Abstract
Glutathione S-transferases (GSTs) have been well documented to be involved in diverse aspects of biotic and abiotic stresses, especially detoxification processes. Whether they regulate plant development remains unclear. Here, we report on our isolation by reverse transcription-polymerase chain reaction of a plant GST, AtGSTU17, from Arabidopsis (Arabidopsis thaliana) and demonstrate that its expression is regulated by multiple photoreceptors, especially phytochrome A (phyA) under all light conditions. Further physiological studies indicated that AtGSTU17 participates in various aspects of seedling development, including hypocotyl elongation, anthocyanin accumulation, and far-red light-mediated inhibition of greening with a requirement of functional phyA. The loss-of-function mutant of AtGSTU17 (atgstu17) resulted in reduced biomass of seedlings and number of lateral roots in the presence of auxin, as well as insensitivity to abscisic acid (ABA)-mediated inhibition of root elongation, with similarity to different phyA mutant alleles. Moreover, the root phenotype conferred by atgstu17 was reflected by histochemical β-glucuronidase staining of AtGSTU17 promoter activity with the addition of auxin or ABA. Further microarray analysis of wild-type Columbia and atgstu17 seedlings treated with far-red irradiation or ABA revealed that AtGSTU17 might modulate hypocotyl elongation by positively regulating some light-signaling components and negatively regulating a group of auxin-responsive genes and modulate root development by negatively controlling an auxin transport protein in the presence of ABA. Therefore, our data reveal that AtGSTU17 participates in light signaling and might modulate various aspects of Arabidopsis development by affecting glutathione pools via a coordinated regulation with phyA and phytohormones.
Glutathione S-transferases (GSTs; E.C. 2.5.1.18) are a superfamily of multifunctional, dimeric enzymes, best known for their role in enzymatic detoxification of endo- and xenobiotics (Moons, 2005). Plant GSTs are divided into six classes, namely, phi (F), tau (U), zeta (Z), theta (T), lamba (L), and dehydroascorbate reductases (Dixon et al., 2002b; Moons, 2005). The large numbers of F and U classes are plant specific and have major roles in herbicide detoxification (Edwards et al., 2000; Dixon et al., 2002b; DeRidder and Goldsbrough, 2006). Recent evidence indicates that GSTs are also involved in endogenous metabolism, including oxidative stress, flavonoid binding, and regulation of apoptosis (Marrs et al., 1995; Alfenito et al., 1998; Mueller et al., 2000; Kilili et al., 2004). Classes Z and T are plant homologs of mammals and other organisms and function in primary metabolism such as isomerization of maleylacetoacetate and detoxification of hydroperoxides formed during oxidative stress (Dixon et al., 2000; Kampranis et al., 2000). Classes L and dehydroascorbate reductases are newly found in plants and function in redox homeostasis (Dixon et al., 2002a; Edwards et al., 2005). Thus, the functions of plant GSTs are diverse and might be due to the ability to conjugate glutathione (GSH) to various targets involved in biotic and abiotic stress.
In addition to having functions in various stresses, plant GSTs appear to be involved in plant growth and development (Gong et al., 2005; Moons, 2005). They have been shown to bind hormones such as auxin and cytokinin (Zettl et al., 1994; Gonneau et al., 1998) and can be induced by a wide variety of phytohormones, including ethylene, auxin, methyl jasmonate, salicylic acid, and abscisic acid (ABA; Wagner et al., 2002; Moons, 2003; Smith et al., 2003). That all these hormones regulate many aspects of plant development implies that plant GSTs may play vital roles in plant growth and development as well. However, evidence to substantiate this role has been limited.
Several plant GSTs have been shown to be induced by different qualities of light (Loyall et al., 2000; Tepperman et al., 2001; Chen et al., 2007). PcGST1 isolated from parsley (Petroselinum crispum) cell cultures by fluorescent differential display was induced by UV-B light and involved in a UV light-dependent signaling pathway to chalcone synthase (Loyall et al., 2000). Tepperman et al. (2001) used a DNA microarray approach to examine the gene expression profiles induced rapidly by far-red (FR) light irradiation. The expression of one GST, AAD32887, increased rapidly but was inhibited by phytochrome A (phyA) mutation (Tepperman et al., 2001). Our previous studies have shown that AtGSTU20 (At1g78370/FIN219-interacting protein1 [FIP1]) can interact in vitro and in vivo with FIN219 (Chen et al., 2007), which functions in a phyA-mediated FR signaling pathway (Hsieh et al., 2000). Moreover, transgenic Arabidopsis (Arabidopsis thaliana) seedlings overexpressing or reducing FIP1 expression exhibited a hyposensitive long-hypocotyl phenotype under continuous FR (cFR) light (Chen et al., 2007). These data indicate that some plant GSTs are regulated by light. However, the functional roles of these GSTs involved in light signaling remain to be elucidated.
To further understand the functional mechanisms of plant GSTs in light signaling pathways, we focused on several candidates affected by phyA or FIN219. Here, we report on functional studies of the GST AAD32887/At1g10370/AtGSTU17 previously detected by microarray assay (Tepperman et al., 2001) and down-regulated by fin219 mutation in FR (H.-J. Chen and H.-L. Hsieh, unpublished data). Our data presented here using transgenic plants and molecular genetic approaches provide further insight into possible functions of AtGSTU17 involved in light signaling, especially phyA-mediated photomorphogenesis, and in the integration of various phytohormones to modulate GSH homeostasis in the regulation of Arabidopsis development.
