Abstract
Photoconvertible fluorescent proteins (FPs) are recent additions to the biologists’ toolbox for understanding the living cell. Like green fluorescent protein (GFP), monomeric EosFP is bright green in color but is efficiently photoconverted into a red fluorescent form using a mild violet-blue excitation. Here, we report mEosFP-based probes that localize to the cytosol, plasma membrane invaginations, endosomes, prevacuolar vesicles, vacuoles, the endoplasmic reticulum, Golgi bodies, mitochondria, peroxisomes, and the two major cytoskeletal elements, filamentous actin and cortical microtubules. The mEosFP fusion proteins are smaller than GFP/red fluorescent protein-based probes and, as demonstrated here, provide several significant advantages for imaging of living plant cells. These include an ability to differentially color label a single cell or a group of cells in a developing organ, selectively highlight a region of a cell or a subpopulation of organelles and vesicles within a cell for tracking them, and understanding spatiotemporal aspects of interactions between similar as well as different organelles. In addition, mEosFP probes introduce a milder alternative to fluorescence recovery after photobleaching, whereby instead of photobleaching, photoconversion followed by recovery of green fluorescence can be used for estimating subcellular dynamics. Most importantly, the two fluorescent forms of mEosFP furnish bright internal controls during imaging experiments and are fully compatible with cyan fluorescent protein, GFP, yellow fluorescent protein, and red fluorescent protein fluorochromes for use in simultaneous, multicolor labeling schemes. Photoconvertible mEosFP-based subcellular probes promise to usher in a much higher degree of precision to live imaging of plant cells than has been possible so far using single-colored FPs.
Multicolored fluorescent proteins (FPs) spanning the entire visible spectrum are considered essential tools for studying gene activity, protein localization, and subcellular interactions in modern biology. Numerous subcellular targeted FP probes have been created for live imaging of plants at the organ, tissue, cell, subcellular, and suborganeller levels. Several dedicated Web-based educational resources have been developed to provide comprehensive and frequently updated information on subcellular targeted FP probes for plants (Mathur, 2007; Held et al., 2008; Mano et al., 2008, 2009).
The routine use of FPs in plant biology has also made us aware of their limitations. The emission spectra of most commonly used FPs span discrete color bands (Shaner et al., 2007); consequently, all targets of a single FP fusion become highlighted in a specific color only. Whereas interactions between dissimilar organelles are readily studied using multicolor labeling with two or more fluorescent proteins (Mathur et al., 2002; Ueda et al., 2004; Kato et al., 2008), single-color labeling becomes a limiting factor when the aim is to understand spatiotemporal aspects of interactions between similar organelles. Further limitations of single-colored FPs become apparent when visualizing local and often transient alterations in the organization of dynamic subcellular elements like the cytoskeleton and endomembranes. Carrying out comparisons for these flexible elements simultaneously is challenging and usually not amenable to quantification. Finally, an issue that plagues most live-imaging approaches is the absence of built-in controls in the cells under observation. For most researchers, the decision of when to stop imaging a cell or a small subcellular region remains empirical rather than one based on a clear imaging parameter. In most studies of living cells, internal controls indicating photodamage are missing, as it is generally assumed that such effects must be minimal. Whereas chlorophyll photobleaching provides a useful visible control in studies involving green tissues, a large proportion of published live-imaging data comes from nongreen cells and tissues in which this internal indicator of cell health cannot be applied. Nevertheless, given the rapid responsiveness of plant cells (Sinclair et al., 2009), internal indicators are extremely important for minimizing artifacts while studying subcellular interactions.
In the majority of transgenic lines created to date, targeted FPs are constitutively expressed and cannot be induced at will. However, as underscored through studies utilizing heat shock and chemically inducible promoters (Ketelaar et al., 2004; Tang et al., 2004; Saidi et al., 2005), FP inducibility is a very desirable trait for studying organelle interactions and protein-protein interactions. It is noteworthy that for most inducible systems developed so far, the induction is global, often involves multiple tissues or entire seedlings, and cannot be easily switched off. Frequently, expression levels fluctuate over time and can lead to cumulative protein overexpression-related artifacts.
At present, the field of imaging subcellular events and interactions in living plants could greatly benefit from monomeric FP probes that combine the favorable properties of existing FPs with rapid, irreversible photoconvertibility. More important, photoconvertible probes should work under existing microscopy infrastructure without requiring additional monetary inputs, be compatible with existing FP probes, and provide quantifiable data. An invaluable quality sought in the new probes should allow their use as internal controls for monitoring cell health during live visualization.
Recently, several new “optical highlighters,” broadly categorized as photoactivatable, photoswitchable, and photoconvertible FPs, have become available (Ai et al., 2006; Shaner et al., 2007; Wiedenmann et al., 2009). In response to specific wavelengths, these proteins undergo structural changes that result in their becoming “switched on” to a bright fluorescent state (photoactivatable FPs; Patterson and Lippincott-Schwartz, 2002) or cause a shift in their fluorescence emission wavelength (photoconvertible FPs; Ando et al., 2002; Wiedenmann et al., 2004; Gurskaya et al., 2006). The use of photoactivatable GFP (PA-GFP) and photoconvertible Dendra and Kaede has been successfully demonstrated for plants (Arimura et al., 2004; Runions et al., 2006; Martin et al., 2009; Brown et al., 2010). Moreover, EosFP, a homolog of Kaede derived from Lobophyllia hemprichii, has been engineered to a monomeric form without loss in fluorescence and photoconversion properties (Wiedenmann et al., 2004; Nienhaus et al., 2005) and utilized for demonstrating clathrin-dependent endocytosis during internalization of PIN auxin efflux carriers (Dhonukshe et al., 2007) and for labeling F-actin (Schenkel et al., 2008) and peroxisomes (Sinclair et al., 2009) in plants. In its unconverted form, monomeric (m)EosFP displays bright green fluorescence that, upon illumination with an approximately 390- to 405-nm waveband, changes irreversibly to red fluorescence (emission maximum of 581 nm).
Here, we present several new mEosFP-based probes targeted to the general cytosol, plasmalemmasomes, endosomes, tonoplast, endoplasmic reticulum, Golgi bodies, peroxisomes, and cytoskeletal elements. The collection includes LIFEACT::mEosFP, a small, most recently developed F-actin marker. The parameters used by us for assessing mEosFP probes include their expression in different cell types under transient and stable conditions and extend to developing optimal and economically feasible conditions for visualizing these probes. We demonstrate their usefulness in tracking single organelles over time, in following membrane and cytoskeletal dynamics, and their compatibility and ease of visualization with other colored FPs while pointing out potential pitfalls and artifacts that might result during visualization. In addition, the discovery of tubular endosomes resulting from mEosFP-based differential coloration of specific vesicles is described. Our detailed characterization of mEosFP-based probes suggests their tremendous potential in advancing spatiotemporal precision during live imaging of plants.
RESULTS AND DISCUSSION
mEosFP-Based Probes: Ease and Specificity of Photoconvertibility
mEosFP (Wiedenmann et al., 2004; Nienhaus et al., 2005) was found to be bright fluorescent green in both transient assays and in transgenic plants (Fig. 1). The protein photoconverted easily to a vivid red color following a 3- to 7-s exposure to violet-blue illumination (Fig. 1). Different microscopy setups (see “Materials and Methods”) could be used for photoconversion. These included a conventional 4′,6-diamidino-2-phenylindole (DAPI) filter (exciter [Ex], 350/50X; CdLP 400; emitter [Em], 460/50 nm; systems 1 and 4, “Materials and Methods”), a violet-blue glass filter (Leica D-filter; Ex, BP 355–425; dichroic 455; long pass [LP] 470 nm; system 2, “Materials and Methods”) on epifluorescence microscopes, and a 50-mW variable power 405-nm laser (system 3, “Materials and Methods”). The automated fluorescence recovery after photobleaching (FRAP) module on a Leica TCS-SP5 confocal laser-scanning microscope was tested and found to provide adequate photoconversion within 3 to 5 s using 70% laser strength. However, practical considerations such as cost constraints associated with purchasing a dedicated 405-nm laser made us assess existing epifluorescence microscopes where photoconversion required manual opening and shutting of the diaphragm. For each probe (Figs. 1–4), a conventional DAPI filter or a violet-blue D-filter allowed us to achieve the same degree of photoconversion as the high-end 405-nm laser-equipped confocal laser-scanning microscope. Since epifluorescence setups usually do not provide beam diameters smaller than 500 μm, we found a simple solution by custom creating smaller pinholes of 100 and 50 μm on our Leica DMRE epifluorescence microscope.
