Abstract
To investigate early salt acclimation mechanisms in a salt-tolerant poplar species (Populus euphratica), the kinetics of molecular, metabolic, and physiological changes during a 24-h salt exposure were measured. Three distinct phases of salt stress were identified by analyses of the osmotic pressure and the shoot water potential: dehydration, salt accumulation, and osmotic restoration associated with ionic stress. The duration and intensity of these phases differed between leaves and roots. Transcriptome analysis using P. euphratica-specific microarrays revealed clusters of coexpressed genes in these phases, with only 3% overlapping salt-responsive genes in leaves and roots. Acclimation of cellular metabolism to high salt concentrations involved remodeling of amino acid and protein biosynthesis and increased expression of molecular chaperones (dehydrins, osmotin). Leaves suffered initially from dehydration, which resulted in changes in transcript levels of mitochondrial and photosynthetic genes, indicating adjustment of energy metabolism. Initially, decreases in stress-related genes were found, whereas increases occurred only when leaves had restored the osmotic balance by salt accumulation. Comparative in silico analysis of the poplar stress regulon with Arabidopsis (Arabidopsis thaliana) orthologs was used as a strategy to reduce the number of candidate genes for functional analysis. Analysis of Arabidopsis knockout lines identified a lipocalin-like gene (AtTIL) and a gene encoding a protein with previously unknown functions (AtSIS) to play roles in salt tolerance. In conclusion, by dissecting the stress transcriptome of tolerant species, novel genes important for salt endurance can be identified.
Excess sodium in soil is a widespread and common stress in natural and agricultural ecosystems (FAO, 2008). High salt concentrations decrease the osmotic potential of the soil solution, thereby lowering the availability of water to plants. Therefore, salinity causes dehydration stress, at least in nonacclimated plants. Furthermore, sodium competes with other cations, thereby affecting plant nutrition and ion homeostasis (Zhu, 2003; Munns and Tester, 2008; Shabala and Cuin, 2008). Since most plant species have only very limited capacities to cope with excess sodium, productivity on saline soil is strongly diminished and plant growth may even become impossible. Globally, salinity is an important environmental problem, because the total area of salt-affected soils, including saline and sodic soils of currently about 831 million ha, is continually increasing (Martinez-Beltran and Manzur, 2005).
The elucidation of salt tolerance mechanisms, therefore, is an important issue and has attracted considerable interest in recent years (Munns, 2005; Vinocur and Altman, 2005; Yamaguchi and Blumwald, 2005; Ma et al., 2006; Munns and Tester, 2008; Chen and Polle, 2010). Comparative analysis of transcriptional profiles of Arabidopsis (Arabidopsis thaliana) and salt cress (Thellungiella halophila) suggested that salt-sensitive and salt-tolerant species have a common set of salt responses whose regulation differs (Taji et al., 2004). Upon sudden salt exposure, both species activated genes involved in ribosomal functions, photosynthesis, cell growth, osmolyte production, and transport activities (Gong et al., 2005), whereas in salt-acclimated T. halophila plants, only a few genes were differentially regulated (Wong et al., 2006).
An intriguing question is how trees, which have a long life span and, therefore, must be able to cope with excess salt for extended periods of time, adapt to high salinity. Populus euphratica is a model for salt tolerance in trees (Chen and Polle, 2010). It withstands sodium concentrations up to 450 mm in nutrient solution (Watanabe et al., 2001). It accumulates high sodium concentrations in leaves, which develop succulence after long-term exposure (Ottow et al., 2005). In a preceding study, a small microarray with 315 cDNAs of transcripts obtained by suppression subtractive hybridization from P. euphratica was used to analyze salt responses, but clear tissues-specific patterns were not found (Gu et al., 2004). Brosché et al. (2005) constructed P. euphratica EST-based microarrays containing 7,342 different ESTs corresponding to about 6,300 different genes. The ESTs were captured from 17 different cDNA libraries of different tissues (roots, leaves, stems) from stress-exposed trees (drought, salt, flooding, cold, ozone). Using microarrays with this collection of stress-responsive genes, Brosché et al. (2005) found that leaves of mature desert-grown P. euphratica trees acclimated to high salinity show only a few differentially expressed genes compared with tap-watered control trees. Salt-acclimated Euphrat poplars show less gene regulation than salt-sensitive poplar species (Ding et al., 2010), probably because transporters important to control excessive salt accumulation and the flavonoid/phenylpropanoid pathway, which may counteract negative salt effects by controlling reactive oxygen species, are constitutively up-regulated in the tolerant poplar species (Janz et al., 2010). However, it is still unclear which early molecular responses are required to adjust the metabolism to life at a new level of osmotic balance and ionic stress.
To obtain insights into the initial events taking place in response to exposure to high salinity in a salt-tolerant tree species, we exposed P. euphratica to salt stress and followed early changes in gene expression, metabolic profiles, and physiological responses to Na+ accumulation and changes in plant water relations. The kinetics of salt accumulation and osmotic adjustment differed between roots and leaves displaying a phase of initial dehydration stress, followed by osmotic adjustment and finally a new level of “high-salinity” homeostasis. These different stages were associated with distinct clusters of coexpressed genes. Comparative in silico analysis with Arabidopsis orthologs was used as a strategy to reduce the number of candidate genes for functional analysis. Using Arabidopsis knockout lines, a lipocalin-like gene (AtTIL) and a gene encoding a protein with previously unknown functions (AtSIS) were shown to play roles in salt tolerance.
RESULTS
The Kinetics of Sodium Accumulation and Recovery of Water Potential Show Distinct Phases in Roots and Leaves of P. euphratica
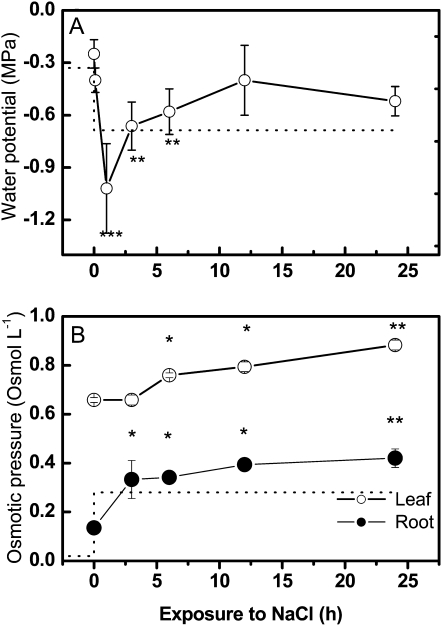
Exposure of plants to elevated salinity imposes stress by a decrease in water potential in the root medium. When young P. euphratica trees were exposed to 150 mm NaCl in the nutrient solution, which corresponds to a drop in osmotic potential of −0.68 MPa, a severe and rapid decrease in the shoot water potential of 0.77 MPa occurred (Fig. 1A). This caused transient wilting of the plants.
Figure 1.
Water potential (A) and osmotic pressure (B) in P. euphratica exposed to salt stress. NaCl at 150 mm or a corresponding water volume was added to the nutrient solution. Water potential was measured on shoots (white circles). Osmotic pressure was measured in pressure sap from roots (black circles) and leaves (white circles). Dotted lines indicate changes in the water potential or osmotic pressure in the nutrient solution. Data indicate means of five plants per treatment and time point ± se. Asterisks indicate significant differences compared with controls.
The shoot water potential recovered almost completely within 24 h (Fig. 1A). Roots and leaves, however, displayed different time courses of osmotic adjustment (Fig. 1B). In roots, acclimation was completed within 3 h, because the osmotic pressure, which increased to ensure water uptake, was adjusted above the osmotic pressure of saline solution in this period of time. Afterward, it increased only marginally (Fig. 1B). In contrast, the osmotic adjustment of leaves started only after roots had reached their new plateau (Fig. 1B).