RESULTS
Expression of AtGSTU17 Is Regulated by Multiple Photoreceptors
To further confirm the expression patterns of FR-regulated AtGSTU17 transcripts, we performed dark-light transition experiments. Wild-type and phyA mutant seedlings were grown in the dark for 2 d, then transferred to FR light for various times; AtGSTU17 expression was examined by RNA gel-blot analysis. AtGSTU17 was induced in 2-d-old wild-type seedlings transferred from the dark to FR light for 1 h, and the level peaked with 6 h FR light; the expression was gradually reduced to constant levels for the remaining FR irradiation periods (Fig. 1A, left section). However, in the phyA mutant seedlings, AtGSTU17 induction by 1 and 6 h FR irradiation was substantially reduced (Fig. 1A, right section), which indicates that AtGSTU17 is indeed induced rapidly by FR, and its expression depends on PHYA.
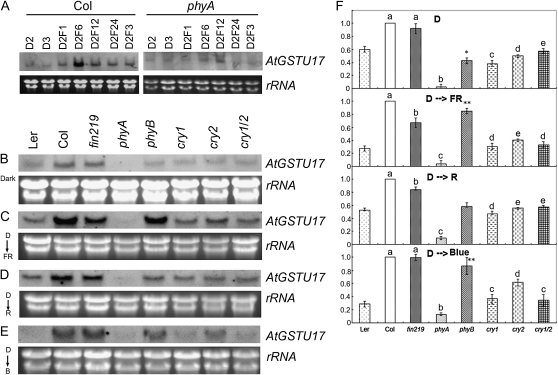
Figure 1.
AtGSTU17 expression is regulated by different photoreceptors. A, RNA gel-blot analysis of AtGSTU17 expression in wild-type Col and the phyA mutant by dark FR light transition. Seedlings of Col and phyA were grown in the dark for 2 d (D2) or 3 d (D3) and then transferred to FR light for 1 h (D2F1), 6 h (D2F6), 12 h (D2F12), 24 h (D2F24) h, or 3 d (D2F3). B to E, RNA gel-blot analysis of AtGSTU17 expression in various photoreceptor mutants under the transition from 3 d dark (B) to 6 h FR light (C), red light (D), and blue light (E). Twenty micrograms of total RNA isolated from treated seedlings were loaded onto each lane and used for RNA gel-blot analyses. The probe is the Dig-labeled 3′ UTR of AtGSTU17. Ribosomal RNA (rRNA) levels in the ethidium bromide-stained gel were used for a loading control. All experiments were repeated twice independently. Light conditions: FR light (1.43 μmol m−2 s−1), red light (16.71 μmol m−2 s−1), and blue light (3.75 μmol m−2 s−1). F, Quantitative representation of AtGSTU17 expression levels shown in B to E. The level of AtGSTU17 expression in Col was set at 1 under the respective conditions. Light conditions: D, dark; R, red; B, blue. Different letters represent statistically different means (P < 0.05). Asterisks indicate significant difference (** at P < 0.01, n = 30; * at P < 0.05, n = 30) compared to wild-type Ler.
To further understand whether AtGSTU17 expression is regulated by other light photoreceptors and different qualities of light, we performed dark-light transition experiments by growing various photoreceptor mutant seedlings in the dark for 3 d, then transferring them to different colors of light for 6 h; the expression of AtGSTU17 transcripts was examined by RNA gel-blot analysis with a gene-specific probe. AtGSTU17 transcripts in the phyA mutant were barely detected under all light conditions, including darkness (Fig. 1, B–F), which implies that AtGSTU17 expression depends strictly on functional PHYA. Moreover, AtGSTU17 expression was also reduced in cry1, cry2, and cry1cry2 (cry1/2) double mutants in the dark (Fig. 1, B and F). In contrast, the level of AtGSTU17 transcripts in fin219 remained comparable to that in Columbia (Col) and was slightly reduced in the phyB mutant as compared with its ecotype Landsberg erecta (Ler) in the dark (Fig. 1, B and F). In the transition of dark-grown seedlings to 6 h FR irradiation, the level of AtGSTU17 transcripts was decreased in fin219, cry1, cry2, and cry1/2 mutants (Fig. 1, C and F) but substantially increased in the phyB mutant under the same conditions (Fig. 1, C and F). In the transition from dark to red light, the level of AtGSTU17 transcripts appeared to be reduced in cry1, cry2, and cry1/2 mutants (Fig. 1, D and F) but remained largely the same in the phyB mutant as in the Ler ecotype, which implies that PHYB under red light may play a lesser role in the regulation of AtGSTU17 expression. As well, the AtGSTU17 transcript level was slightly lower in fin219 than in Col (Fig. 1, D and F). In the transition from dark to blue light, the AtGSTU17 transcript level was decreased in cry1, cry2, and cry1/2 double mutants, with a substantial reduction in cry1 and cry1/2 (Fig. 1, E and F). Intriguingly, the level was markedly higher in the phyB mutant than in the Ler ecotype, in which AtGSTU17 transcripts were barely detected. In addition, AtGSTU17 expression was not affected by fin219 under the blue light transition (Fig. 1, E and F). Taken together, these expression data reveal that AtGSTU17 is differentially regulated by various photoreceptors and highly controlled by functional phyA.
AtGSTU17 Regulates Hypocotyl Elongation Mainly in FR Light
To further understand the functions of AtGSTU17 involved in light signaling, we cloned it by reverse transcription (RT)-PCR on the basis of sequence information deposited in the National Center for Biotechnology Information database. The full-length complementary DNA (cDNA) encodes a 227 amino acid protein and shares 74% identity at the amino acid level with AtGSTU18 (At1g10360) within the same U class (Supplemental Fig. S1). The recombinant proteins generated from an Escherichia coli expression system showed enzymatic activities to the substrates GSH and 1-chloro-2,4-dinitrobenzene (CDNB), with Km value 0.285 mm for GSH at 1.0 mm CDNB and 1.400 mm for CDNB at 1.0 mm GSH (Supplemental Fig. S1), which indicates that AtGSTU17 has higher affinity to both substrates than do other plant GSTs reported in the literature (23). Thus, AtGSTU17 has GST activities.