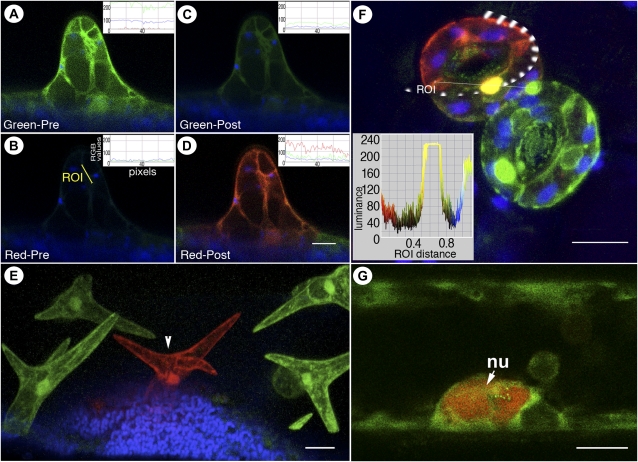
Figure 1.
Visualization of green and red forms of mEosFP distinct from chlorophyll fluorescence (false-colored blue) in transgenic Arabidopsis plants expressing mEosFP-cytosolic. A to D, Differential labeling of a single-celled trichome at an early developmental stage using nontargeted mEosFP-cytosolic expressed under the control of a trichome-specific GLABRA2 promoter. The inset RGB fluorescence intensity line traces in each panel depict the prephotoconversion and postphotoconversion values of red, green, and blue per pixel along the diagonal depicted in B. The RGB values are calculated on a standard eight-bit scale of 0 to 255 stretching across the region of interest (ROI). Note the residual green fluorescence of mEosFP in C. E, Mature trichomes on a transgenic Arabidopsis leaf visualized 48 h after mEosFP-cytosolic had been photoconverted in the central trichome cell (arrowhead) show the stability of the red form of mEosFP following its photoconversion. Note that simultaneous visualization of both green and red forms provides the comparative controls within a single scanned image. F, Two pairs of guard cells developing in an aberrant pattern on a transgenic mEosFP-cytosolic plant demonstrate the major colors that can be obtained following epifluorescent photoconversion followed by simultaneous visualization. The top guard cell (red) contains fully photoconverted mEosFP-cytosolic (region of illumination ringed by the broken line); a partially photoconverted nucleus appears yellow, while cytoplasmic aggregates in the lower pair of guard cells retain the unconverted G-mEosFP form. The inset depicts color luminance along the marked region of interest, which is helpful in quantifying and interpreting color overlap following photoconversion. G, A nucleus (nu) in a hypocotyl epidermal cell of a light-grown seedling exhibits the R-mEosFP form without an intentional photoconversion and suggests that the red form might accumulate within the nucleus. All images were taken using microscopy system 2. Bars = 25 μm (A–E), 10 μm (F), and 5 μm (G).
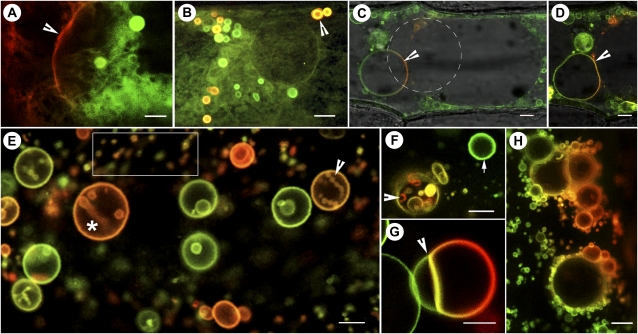
Figure 2.
mEosFP-based probes targeted to different membrane compartments. A, A region lying between two onion epidermal cells transiently expressing the mEosFP::PIP1 probe shows the nonconverted (green) and the photoconverted (red) labeling of tubular-vesicular membranes. The arrowhead points to the plasma membrane. (See also Supplemental Fig. S1.) B, Membrane tubules and multilamellar vesicles (arrowhead) in a mEosFP::PIP1-expressing cell. Vesicles are motile, and the red (photoconverted) mix readily with nonconverted (green) vesicles within minutes. The merged image acquired from both green and red channels is a single scan along the xy axis. Due to the rapid motility of vesicles and tubules, separate preconversion and postconversion images from green and red channels do not exhibit the vesicles observed here and can only inform about the nonvisibility of the probe in the red channel prior to its photoconversion. C and D, An onion epidermal cell transiently expressing the mEosFP::α-TIP1 probe shows vacuolar membranes being highlighted. The photoconverted portion of membrane (arrowheads) delimiting a minivacuole appears red and shows a regular spherical shape (C) that changes into an ellipsoid form in a subsequent scan (D). The merged images are sufficient to show dynamic changes in the highlighted compartment like conventional FPs, but through sequential time-lapse imaging they have the potential to inform about membrane dynamics within the vacuole compartment. E, Differential labeling of various PI(3)P-enriched vesicles following transient expression of mEosFP::2xFYVE in an onion epidermal cell. Based on their size, the smaller vesicles (e.g. boxed area) qualify as endosomes, while the larger vesicles (asterisk) are considered prevacuoles and vacuoles. The number of endosomes is fairly representative of onion cells exhibiting active cytoplasmic streaming. A number of prevacuoles (e.g. arrowhead and asterisk) display internalized vesicles. Full photoconversion labels vesicles red, whereas nonconverted vesicles remain green. Partial conversion results in labeling hues ranging from green to red. F, A single mEosFP::2xFYVE-labeled vesicle (arrowhead) illuminated with a 3-s pulse of violet-blue light exhibits internalized vesicles of different colors. This suggests that sequential photoconversion pulses may be used to determine the relative differences in protein content or membrane labeling between vesicles. The arrow points to an unconverted vesicle used as a control. G, Rapid membrane fusion observed along a line of contact between photoconverted and nonphotoconverted vesicles (arrowhead) achieved through transient salt-induced plasmolysis suggests the usefulness of the mEosFP::2xFYVE probe for understanding homotypic vesicle fusion. H, A PEG-treated cell exhibiting aggregation of numerous small mEos::2xFYVE vesicles around the larger ones. This state is maintained until PEG is removed, when rapid vesicle fusion occurs. All images were acquired using microscopy system 2. Bars = 5 μm (A–D and F–H) and 1 μm (E).
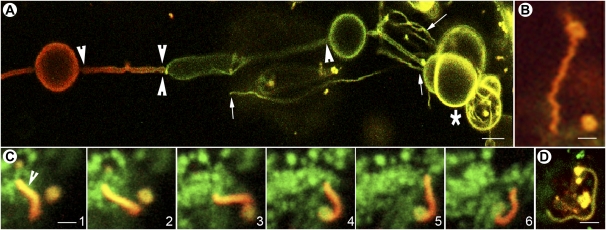
Figure 3.
Tubular projections from mEosFP::2xFYVE highlighted PI(3)P-enriched vesicles. A, An onion epidermal cell transiently expressing mEosFP::2xFYVE exhibits differentially colored vesicles, and associated tubules of varying diameters range from very thin (small arrows) to thick (large arrowheads) after a 5-s exposure to violet-blue light. Fused vesicles at bottom right (asterisk) extend multiple, thin tubules from their surface. The facing arrowheads depict a junction where two tubules extended from different vesicles appear to meet. Photoconverted protein does not extend beyond the junction, suggesting that despite their proximity the tubules maintain closed ends. B, A single vesicle exhibiting a tubule and an oscillatory movement resulting in the zigzag shape of the tail. C, Six frames from a time-lapse sequence taken over 24 s show a single photoconverted tubule and its contortions. Note that the tubule appears to snare a vesicle. (See also Supplemental Movie S1.) D, A single tubule that has ensnared many vesicles within a cup-shaped structure. All images were acquired using microscopy system 2. Bars = 5 μm (A) and 2.5 μm (B–D).
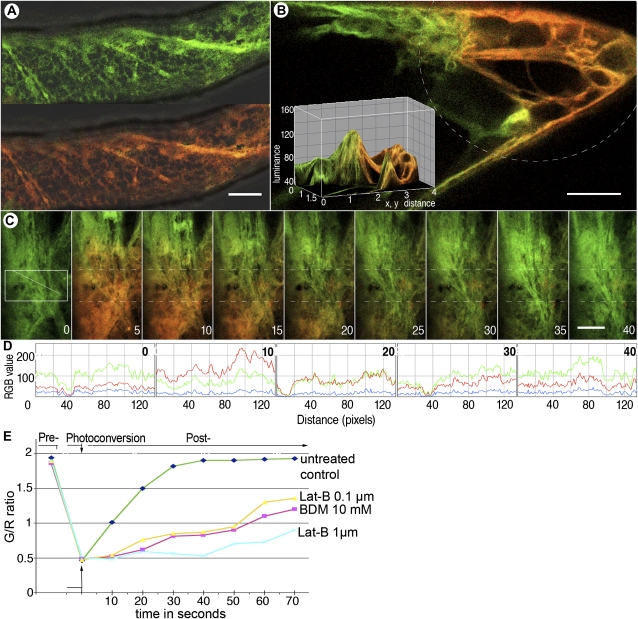
Figure 4.
