pressure to inactivate p53 in Pten deficient leukemias, leukemias should arise more quickly from p53+/− mice as compared to p53+/+ mice and we would expect the leukemias to delete the wild-type p53 allele. Young adult wild-type, p53+/−, Ptenfl/fl;Mx-1-Cre+;p53+/+, and Ptenfl/fl;Mx-1-Cre+;p53+/− mice were treated with pIpC and monitored. Wild-type and p53+/− mice all survived for the duration of the experiment (50 days) with no signs of leukemia, while Ptenfl/fl;Mx-1-Cre+;p53+/+ mice died 21 to 45 days after pIpC treatment with T-ALL and MPD (Fig. 7A). Ptenfl/fl;Mx-1-Cre+;p53+/− mice died significantly (p=0.0008) more quickly, 8 to 39 days after ending pIpC treatment, again with T-ALL and MPD (Fig. 7A). Heterozygosity for p53 therefore accelerates leukemogenesis.
Figure 7. Some Pten-deficient leukemias inactivate p53.
(A) Wild-type, p53+/−, Ptenfl/fl;Mx-1-Cre+;p53+/+, and Ptenfl/fl;Mx-1-Cre+;p53+/− mice 5 to 6 weeks of age were treated with three doses of pIpC then monitored. Ptenfl/fl;Mx-1-Cre+;p53+/− mice died significantly more quickly than Ptenfl/fl;Mx-1-Cre+;p53+/+ mice (p=0.0008 by log-rank test). (B) Western blotting of thymocytes from Ptenfl/fl;Mx-1-Cre+;p53+/+ and Ptenfl/fl;Mx-1-Cre+;p53+/− mice with T-ALL showed strongly diminished expression of p53 in the Ptenfl/fl;Mx-1-Cre+;p53+/− cells. (C) Genomic PCR analysis of thymocytes from Ptenfl/fl;Mx-1-Cre+;p53+/− mice showed a significantly diminished band corresponding to the wild-type p53 allele suggesting loss of heterozygosity. The weak wild-type p53 band may reflect residual normal thymocytes or thymic stromal cells. 9–12 mice/genotype were used in this experiment.
We next examined p53 expression in the thymus of mice that developed leukemias. In each case, the thymus was enlarged, completely effaced, and appeared to consist mainly of leukemic lymphoblasts. Whereas thymocytes from Ptenfl/fl;Mx-1-Cre+;p53+/+ mice showed robust expression of p53 (6 of 6 samples), thymyocytes from Ptenfl/fl;Mx-1-Cre+;p53+/− mice showed little or no p53 expression (4 of 4 samples; Fig. 7B). PCR amplification of genomic DNA showed that thymic lymphoblasts from all four Ptenfl/fl;Mx-1-Cre+;p53+/− mice had deleted the wild-type allele of p53 (Fig. 7C). Thus, Pten deletion induces a strong selective pressure to inactive the tumor suppressor response in leukemogenic cells.
DISCUSSION
Pten deletion increased Akt, mTORC1, and S6 kinase activation in HSCs (Fig. 1B, D, E) but we could find no evidence for reduced FoxO1 or FoxO3a expression or cytoplasmic sequestration (Fig. 1B, F–H; Fig. S2). We observed a clear increase in ROS levels within thymocytes after Pten deletion but not in HSCs (Fig. 2). Consistent with this, NAC treatment attenuated the increase in ROS levels in thymocytes but did not rescue the changes in hematopoiesis, HSC frequency (Fig. 2), or HSC reconstituting capacity (Fig. 3A) after Pten deletion. This contrasted with results from FoxO1/3/4-deficient mice in which ROS levels clearly increased within HSCs and NAC treatment at least partially rescued HSC depletion (Tothova et al., 2007). Pten deletion and FoxO1/3/4 deletion thus lead to the depletion of HSCs by different mechanisms. HSC depletion after Pten deletion is mediated largely by mTOR activation with no evidence so far for an important contribution by oxidative stress.
Pten deletion induced a tumor suppressor response in hematopoietic cells, characterized by increased expression of p19Arf and p53 in splenocytes (Fig. 4A, B) and increased expression of p16Ink4a and p53 in HSCs (Fig. 4C–F). p16Ink4a/p19Arf deficiency, p19Arf deficiency, or p53 deficiency significantly accelerated leukemogenesis after Pten deletion (Fig. 5A, B, D). p16Ink4a deficiency did not significantly affect the rate of leukemogenesis after Pten deletion, though it did suppress the generation of histiocytic sarcomas (Fig. 5C). This suggests hematopoietic cells mainly rely upon the p53 pathway to suppress leukemogenesis after Pten deletion. This is consistent with results from mouse prostate in which Pten deletion induces p53-dependent senescence (Chen et al., 2005). Interestingly, this senescence response is p19Arf-independent in prostate (Chen et al., 2009) but p19Arf did suppress leukemogenesis after Pten deletion, indicating tissue-specific functions for p19Arf in tumor suppression.