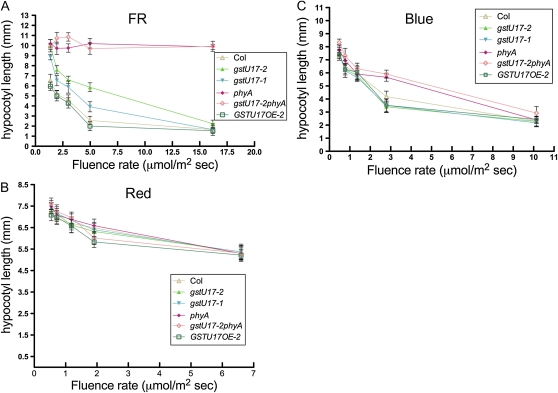
To further elucidate the functional regulation between AtGSTU17 and phyA, we used two T-DNA-inserted null mutants, SALK_139615 and SALK_025503, named atgstu17-1 and atgstu17-2 from the Arabidopsis Biological Resource Center (ABRC; Supplemental Fig. S2), a new phyA allele in a Col background (Supplemental Fig. S4), the double mutant gstu17-2phyA, and AtGSTU17-overexpressed transgenic lines in Col (GSTU17OE-2) for phenotypic examination under various light conditions. The atgstu17 mutants atgstu17-1 and atgstu17-2 exhibited a hyposensitive long-hypocotyl phenotype as compared with Col under low fluences of FR light (Figs. 2A and 3A) but not other light conditions (Figs. 2, B and C, and 3A). In contrast, GSTU17OE-2 displayed a short-hypocotyl phenotype comparable to that of Col under all light conditions examined (Figs. 2 and 3A). The double-mutant atgstu17phyA showed slightly longer hypocotyls than did phyA in the low FR fluence (<5 μmol m−2 s−1) and was much longer than atgstu17 under FR light (Figs. 2A and 3A) but did not display a significant hypocotyl phenotype under other light conditions, including darkness (Figs. 2, B and C, and 3A). The phenotype examination indicated that AtGSTU17 might have a function in the control of hypocotyl elongation in response to FR irradiation.
Figure 2.
The atgstu17 mutant displays a long-hypocotyl phenotype specifically in cFR light. Seedlings of wild-type Col, atgstu17 mutants (gstu17-1 and gstu17-2), phyA, the double mutant gstu17-2phyA, and AtGSTU17 overexpression line (GSTU17OE-2) were grown in different fluence rates of FR (A), red (B), and blue (C) light for 3 d, then hypocotyl lengths were measured. Mean hypocotyl lengths were calculated from at least 40 seedlings. Error bar represents sd from two independent experiments.
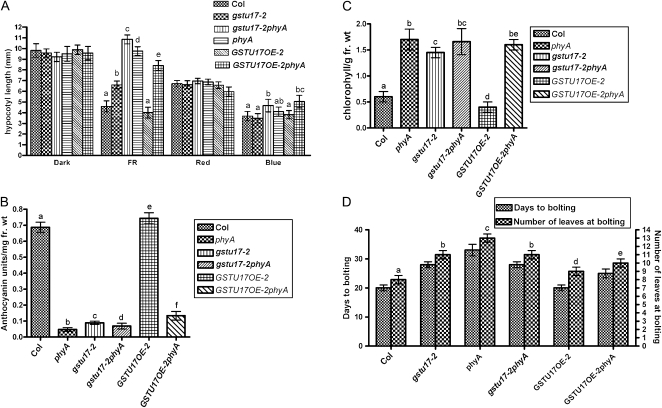
Figure 3.
AtGSTU17 shows a requirement for functional phyA in the regulation of various aspects of Arabidopsis development. A to C, Seedlings of Col, atgstu17 mutant (gstu17-2), the double mutant gstu17-2phyA, phyA, GSTU17OE-2, and GSTU17OE-2 in a phyA mutant background (GSTU17OE-2phyA) were grown in a low FR fluence rate (2 μmol m−2 s−1) for 3 d, then underwent measurement of hypocotyl lengths (A), anthocyanin accumulation (B), or chlorophyll content after 2 d exposure to white light (C). D, The same materials were grown in white light under long-day conditions until the inflorescent stem appeared. Days from seedlings sown in the soil to appearance of the first inflorescence and leaf number at bolting were recorded. All these experiments were replicated at least once. Error bar represents sd. Different letters represent statistically different means (P < 0.05).