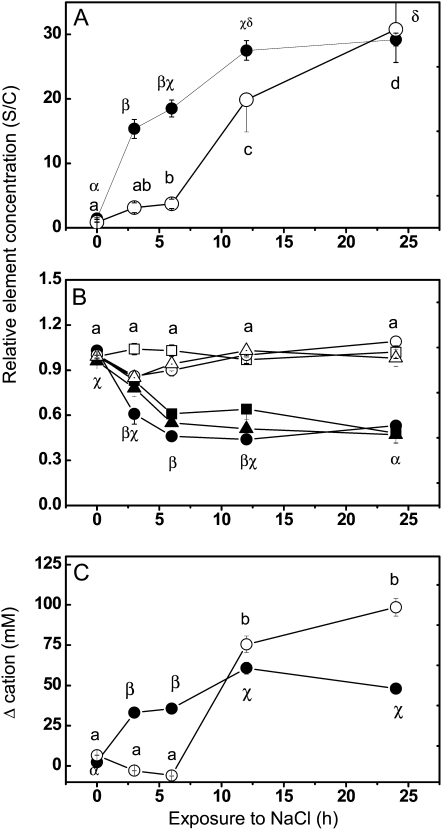
Since changes in osmotic potential are the result of changes in solute concentrations in plant tissues, we determined profiles of nutrient elements and organic metabolites. In roots, salt exposure caused 28-fold increases in Na+ concentrations within 12 h and simultaneously about 2-fold decreases in major cations (Ca2+, K+, and Mg2+; Fig. 2, A and B). Afterward, elevated Na+ and decreased cation levels were maintained in roots (Fig. 2, A and B). In leaves, an about 30-fold increase in Na+ was also observed; however, accumulation started with a delay of about 6 h compared with roots and reached levels similar to those of roots after 24 h of sodium exposure (Fig. 2A), whereas the major cations in leaves remained unaffected (Fig. 2B). Other nutrient elements, such as phosphorus (P) and sulfur (S), were neither influenced in roots nor in leaves of salt-stressed poplar [means across all time points and controls and salt-exposed plants: P(roots) = 6.50 ± 0.27 mg g−1 dry weight, S(roots) = 4.45 ± 0.06 mg g−1 dry weight, P(leaves) = 7.12 ± 0.10 mg g−1 dry weight, S(leaves) = 7.97 ± 0.05 mg g−1 dry weight].
Figure 2.
Salt-induced changes in element concentrations in leaves and roots of P. euphratica. Element concentrations of leaves (white symbols) or roots (black symbols) after salt treatment (150 mm) were divided by mean concentrations of controls (S/C). A, Sodium. B, Magnesium (triangles), potassium (circles), and calcium (squares). C, Change in cation concentrations (mm). Data indicate means of 15 plants ± se. Different letters indicate significant differences at P ≤ 0.05, with Greek letters for roots and Roman letters for leaves. Control plants contained the following ion concentrations (mg g−1 dry mass ± se) in roots: sodium, 1.43 ± 0.37; potassium, 1.00 ± 0.11; calcium, 1.03 ± 0.07; and magnesium, 0.96 ± 0.05. Control plants contained the following ion concentrations (mg g−1 dry mass ± se) in leaves: sodium, 0.16 ± 0.01; potassium, 37.85 ± 0.98; calcium, 14.4 ± 0.91; and magnesium, 5.75 ± 0.32.
Differences between sodium uptake by roots and leaves affected the overall pattern of cation accumulation, leading to earlier but less pronounced increases in total cation concentrations in roots compared with leaves (Fig. 2C). The changes in water contents were small, −3.2% and −1.2% in leaves (control, 85.4%) and roots (control, 94.9%), respectively.
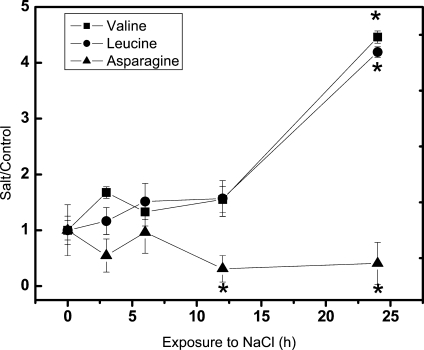
Metabolite profiling was conducted for soluble carbohydrates, sugar alcohols, organic acids, and amino acids in leaves and did not show significant changes for most compounds analyzed (Supplemental Table S1). Exceptions were Asn, whose levels were decreased after 12 h, and Val and Ile, which showed significant increases after 24 h of salt exposure (Fig. 3). However, the estimated concentrations of Val and Ile of 7.4 and 3.3 μg g−1 dry weight contributed only 0.03% and 0.01%, respectively, to the osmotic potential imposed by sodium. Thus, the changes of these amino acids were too low to affect the total osmotic potential of leaves significantly. Other compounds such as Thr and citric acid showed only transient changes after 12 and 3 h, respectively. Overall, osmotic recovery in P. euphratica was thus mainly achieved by sodium accumulation.
Figure 3.
Salt-induced changes in amino acids in leaves of P. euphratica. Data indicate means ± se (n = 5). Asterisks indicate significant changes compared with controls.
Transcriptional Profiling Reveals Groups of Coregulated Genes
As changes in the ion balance were completed in roots (0–12 h) before adjustment in leaves started (12–24 h), we took advantage of these distinct phases to dissect transcriptional responses related to water deficit and/or high sodium concentrations in leaves and roots, respectively. We used previously developed microarrays containing ESTs of about 6,340 genes from P. euphratica (Brosché et al., 2005). To identify tightly stress-responsive genes, stringent settings of statistical analysis of microarrays (SAM; Δ = 0.189–0.269) were applied to obtain low false discovery rates (0–0.1).
Using these criteria, we identified 56 and 48 ESTs in leaves and roots, respectively, which showed significant changes of transcript levels in response to salinity (Supplemental Table S2). These numbers correspond to 0.9% and 0.8% of all ESTs present on the array. About 22% and 41% of these transcripts in leaves and roots, respectively, did not show homology with genes of known functions. Only three genes (dehydrin [Dhn1 protein; AJ774829], osmotin-like protein [AJ779386], Suc synthase [AJ778233, AJ778234]) responded with significant changes in transcript abundance in both roots and leaves, respectively. Therefore, the total number of stress-responsive ESTs was 101, corresponding to 1.7% of the ESTs on the array. The total number of unique salt-responsive genes was even lower, because some of the identified transcripts (e.g. those for Rubisco activase, PsbA, and ribosomal RNA) were represented by multiple ESTs on the array (Supplemental Table S2).
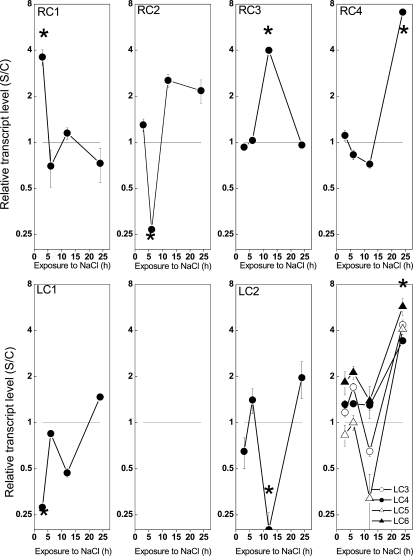
At a given time point, all responsive genes had either only increased or decreased expression. For some selected genes, the response was confirmed by quantitative real-time PCR (Supplemental Table S3). Cluster analysis based on the transcript levels across all time points revealed groups of coexpressed genes (Fig. 4). In roots, four clusters were obtained, one for each time point (Fig. 4, RC1–RC4; for genes included in each cluster, see Supplemental Table S2). In leaves, six clusters were obtained, because transcriptional responses after 24 h of salt exposure were more diverse than at other time points (Fig. 4).