p16Ink4a deficiency, p16Ink4a/p19Arf deficiency, or p53 deficiency all significantly prolonged the ability of Pten-deficient HSCs to give multilineage reconstitution in irradiated mice (Fig. 6). p19Arf deficiency did not prolong the reconstituting capacity of Pten-deficient HSCs (Fig. 6B). Thus p19Arf is critical for the suppression of leukemogenesis but not for HSC depletion after Pten deletion. In contrast, p16Ink4a is critical for HSC depletion but plays a limited role suppressing leukemogenesis. One possible explanation for this distinction is that some leukemias may arise from cells other than HSCs and this process could be inhibited by p19Arf expression in those cells.
Our results thus indicate that Pten deletion induces an mTOR mediated tumor suppressor response in hematopoietic cells, suppressing leukemogenesis and depleting HSCs. This suggests that leukemias likely arise from rare clones of Pten-deficient hematopoietic cells that acquire secondary mutations that attenuate the tumor suppressor response. Consistent with this, we observed loss of p53 heterozygosity in leukemias that arose from Ptenfl/fl;Mx-1-Cre+;p53+/− mice, indicating that Pten deletion imposes a strong selection against the tumor suppressor response (Fig. 7). It is also important to note that we do not know which hematopoietic cells are transformed after Pten deletion. Therefore, the tumor suppressors may act in HSCs themselves to suppress leukemogenesis or they may act in downstream cells.
Although p16Ink4a and p19Arf are both encoded at the Cdkn2a locus, they are regulated by different promoters, have no sequence homology, and different molecular functions (Sherr, 2001). The mechanisms by which Pten deletion or other oncogenic stimuli induce these tumor suppressor expression are not understood. While it is well established that oncogenic stresses such as c-Myc or E1A expression also activate the p19Arf-p53 pathway (de Stanchina et al., 1998; Zindy et al., 1998), the mechanisms behind this activation remain unclear. The mechanisms behind p16Ink4a and p53 activation in response to Ras activation also remain unknown (Serrano et al., 1997).
The best-characterized consequences of p53 or p16Ink4a activation are senescence and apoptosis. However, these responses have only been characterized in certain non-stem cell populations, and it is possible that p53 or p16Ink4a activation may have other effects on stem cells. We did not detect any evidence that hematopoietic cells underwent senescence or cell death after Pten deletion (Fig. S7). However, HSCs are asynchronously depleted over a 4 to 8 week period after Pten deletion (Fig. 6). This raises the formal possibility that HSCs asynchronously undergo cell death or senescence over 4 to 8 weeks, such that very few HSCs express markers of cell death or senescence at any single time point, rendering it undetectable. Nonetheless, the simplest interpretation of our data is that p16Ink4a and p53 expression cause HSCs to prematurely exit the stem cell pool, perhaps by maturing to transit amplifying MPPs. As this would occur asynchronously over time, the number of HSCs that prematurely exit the stem cell pool at any single time point would be imperceptibly small but the cumulative effect of premature maturation over a period of weeks would deplete HSCs.
Consistent with this model, deficiency for p16Ink4a, p19Arf, and p53 dramatically expands the frequency of long-term multilineage reconstituting cells by conferring long-term self-renewal potential to MPPs which normally only give transient multilineage reconstitution (Akala et al., 2008; Kiel et al., 2008). These tumor suppressors thus play a physiological role promoting the transition from HSCs to MPPs and negatively regulating the self-renewal potential of multipotent cells. Increased expression of p16Ink4a and p53 in dividing HSCs after Pten deletion may accelerate the normal maturation of cells out of the HSC pool, leading to HSC depletion.
The ability to rescue the hematopoietic phenotypes in Pten-deficient mice with rapamycin suggests these phenotypes are driven by mTORC1 activation. However, our data indicate only that increased mTORC1 activation is required for HSC depletion and leukemogenesis, not that it is sufficient. This may explain why other genetic backgrounds that activate mTORC1, such as Tsc1 deletion (Chen et al., 2008; Gan et al., 2008), do not necessarily lead to leukemogenesis. mTORC1-independent pathways downstream of Pten presumably also contribute to leukemogenesis. Since rapamycin can indirectly inhibit mTORC2 in addition to mTORC1 (Sarbassov et al., 2006) and mTORC2 is required for the development of prostate cancer after Pten deletion (Guertin et al., 2009), mTORC2 may mediate some of the effects of Pten deletion on HSCs and other hematopoietic cells.