CX::mEosFP demonstrates potential for estimating membrane flow through the analysis of initial color recovery after photoconversion using microscopy system 2. A, A cell from a transgenic Arabidopsis seedling expressing CX::mEosFP shows unconverted (green, top portion) and photoconverted (red, bottom portion) labeling of cortical ER membranes. B, Locally photoconverted CX::mEosFP (broken line) in a cell reveals a quantifiable color gradient (depicted in the inset) due to the rapid mobility and intermixing of red- and green-highlighted ER membranes. This observation formed the basis for the color tracking shown in C to E. C, Time-lapse sequence over 40 s shows the initial (nonconverted) green state of CX::mEosFP (panel 1 at 0 s). Photoconversion carried out using a circular beam from a violet-blue D filter for 5 s made a subcellular region fluoresce red (panel 2 at 5 s). Subsequent recovery of the green color in the delineated region of interest lying along the diagonal line within the photoconverted area (box in panel 1 and hatched line in subsequent time panels) is shown taking place over the next seven steps. Bars = 10 μm. D, Line traces of diagonals in successive panels from C subjected to color quantification using a standard 0 to 255 RGB scale depict the change in green and red levels and the nearly complete recovery of green fluorescence over time (e.g. time point 0 compared with 40 s). E, A comparison of CX::mEosFP movement over 70 s in untreated control cells and cells treated with 0.1 or 1.0 μm latrunculin B (Lat-B) and 10 mm BDM following a photoconversion step of 5 s. Green/red (G/R) ratios (averaged from three separate experiments) plotted against time demonstrate that the relative green fluorescence recovery time varies with cellular conditions. Therefore, this method can be applied to analyze and compare protein and membrane mobility in cells.
Thus, where available, the 405-nm laser should be considered the preferred mode for photoconversion, as it allows a high degree of precision through associated automated controls. However, lack of the 405-nm laser does not limit the use of these probes, as conventional filters on epifluorescence microscopes can be used for photoconversion. To allow complete reproducibility, all the photoconversions shown in the images presented here have been carried out using glass filters on epifluorescence microscopes.
Properties, Uses, and Caveats Associated with Various mEosFP Probes in Plants
A number of mEosFP fusion probes targeted to different subcellular compartments and structures are reported (Table I). While demonstrating photoconvertibility for each probe, we have utilized some of them for establishing the novel properties, associated caveats, and potential uses of mEosFP in plant research.
Table I. Photoconvertible mEosFP-based probes targeted to different subcellular compartments.
| Name of Probe | Target | Sequence Used for Targeting/Base Reference |
| mEosFP-cytosolic | Cytosol | Nontargeted mEosFPa |
| mEosFP::PIP1 | Plasma membrane | At3g61430: CDS plasma membrane intrinsic protein 1, ATPIP1/Fetter et al. (2004) |
| mEosFP:: α-TIP1 | Vacuolar membrane | At1g73190: CDS-α tonoplast intrinsic protein/Hunter et al. (2007) |
| mEosFP::ER membrane | ER membrane | At5g61790: membrane-targeting sequence of calnexin 1a/Runions et al. (2006) |
| Mito-mEosFP | Mitochondria | First 261-bp of the Nicotiana plumbaginifolia mitochondrial ATP2-1 coding sequence (GenBank accession no. X028680) |
| mEosFP-2xFYVE | Endosomes/prevacuolar compartment | 2xFYVE domain from mouse HGF-regulated Tyr kinase substrate proteina/Voigt et al. (2005) |
| mEosFP::GONST1 | Golgi bodies | At2g13650: CDS GONST1/Baldwin et al. (2001) |
| mEosFP::PTS1 | Peroxisome matrix | C-terminal tripeptide SKL (PTS1)ab/Mathur et al. (2002); Sinclair et al. (2009) |
| mEosFP::MBD-MAP4 | Microtubules | Microtubule-binding domain mammalian MAP-4a/Marc et al. (1998) |
| LIFEACT::mEosFP | F-actin | 17-amino acid peptide from yeast Abp140pa/Riedl et al. (2008) |
| mEosFP::FABD-mTalin | F-actin | F-actin-binding domain of mammalian Talinab/Kost et al. (1998); Schenkel et al. (2008) |
Transgenic lines created in Arabidopsis.
Previously reported on the basis of transient assays.
Transgenic Arabidopsis (Arabidopsis thaliana) plants expressing the mEosFP-cytosolic probe (Fig. 1) were used for assaying the stability of both colored forms of mEosFP. Observations on more than 70 different transgenic lines harboring mEosFP-cytosolic did not reveal growth and reproductive abnormalities, suggesting that the protein is well tolerated by plants. Photoconversion of single epidermal cells provided clear differentiation between the green and red forms of mEosFP (Fig. 1, A–F). Differences were also seen between the red form of the protein (R-mEosFP; Fig. 1 D) and chlorophyll autofluorescence (false-colored blue; Fig. 1D) by collecting emission spectra between 570 and 620 nm and between 626 and 763 nm, respectively. Both sequential as well as simultaneous use of 488-nm and 543-nm lasers during confocal visualization allowed constant monitoring of the nonphotoconverted green fluorescent form (G-mEosFP) of mEosFP within a cell. This provided the desirable internal control during live cell imaging, since photobleaching of G-mEosFP from nonphotoconverted regions was an indication of compromised cellular health. In all subsequent visualizations reported here using different mEosFP-based probes, this internal control was rigorously maintained. It is noteworthy that in cells that have high levels of mEosFP expression, a short photoconversion period will allow color change to be readily visible but will invariably leave a certain residual green fluorescent form of mEosFP (Fig. 1, A versus C). Care must be taken to record this residual form in the first scan after photoconversion. This is important, as in subsequent scans with a 488-nm laser, depending upon the laser strength, this partially activated form can either become photobleached or fully photoconverted. A fluorescence intensity line tracing of red, green, blue (RGB) values within the photoconverted region (Fig. 1, A–D, insets) usually helps to resolve whether the change involves photoconversion or photobleaching. In contrast to the situation in Figure 1D, during photobleaching the decrease in green fluorescence is not accompanied by a concomitant increase in red fluorescence. If a slight increase is observed in chlorophyllous cells, it is usually attributable to photobleached chloroplasts. In general, a minimal 488-nm laser power (1%–5%) and a high (approximately 60%–80%) 543-nm laser power allowed us to avoid extra photoconversion as well as photobleaching while providing a high green-to-red contrast. In symplastically isolated epidermal cells such as mature trichomes (Fig. 1E) and guard cells (Fig. 1F), the R-mEosFP is very stable and can be easily detected even after 48 h.
While observing photoconverted cells in mEosFP-cytosolic plants, a general artifact that relates equally to all photoconvertible probes became apparent. The diameter of an excitation beam notwithstanding, the illumination and consequent photoconversion of G-mEosFP occurs in a concentric manner where the intensity of the excitation wavelength disperses around the beam focal point. Thus, under short exposure time, G-mEosFP molecules at the periphery might be photoconverted only partially in comparison with those lying on the precise focal point. This slight variation in the distribution of excitation intensity results in mEosFP hues ranging from yellow to orange-red at the periphery (Fig. 1F). Therefore, it is important to recognize the minor variability in shades of red that occurs during green-to-red photoconversion and use the maximal red as a point of reference. Figure 1F, where the photoconversion was carried out on the top guard cell using a circular beam (broken white lines), demonstrates this color variability. A green fluorescent nucleus unexposed to the maximal intensity of photoconverting light did photoconvert completely and therefore appears yellow rather than red or green (Fig. 1F). A histogram of RGB values (Fig. 1F, inset) across the red-to-green region (the region of interest) underscores this point, while the color bar on the side clearly depicts the mixing of red and green colors to produce a variety of hues. Awareness of this artifact is especially important when using mEosFP in studies involving protein colocalization, since a partial photoconversion as well as full colocalization will both result in similar yellow hues. However, when 488- and 543-nm laser lines are used together, the green moiety in partially photoconverted cells usually photobleaches faster (within a few scans) and the yellow color changes to a stable red. Therefore, protein colocalization should be inferred only if a yellow signal persists after multiple laser scans.
Another artifact relating to the use of mEosFP became apparent in mEosFP-cytosolic plants grown in bright fluorescent white light (approximately 80–100 μmol m−2 s−1), where up to 25% of hypocotyl epidermis cells contained red nuclei (Fig. 1G) even without intentional photoconversion. Notably, mEosFP (226 amino acids; approximately 25.8 kD) is slightly smaller than monomeric GFP (238 amino acids; approximately 26.9 kD). Like nontargeted GFP (Haseloff et al., 1997), mEosFP highlights the entire cytosol and apparently diffuses freely in and out of the nucleus (Fig. 1, A and B). The artifact might thus be attributed to a combination of a high violet-blue component in the fluorescent white light spectrum and a possible higher concentration of mEosFP within the nucleus of certain endoreduplicating hypocotyl epidermal cells (Gendreau et al., 1998). Similarly, cells transiently expressing mEosFP-cytosolic and injured cells on transgenic plants were often found to contain bright fluorescent nuclear aggregates. Thus, care must be taken to check cells under both GFP/fluorescein isothiocyanate and red fluorescent protein (RFP)/tetramethyl rhodamine isothiocyanate filters before photoconversion. Additional care is required in interpreting the nontargeted nuclear localization of mEosFP-cytosolic, as after photoconversion its bright red nuclear fluorescence often overshadows the fainter cytoplasmic fluorescence.