Figure 4.
Salt-induced changes in transcript levels in roots and leaves of P. euphratica. Data indicate mean transcript levels of genes assigned to root clusters (RC) and leaf clusters (LC) according to Supplemental Table S2. Asterisks indicate significant changes compared with controls.
Root Genes Responsive to Different Phases of Osmotic Stress and Recovery
We exploited the different phases of salinity stress (Figs. 1 and 2) to assign genes forming transcriptional clusters to different physiological stages: dehydration stress, osmotic adjustment, and homeostasis associated with high tissue salt concentrations.
Root cluster 1 (RC1) was associated with the initial dehydration phase. This cluster was small, containing only two members both of the dehydrin family (Fig. 4; Supplemental Table S2).
RC2, which characterized the major phase of osmotic readjustment of roots, was formed by a group of genes with decreased transcript levels (Fig. 4) and contained three putative aquaporins. Furthermore, several genes involved in protein metabolism and genes that may play roles in stress sensing and signal transduction (a putative Gly-rich RNA-binding protein, a homolog to a metallothionein MT1a, and a Suc synthase; Supplemental Table S2) were also found.
Genes forming RC3 were associated with a physiological stage in which osmotic recovery of the roots was accomplished but in which most likely metabolic adjustment to the new ionic relationships was still ongoing (Fig. 2A). This cluster contained the highest proportion of genes with unknown functions or no hit in the databases (74%). The remaining transcripts indicated cell wall changes (polygalacturonase-like protein), stress and stress signaling (a cyclophilin, a putative PR protein, and a putative Ser/Thr kinase), the involvement of glycolysis (malate dehydrogenase), and remodeling of protein metabolism (tRNA for Gln synthase, Ser hydroxymethyltransferase, and a proteasome AAA-ATPase subunit; Supplemental Table S2). However, salt-induced changes in protein content were not found (Supplemental Fig. S2).
RC4 contained only two genes, both belonging to the osmotin family (Fig. 4; Supplemental Table S2). The different time courses of dehydrins (RC1) and osmotins (RC4) suggest that dehydrins are important for the primary adaptation processes in roots, whereas osmotins seem to be required in the homeostatic phase to maintain cell functions at low osmotic potentials and high ionic stress.
Leaf Genes Responsive to Different Phases of Osmotic Stress and Recovery
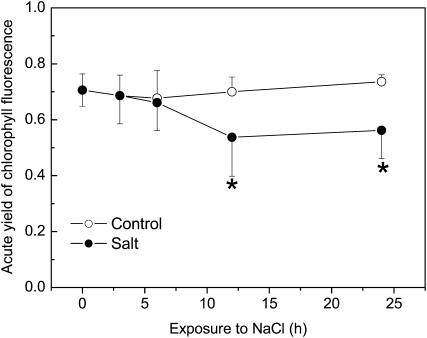
In leaves, a higher number of genes (leaf cluster 1 [LC1]) were responsive to the initial phase of dehydration stress than in roots (Supplemental Table S2). This was remarkable because at this early time point, leaves were not yet exposed to elevated sodium concentrations (Fig. 2A) but the plants showed wilting, which recovered during the further time course of sodium exposure. LC1 was composed of six transcripts representing Rubisco activase and one of a Clp protease (heat shock protein) with putative chloroplast localization (Supplemental Table S2). This suggests a massive influence of the initial dehydration phase on chloroplast metabolism. To corroborate this assumption, we determined PSII activity by measuring chlorophyll fluorescence. However, at this early time point, PSII activity was not yet affected (Fig. 5), although photosynthetic CO2 assimilation and stomatal conductance were decreased (Wang et al., 2007). Furthermore, LC1 contained mitochondrial catalase, cytochrome P450 monooxygenase, and glyoxalate oxidase (Supplemental Table S2). Decreased transcript abundance of these genes suggests compromised mitochondrial hydrogen peroxide metabolism and the formation of methylglyoxalates, which are generated as toxic oxidative side products of glycolysis. Suc synthase transcripts were also decreased. However, correlations of transcriptional changes of this gene with carbohydrate concentrations were not found (Supplemental Table S1). The maintenance of soluble carbohydrate concentrations might have been achieved by the activity of other enzymes whose transcript abundance remained unchanged.
Figure 5.
Quantum yield of photochemistry in irradiated leaves of control and salt-exposed P. euphratica plants. Data indicate means ± se (n = 5). Asterisks indicate significant changes compared with controls.
It is notable that in leaves after 6 h of salt exposure, when the initial dehydration stress was relieved (Fig. 1B) but sodium accumulation had not yet occurred (Fig. 2B), no changes in transcripts were found.
After 12 h, at the stage when massive sodium accumulation occurred, only genes with significantly decreased transcript levels (LC2) were identified (Fig. 4). Five members of LC2 were of chloroplast origin (three psbA-related proteins, D2 protein, Rubisco large subunit). At this stage, PSII activity was also decreased (Fig. 5). Furthermore, LC2 contained genes for plastidic and nuclear protein synthesis (26S and 60S ribosomal RNA and a translation initiation factor), genes with ill-defined or undefined functions (putative glycosyl transferase, predicted GTP-binding protein, unknowns), and genes pointing to glycolysis (oxoglutarate dehydrogenase) and stress (a putative glutathione-S-transferase and COBRA protein precursor; Supplemental Table S2).
Increased transcript levels in leaves were only found after 24 h, when the water potential had been restored and the tissues were exposed intrinsically to high salinity (Figs. 1A, 2A, and 4). Time courses and relative transcript levels differed among each other, yielding four clusters (Fig. 4). Two of these clusters (LC3, LC5) contained genes whose transcript levels showed time courses similar to those of LC2, with a decrease at 12 h of salt exposure, although stronger increases after 24 h that those of LC2. LC3 contained calcineurin-like protein CLB10, which is known to play a role in the response to salinity, and 1-aminocyclopropane-1-carboxylic acid oxidase, suggesting the recruitment of ethylene-related pathways (Ottow et al., 2005). LC5 contained mainly organelle-specific genes (psbA, mitochondrial ATPase) and the osmotin-like protein that also had increased expression in roots.
In contrast to LC3 and LC5, transcript levels of genes in clusters LC4 and LC6 did not change during the initial phases of salt exposure, suggesting that these groups must be specific for acclimation to high cellular solute concentrations. Relative transcript levels of LC6 were higher than those of LC4, but otherwise no differences between these two clusters were found. LC4 was composed of a GA-responsive gene (GASA3), a dehydrin-like protein, an aldehyde dehydrogenase, a homeodomain transcription factor with homology to ATHB-7, a lipocalin-like protein, two metallothioneins (MT2a, MT3), and genes with unknown functions. LC6 contained genes for a putative Kunitz trypsin inhibitor, Asn synthase, GA-responsive protein, and a metallothionein MTa-like protein. Among all clusters, LC4 and LC6 were characterized by the highest abundance of stress-related genes and transcription factors. Since osmotic recovery was almost accomplished at this stage (Fig. 1A), these genes must play roles in the adaptation to high salinity or in maintaining cellular metabolism at a new level of osmotic pressure.