AtGSTU17-Regulated Photomorphogenic Responses Require Functional phyA
Because AtGSTU17 is regulated by phyA and its loss-of-function or gain-of-function mutants show altered hypocotyl response to FR irradiation, we further examined whether changes in AtGSTU17 expression affected phyA-regulated responses. GSTU17OE-2 in the phyA mutant background (GSTU17OE-2phyA) showed a slightly shorter long-hypocotyl phenotype as compared with phyA but longer hypocotyls than atgstu17-2 under FR light (Fig. 3A); in contrast, GSTU17OE-2 in Col under the same condition exhibited a phenotype comparable to that of Col, which implies that AtGSTU17 function requires functional phyA to regulate hypocotyl elongation in response to FR light. Moreover, atgstu17 showed reduced anthocyanin accumulation and defects in FR-mediated blockage of greening regulated by phyA (Fig. 3, B and C). In contrast, GSTU17OE-2 in Col showed opposite effects on anthocyanin levels and chlorophyll content (Fig. 3, B and C). The double-mutant atgstu17phyA showed levels of anthocyanin and chlorophyll comparable to that of the respective single mutants atgstu17 and phyA (Fig. 3, B and C). GSTU17OE-2phyA showed defects in chlorophyll content and anthocyanin accumulation that were substantially different from those of GSTU17OE-2 in Col (Fig. 3, B and C), which suggests that the effects of AtGSTU17 on FR blockage of greening and anthocyanin accumulation require functional phyA. In addition, atgstu17 showed a delayed flowering phenotype similar to that of phyA (Fig. 3D). The double-mutant atgstu17phyA showed a flowering time similar to that of atgstU17. In contrast, GSTU17OE-2 exhibited a flowering time similar to that of wild-type Col, but GSTU17OE-2phyA showed a delayed flowering phenotype under long-day conditions (Fig. 3D). Thus, AtGSTU17 participates in the control of hypocotyl elongation, anthocyanin accumulation, FR blockage of greening, and flowering in a phyA-dependent manner.
AtGSTU17 Mutants Show Defects in Auxin- and ABA-Regulated Root Development
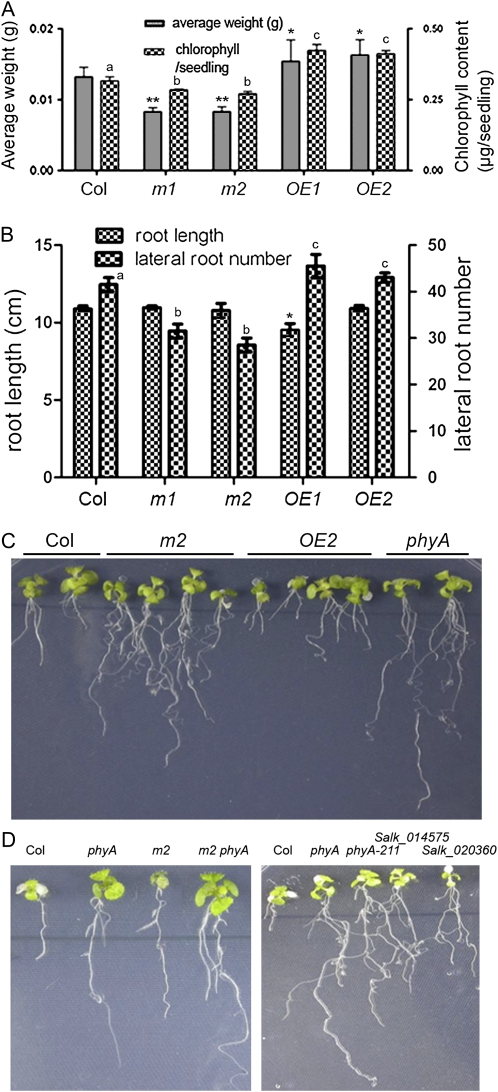
GSTs are induced by stresses from biotic and abiotic factors, including herbicides, chemical toxins, osmotic stress, and plant hormones (Marrs, 1996; Smith et al., 2003; DeRidder and Goldsbrough, 2006). Our studies by RNA gel-blot analysis also revealed AtGSTU17 transcripts up-regulated by ABA, 2,4-dichlorophenoxyacetic acid (2,4-D), and jasmonic acid (JA) treatment (Supplemental Fig. S3). Intriguingly, the induction of AtGSTU17 expression by 2,4-D and JA was enhanced in white light (Supplemental Fig. S3), which implies that AtGSTU17 function may involve interactions between light and phytohormones such as auxin and JA. In addition, atgstu17 mutant seedlings showed less biomass and chlorophyll content than those of Col in the presence of auxin (Fig. 4A); however, AtGSTU17 overexpression (OE1 and OE2) resulted in an opposite phenotype, with increased biomass and chlorophyll content under the same condition (Fig. 4A), which implies that AtGSTU17 is able to affect seedling development in response to auxin stimulation. Moreover, AtGSTU17 affected the generation of lateral roots in the presence of auxin, with reduced number of lateral roots in atgstu17 mutants and more lateral roots in OE1 and OE2, but did not largely influence the length of primary roots (Fig. 4B).
Figure 4.
Loss- and gain-of-function mutants of AtGSTU17 show changes in physiological responses and root development in response to auxin and ABA. A, Three-day-old seedlings of wild-type Col, atgstu17 mutants (m1 and m2), and AtGSTU17 overexpression lines (OE1 and OE2) grown in GM plates were transferred to vertical plates with 0.5 μm 2,4-D and incubated for another 10 d for measurement of biomass and chlorophyll content. Error bar represents sd. Different letters represent statistically different means (P < 0.05). Asterisks mark significantly different data at P < 0.001. B to D, Seedlings of 7-d-old Col, m1, m2, OE1, OE2, the double mutant atgstu17phyA, and different phyA alleles were transferred to vertical plates containing 0.01 μm 2,4-D (B) or 30 μm ABA (C and D) and incubated for 10 more days in white light for measurement of primary root length and number of lateral roots. Different letters represent statistically different means (P < 0.05). Asterisks indicate significantly different data at P < 0.05. n = 10.