All components in a living plant cell show dynamic behavior. While observations on organelles such as mitochondria, plastids, peroxisomes, and Golgi bodies are aided to a great extent by their well-defined morphology, the shape of endomembrane compartments is not fixed. In striving to adjust the subcellular milieu to the external environment, endomembranes are in constant flux. Endomembrane compartments frequently form transient contacts, glide over each other, and often form tubules and vesicles of assorted shapes (Harris, 1986). Selective labeling and tracking of endomembrane compartments presents a major challenge for live imaging. A number of probes described here (Table I), therefore, have been targeted to endomembrane compartments for assessing the utility of mEosFP for tracking vesicles and estimating changes in membrane dynamics.
mEosFP Probes Allow Highly Specific Differential Labeling of Membranes and Vesicles
The plasma membrane is the outer limiting membrane of the plant cell but maintains high connectivity with the cell interior through tubulovesicular compartments known as plasmalemmasomes or plasmatubules (Robinson et al., 1996). The aquaporin PIP1 (for plasma membrane intrinsic protein 1; Fetter et al., 2004) has been shown to be enriched on paramural plasma membrane invaginations (Robinson et al., 1996) and was used for generating the probe for plasmalemmasomes/plasmatubules. Transient expression of the mEosFP::PIP1 probe in onion (Allium cepa) epidermal cells highlighted a convoluted plasma membrane as well as internal tubular-vesicular compartments, including numerous multilamellar vesicles of different sizes (Fig. 2A). Morphologically, the highlighted compartments closely match the electron micrographs of plasmalemmasomes (Harris et al., 1982; Robinson et al., 1996). The presence of multilamellar vesicles is also consistent with the observations of Marchant and Robards (1968), who had considered plasma membrane evaginations as precursors for multivesicular bodies possibly associated with cell wall formation. Individual mEosFP::PIP1 vesicles were photoconverted and maintained their color-differentiated status over 12 h of visualization.
From the cell interior perspective, vacuoles are able to maintain an optimal water homeostasis within the cell through constant communication with the cell boundary. Aquaporins belonging to the TIP1 (for tonoplast intrinsic protein 1) subfamily of major intrinsic proteins have been shown to localize to the vacuolar membrane (Cutler et al., 2000; Ma et al., 2004; Hunter et al., 2007). The localization of mEosFP::α-TIP1 matched the observations of Hunter et al. (2007), as it labeled the tonoplast of large central vacuoles in onion epidermal cells (Fig. 2, C and D). However, in our transient expression assays, endomembranes highlighted by mEosFP::α-TIP1 included smaller vacuoles (Fig. 2C, arrowhead) and tubular strands. Photoactivation of small domains on vacuoles followed by time-lapse observations showed the flexible nature of the vacuolar compartment but also raised the possibility that α-TIP1 localization might be similar to that reported for AtTIP1;1, a γ-TIP that localizes to the tonoplast junctions (Beebo et al., 2009). Transgenic plants required for further investigating this possibility are being created.
Different early and late endosomes/multivesicular bodies and prevacuolar vesicles traffic between the cell boundary and large vacuoles (Bethke and Jones, 2000; Lam et al., 2007; Robinson et al., 2008). A third probe with high potential for investigating relationships between different endomembrane compartments was created by fusing mEosFP to two tandem FYVE domains. The FYVE domain specifically binds phosphatidylinositol 3-phosphate [PI(3)P], a key player in membrane trafficking in animals, yeast, and plants (Burd and Emr, 1998; Gaullier et al., 1998; Gillooly et al., 2000). As observed with other FP::2xFYVE fusions (Voigt et al., 2005; Vermeer et al., 2006), the mEosFP::2xFYVE probe highlighted numerous vesicles ranging in size from 200 nm to 25 μm in diameter (Fig. 2E). Based on prior publications (Kim et al., 2001; Voigt et al., 2005; Lam et al., 2007) and our unpublished colocalization (data not shown) with the commonly used endosome-labeling steryl dye FM4-64 (Vida and Emr, 1995), the smaller vesicles (Fig. 2E, box) were considered to be endosomes. As suggested by Vermeer et al. (2006), the larger vesicles were interpreted as prevacuoles or vacuoles (Fig. 2E). Since small vesicles move rapidly within the cell, a 3-s exposure resulted in varying degrees of photoconversion. Figure 2E illustrates the multicolor labeling that was achieved in this manner. The observation suggests a correlation between FP content and the color of a vesicle. Exposure of prevacuoles containing one or more internalized vesicles lent further support to this conjecture. A single exposure produced multicolored vesicles inside a larger vesicle (Fig. 2F, arrowhead). These observations suggest that short pulses of photoconverting wavelength can be used to estimate the relative concentrations of proteins in vesicles in living cells. Alternatively, as single vesicles of more than 2 μm could be easily photoconverted separately (Fig. 2G), we wondered whether the mEosFP::2xFYVE probe could be used for assessing homotypic vesicle fusion. Vesicle fusion takes place under normal conditions and is responsible for the growth and development of large vacuoles. The process is greatly augmented during stress. mEosFP::2xFYVE-labeled vesicles were photoconverted within a small region of the cell and allowed to mix for 5 min before the cell was challenged with 500 mm NaCl. As shown (Fig. 2G, arrowhead), membrane fusion occurred along lines of contact during the salt-induced plasmolysis. Interestingly, a treatment involving 10% polyethylene glycol 6000 (PEG) resulted in the aggregation of small vesicles around larger ones (Fig. 2H), with rapid fusion taking place upon deplasmolysis following PEG removal. These observations suggest that both forms of mEosFP are stable under low-hydration conditions and that mEosFP::2xFYVE can be used as an experimental tool for understanding membrane fusion.
Although different XFP::2xFYVE probes have been reported to label endosomes and vacuoles (Kim et al., 2001; Voigt et al., 2005; Vermeer et al., 2006), there are no reports of their highlighting long membrane tubules. In our observations on onion cells transiently expressing the mEosFP::2xFYVE probe, we invariably observed tubular connectivity between large vesicles. Figure 3A shows tubules of varying diameters (ranging from 0.25 to 1.5 μm) extending between large prevacuolar vesicles. Since their initial observation in onion epidermal cells, we have been able to confirm similar tubules extending over the vacuolar surface in transgenic Arabidopsis plants expressing GFP/RFP/mEosFP::2xFYVE probes. Photoconversion of single vesicles allowed an associated tubule to achieve the same color within 10 to 15 s. However, tubules that appeared to be continuous maintained distinct colors (Fig. 3A, facing arrowheads), suggesting that their ends are sealed and do not create a continuous compartment.
mEosFP-Aided Vesicle Tracking Reveals a Unique FYVE-Labeled Compartment
The ability to differentially highlight a few vesicles among hundreds of similar vesicles led to a serendipitous observation. Sporadically, a few mEosFP::2xFYVE-labeled vesicles displayed an aberrant oscillatory-rotatory motion. These aberrantly behaving vesicles were photoconverted and found to extend tubular projections. The asymmetrically shaped vesicles rapidly elongated into narrow tubules with lengths ranging between 5 and 15 μm and a diameter of 0.6 ± 0.13 μm (Fig. 3B). The photoconverted tubules could be easily tracked between nonphotoconverted mEosFP::2xFYVE-labeled vesicles. Tubules frequently formed loops and appeared to snare other vesicles into loose aggregates (Fig. 3, C and D). Ensnared vesicles finally fused with the tubules. The persistence of tubules within a cell varied considerably, ranging from a few minutes to up to 4 h. New tubules were constantly generated, while all tubules invariably recircularized to form spherical vesicles again. Although our observations do not provide an immediate explanation for these tubular compartments and their snaring of vesicles, it is noteworthy that in animal cells, early and recycling endosomes are often described as tubulovesicular compartments that accumulate internalized cargo and display tubular extensions (Tooze and Hollinshead, 1991; Gruenberg, 2001; Miaczynska and Zerial, 2002). Similar membrane extensions from either endosomes or prevacuolar compartments have not been reported in plant cells so far (Otegui and Spitzer, 2008). The transient and sporadic occurrence of these membrane extensions may explain why they have not been reported in earlier studies involving FP fusions with different SNAREs, Rab GTPases, and plasma membrane proteins that have formed the basis of our views on the endosomal compartments in plants (Sönnichsen et al., 2000; Geldner et al., 2009). As an alternative explanation, some of the tubular structures appear to resemble the cup- or bulb-shaped structures observed by Beebo et al. (2009). However, the majority of tubules reported here do not appear to fall into similar categories. The oscillatory movement that initially drew attention to a few vesicles and made us photoconvert them probably resulted from a growing asymmetry in vesicle shape due to localized tubular projections. Despite the asymmetry-driven aberrant motility, these vesicles continued to provide a generally circular profile that would have been easily missed among the hundreds of similarly fluorescent vesicles in a cell. Photoconversion also confirmed that the vesicles carried the mEosFP::2xFYVE label and are not imaging artifacts. Since their initial visualizations in onion cells, we have been able to observe similar short-lived tubules in transgenic Arabidopsis plants expressing XFP::2xFYVE and to confirm that these are not artifacts of transient expression. While an element of serendipity is apparent in our observations, the discovery of tubular endosomal compartments in plant cells can directly be attributed to our ability to differentially highlight a subpopulation of green fluorescent vesicles and follow them over time. Photoconvertible mEosFP clearly has tremendous potential for unveiling new and novel subcellular compartments and activities through differential highlighting. Follow-up studies on these transient tubular compartments are being pursued in stable transgenic Arabidopsis plants expressing mEosFP::2xFYVE and will be reported separately (J. Mathur, R. Radhamony, E. Roach, and N. Mathur, unpublished data).