Identification of Novel Salt-Resistant Genes by Comparative Analysis of Arabidopsis Orthologs of P. euphratica Genes and Functional Characterization of Knockout Mutants
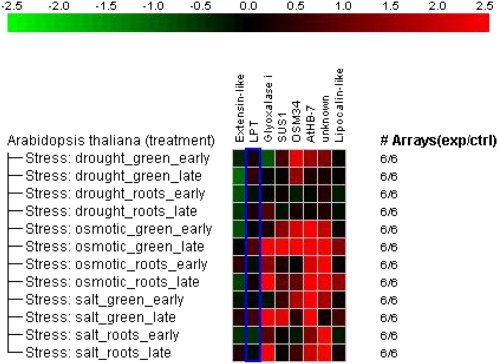
To investigate the significance of the stress-responsive genes identified by transcriptional profiling in P. euphratica in a wider context, we BLASTed the poplar ESTs against The Arabidopsis Information Resource database to extract Arabidopsis genes with significant homology. For the leaf set, 39 significant matches, and for the root set, 27 significant matches with Arabidopsis genes were obtained (Supplemental Table S2). Some of these genes were retrieved in both tissues and some were represented by different ESTs, resulting in a total of 49 different Arabidopsis homologs. This condensed data set was used to search Genevestigator (Hruz et al., 2008) for genes that were silent during normal vegetative development. Appling this criterion, we obtained a short list of eight stress-specific genes out of the total 49 stress-responsive genes (Supplemental Fig. S1). Two of these Arabidopsis orthologs were found in both roots and leaves of P. euphratica (osmotin [At4g11650], Suc synthase [At5g20830]), three genes were only responsive in roots (glyoxalase [At1g80160], extensin [At2g27380], seed storage/lipid transfer protein [At5g05960]), and three were only responsive in leaves (ATBH-7 homeodomain transcription factor [At2g46680], a lipocalin-like gene [At5g58070], a gene with unknown functions [At5g02020]). The latter three genes and osmotin belonged to the group of genes induced in P. euphratica leaves after 24 h of salt stress. With the exception of the lipocalin-like gene, all other leaf genes were also responsive to salt, drought, and osmotic stress in Arabidopsis, whereas the lipocalin-like gene was only marginally induced in response to osmotic and salt stress (Fig. 6).
Figure 6.
Analysis of changes in transcript levels in response to salinity, drought, and osmotic stress in Arabidopsis orthologs of eight selected poplar salt-responsive genes. Analysis was conducted with Genevestigator.
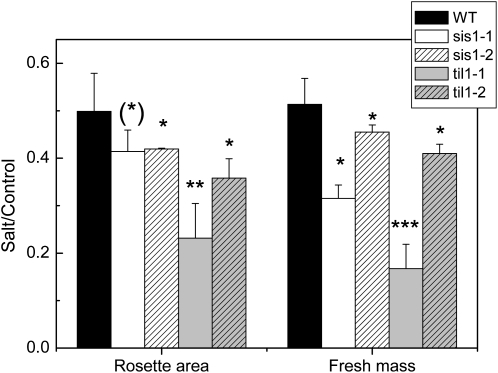
The Arabidopsis lipocalin-like gene (At5g58070) and the gene with unknown functions (At5g02020) have no established roles in salt tolerance. Since the unknown gene was induced by salt in Arabidopsis as well as in poplar (Fig. 6; Supplemental Table S2) and contained an overabundance of Ser residues in its amino acid sequence (Fayyaz, 2008), we termed the gene At5g02020 as Salt-Induced Ser-rich (AtSIS). The lipocalin-like gene (At5g58070) belongs to a family of previously annotated temperature-induced lipocalin-like genes (Flower et al., 2000) and, therefore, was termed AtTIL. To find out whether these candidate genes contribute to salt resistance, homozygous T-DNA insertion mutants Attil1-1, Attil1-2, Atsis1-1, and Atsis1-2 were isolated and exposed to salinity stress. Growth assays in soil supplemented with increasing NaCl concentrations revealed a significantly higher sensitivity of leaf expansion of Attil1-1 and Attil1-2 to excess salt than that of the wild type (Fig. 7). Atsis1-1 and Atsis1-2 showed intermediate behavior (Fig. 7). All knockout mutants produced significantly less biomass under salt stress than the wild type, showing that these genes play roles in salt tolerance (Fig. 7).
Figure 7.
Performance of Arabidopsis T-DNA insertion lines Attil1-1, Attil1-2, Atsis1-1, and Atsis1-2 in response to salinity. Plants were grown in soil and exposed to increasing concentrations of NaCl (25, 50, and 250 mm). Rosette leaf area and biomass were determined 20 d after starting salt exposure (n = 7–10; values are means ± se). WT, Wild type.
DISCUSSION
Different Transcriptional Timetables of Salt Responses in Roots and Leaves Accommodate Different Physiological Needs
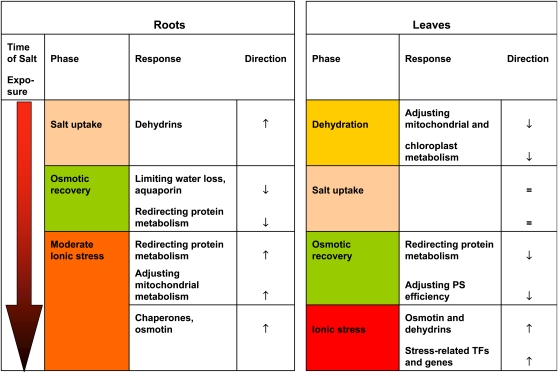
In this study, we dissected the physiological responses of the salt-tolerant Euphrat poplar into distinct phases and showed that these phases were characterized by suppression or activation of clusters of coregulated genes. The major events have been compiled in an overview (Fig. 8). By these kinetic analyses, we gained insights into the molecular responses of salt-acclimation processes associated with an initial dehydration phase in leaves, subsequent salt accumulation, and finally restored osmotic homeostasis at a new level of ionic stress. The timing of adaptive responses differed in aboveground and belowground plant tissues, because transport through the stem delays salt accumulation in leaves compared with roots. Furthermore, our study highlights important differences between poplar roots and leaves regarding early salt effects, with little overlap of the genes involved (three of 101). Differences between early and late transcriptional responses as well as between roots and leaves have also been reported for Arabidopsis (Ma et al., 2006). However, in those previous studies, the link to physiological differences in the tissues was lacking. Our study shows that ionic stress is immediately high in roots. In contrast, leaves suffer initially from dehydration stress by water loss but not from shifts in the ion balance. With continued sodium uptake, roots can avoid excessive ion accumulation by compensatory decreases in other cations. The existing leaves must accumulate sodium on top of the cations already present, thereby experiencing the strongest ionic stress (Fig. 2C).
Figure 8.
Overview of the different phases of salt stress in roots and leaves of P. euphratica and the molecular responses. Time of salt exposure was 0 to 24 h. TF, Transcription factors.
Notably, only two genes, osmotins and dehydrins, were commonly induced in roots and leaves, albeit with different time courses. This finding points to functional differences of chaperones in stress acclimation of Euphrat poplar. Dehydrins that occur immediately in roots but only late in leaves have been shown to maintain a highly disordered status under high solute concentrations where globular proteins would collapse (Mouillon et al., 2006). They may also act as protein cryoprotectants (Hara et al., 2001; Seki et al., 2002) and contribute to protection from drought stress (Rinne et al., 1999; Wang et al., 2004). The time courses observed here suggest a protective function of dehydrins against ionic disorders (Mouillon et al., 2008), whereas the delayed activation of osmotins, another class of well-known stress-activated genes (Shinozaki and Yamaguchi-Shinozaki, 2007), indicates that these proteins must be involved in helping the plant to cope with life at a new level of cellular homeostasis.