Because AtGSTU17 was highly induced at 1 h after ABA treatment (Supplemental Fig. S3A), we wondered about a phenotypic response on exposure to ABA. Surprisingly, atgstu17 (m2) was less sensitive to ABA-mediated inhibition of root elongation as compared with Col (Fig. 4C). By contrast, GSTU17OE-2 (OE2) displayed a root length comparable to that of Col. Intriguingly, the phyA mutant showed insensitivity to ABA-mediated inhibition of root elongation as well (Fig. 4C). To substantiate the insensitivity of atgstu17 and phyA responses to ABA, the double-mutant atgstU17phyA and different phyA alleles, including phyA-211, Salk_014575, and Salk_020360, were included for further examination. atgstu17phyA (m2phyA) showed even longer primary roots and lateral roots than those in the respective single mutants (Fig. 4D, left section). As well, all the phyA alleles examined exhibited insensitivity to ABA inhibition of root elongation (Fig. 4D, right section). Thus, AtGSTU17 and PHYA might play a critical role in the control of root development regulated by auxin and ABA.
AtGSTU17 Mutants Affect GSH Homeostasis in Response to Auxin and FR Light Treatment
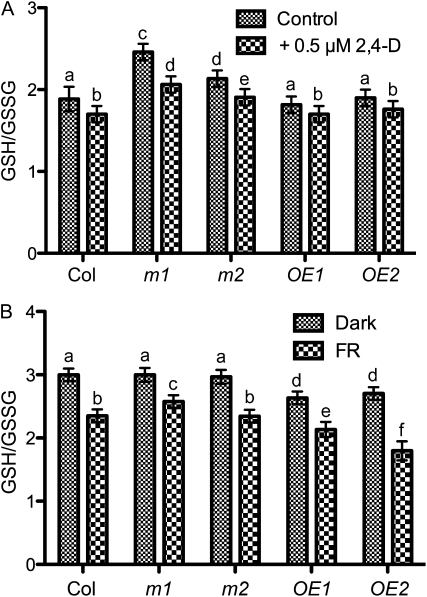
AtGSTU17 proteins showed enzymatic activity to GSH (Supplemental Fig. S1), which indicates that AtGSTU17 may regulate GSH pools in Arabidopsis under light and various hormone conditions. The reduced (GSH) and oxidized forms (GSSG) of GSH, as well as its redox state, play critical roles in the control of plant development in response to changes in light and environmental signals (Loyall et al., 2000; Belmonte et al., 2005; Foyer and Noctor, 2009). To associate the GSH/GSSG ratio with the atgstu17 mutant phenotype under cFR light and auxin treatment, we determined the GSH/GSSG ratio in seedlings of atgstu17 (m1 and m2) and GSTU17OE (OE1 and OE2). The GSH/GSSG ratio was higher in the atgstu17 mutant than in the wild-type Col in the absence of auxin; however, the GSH/GSSG ratio was decreased in atgstu17 in the presence of 0.5 μm 2,4-D but was still higher than that in Col under the same condition (Fig. 5A), which suggests that AtGSTU17 may act as a key component to regulate a GSH pool for fine tuning the GSH/GSSG ratio in cells. In contrast, the GSH/GSSG ratio in GSTU17OE (OE1 and OE2) was similar to that in Col in the control treatment and in the presence of auxin. Thus, AtGSTU17 is able to affect GSH/GSSG homeostasis in response to the phytohormone auxin.
Figure 5.
Ratio of GSH/GSSG in AtGSTU17 mutant and overexpressed seedlings. A, Seedlings of wild type (Col), atgstu17 mutants (m1 and m2), and AtGSTU17 overexpression lines (OE1 and OE2) grown in GM plates for 3 d were transferred to vertical plates without (Control) or with 0.5 μm 2,4-D and incubated for another 10 d for measurement of total GSH and GSSG. Error bar represents sd based on two biological replicates. Different letters represent statistically different means (P < 0.05). B, Wild type (Col), atgstu17 mutants (m1 and m2), and AtGSTU17 overexpression lines (OE1 and OE2) were grown in GM plates under darkness or FR light for 3 d, then used for measurement of total GSH and GSSG. Error bar represents sd based on two biological replicates. Different letters represent statistically different means (P < 0.05).
In addition, wild-type seedlings grown in cFR versus the dark control showed a significant reduction in GSH/GSSG ratio, which corresponds to a short-hypocotyl phenotype; however, atgstu17 (m1) showed slightly increased GSH/GSSG ratio as compared to Col in cFR. GSTU17OE (OE1 and OE2) showed a substantial reduction of GSH/GSSG ratio under the same condition (Fig. 5B). These data are consistent with a role of AtGSTU17 induced by FR irradiation and leading to a decrease in GSH/GSSG ratio. By contrast, the GSH/GSSG ratio for atgstu17 grown in the dark did not differ from that of Col, but GSTU17OE showed a reduction in GSH/GSSG ratio (Fig. 5B). Thus, AtGSTU17 is highly regulated by the FR light photoreceptor phyA (Fig. 1) and may play a more prominent role in the regulation of GSH pools in FR light than in the dark.
AtGSTU17 Is Highly Expressed in the Initiation Site of Lateral Roots in Response to ABA
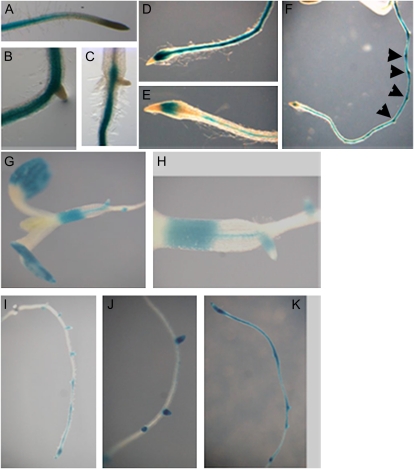
Further examination of the promoter activities of AtGSTU17 by histochemical GUS staining revealed that AtGSTU17 was mainly expressed in the maturation zone of roots and the basal region of hypocotyls (Fig. 6, A–C). In the presence of auxin, the staining pattern was extended to the meristematic zone with enhanced stains in the initiation sites of lateral roots (Fig. 6, D–F). By contrast, in the presence of ABA, GUS staining was detected in cotyledons, the elongation region (upper portion) and vascular tissue of hypocotyls, the profound initiation sites of lateral roots, and the elongation zone, as well as the meristematic zone of roots (Fig. 6, G–K). Thus, AtGSTU17 expression patterns appear to be associated with phenotypic responses to both auxin and ABA.