The depletion of HSCs (and other hematopoietic progenitors) after Pten deletion may explain why few leukemias exhibit Pten deletion (Aggerholm et al., 2000; Chang et al., 2006; Sakai et al., 1998) other than T-ALL (Gutierrez et al., 2009). Rare clones of Pten-deficient hematopoietic stem/progenitor cells would be unlikely to have the opportunity to acquire secondary mutations before being depleted and therefore would be unlikely to progress to leukemia. Leukemias may be more likely to hyper-activate the PI-3kinase pathway by other types of mutations that are better tolerated by hematopoietic cells. Additional studies of the PI-3kinase pathway in stem cells will provide additional insights into stem cell regulation and the development of cancer.
MATERIALS AND METHODS
Mice
C57BL/Ka-Thy-1.1 (CD45.2) and C57BL/Ka-Thy-1.2 (CD45.1) mice were used in hematopoietic reconstitution experiments. Mx-1-Cre+ mice (Kuhn et al., 1995), Ptenfl/fl mice (Groszer et al., 2001), Ink4a+/− mice (Sharpless et al., 2001), Arf+/− mice (Kamijo et al., 1997), Ink4a/Arf+/− mice (Serrano et al., 1996), and p53+/− mice (Jacks et al., 1994) were backcrossed for at least ten generations onto a C57BL/Ka background. All mice were housed in the Unit for Laboratory Animal Medicine at the University of Michigan.
pIpC, rapamycin, and N-Acetyl-L-cysteine administration
Polyinosine-polycytidine (pIpC) and rapamycin were administered as previously described (Yilmaz et al., 2006). Briefly, pIpC from Sigma (St. Louis, MO) or from Amersham (Piscataway, NJ) was resuspended in Dulbecco’s phosphate-buffered saline (D-PBS) at 2 mg/ml (for Sigma) or 50 μg/ml (for Amersham), and mice were injected intraperitoneally (IP) with 25 μg/gram (for Sigma) or 0.5 μg/gram (for Amersham) of body mass every other day for 6–14 days. Rapamycin (LC Laboratories, Woburn, MA) was dissolved in absolute ethanol at 10 mg/ml and diluted in 5% Tween-80 (Sigma) and 5% PEG-400 (Hampton Research, Aliso Viejo, CA) before being administered daily by IP injection at a dose of 4 mg/kg. N-Acetyl-L-cysteine (NAC; Sigma) was administered daily by subcutaneous injection at 100 mg/kg (pH 7.0) starting one day after the final dose of pIpC.
Flow cytometry and HSC isolation
Bone marrow cells were obtained by crushing the long bones (tibias and femurs), pelvic bones, and vertebrae in a mortar and pestle with Hank’s buffered salt solution without calcium or magnesium, supplemented with 2% heat-inactivated calf serum (GIBCO, Grand Island, NY; HBSS+). Cells were triturated and filtered through nylon screen (45 μm, Sefar America, Kansas City, MO) to obtain a single-cell suspension. For isolation of c-kit+Flk2 Lin Sca-1+CD48 cells, whole bone marrow cells were incubated with FITC-conjugated monoclonal antibodies to lineage markers including B220 (6B2), CD3 (KT31.1), CD4 (GK1.5), CD5 (53-7.3), CD8 (53-6.7), Gr-1 (8C5), Mac-1 (M1/70), and Ter119 (TER-119) in addition to APC-conjugated anti-Sca-1 (Ly6A/E; E13-6.7), biotin-conjugated anti-c-kit (2B8), and PE/Cy5-conjugated anti-Flk-2 (A2F10.1). Biotin-conjugated c-kit staining was visualized using streptavidin APC-Cy7.
For isolation of CD150+CD48−CD41−Lin−Sca-1+c-kit+ HSCs, bone marrow cells were incubated with PE-conjugated anti-CD150 (TC15-12F12.2; BioLegend, San Diego, CA), FITC-conjugated anti-CD48 (HM48-1; BioLegend), FITC-conjugated anti-CD41 (MWReg30; BD PharMingen, San Diego, CA), APC-conjugated anti-Sca-1 (Ly6A/E; E13-6.7), and biotin-conjugated anti-c-kit (2B8) antibody, in addition to antibodies against the following FITC-conjugated lineage markers: CD2 (RM2–5), B220 (6B2), CD3 (KT31.1), CD5 (53-7.3), CD8 (53-6.7), Gr-1 (8C5), and Ter119 (TER-119). Biotin-conjugated c-kit was visualized using streptavidin-conjugated APC-Cy7. HSCs were sometimes pre-enriched by selecting c-kit+ cells using paramagnetic anti-biotin microbeads and autoMACS (Miltenyi Biotec, Auburn, CA). Dead cells were excluded using the viability dye 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml).