Surprisingly, Suc synthase was also salt responsive in both roots and leaves, but its transcript levels decreased under stress (Gu et al., 2004; Bogeat-Triboulot et al., 2007; this study). In contrast, its homolog AtSUS1 increases under stress (Ma et al., 2006). AtSUS1 also responds to elevated carbohydrate concentrations (Bieniawska et al., 2007). However, our metabolite profiling did not provide evidence for strong redirecting of carbohydrate metabolism during salt stress. As PeSUS1 was decreased in the early stage of leaf dehydration stress, it may be responsive to changes in cell turgor that lead to physiological increases in solutes.
Stress Acclimation Involves a Redirection of Protein Metabolism in Roots and Leaves But Different Stress Pathways
Although there was little overlap between stress-responsive genes in roots and leaves, both tissues showed similar patterns of regulation of pathways involved in protein and amino acid metabolism. When tissues were exposed to an increment in sodium (6 h in roots, 12 h in leaves), decreased transcript abundance of genes for protein biosynthesis suggested that this metabolic path was transiently suppressed in both roots and leaves. The recovery correlated with increased transcript abundance for genes involved in amino acid biosynthesis (12 h in roots, 24 h in leaves). Indeed, long-term salt stress leads to an accumulation of free amino acids in both salt-sensitive and salt-tolerant poplar species (Ottow et al., 2005; Ehlting et al., 2007). It is likely that amino acids, or at least certain amino compounds, have significance for salt stress compensation, because functional links between Gln synthetase and drought and salt tolerance exist (Hoshida et al., 2000; El-Khatib et al., 2004).
Overall, there was surprisingly little overlap between the stress transcriptomes of leaves and roots. Our analysis suggested that the restoration of the water potential in roots involved aquaporins (Supplemental Table S2). These genes are important for the regulation of plant internal water transport (Chaumont et al., 2005) and respond to salt and drought stress in different species (Boursiac et al., 2005; Bogeat-Triboulot et al., 2007) as well as to salt stress in the field (Brosché et al., 2005). Their suppression at early stages may help to prevent water loss. As sodium uptake of roots progressed, cyclophilins were transiently induced. This is further support for the idea that chaperones play a major role in protecting metabolism when plants use Na+ as an osmolyte.
A putative Ser/Thr kinase was increased when salt adjustment in roots was almost accomplished. Kinases are involved in signal transduction in response to stress (Zhu, 2002), but the relatively long delay between stimulus and response does not suggest a primary role of this kinase in salt signaling. An involvement in oxidative stress signaling is possible, because we also observed a transient increase in transcripts for glyoxalases (syn. lactoyl-glutathione lyase) that reduce toxic glyoxals to glycolate (Shangari et al., 2003).
In leaves, the early phase of dehydration stress was characterized by the suppression of transcripts encoding chloroplastic or mitochondrial enzymes, suggesting cross talk between these organelles. Rapid suppression of Rubisco activase and chloroplastic Clp protease, which was also observed under natural salt stress in P. euphratica (Brosché et al., 2005), may be required for the down-regulation of photosynthetic CO2 assimilation and the stress adaptation of the photosynthetic apparatus, respectively (Zheng et al., 2006; Wang et al., 2007; Stanne et al., 2009). Decreased PSII yield occurred only after the leaves started to accumulate sodium and was probably caused by suppression of the biosynthesis of PSII D1 protein (psbA), Qa protein, and Rubisco large subunit. This indicates the necessity to establish a new level of homeostasis for photosynthetic electron transport. Since the decrease in fluorescence yield was reversible in darkness, the overall electron transport capacity was not injured but down-regulated. This probably reflects an accommodation for decreases in assimilation. Notably, similar responses have also been found in salt cress in response to excess salinity (Wong et al., 2006). Comparisons of stress transcriptomes of Arabidopsis also support the notion that down-regulated clusters mainly contain genes for developmental processes and metabolic adjustment (Ma and Bohnert, 2007).
In contrast to roots, sodium accumulation activated some of the well-known stress and signal transduction pathways in leaves, including transcripts for calcineurin B-like protein and 1-aminocyclopropane carboxylate oxidase, pointing to the involvement of calcium and ethylene. Salt exposure generally leads to increased abscisic acid levels, but in Euphrat poplar more rapidly than in salt-sensitive poplar species (Chen et al., 2001; Luo et al., 2009). Here, activation of a putative homeodomain transcription factor (homolog to ATHB-7) and aldehyde dehydrogenase was observed, which are known to be regulated by abscisic acid and to be responsive to salinity and drought (Söderman et al., 1996; Seki et al., 2002; Olsson et al., 2004; Sunkar et al., 2003; Kotchoni et al., 2006). The late response of these genes, which occurred only when the leaves were exposed to a high salt level, suggests that they are necessary to afford tissue protection from excessive ion accumulation or high osmotic pressure. Notably, transcript levels of aldehyde dehydrogenase, osmotin, metallothioneins, as well as an unknown gene (AJ773239) were also salt affected under field conditions (Brosché et al., 2005). The late response observed here indicates that they are not involved in the primary recognition and signaling of salt stress. As metallothioneins, GA-responsive genes, and several genes with unknown functions were present in the same regulon together with well-established stress-regulated genes, they are likely candidates for mediating salt stress protection.
Lipocalin-Like TIL and Ser-Rich Salt-Induced SIS Are Required for Salt Endurance
Transcriptional profiling of salt-tolerant species holds the promise of revealing candidate genes involved in stress adaptation. In our study, about 25% of the salt-responsive genes had unknown functions and, furthermore, about 10% had no significant hit with GenBank, suggesting that we do not yet fully understand the molecular events taking place to adjust the metabolism to excess salt and that P. euphratica contains a number of unique salt-responsive genes. By in silico analysis, we identified eight Arabidopsis orthologs among the salt-responsive genes of Euphrat poplar that were silent during most stages of vegetative plant development, but some of them were activated during seed formation or germination (Supplemental Fig. S1). This suggests a role of these genes in osmotic adaptation. However, not all of the eight Arabidopsis genes showed the same responses to salt, drought, or osmotic stress as poplar genes, pointing to differences in stress regulation. Comparative analysis of salt cress and Arabidopsis transcriptomes also supported the idea that the regulation of some key genes may be different in salt-tolerant and salt-sensitive species (Gong et al., 2005; Wong et al., 2006), and our study is strong experimental evidence for this suggestion.
In our study, differences in regulation between poplar and Arabidopsis were particularly obvious for genes encoding a putative lipocalin-like protein (TIL), a lipid transfer protein, and SUS1. We found the lipocalin-like protein particularly intriguing, because it was not regulated in salt-stressed or osmotically stressed Arabidopsis (Kilian et al., 2007), whereas other members of this gene family are stress responsive, especially under low temperatures (Frenette Charron et al., 2005). Lipocalins are most likely localized in the plasma membrane (Kawamura and Uemura, 2003), bind small hydrophobic ligands, act as transporters, and protect plants from oxidative stress (Frenette Charron et al., 2002; Charron et al., 2008). We used loss-of-function mutants of Arabidopsis to test the involvement of TIL and SIS, a salt-regulated unknown gene in salt tolerance. Data mining showed that SIS expression was typically regulated by salt treatment (Kilian et al., 2007; Luo et al., 2009). We show that suppression of its expression renders Arabidopsis knockout mutants more salt susceptible. Attil mutants were also more salt susceptible than the wild type, although this gene was not identified in previous salt screens. Our results indicate that both AtTIL and AtSIS play roles in salt tolerance, probably by increasing the protection of membranes.