Figure 6.
Histochemical GUS staining of AtGSTU17 promoter activities. Transgenic seedlings harboring the construct of AtGSTU17 promoter fused with GUS were grown in GM plates without 2,4-D (A–C) or with 1 μm 2,4-D (D–F) or 30 μm ABA (G–K) under white light for 1 d and then subjected to GUS staining. Arrowheads indicate the initiation sites of lateral roots.
AtGSTU17 Mutation Results in Misregulation of Light-Responsive Genes, Auxin-Induced Genes, and Transcription Factors
To reveal the molecular mechanisms underlying atgstu17 insensitivity to FR and ABA responses, we performed microarray analysis using wild-type Col and mutant seedlings grown under cFR or ABA treatment. The mutant atgstu17 in cFR light showed altered expression of 1,332 genes, with at least 2-fold difference in expression as compared with Col; example genes included some light-regulated genes such as positive-regulator FAR1-related Sequence (FRS8; At1g80010), negative-regulator Suppressor of PHYA-105 (SPA1; At2g46340) of photomorphogenesis, Chalcone Isomerase, and key regulators of flowering time, AGAMOUS (At4g18960) and FLOWERING LOCUS C (FLC; At5g10140). Surprisingly, the expression of phototropism-related genes such as Non-phototropic Hypocotyl3 (NPH3; At3g08660) and NPH1 (PHOT1; At3g45780) was affected in atgstu17 under cFR (see Supplemental Table S1). As well, more than 20 auxin-responsive genes, including IAA5 (At1g15580), IAA6 (At1g52830), and SAURs (At3g03830; At4g13790; At4g38850) were up-regulated in atgstu17 under cFR light, which implies that AtGSTU17 negatively regulates the expression of these auxin-responsive genes under cFR light. In addition, atgstu17 showed altered positive or negative expression of more than 15 transcription factors (Supplemental Table S1). Moreover, atgstu17 under ABA treatment showed altered levels of 1,850 genes, with at least 2-fold difference in expression from that in Col; examples included greatly reduced expression of florigen gene FT (At1g65480), AGAMOUS (At4g18960), and several light-regulated receptor kinases (Supplemental Table S2A). In contrast, SPA1 was up-regulated in atgstu17 under ABA treatment (Supplemental Table S2B). Some auxin-responsive genes, including auxin transport protein PIN7 (At1g23080), were also affected in ABA-treated atgstu17. Intriguingly, 12 Myb-related transcription factors, one WRKY, and two bHLH transcription factors were down-regulated, and four Myb and three bZIP transcription factors were up-regulated in atgstu17 under ABA treatment. As well, the negative regulator of flowering time, FLC, and MADS-box proteins AGL3 and AGL13 were up-regulated in atgstu17 in the presence of ABA (Supplemental Table S2B). Intriguingly, 13% of the genes were the common genes affected by atgstu17 mutation in both FR and ABA conditions account. Thus, AtGSTU17 may function in upstream signaling networks to modulate various regulators, including transcription factors, in the control of diverse aspects of plant development via integration of FR light and ABA.
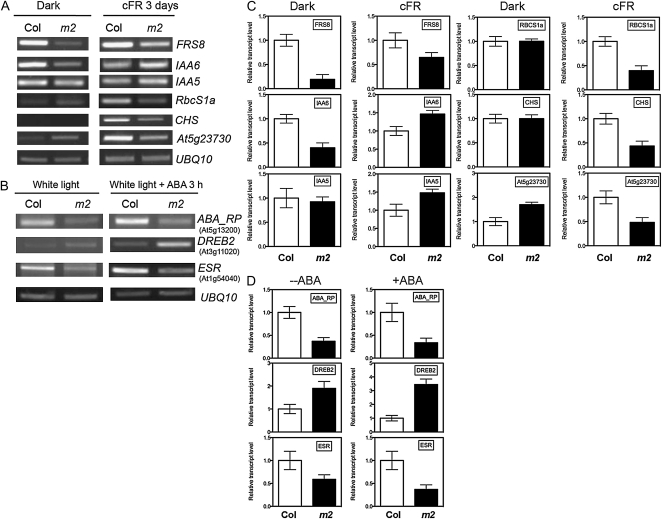
To validate the expression of the genes affected in atgstu17 revealed by microarray analysis, several light-regulated genes, including FRS8 (At1g80010), RbcS1a, CHS, and COP1 like (At5g23730); auxin-responsive genes such as IAA6 (At1g52830) and IAA5 (At1g15580); and ABA-regulated genes, including ABA_RP (At5g13200), DREB2B (At3g11020), and ESR (At1g54040), were selected for confirmation of transcript levels under FR or ABA treatment. The genes examined by RT-PCR showed expression patterns consistent with those from microarray analysis (Fig. 7), which indicates that AtGSTU17 can regulate a group of light-responsive genes positively and auxin-regulated genes negatively under FR light, as well as ABA-regulated genes, to modulate seedling development.
Figure 7.
Validation of the expression of some light-, auxin-, and ABA-responsive genes derived from microarray analysis. A and B, Seedlings of Col and atgstu17 were grown in the dark and cFR for 3 d (A) or white light with or without ABA (B) for 3 h and then underwent RT-PCR analysis with gene-specific primers for the genes shown in the figures. UBQ10, ubiquitin10 gene used for the internal control. C and D, Quantitative representation of the expression of the genes shown in A and B, respectively. The level of the indicated gene in Col was set at 1 under the respective conditions.