Endoplasmic Reticulum-Targeted mEosFP Provides a Mild Method for Assessing Endomembrane Dynamics
The cortical endoplasmic reticulum (ER) is composed of labile extending and retracting, anastomosing membrane tubules, fenestrated sheets, as well as compact spindle-shaped ER bodies (Matsushima et al., 2003; Hara-Nishimura et al., 2004). The dynamic nature of the ER suggested a way of using mEosFP for assessing rapid membrane flow and reorganization. The mGFP-ER construct (Haseloff et al., 1997) has been one of the most used probes for plants. Using similar constructs, the ER has also been successfully visualized in other colors (Sinclair et al., 2009). However, ss::mEosFP-HDEL is retained within the ER lumen and does not allow the ER membrane to be visualized specifically. The PA-GFP-ER construct consisting of transmembrane domains of Arabidopsis calnexin (Huang et al., 1993) fused to a photoactivatable GFP has been reported by Runions et al. (2006) to efficiently label the ER membrane. It formed the basis for creating the CX::mEosFP probe. Unlike PA-GFP-ER, which is nearly nonfluorescent prior to its photoactivation, the nonphotoconverted CX::mEosFP is bright, fluorescent green (Fig. 4A, top). It thus provides a significant advantage over the parental probe. CX::mEosFP photoconverts readily (Fig. 4A, bottom) and efficiently highlights rapidly moving membrane vesicles, fenestrated membrane sheets, and cortical ER tubules while negatively highlighting ER bodies and the nucleus. In general, the green form of mEosFP is irreversibly photoconverted to the red form (Wiedenmann et al., 2004; Fig. 1E), and thus color dilution can be attributed to protein dispersal, membrane flow, and reorganization. In healthy plant cells, the rapid movement of CX::mEosFP creates a quick merging of green and red fluorescence. As shown in Figure 4B, the localized photoconversion of CX::mEosFP creates a color range between green and red. CX::mEosFP was used to estimate membrane flow from one subcellular locale to another by analyzing the ratio of unconverted, green CX::mEosFP to its red form (Fig. 4, C and D). Recovery of green fluorescence in the same region can be used to estimate membrane and fusion protein dynamics (Fig. 4D). The applicability of the method for providing correlative measurements is further demonstrated using 2,3-butanedione monoxime (BDM) and latrunculin B-aided inhibition of ER motility (Fig. 4E). Acquisition of similar data can be useful for interpreting the mobility and behavior of other organelles. The method of using mEosFP probes and assessing color recovery after photoactivation follows the same principle as the commonly used FRAP (Reits and Neefjes, 2001) method for measuring protein mobility and activity in living cells. However, it promises a significant advantage over FRAP, since the harsh photobleaching step is replaced by the much gentler photoconversion of mEosFP. A major point to be considered in the color-based approach relates to color conversion efficiency (CCE). This may be defined as the ratio of green and red forms of mEosFP obtained following a colorimetric RGB scale of 0 to 255 values plotted over time. The CCE depends upon the initial amount of fusion protein in a cell as well as its turnover over a specific period. Thus, the photoconversion of protein accumulated within a small compartment might be quite rapid, but depending upon the rate of turnover, its subsequent recovery might take a long time. For certain proteins, the cytoplasmic diffusion rates might be quite considerable and would need to be taken into account. CCE will also vary with the photoconversion filters/laser and the focal intensity of the excitation beam. Nevertheless, as long as observations are made under the same conditions, the method should allow direct comparisons of digitized images.
Small mEosFP-Based Cytoskeletal Probes
Both actin filaments and microtubules create very flexible and highly responsive arrangements on the cell cortex. In recent years, several probes have been created for labeling F-actin, and each new probe has provided a better appreciation of the dynamic nature of the actin cytoskeleton. We have reported a mEosFP::FABD-mTalin probe previously and demonstrated the successful labeling and photoconversion of F-actin in living plant cells (Schenkel et al., 2008). Nevertheless, smaller probes that result in minimal interference with actin dynamics are still sought. A 17-amino acid peptide called LIFEACT (amino acid sequence MGVADLIKKFESISKEE) from the yeast Abp140p fused to a GFP or RFP has emerged as the smallest live probe for labeling the actin cytoskeleton (Riedl et al., 2008). LIFEACT::XFP has already been tested in several systems, including plants, and reportedly does not interfere with actin polymerization or depolymerization. Reportedly, its use does not influence the major actin-dependent processes of polar tip growth in lower and higher plants (Era et al., 2009; Vidali et al., 2009). The LIFEACT::mEosFP fusion presented here (Fig. 5, A and B) is only 246 amino acids (approximately 28 kD), as compared with LIFEACT::GFP (263 amino acids, approximately 30 kD; Riedl et al., 2008), GFP-mTalin (447 amino acids, approximately 49 kD; Kost et al., 1998), and ABD2::GFP (70 kD; Sheahan et al., 2004; Hofmann et al., 2009). It is much smaller than the other photoconvertible F-actin-highlighting probe mEosFP::FABDmTalin (425 amino acids, approximately 47 kD; Schenkel et al., 2008) and thus combines two highly desirable properties. The potential of LIFEACT-mEosFP for estimating localized alterations in F-actin organization are evident from Figure 5E. Whereas the change in morphology of F-actin strands might reflect their rearrangement during cytoplasmic streaming, the ability to quantify changes within an actin strand is a powerful method to understand local actin dynamics (Watanabe and Mitchison, 2002; Ponti et al., 2003). As shown in Figure 5F, changes in the green-to-red ratio within an actin cable can be observed and quantified. Progressive quantification of a green or red region suggests a viable method for in vivo speckle labeling and determining actin filament polarity (Fig. 5F). Although the LIFEACT marker can readily release from actin filaments and rebind at other positions (Riedl et al., 2008), it is suggested that with refined controls, LIFEACT-mEosFP can become a useful addition to the existing fluorescent speckle microscopy methodology (Danuser and Waterman-Storer, 2006). However, these are early stages of LIFEACT use in plants, and there could be potential pitfalls that will become apparent only upon wider usage.
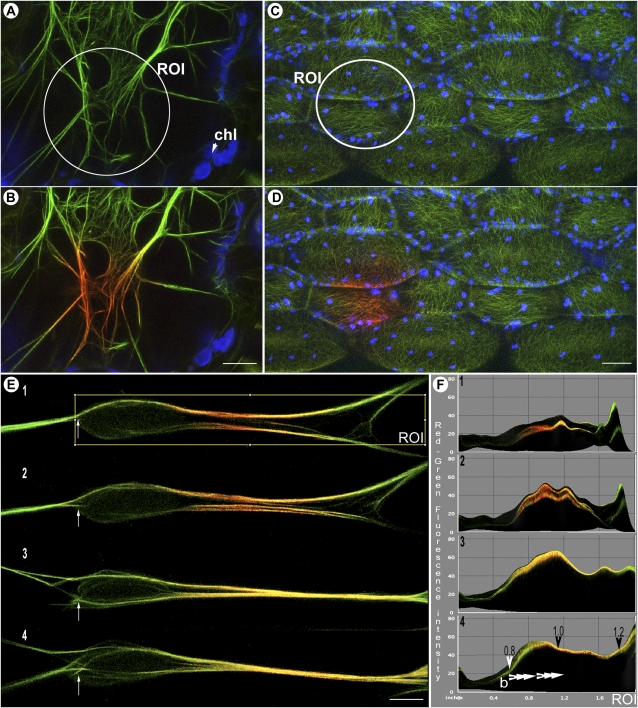
Figure 5.