CONCLUSION
In this study, we report the molecular timetable of the early salt transcriptome of the salt-tolerant Euphrat poplar. Molecular chaperones, especially dehydrins and osmotin, which help the cell to keep their proteins in an active status, appear to be key factors for coping with elevated salinity and ionic stress. Together with Suc synthase, these two chaperones were the only overlapping salt-responsive genes in roots and leaves. Adjustment of cellular metabolism involved remodeling of protein and amino acid biosynthesis in both roots and leaves, albeit employing different genes. In roots, decreases in aquaporins occurred, which might have limited water loss. In leaves, flexible down-regulation and well-orchestrated adjustment of energy metabolism involving mitochondrial and photosynthetic genes were among the initial responses to salinity and not, as one might have expected, up-regulation of typical stress-related enzymes. Induction of known stress-associated genes occurred only at late stages in leaves, when leaves had accumulated excessive sodium concentrations. Roots and leaves perceived physiologically different stress situations, and accommodation obviously required the activation of different stress transcriptomes.
To identify the stress-specific genes within the salt transcriptome, the Arabidopsis orthologs of poplar genes were used for in silico analyses to extract genes that were silent during vegetative development. This strategy resulted in eight candidates (Suc synthase, osmotin, glyoxalase, extensin, seed storage/lipid transfer protein, homeodomain transcription factor AtHB-7, a lipocalin-like gene [TIL], and a gene with unknown functions [SIS]), among which the lipocalin-like protein (TIL) was unique because it did not show early responses to salt or osmotic stress in Arabidopsis and SIS was unique because it was found in all salt experiments analyzed. Knockout mutants of Arabidopsis for TIL or SIS showed salt-sensitive phenotypes. In conclusion, the stress regulon of the salt-tolerant Euphrat poplar contains genes previously not known for functions in salt tolerance, such as TIL and SIS. In a wider context, these findings imply that salt-susceptible plants harbor genes important for salt tolerance that cannot be identified by conventional salt screens relying on differential gene expression but by comparison of sensitive and tolerant species.
MATERIALS AND METHODS
Plant Material
Populus euphratica (clone B2 from the Ein Avdat Valley in Israel; Hebrew University of Jerusalem) was multiplied by micropropagation (Leplé et al., 1992). Rooted plantlets were transferred to aerated hydroculture with Long Ashton nutrient solution (Hewitt and Smith, 1975) and grown for 3.5 months at 22°C, 150 μmol m−2 s−1 photosynthetically active radiation, with a photoperiod of 16 h of light. After reaching an average height of 0.83 ± 0.02 m, the medium was supplemented with 150 mm NaCl. Roots and leaves of six plants per treatment were harvested after 0, 3, 6, 12, and 24 h of salt stress and of nonstress treatment for controls. Plants were removed from the nutrient solution, immersed for 10 s with the roots in distilled water, and divided into roots and shoots. Root and leaf tissues were immediately frozen in liquid nitrogen. During the experimental phase, light was continuously supplied to avoid interference of light/dark transitions. The experiment was performed three times corresponding to three biological replications.
Ecophysiological Measurements
Chlorophyll fluorescence was measured by a pulse-modulated MINI-PAM (Walz) on adult leaves of stressed plants and controls under ambient light (Maxwell and Johnson, 2000) and used to calculate the actual yield of photochemistry: Φlight = (Fm′ − Fo′)/Fm′. A separate set of plants was used to measure the fluorescence of dark-adapted leaves to determine the maximum yield of photochemistry: Φdark = (Fm – Fo)/Fm.
Water potential was determined on 30-cm-long main shoots of plants (measured from the apex) at the indicated time points using a Scholander pressure chamber (Scholander et al., 1965; Cochard et al., 2001).
To determine osmotic pressure, leaves and roots that had been shock frozen in liquid nitrogen and stored at −80°C were thawed, and pressure sap was gained with a home-built pressing device. The sap was kept on ice and centrifuged (5 min, 5,000g), and the osmolality (osmol kg−1) of the supernatant was determined in a Cryo-osmometer (Osmomat 030; Ganotec). To convert differences in water potential into osmotic pressure, the following equation was used: 1 osmol = 0.00832 × Tabs MPa, where Tabs = 293.15 K.
Element Analysis
Plant tissues were dried at 70°C and subsequently digested by using the nitric acid pressure system according to Heinrichs et al. (1986). Quantification of elements was carried out by inductively coupled plasma-optical emission spectrometry (Spectro Analytical Instruments) at λ = 559 nm.
To determine cation solute concentrations, the measured concentrations of Na+, K+, Ca2+, and Mg2+ were summed up and expressed on the basis of the water content of the tissue (mm). The water content was determined as follows: (fresh mass – dry mass)/dry mass (mL g−1).
Analysis of Leaf Metabolites by Gas Chromatography-Mass Spectrometry
The extraction protocol was modified from Roessner-Tunali et al. (2003). Briefly, frozen leaf tissue powder (100 mg dry mass) was extracted in 1.4 mL of 80% (v/v) aqueous methanol in a 2.0-mL microcentrifuge tube for 2 h by shaking (200 rpm) at ambient temperature. A total of 120 μL of ribitol (0.2 mg mL−1 water) was added as an internal standard prior to incubation. The mixture was extracted for 15 min at 70°C. The extract was vigorously mixed with 1,500 μL of water and subsequently centrifuged for 15 min at 4,000 rpm. Aliquots of the methanol/water supernatant (150 μL) were dried in vacuum overnight. The dry residue was modified for gas chromatography-mass spectrometry analysis according to Fiehn et al. (2000). Residues after reduction were redissolved and derivatized for 90 min at 37°C (in 60 μL of 30 mg mL−1 methoxyamine hydrochloride in pyridine) followed by a 30-min treatment with 120 μL of N-methyl-N-[trimethylsilyl]trifluoroacetamide at 37°C. A total of 10 μL of a retention time standard mixture (0.029% [v/v] n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotracontane, and n-hexatriacontane dissolved in pyridine) was added before trimethylsilylation. Sample volumes of 1 μL were then injected onto the gas chromatograph column on a splitless mode.
The gas chromatography-mass spectrometry system was composed of a Pal autosampler (Ophir Analytic), a TRACE GC 2000 gas chromatograph, and a TRACE DSQ quadrupole mass spectrometer (Bargal Analytics). The mass spectrometer was tuned according to the manufacturer’s recommendations. Gas chromatography was performed on a 30-m Rtx_5Sil MS column with 0.25-μm film thickness (Restek). The injection temperature was set at 230°C, the interface at 250°C, and the ion source adjusted to 200°C. Helium was used as the carrier gas at a flow rate of 1 mL min−1. The analysis was performed under the following temperature program: 5 min of isothermal heating at 70°C, followed by a 5°C min−1 oven temperature ramp to 350°C, and a final 5-min heating at 330°C. The system was then temperature equilibrated for 1 min at 70°C before injection of the next sample. Mass spectra were recorded at two scans per second with a scanning range of 50 to 600 mass-to-charge ratio. Both chromatograms and mass spectra were evaluated using the XCALIBUR version 1.3 program (ThermoFinnigan). A retention time and mass spectral library for automatic peak quantification of metabolite derivatives was implemented within the XCALIBUR method format. Substances were identified by comparison with authentic standards, as described by Roessner-Tunali et al. (2003).
Identified peak areas were normalized based on the internal standard and dry weight and analyzed statistically with the Tukey multiple comparison algorithm (P = 0.05) incorporated into the JMP statistical software (Statistics and Graphics Guide: Version 3; SAS Institute).