To measure ROS levels, bone marrow cells were isolated and stained as above except that the HSC stain was modified to make the FITC channel available for DCFDA (2′-7′-dichlorofluorescein diacetate, Molecular Probes, Eugene, OR) staining. Antibodies for HSC isolation were as described above except for the following antibodies: PE/Cy5-conjugated anti-CD150 (TC15-12F12.2; BioLegend), PE-conjugated anti-CD48 (HM48-1; BioLegend), PE-conjugated anti-CD41 (MWReg30; BD PharMingen), and PE-conjugated antibodies against lineage markers. After antibody staining, thymus cells, whole bone marrow cells, or c-kit+ enriched cells were incubated with 5 μM DCFDA for 15 min at 37°C followed by flow cytometry.
For details regarding immunofluorescence assays, western blotting and quantitative PCR see Supplementary Materials.
HIGHLIGHTS.
Oxidative stress is not a major driver of HSC depletion after Pten deletion
mTOR activation after Pten deletion induces tumor suppressors that deplete HSCs
p53 and p16Ink4a suppress leukemia and promote HSC depletion after Pten deletion
Pten-deficient leukemias acquire mutations that attenuate the tumor suppressors
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute. J.Y.L was supported by predoctoral fellowships from the University of Michigan (UM) Biology of Aging Training Grant and the Medical Scientist Training Program. D.N. was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science. Flow-cytometry was partially supported by the UM-Comprehensive Cancer NIH CA46592. Thanks to David Adams and Martin White for flow-cytometry. Thanks to Chris Mountford and Sara Grove for mouse colony management and to Michael Smith and Mayya Malakh for help with genotyping.
Footnotes
AUTHOR CONTRIBUTIONS
J.Y.L. and D.N. performed all experiments and participated in the design and interpretation of experiments. O.H.Y. initiated the project and generated the compound mutant mice. M.S.L. analyzed mouse pathology with help from J.Y.L. Z.T. and D.G.G. participated in the conception and interpretation of experiments. S.J.M. participated in the design and interpretation of Pten experiments and wrote the paper with J.Y.L and D.N.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggerholm A, Gronbaek K, Guldberg P, Hokland P. Mutational analysis of the tumour suppressor gene MMAC1/PTEN in malignant myeloid disorders. European journal of haematology. 2000;65:109–113. doi: 10.1034/j.1600-0609.2000.90181.x. [DOI] [PubMed] [Google Scholar]
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Chang H, Qi XY, Claudio J, Zhuang L, Patterson B, Stewart AK. Analysis of PTEN deletions and mutations in multiple myeloma. Leukemia research. 2006;30:262–265. doi: 10.1016/j.leukres.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Carracedo A, Lin HK, Koutcher JA, Behrendt N, Egia A, Alimonti A, Carver BS, Gerald W, Teruya-Feldstein J, et al. Differential p53-independent outcomes of p19(Arf) loss in oncogenesis. Science signaling. 2009;2:ra44. doi: 10.1126/scisignal.2000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes & development. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorian C, Nakashima J, Le Belle J, Ohab J, Kim R, Liu A, Smith KB, Groszer M, Garcia AD, Sofroniew MV, et al. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J Neurosci. 2009;29:1874–1886. doi: 10.1523/JNEUROSCI.3095-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, Zack JA, Geschwind DH, Liu X, Kornblum HI, Wu H. PTEN negatively regulates neural stem cell self-renewal by modulating G0–G1 cell cycle entry. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science (New York, NY) 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, Mullholland DJ, Magnuson MA, Wu H, Sabatini DM. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, Dahlberg S, Neuberg D, Moreau LA, Winter SS, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. The Journal of cell biology. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kharas MG, Okabe R, Ganis JJ, Gozo M, Khandan T, Paktinat M, Gilliland DG, Gritsman K. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115:1406–1415. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Morrison SJ. CD150− cells are transiently reconstituting multipotent progenitors with little or no stem cell activity. Blood. 2008;111:4413–4414. doi: 10.1182/blood-2007-12-129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science (New York, NY) 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes & development. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell death and differentiation. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- Sakai A, Thieblemont C, Wellmann A, Jaffe ES, Raffeld M. PTEN gene alterations in lymphoid neoplasms. Blood. 1998;92:3410–3415. [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nature Reviews Molecular Cell Biology. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5-triphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yalcin S, Zhang X, Luciano JP, Mungamuri SK, Marinkovic D, Vercherat C, Sarkar A, Grisotto M, Taneja R, Ghaffari S. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. The Journal of biological chemistry. 2008;283:25692–25705. doi: 10.1074/jbc.M800517200. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & development. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.