mEosFP-based probes for cytoskeletal elements. A and B, LIFEACT::mEosFP represents the smallest live imaging probe for F-actin. Actin filaments in transgenic Arabidopsis plants are efficiently highlighted in green (region of interest [ROI] in A) before and in red (B) after photoconversion. Chloroplasts (chl; false-colored blue) are covisualized with both forms of mEosFP. C and D, Transgenic Arabidopsis plants expressing mEosFP::MBD-MAP4 highlight cortical microtubules (C), which have been photoconverted locally (D) in a specific region of interest (shown in C). Covisualized chloroplasts are depicted in blue. E, Tracking changes in cytoskeletal elements is possible using mEosFP labeling. Shown in E1 to E4 are images taken 10 s apart to show a change in F-actin organization as filaments (E1) bundle together (E4). The directional dispersal of the photoactivated red form of mEosFP is apparent between the four images and provides a means of assessing F-actin polarity in living cells. An independent positional marker on the sequential images (e.g. arrows in E1–E4) may minimize interpretational errors arising from a cytoplasmic streaming-induced drift in the actin cable. F, A post-image-acquisition three-dimensional surface plot (ImageJ) of the region of interest depicted in E1 (and subsequent images) provides quantification of red and green fluorescence values existing in an image on a standard RGB scale (values between 0 and 255). Using these values, a red-green ratio of 1 in the acquired image suggests equal green and red molecules, while ratios higher than 1 suggest increased red levels. F1 to F4 depict the shift in red fluorescent molecules toward the right side of the time-lapse images. Alterations in the red-green ratio (arrowheads in F4) suggest that new, unphotoconverted monomers are being added from one end (left side of images) and therefore cause the photoconverted F-actin region to shift toward the right. As indicated by the arrowheads (F4), the net fast-growing, barbed ends of the actin filaments lie on the left. All images were acquired using microscope system 2. Bars = 5 μm (A, B, and E) and 20 μm (C and D).
The fusion of different FPs to the microtubule-binding domain of the mammalian MAP4 gene has been very useful for observing cortical microtubules and their dynamics in plants (Marc et al., 1998; Mathur and Chua, 2000). Due to the smaller size of mEos, the mEosFP::MBD-MAP4 version (Fig. 5, C and D) is slightly smaller than similar cyan fluorescent protein (CFP)/GFP/yellow fluorescent protein (YFP)/RFP probes and has the added feature of photoconvertibility. Whereas a number of transgenic lines expressing GFP-MDB-MAP4 exhibit microtubule bundling (Marcus et al., 2001), the transgenic Arabidopsis plants expressing mEosFP::MBD-MAP4 do not exhibit this defect so far.
Using mEosFP to Understand Organelle Behavior and Interactions
The probes mito-mEosFP (Fig. 6, A, a, B, and b) and mEos::GONST1 (Fig. 6, C and c) target to mitochondria and Golgi bodies, respectively. While probes with similar targets have been created using KaedeFP, a tetrameric homolog of EosFP (Arimura et al., 2004; Brown et al., 2010), the two probes developed by us use the monomeric form.
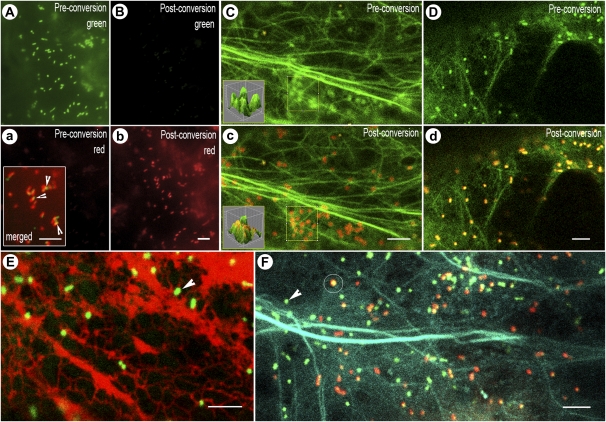
Figure 6.
Visualizing motile organelles using mEosFP alone and in combination with other FPs. A, a, B, and b, Mito-mEosFP transiently expressed in tobacco leaves by means of agroinfiltration efficiently targets to mitochondria (A) and is near fully photoconverted (b) after a 30-s exposure to 365/50-nm wavelength. Microscope system 4 was used for acquiring these images. Note that while preconversion fluorescence is barely detectable in the red channel (a) following photoconversion, it is not picked up in the green channel (B). The exposure times for images A and B are identical, as are those for images a and b. The inset in a is a merged image of the green and red channels postconversion using identical exposure times and demonstrates the slight shift and resultant artifact (arrowheads) that occurred during the time lapse between capture of the two sequential images, which could be misinterpreted as an absence of colocalization. C and c, Golgi bodies highlighted by mEosFP::GONST1 and F-actin (highlighted using GFP::mTalin) covisualized before (C) and after photoconversion (c). The rapid motility of Golgi bodies does not allow direct comparisons to be drawn between preconversion and postconversion images in separate channels; therefore, merged images acquired in both channels are presented. The inset three-dimensional surface plot clearly shows quantifiable changes in the color of Golgi bodies from their prephotoconversion to their postphotoconversion state (for more details, see Supplemental Fig. S2). D and d, Peroxisomes labeled with mEosFP-PTS1 and F-actin labeled using YFP::mTalin covisualized before (D) and after photoconversion (d). E, Peroxisomes labeled with mEosFP-PTS1 can be clearly discriminated (arrowhead) from RFP-labeled ER during prolonged covisualization using 488- and 543-nm lasers. Unintended photoconversion of mEosFP does not occur. F, Both the nonphotoconverted (arrowhead, green) and photoconverted (circle, red) forms of mEosFP-PTS1 covisualized with CFP::mTalin targeted to F-actin. Photoconversion was carried out separately, since the 458-nm argon laser line does not cause mEosFP to change color. Images in C to F were acquired using microscope system 2. Bars = 5 μm.
However, a potential artifact must be pointed out when using photoconvertible FPs to understand organelle interactions. Among other parameters, such studies rely heavily on demonstrating colocalization between two organelles (Arimura et al., 2004; Fig. 6, A, a, B, and b). Notably, in living cells, organelles like mitochondria and peroxisomes are highly motile; thus, even a minimal lapse in the timing of the capture of sequential red or green images (caused by single-band cube switching when using broad-spectrum illumination) can lead to the appearance of a lack of colocalization, as a result of a given organelle moving in the x, y, or z plane between exposures. As depicted in the inset in Figure 6a, merged images may produce a green signal that could be interpreted as a lack of colocalization, when in fact there may be complete colocalization. Laser scanning acquisition allows simultaneous fluorescence excitation, but depending on the method of image capture, a single fast-moving organelle may appear multiple times in the same image. Unambiguous imaging of multicolored fast-moving organelles therefore necessitates either the use of a color camera or the use of a beam splitter.
The GONST1 gene of Arabidopsis encodes a Golgi-localized GDP-Man transporter (Baldwin et al., 2001), and its full-length cDNA was used to create mEosFP::GONST1. As shown for the GONST1::YFP fusion (Baldwin et al., 2001), the mEosFP::GONST1 probe labeled motile punctate bodies (Fig. 6C) that photoconverted readily (Fig. 6C). Recently, Golgi-targeted fusions with a tetrameric Kaede protein have been reported (Brown et al., 2010). Overexpression of oligomeric proteins usually results in aggregates ranging in size from 0.5 to 2.5 μm and thus limits their suitability for visualizing small organelles, which fall within the same size range (Wiedenmann et al., 2004). Whereas intuitively, the monomeric form of a protein should be considered superior to its tetrameric form, for live imaging purposes, the relative pros and cons of using mEosFP probes or Kaede-based probes require further investigation.
mEosFP Is Fully Compatible with Other Colored FPs
For mEosFP-based probes to be truly useful in multicolor live imaging, they should be compatible with FP of other colors such as CFP, GFP, YFP, and RFP. This was tested using mEosFP probes targeted to Golgi bodies and the previously described mEosFP-PTS1 probe targeted to peroxisomes (Sinclair et al., 2009). The simultaneous visualization of mEosFP::GONST1 with GFP-mTalin (which labels F-actin; Fig. 6, C and c) and mEosFP-PTS1 with YFP::mTalin (Fig. 6, D and d) demonstrates mEosFP compatibility with GFP and YFP, respectively. In order to assess whether nonphotoconverted mEosFP can be used safely with a RFP, the mEosFP-PTS1 was covisualized with RFP-ER using 488- and 543-nm lasers (Fig. 6E). Simultaneous visualization of both probes demonstrated that the green form is stable and not prone to spontaneous photoconversion. We then tested whether CFP, excited by the 458-nm laser, could also be covisualized with mEosFP. Both the green mEosFP-PTS1 and its red form could be visualized with CFP-labeled actin filaments. Covisualization with CFP confirmed that mEosFP does not get photoconverted using the 458-nm laser and once again attested to the stability of both forms of mEosFP under simultaneous illumination by 458- and 543-nm lasers (Fig. 6F).