Statistical Analyses of Physiological Data
Data are shown as means ± se. Six plants were analyzed per time point. Statistical analysis (ANOVA) was performed using Statgraphics (Centurion XV). Significant differences at P ≤ 0.05 were obtained after ANOVA by a least significant difference test. Asterisks in figures are as follows: * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
Microarray Analyses
RNA was isolated from P. euphratica leaves or roots using the method described by Chang et al. (1993). RNA samples from three biological replicates were used; each of the independent biological samples consisted of leaves or roots from six plants. To avoid bias in the microarray evaluation as a consequence of dye-related differences in labeling efficiency and/or differences in recording fluorescence signals, dye labeling for each paired sample was reversed in two individual hybridizations on microarrays containing 7,662 different Populus ESTs, among them 6,320 ESTs from P. euphratica. The EST libraries were constructed from different tissues and stressed plants and used to produce microarrays on glass slides as described by Brosché et al. (2005). A total of six hybridizations per time point were obtained. The complete protocols for probe labeling and hybridization, normalized data, and raw data files are available from the ArrayExpress database (www.ebi.ac.uk/arrayexpress/) under the accessions of E-MEXP-2095. By the use of P. euphratica arrays, problems that may arise by low hybridization efficiencies caused by sequence divergence are avoided (Janz et al., 2010).
Images were analyzed in GenePixPro 5.1 (Axon Instruments). Visually bad spots or areas on the array and low-intensity spots were excluded. Low-intensity spots were determined as spots where less than 55% of the pixels had intensity above the background + 1 sd in either channel. Slides with hybridized microarrays were scanned sequentially for Cy3- and Cy5-labeled probes. Raw expression data were normalized for sources of systematic variation using a normalization model based on Wolfinger et al. (2001), which is a modified version of the global ANOVA model of Kerr and Churchill (2001). The log2-transformed raw data are represented by y and subjected to the normalization model ygijk = μ + Ti + Aj + Dk + (TA)ij + (DA)ij + εgijk, where μ represents an overall mean value, T the fixed treatment effect, A the random array effect, D the fixed dye effect, TA the random interaction effect of arrays and treatments, DA the random interaction effect of arrays and dyes, and ε the residual stochastic error. Normalization was carried out using SAS. For each gene g, the empirical residuals calculated by subtraction of the fitted effects from the observed ygijk were used as normalized data in the subsequent significance analysis with SAM (Tusher et al., 2001). The SAM algorithm was obtained as a Microsoft Excel Add-in from http://www-stat.stanford.edu/~tibs/SAM/. It uses the false discovery rate and q value method presented by Storey (2002).
SAM was used with the option “paired data,” the Δ value was chosen as the false discovery rate became 0, and the limit of fold change was set to 3. The fold changes in gene expression between the control and salt-treated samples for each harvesting time point were estimated after normalization as the ratio of the mean signal intensities.
Six arrays were used for each time point (except for root after 12 h, where two technical replicates were missing) and probed with samples of the three biological replications.
The normalization method described above led to homogenous array means of log2-transformed raw data, and not all effects in the normalization model could be tested because of missing degrees of freedom. Therefore, the simpler normalization model ygijk = μ + Ti + Aj + Dk + εgijk was tested. In this case, all effects were significant at the 5% error level, but array means were no longer homogenous after normalization. The sets of genes recognized as significant by the SAM algorithm for the normalization model with interactions were very similar to those based on the simpler normalization model. They differed negligibly in the number of significant genes, and they always were subsets of each other. Therefore, we always refer to the results based on the normalization model.
Expression Analysis by Quantitative Real-Time PCR
The transcript levels of five genes, which showed significant changes in response to salt stress in the microarray experiments, were confirmed by quantitative real-time PCR. RNA was isolated according to the protocol of Chang et al. (1993). DNA was removed with DNase I (Fermentas), and first-strand cDNA was reverse transcribed from 5 μg of RNA using reverse transcriptase (Fermentas) according to the manufacturer’s recommendations. Gene-specific primers were designed by using the Primer3 software (Rozen and Skaletsky, 2000). The relative transcript abundance was detected by the iCycler using iQ SYBR Green Supermix (Bio-Rad). The tubulin amplicon was used as an internal control for normalization.
Analysis of Arabidopsis T-DNA Insertion Lines
Arabidopsis (Arabidopsis thaliana) lines with T-DNA insertions in the genes At5g58070.1 (Salk_136775 = Attil1-1, SALK_150259 = Attil1-2) and At5g02020 (Salk_146631 = Atsis1-1, Salk_064028 = Atsis1-2) were obtained from the Nottingham Arabidopsis Stock Center. Seeds of the segregating T3 generation were sown on half-strength Murashige and Skoog medium supplemented with 50 μg mL−1 kanamycin as the selectable marker. DNA of kanamycin-resistant plants was isolated with the DNeasy Kit (Qiagen) according to the protocol of the manufacturer. Homozygous lines were identified by PCR analysis using gene-specific primers (RP-TIL, LPTIL, RP-SIS, LP-SIS) and the left border primer LBa1 (Supplemental Table S4). PCR was performed with a 5-min initial denaturation step at 94°C and 40 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C. PCR analysis revealed the T-DNA insertion into each target gene. Using the gene-specific primers exp_RP and exp_LP did not result in a PCR product, indicating that the selected lines Attil1-1, Attil1-2, Atsis1-1, and Atsis2-1 were homozygous for the T-DNA insertion.
To test salt responses of soil-grown plants, Arabidopsis wild-type and T-DNA lines were germinated for 10 d on water-agar (16 h of light at 150 μmol m−2 s−1 photosynthetically active radiation, 25°C, 60% relative air humidity). Uniform seedlings were planted into soil (T25 Feinerde; Frühstorfer Erde) and grown in a climatized cabinet under short-day conditions (8 h light at 115 μmol m−2 s−1 photosynthetically active radiation/16 h of darkness, 20°C air temperature, 60% relative air humidity). After 10 d, the plants were gradually exposed to NaCl (50 mm for 5 d, 100 mm for 5 d, and 200 mm subsequently) and harvested after 10 d of exposure to 200 mm NaCl.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genevestigator analyses (development, anatomy) of orthologs in Arabidopsis of P. euphratica salt-responsive genes in roots and leaves, respectively.
Supplemental Figure S2. Changes in protein content in response to salt in leaves and roots of P. euphratica.
Supplemental Table S1. Changes in metabolites in response to salt exposure.
Supplemental Table S2. Genes with significantly changed transcript abundance in salt-exposed compared with control roots of poplar.
Supplemental Table S3. Comparison of quantitative real-time PCR of selected genes with their fold changes on the microarrays, and primers used for quantitative real-time PCR.
Supplemental Table S4. Primers used for the analysis of Arabidopsis T-DNA insertion mutants of TIL and SIS.