DISCUSSION
GSTs belong to a large gene family of 53 members in Arabidopsis and have diverse functions such as detoxification, metabolism, and defense in plants (Dixon et al., 2002b). Here we report that AtGSTU17, a member of the U class in Arabidopsis, is mainly involved in FR light signaling and is regulated by various photoreceptors, especially phyA, under all light conditions examined. Moreover, its function appears to require functional phyA in hypocotyl elongation, FR-mediated inhibition of greening (Barnes et al., 1996), anthocyanin accumulation, and flowering time (Fig. 3). As well, the double mutant atgstu17phyA showed defects in physiological responses, including slightly longer hypocotyls in low FR, reduced anthocyanin levels, and insensitivity in FR-mediated inhibition of greening, which suggests that AtGSTU17 might act in a parallel manner to or interact with phyA to modulate some phyA-mediated responses. Furthermore, microarray analysis revealed atgstu17 with up-regulated expression of at least 20 auxin-responsive genes and down-regulated expression of the FR signaling component FRS8 under FR conditions, which may contribute to combined effects on the long-hypocotyl phenotype of atgstu17 in FR. In addition, a suppressor of phyA-105 1 (SPA1; Hoecker et al., 1998) was up-regulated in atgstu17 under cFR light, which implies that phyA, AtGSTU17, and SPA1 might work in a feedback inhibition manner to maintain a dynamic balance for the regulation of photomorphogenesis.
In addition to its expression being regulated by PHYA, AtGSTU17 was affected positively by the blue-light photoreceptors cry1 and cry2 under all light conditions, especially blue light, in which the cry1 mutant showed greatly reduced AtGSTU17 mRNA levels (Fig. 1E). Thus, various photoreceptors, including phyB, may have specific effects on AtGSTU17 functions in addition to their combined regulation. The physiological significance of AtGSTU17 regulation by different photoreceptors might be related to the adaptation of certain stress conditions, as was revealed by Xu et al. (2009), who showed that wheat (Triticum aestivum) cryptochromes are involved in osmotic stress responses (Xu et al., 2009).
Plant GSTs are involved in a number of biotic and abiotic responses, which might be related to GSTs being able to conjugate GSH to different targets, including phytohormones, that regulate various aspects of plant development and thus may affect homeostasis of phytohormones such as auxin (Zettl et al., 1994) or cytokinin (Gonneau et al., 1998) and GSH within the cells or tissues. Changes in GSH levels have been implicated as affecting plant flowering (Ogawa et al., 2001), root elongation (Vernoux et al., 2000; Espunya et al., 2006), chloroplast transcription (Baena-González et al., 2001), and responses to pathogen infection (Gullner et al., 1999). The ROOT MERISTEMLESS1 (RML1) gene encoding the first enzyme of GSH biosynthesis regulates the establishment of an active postembryonic meristem in the root apex. The rml1 mutant is deficient in γ-glutamylcysteine synthetase activity and results in almost no GSH, thus leading to a very short root phenotype (Vernoux et al., 2000). GSH depletion by the inhibitor of its synthesis, buthionine sulfoximine, specifically reduces primary root growth and results in loss of the auxin efflux carriers PIN1, PIN2, and PIN7 from the root tips of primary roots, thus affecting auxin transport (Wiśniewska et al., 2006; Koprivova et al., 2010). Bashandy et al. (2010) reported that mutations of two key thiol reduction pathways, thioredoxin and GSH (the ntra ntrb cad2 mutant), resulted in a loss of auxin-regulated phenotypes such as apical dominance, vasculature, and secondary root production, and auxin transport and auxin levels, which implies that the GSH pool within cells is linked to auxin homeostasis. Moreover, alterations of the GSH redox state can improve apical meristem structure and somatic embryo quality in white spruce (Picea glauca; Belmonte et al., 2005). AtGSTU17, encoding a U class member of GST, has GST activity (Supplemental Fig. S1). It can be induced by ABA (Fig. 6, G–K; Supplemental Fig. S3), and the loss-of-function mutant exhibits a less-sensitive root phenotype to ABA-mediated inhibition of root elongation than does the wild type (Fig. 4, C and D). This phenotype might be due to high levels of GSH accumulating in the roots of the loss-of-function atgstu17 mutant as compared with the wild type under ABA treatment (Supplemental Fig. S5), which is consistent with high levels of endogenous GSH enhancing cell division in the root meristematic region leading to root elongation (Vernoux et al., 2000). Because promoter activity assay revealed AtGSTU17 expression in roots during seedling development (Fig. 6, A–C), AtGSTU17 expression was even enhanced in the meristematic zone and the initiation sites of lateral roots under auxin and ABA treatments (Fig. 6, D–F and I–K), which is consistent with high levels of GSH in atgstu17 seedlings treated with auxin or ABA (Supplemental Fig. S5). Intriguingly, microarray data revealed that atgstu17 showed substantially increased levels of PIN7 transcript (Wiśniewska et al., 2006; Supplemental Table S2), thus leading to more auxin flow toward the initiation sites of lateral roots under ABA treatment, which may lead to more lateral roots in the atgstu17 mutant in the presence of ABA (Fig. 4, C and D). This speculation is consistent with very recent reports indicating that the GSH level is linked to PIN proteins such as PIN7, and accordingly auxin transport (Bashandy et al., 2010; Koprivova et al., 2010). Surprisingly, the phyA mutant also showed an insensitive long-root phenotype in response to ABA-mediated inhibition of root elongation. Other phyA alleles, including phyA-211 and SALK_014575 (T-DNA insertion line) and SALK_020360, were also less sensitive to ABA inhibition of root elongation (Fig. 4, C and D). Moreover, phytochrome mutants were reported to differentially affect lateral root production and showed up-regulated expression of PIN3 and PIN7 transcripts (Devlin et al., 2003; Salisbury et al., 2007). Intriguingly, Sung et al. (2007) reported that a genetic screen identified an arsenic-tolerant double mutant with a single T-DNA insertion in the PHYA gene, which exhibited elevated levels of the thiols Cys, γ-glutamylcysteine, and GSH. This finding indicates that PHYA negatively regulates thiol synthesis (Sung et al., 2007), which is consistent with our data showing that AtGSTU17 transcripts are highly down-regulated in the phyA mutant (Fig. 1), corresponding to increased levels of GSH in the atgstu17 mutants. Thus, phyA might regulate root development by integrating with AtGSTU17 to fine tune GSH homeostasis in response to ABA.