CONCLUSION
The observations presented here demonstrate that mEosFP-based probes retain all the qualities of single-colored fluorescent proteins while providing the additional capability of photoconversion. The major strength of mEosFP probes lies in their ability to create color-based differentiation within and between organelle populations and membranes. Both green and red fluorescent forms of mEosFP are stable and thus provide the highly desirable intracellular controls during prolonged live imaging. These ready-to-use probes greatly enhance spatiotemporal precision during live imaging of plant cells and, as demonstrated, can be used to label single cells or small regions within a cell, track single organelles, and allow an analysis of membrane and cytoskeletal dynamics. All mEosFP-based probes are amenable to differential color quantification methods. As demonstrated by the discovery of tubular PI(3)P-enriched compartments, the new probes possess tremendous potential for further discovery within the plant cell.
MATERIALS AND METHODS
Molecular Methods
Supplemental Table S1 provides a list of PCR primers used for constructing different mEosFP fusion constructs. All PCR products were cloned into pGEM-T-Easy vector before excising relevant fragments and inserting them into an intermediate vector carrying a pCaMV-35S promoter and a nos terminator sequence. The binary pCAMBIA 1300 base vector (http://www.cambia.org.au) to which a pCaMV-35S promoter and a nos terminator sequence had been added was used for generating plant transformation-competent mEosFP-cytosolic, mEosFP-PTS1, p35S-CX::mEosFP, LIFEACT::mEosFP, and mEosFP::MBD-MAP4. An additional version of mEosFP-cytosolic was driven by a GLABRA2 promoter, which is strongly active in trichomes (Szymanski et al., 1998). For the mito-mEosFP construct, Eos was PCR amplified from a pcDNA3.1 clone (Riedl et al., 2008) using relevant primers (Supplemental Table S1). The PCR product was purified and digested with SpeI and SacI. The backbone for this construct was pBINmgfp5-atpase (Logan and Leaver, 2000) cut with SpeI and SacI to remove mGFP5. The 5′-SpeI-Eos-3′-SacI was then ligated in to replace mGFP5. Standard molecular biology protocols were followed (Sambrook et al., 1989).
Expression in Plant Cells
Transient expression of different mEosFP probes was carried out in onion (Allium cepa) epidermal cell and 8- to 12-d-old Arabidopsis (Arabidopsis thaliana) seedlings. Gold particle coating with DNA and bombardment using a biolistic particle delivery system (Bio-Rad PDS-1000/He; http://www.bio-rad.com/) was carried out following the manufacturer’s instructions. mEosFP expression was assessed between 16 and 20 h after biolistic particle bombardment. For mito-mEosFP, the transient expression was carried out using agroinfiltration (strain GV3101 at an optical density at 600 nm of 0.1) of tobacco (Nicotiana tabacum) leaves according to Sparkes et al. (2006). The images of mito-mEosFP expression were taken 6 d after infiltration.
Stable transgenic lines for CX::mEosFP, mEosFP-PTS1, mEosFP-cytosol, LIFEACT::mEosFP, and mEosFP::MBD-MAP4 were generated using Agrobacterium tumefaciens (strain GV3101)-mediated floral dip transformation (Clough and Bent, 1998). Seeds were grown on 1% agar-gelled Murashige and Skoog (1962) medium supplemented with 3% Suc and with pH adjusted to 5.8. Plants were grown in petri dishes in a growth chamber maintained at 21°C ± 2°C and a 16-h/8-h light/dark regime using cool-white light at approximately 80 to 100 μmol m−2 s−1.
Microscopy and Drug Treatments
For live imaging, plant tissue and seedlings were placed in a depression slide in distilled water under a glass coverslip. Four different microscope systems were used.
System 1 consisted of an upright epifluorescence microscope (Nikon Eclipse 80i) equipped with a 40× Nikon Plan Apochromat lens with numerical aperture 0.95, and a 12-bit 3CCD color digital camera (Qicam fast 1394; Qimaging; http://www.adept.net.au/cameras/qimaging/) was used for epifluorescence microscopy. For this setup, the chroma filter sets used were Endow GFP-LP filter set 41018 (Ex, HQ 470/40X; dichroic, Q495LP; Em, HQ500LP), tetramethyl rhodamine isothiocyanate filter set 41002c (Ex, HQ545/30X; dichroic, Q570LP; Em, HQ 620/60), and the DAPI/Hoechst/aminomethylcoumarin acetate filter set 31000V2 (Ex, 350/50X; central dichroic, LP 400; Em, 460/50). Photoconversion was performed manually using epifluorescent lighting through the DAPI filter cube. Images were captured and processed using SIMPLE PCI software (Compix, Inc.; http://www.cimaging.net).
System 2 consisted of a Leica TCS-SP5 confocal laser-scanning microscope equipped with a 488-nm argon laser and a 543-nm helium-neon laser. The epifluorescence setup consisted of a Leica DM6000CS microscope equipped with a 40× water-immersion lens (numerical aperture 0.80). Images were obtained in a 1,024- × 512-pixel format in x/y/z and x/y/time dimensions and processed using proprietary Leica software. Unless stated otherwise, the time lapse between x/y/time scans was maintained at 1.37 s. Sequential images had a 1-μm (z axis) distance between them for x/y/z mode acquisition. Fluorescence emission collection was at 490 to 510 nm for GFP, 500 to 522 nm for YFP, 570 to 620 nm for RFP, and 626 to 763 nm for chlorophyll. For visualizing CFP-mTalin (Fig. 6E), the probe was excited using a 458-nm laser, and emission was collected between 493 and 510 nm for CFP, between 511 and 540 nm for GFP, and between 568 and 600 nm for RFP.
Photoconversion was performed manually by controlling the diaphragm. The diaphragm on the Leica DM6000CS microscope was modified to achieve 50- and 100-μm apertures. Closing down the iris, or moving the stage so that only a small part of the cell was exposed to the beam, achieved photoconversion of an organelle subpopulation. Epifluorescent lighting was through a D filter cube (Leica UV/violet; Ex, BP 355–425; dichroic, 455; Em, LP 470 nm) and a 40× water-immersion lens. For imaging the photoconverted probe simultaneously with nonconverted FP, the 488-nm argon laser and the 543-nm helium-neon lasers were used at approximately 10% and 80% power, respectively. Fluorescence bleedthrough was minimized by adjusting the pinhole and photomultiplier tubes and confirmed through sequential laser scans. However, for most visualization, both argon and helium-neon lasers were used simultaneously.
System 3 comprised a Leica TCS-SP5 confocal laser-scanning microscope and multiphoton imaging setup on an upright Leica 6000B microscope with 63× (numerical aperture 0.90) water-immersion lens. This system has a Radius 50-mW, 405-nm laser in addition to the argon and helium-neon lasers. This setup was used only for checking probe conversion and utilized Leica proprietary FRAP software for controlling 405-, 488-, and 543-nm lasers. It was not used routinely in the experiments presented here.
System 4 was used specifically for observing the mito-mEosFP-infiltrated tobacco leaves and consisted of a Zeiss Axioimager-Z1 microscope with light from a HXP120 mercury lamp, an Apochromat 63× oil-immersion lens (numerical aperture 1.4), a Zeiss 38HE GFP cube (Ex, 470/40; dichroic, 495; Em, 525/50), and a Zeiss 43HE Red cube (Ex, 550/25; dichroic, 570; Em, 605/70). A Zeiss-49 (DAPI/Hoechst; Ex, 365/50; dichroic, 395; Em, 445/50) filter was used for photoconversion.
For drug treatments, both latrunculin B and BDM were purchased from Molecular Probes (Invitrogen), dissolved in 30% dimethyl sulfoxide, and used at the concentrations shown (Fig. 4). Plants used in the control experiment depicted in Figure 4 were placed in water containing an equivalent amount of dimethyl sulfoxide.
Postacquisition Image Processing and Quantification
All images were cropped and processed for brightness/contrast as complete montages using Adobe Photoshop CS3 (http://www.adobe.com). The layer function in Photoshop was used to introduce text, regions of interest, and color overlays. Green and red images for mito-mEosFP (Fig. 6A, inset) were merged after acquisition using ImageJ software (National Institutes of Health). Images acquired using the Leica confocal microscope were processed directly using the proprietary fluorescence intensity quantification tools in various regions of interest. Alternatively, the histogram analysis tool that provides RGB data on an eight-bit value scale of 0 to 255 (Cowlishaw, 1985), Interactive 3D surface plot version 2.22 (http://rsbweb.nih.gov/ij/plugins/surface-plot-3 d.html), which uses image luminance for plot height, Color inspector 3D version 2, which shows color distribution within a three-dimensional color space and allows color cell frequency to be presented in histograms (http://www.f4.fhtw-berlin.de/∼barthel/ImageJ/ColorInspector/help.htm), and the RGB profiler (http://rsbweb.nih.gov/ij/plugins/rgb-profiler.html) plug-ins from ImageJ version 1.40g (http://rsbweb.nih.gov/ij/) were used. Color quantification (e.g. color bar in Fig. 1C) followed the International Color Consortium-compliant Adobe Photoshop color coding as described (Schenkel et al., 2008).