Supplementary Material
Acknowledgments
C. Kettner, S. Elend, and T. Klein (Laboratory for Radioisotopes) provided excellent technical assistance.
References
- Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Bogeat-Triboulot MB, Brosché M, Renaut J, Jouve L, Le Thiec D, Fayyaz P, Vinocur B, Witters E, Laukens K, Teichmann T, et al. (2007) Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol 143: 876–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C. (2005) Early effects of salinity on water transport in Arabidopsis roots: molecular and cellular features of aquaporin expression. Plant Physiol 139: 790–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosché M, Vinocur B, Alatalo ER, Lamminmäki A, Teichmann T, Ottow EA, Djilianov D, Afif D, Bogeat-Triboulot MB, Altman A, et al. (2005) Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol 6: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Purear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Charron JB, Ouellet F, Houde M, Sarhan F. (2008) The plant apolipoprotein D ortholog protects Arabidopsis against oxidative stress. BMC Plant Biol 8: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Moshelion M, Daniels MJ. (2005) Regulation of plant aquaporin activity. Biol Cell 97: 749–764 [DOI] [PubMed] [Google Scholar]
- Chen SL, Li J, Wang S, Hüttermann A, Altman A. (2001) Salt nutrient uptake and transport, and ABA of Populus euphratica, a hybrid in response to increasing soil NaCl. Trees (Berl) 15: 186–194 [Google Scholar]
- Chen SL, Polle A. (2010) Salinity tolerance of Populus. Plant Biol (Stuttg) 12: 317–333 [DOI] [PubMed] [Google Scholar]
- Cochard H, Forestier S, Améglio T. (2001) A new validation of the Scholander pressure chamber technique based on stem diameter variations. J Exp Bot 52: 1361–1365 [PubMed] [Google Scholar]
- Ding MQ, Hou PC, Shen X, Wang MJ, Deng SR, Sun J, Xiao F, Wang RG, Zhou XY, Lu CF, et al. (2010) Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol Biol 73: 251–269 [DOI] [PubMed] [Google Scholar]
- Ehlting B, Dluzniewska P, Dietrich H, Selle A, Teuber M, Hänsch R, Nehls U, Polle A, Schnitzler JP, Rennenberg H, et al. (2007) Interaction of nitrogen nutrition and salinity in grey poplar (Populus tremula × alba). Plant Cell Environ 30: 796–811 [DOI] [PubMed] [Google Scholar]
- El-Khatib RT, Hamerlynck EP, Gallardo F, Kirby EG. (2004) Transgenic poplar characterized by ectopic expression of a pine cytosolic glutamine synthetase gene exhibits enhanced tolerance to water stress. Tree Physiol 24: 729–736 [DOI] [PubMed] [Google Scholar]
- FAO (2008) FAO Land and Plant Nutrition Management Service. http://www.fao.org/ag/agl/agll/spush (October 29, 2010)
- Fayyaz P. (2008) Effects of salt stress on ecophysiological and molecular characteristics of Populus euphratica Oliv., Populus x canescens (Aiton) Sm. and Arabidopsis thaliana L. PhD thesis Cuivillier Verlag, Goettingen, Germany [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. (2000) The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta 1482: 9–24 [DOI] [PubMed] [Google Scholar]
- Frenette Charron JB, Breton G, Badawi M, Sarhan F. (2002) Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett 517: 129–132 [DOI] [PubMed] [Google Scholar]
- Frenette Charron JB, Ouellet F, Pelletier M, Danyluk J, Chauve C, Sarhan F. (2005) Identification, expression, and evolutionary analyses of plant lipocalins. Plant Physiol 139: 2017–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong QQ, Li PH, Ma SS, Indu Rupassara S, Bohnert HJ. (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Gu R, Fonseca S, Puskás LG, Hackler L, Jr, Zvara A, Dudits D, Pais MS. (2004) Transcript identification and profiling during salt stress and recovery of Populus euphratica. Tree Physiol 24: 265–276 [DOI] [PubMed] [Google Scholar]
- Hara M, Terashima S, Fukaya T, Kuboi T. (2001) Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. Plant Physiol 158: 1333–1339 [Google Scholar]
- Heinrichs H, Brumsack HJ, Loftfield N, König N. (1986) Verbessertes Druckaufschlussystem für biologische und anorganische Materialien. Z Pflanzenernaehr Bodenkd 149: 350–353 [Google Scholar]
- Hewitt EJ, Smith TA. (1975) Plant Mineral Nutrition. English Universities Press, London [Google Scholar]
- Hoshida H, Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Takabe T, Takabe T. (2000) Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol Biol 43: 103–111 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz D, Behnke K, Schnitzler J-P, Kanawati B, Schmitt-Kopplin P, Polle A. (2010) Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biol 10: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Uemura M. (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36: 141–154 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. (2001) Statistical design and the analysis of gene expression microarray data. Genet Res 77: 123–128 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D. (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Brasiliero A, Michel MF, Delmotte F, Jouanin L. (1992) Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep 11: 137–141 [DOI] [PubMed] [Google Scholar]
- Luo ZB, Janz D, Jiang XN, Göbel C, Wildhagen H, Tan YP, Rennenberg H, Feussner I, Polle A. (2009) Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol 151: 1902–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ. (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. (2006) Dissecting salt stress pathways. J Exp Bot 57: 1097–1107 [DOI] [PubMed] [Google Scholar]
- Martinez-Beltran J, Manzur CL. (2005) Overview of salinity problems in the world and FAO strategies to address the problem. Proceedings of the International Salinity Forum. Riverside Publishing, Riverside, CA, pp 311–315 [Google Scholar]
- Maxwell K, Johnson GN. (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Mouillon JM, Eriksson SK, Harryson P. (2008) Mimicking the plant cell interior under water stress by macromolecular crowding: Disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol 148: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon JM, Gustafsson P, Harryson P. (2006) Structural investigation of disordered stress proteins: comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol 141: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. (2005) Genes and salt tolerance: bringing them together. New Phytol 167: 645–663 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Olsson ASB, Engström P, Söderman E. (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55: 663–677 [DOI] [PubMed] [Google Scholar]
- Ottow EA, Brinker M, Teichmann T, Fritz E, Kaiser W, Brosché M, Kangasjärvi J, Jiang X, Polle A. (2005) Populus euphratica displays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress. Plant Physiol 139: 1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne PL, Kaikuranta PL, van der Plas LH, van der Schoot C. (1999) Dehydrins in cold-acclimated apices of birch (Betula pubescens Ehrh.): production, localization and potential role in rescuing enzyme function during dehydration. Planta 209: 377–388 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Krawetz S, Misener S, , Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Scholander PF, Bradstreet ED, Hemmingsen EA, Hammel HT. (1965) Sap pressure in vascular plants: negative hydrostatic pressure can be measured in plants. Science 148: 339–346 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA. (2008) Potassium transport and plant salt tolerance. Physiol Plant 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Shangari N, Bruce WR, Poon R, O’Brien PJ. (2003) Toxicity of glyoxals: role of oxidative stress, metabolic detoxification and thiamine deficiency. Biochem Soc Trans 31: 1390–1393 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Söderman E, Mattsson J, Engström P. (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J 10: 375–381 [DOI] [PubMed] [Google Scholar]
- Stanne TM, Sjögren LLE, Koussevitzky S, Clarke AK. (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc B 64: 479–498 [Google Scholar]
- Sunkar R, Bartels D, Kirch HH. (2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 35: 452–464 [DOI] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinocur B, Altman A. (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16: 1–10 [DOI] [PubMed] [Google Scholar]
- Wang R, Chen S, Deng L, Fritz E, Hüttermann A, Polle A. (2007) Leaf photosynthesis, fluorescence response to salinity and the relevance to chloroplast salt compartmentation and anti-oxidative stress in two poplars. Trees (Berl) 21: 581–591 [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Katsumi K, Yuji I, Sasaki S. (2001) Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tissue Organ Cult 63: 199–206 [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, Guevara D, Nuin P, Whitty B, Diaz C, Golding GB, Gray GR, Weretilnyk EA, et al. (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Blumwald E. (2005) Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10: 615–620 [DOI] [PubMed] [Google Scholar]
- Zheng B, MacDonald TM, Sutinen S, Hurry V, Clarke AK. (2006) A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224: 1103–1115 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.