All experiments reported here were replicated at least four times. Where applicable, speed measurements are provided as means ± se followed by the total number (n) of observed cells or organelles.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transient expression and photoconversion of mEosFP::PIP1 probe.
Supplemental Figure S2. Magnified view of Golgi bodies highlighted by mEosFP::GONST1 and F-actin.
Supplemental Table S1. Primers used for different mEosFP-based probes.
Supplemental Movie S1. Time-lapse imaging of a PI(3)P-enriched tubule.
Supplementary Material
Acknowledgments
We thank J. Wiedenmann (University of Southampton) for mEosFP, G. Patterson and J. Lippincott-Schwartz (National Institutes of Health) for PA-GFP, J. Runions and C. Hawes (Oxford Brookes University) for the PA-GFP-ER construct, T. Timmers (INRA) for the GFP::2xFYVE construct, R. Wedlich-Soldner (Max Planck Institute of Biochemistry) for the LIFEACT probe, and M. Otgui (University of Wisconsin) and N. Geldner (University of Lausanne) for comments and discussions on the endosomal compartment.
References
- Ai HW, Henderson JN, Remington SJ, Campbell RE. (2006) Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J 400: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. (2002) An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA 99: 12651–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N. (2004) Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA 101: 7805–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin TC, Handford MG, Yuseff MI, Orellana A, Dupree P. (2001) Identification and characterization of GONST1, a Golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13: 2283–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebo A, Thomas D, Der C, Sanchez L, Leborgne-Castel N, Marty F, Schoefs B, Bouhidel K. (2009) Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol Biol 70: 193–209 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Jones RL. (2000) Vacuoles and prevacuolar compartments. Curr Opin Plant Biol 3: 469–475 [DOI] [PubMed] [Google Scholar]
- Brown SC, Bolte S, Gaudin M, Pereira C, Marion J, Soler MN, Satiat-Jeunemaitre B. (2010) Exploring plant endomembrane dynamics using the photoconvertible protein Kaede. Plant J 63: 696–711 [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. (1998) Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell 2: 157–162 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cowlishaw MF. (1985) Fundamental requirements for picture presentation. Proc Soc Information Display 26: 101–107 [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser G, Waterman-Storer CM. (2006) Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct 35: 361–387 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Era A, Tominaga M, Ebine K, Awai C, Saito C, Ishizaki K, Yamato KT, Kohchi T, Nakano A, Ueda T. (2009) Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter K, Van Wilder V, Moshelion M, Chaumont F. (2004) Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 16: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. (1998) FYVE fingers bind PtdIns(3)P. Nature 394: 432–433 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Höfte H, Grandjean O, Brown S, Traas J. (1998) Phytochrome controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl. Plant J 13: 221–230 [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19: 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. (2001) The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol 2: 721–730 [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24: 461–465 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R, Shimada T, Nishimura M. (2004) Diversity and formation of endoplasmic reticulum-derived compartments in plants: are these compartments specific to plant cells? Plant Physiol 136: 3435–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N. (1986) Organization of the endomembrane system. Annu Rev Plant Physiol 37: 73–92 [Google Scholar]
- Harris N, Oparka KJ, Walker-Smith DJ. (1982) Plasmatubules: an alternative to transfer cells? Planta 156: 461–465 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Boulaflous A, Brandizzi F. (2008) Advances in fluorescent protein-based imaging for the analysis of plant endomembranes. Plant Physiol 147: 1469–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Niehl A, Sambade A, Steinmetz A, Heinlein M. (2009) Inhibition of tobacco mosaic virus movement by expression of an actin-binding protein. Plant Physiol 149: 1810–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Franklin AE, Hoffman NE. (1993) Primary structure and characterization of an Arabidopsis thaliana calnexin-like protein. J Biol Chem 268: 6560–6566 [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L. (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Reynolds D, Brown ML, Boisdore M, Fujikawa Y, Morales A, Meisel LA. (2008) Multidimensional fluorescence microscopy of multiple organelles in Arabidopsis seedlings. Plant Methods 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Anthony RG, Hussey PJ. (2004) Green fluorescent protein-mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor’s actin depolymerizing activity in vitro. Plant Physiol 136: 3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Lam SK, Tse YC, Miao Y, Li HY, Wang J, Lo SW, Jiang L. (2007) Molecular characterization of plant prevacuolar and endosomal compartments. J Integr Plant Biol 49: 1119–1128 [Google Scholar]
- Logan DC, Leaver CJ. (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Ma S, Quist TM, Ulanov A, Joly R, Bohnert HJ. (2004) Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. Plant J 40: 845–859 [DOI] [PubMed] [Google Scholar]
- Mano S, Miwa T, Nishikawa S, Mimura T, Nishimura M. (2008) The Plant Organelles Database (PODB): a collection of visualized plant organelles and protocols for plant organelle research. Nucleic Acids Res 36: D929–D937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Miwa T, Nishikawa S, Mimura T, Nishimura M. (2009) Seeing is believing: on the use of image databases for visually exploring plant organelle dynamics. Plant Cell Physiol 50: 2000–2014 [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ. (1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant R, Robards AW. (1968) Membrane systems associated with the plasmalemma of plant cells. Ann Bot (Lond) 32: 457–471 [Google Scholar]
- Marcus AI, Moore RC, Cyr RJ. (2001) The role of microtubules in guard cell function. Plant Physiol 125: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Mathur J. (2007) The illuminated plant cell. Trends Plant Sci 12: 506–513 [DOI] [PubMed] [Google Scholar]
- Mathur J, Chua NH. (2000) Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Hülskamp M. (2002) Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol 128: 1031–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Yamada K, Shimada T, Nishimura M, Hara-Nishimura I. (2003) The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol 44: 661–666 [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Zerial M. (2002) Mosaic organization of the endocytic pathway. Exp Cell Res 272: 8–14 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. (2005) Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc Natl Acad Sci USA 102: 9156–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Spitzer C. (2008) Endosomal functions in plants. Traffic 9: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. (2002) A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297: 1873–1877 [DOI] [PubMed] [Google Scholar]
- Ponti A, Vallotton P, Salmon WC, Waterman-Storer CM, Danuser G. (2003) Computational analysis of F-actin turnover in cortical actin meshworks using fluorescent speckle microscopy. Biophys J 84: 3336–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits EA, Neefjes JJ. (2001) From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol 3: E145–E147 [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008) Lifeact: a versatile marker to visualize F-actin. Nat Methods 5: 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Jiang L, Schumacher K. (2008) The endosomal system of plants: charting new and familiar territories. Plant Physiol 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Sieber H, Kammerloher W, Schaffner AR. (1996) PIP1 aquaporins are concentrated in plasmalemmasomes of Arabidopsis thaliana mesophyll. Plant Physiol 111: 645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runions J, Brach T, Kühner S, Hawes C. (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57: 43–50 [DOI] [PubMed] [Google Scholar]
- Saidi Y, Finka A, Chakhporanian M, Zrÿd JP, Schaefer DG, Goloubinoff P. (2005) Controlled expression of recombinant proteins in Physcomitrella patens by a conditional heat-shock promoter: a tool for plant research and biotechnology. Plant Mol Biol 59: 697–711 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schenkel M, Sinclair AM, Johnstone D, Bewley JD, Mathur J. (2008) Visualizing the actin cytoskeleton in living plant cells using a photo-convertible mEos:FABD-mTn fluorescent fusion protein. Plant Methods 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. (2007) Advances in fluorescent protein technology. J Cell Sci 120: 4247–4260 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW. (2004) A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J. (2009) Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J 59: 231–242 [DOI] [PubMed] [Google Scholar]
- Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. (2000) Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol 149: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD. (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Tang W, Collver H, Kinken K. (2004) Dexamethasone-inducible green fluorescent protein gene expression in transgenic plant cells. Genomics Proteomics Bioinformatics 2: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M. (1991) Tubular early endosomal networks in AtT20 and other cells. J Cell Biol 115: 635–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A. (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW, Jr, Munnik T. (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Rounds CM, Hepler PK, Bezanilla M. (2009) Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Samaj J, Hlavacka A, Ueda T, Preuss M, Nielsen E, Mathur J, Emans N, Stenmark H, et al. (2005) Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur J Cell Biol 84: 609–621 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. (2002) Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science 295: 1083–1086 [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler KD, Nienhaus GU. (2004) EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA 101: 15905–15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann J, Oswald F, Nienhaus GU. (2009) Fluorescent proteins for live cell imaging: opportunities, limitations, and challenges. IUBMB Life 61: 1029–1042 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.