As well, atgstu17 exhibited a long-hypocotyl phenotype under cFR and showed a slight increase of GSH/GSSG ratio compared to wild-type Col; furthermore, AtGSTU17 overexpression resulted in a significant reduction of GSH/GSSG ratio (Fig. 5B), which implies that AtGSTU17 can contribute to the regulation of the GSH/GSSG ratio; a high GSH/GSSG ratio is essential for cell division (Belmonte et al., 2005). However, AtGSTU17 in the dark may have a redundant effect with other GST members on GSH/GSSG ratio for hypocotyl elongation, which led to atgstu17 having a GSH/GSSG ratio similar to that of Col (Fig. 5B). In addition to the FR effect on GSH/GSSG ratio, application of exogenous auxin could decrease the GSH/GSSG ratio in the wild-type Col (Fig. 5A), which is consistent with data showing that AtGSTU17 is induced by auxin and has an enzymatic activity to the substrate GSH (Supplemental Figs. S1 and S3). In contrast, the atgstu17 mutant showed an increase of GSH/GSSG ratio in the absence or presence of auxin (Fig. 5A), which resulted in a significant decrease of biomass and chlorophyll content (Fig. 4A). This finding is probably due to excess oxidant status caused by higher concentration of auxin, such as 0.5 μm 2,4-D, which results in a redox imbalance toward oxidation. This speculation is supported by our observation of the atgstu17 mutant with more anthocyanin accumulation under the same condition (data not shown).
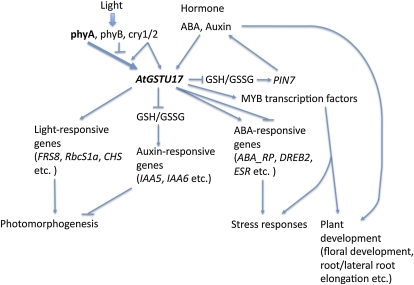
In addition, the atgstu17 mutant in the presence of ABA showed down-regulated levels of 12 MYB genes (Supplemental Table S2), which mainly participate in the regulation of the architecture of Arabidopsis inflorescence (Kirik et al., 1998), floral development (Baumann et al., 2007; Zhou et al., 2008; Zhang et al., 2009), phosphate starvation response (Devaiah et al., 2009), epidermal cell outgrowth (Jakoby et al., 2008), cell wall biosynthesis (Zhong et al., 2008), ABA signaling, and drought stress (Urao et al., 1996; Abe et al., 2003). In conclusion, AtGSTU17 regulated by various photoreceptors, especially phyA, and different phytohormones, including auxin and ABA, may have complicated functions regulating gene expression and plant development such as hypocotyl and root elongation, as well as stress responses by fine tuning GSH homeostasis and redox status in response to changes in light and environmental signals (Fig. 8).
Figure 8.
A model illustrates the possible functions of AtGSTU17 in Arabidopsis. Arabidopsis AtGSTU17 is differentially regulated by multiple photoreceptors, especially phyA under various light conditions, and by different phytohormones, including ABA and auxin. AtGSTU17 can fine tune GSH homeostasis and GSH/GSSG ratio to regulate the expression of PIN proteins such as PIN7 and auxin-responsive genes, as well as light-responsive genes, ABA-responsive genes, and a number of transcription factors, including MYBs for the modulation of photomorphogenesis, Arabidopsis development such as hypocotyl and root elongation, and stress responses. Bold arrow, highly positive effect; regular arrow, positive effect; inverted T, negative effect. [See online article for color version of this figure.]
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Col, Ler, Arabidopsis (Arabidopsis thaliana), and all mutants in the Col background, except for phyB in the Ler background, were used in this study. The T-DNA insertion lines atgstu17-1 (SALK_139615) and atgstu17-2 (SALK_025503) were obtained from ABRC. Wild-type Col, Ler, and mutant seeds were surface sterilized, sown onto the GM (Murashige and Skoog salts plus vitamins, 0.5 g/L MES buffer, pH 5.7, and 0.3% Suc) plates, and treated with various light wavelengths as described (Hsieh et al., 2000). Light fluence rates used in specific light qualities are in the figure legends. As well, homozygous atgstu17-1 and atgstu17-2 mutants were crossed to phyA to generate the homozygous double mutants atgstu17-1phyA and atgstu17-2phyA. For transgenic studies, at least T3 homozygous lines of seeds were grown for 3 d under different light conditions for phenotypic examination and RNA gel-blot analyses.
Generation of the Double Mutant atgstu17phyA
To generate the double mutant atgstu17phyA, T-DNA insertion lines of AtGSTU17, atgstu17-1, and atgstu17-2 were crossed with the phyA mutant. The resulting F2 progenies were screened for the fast-greening phenotype in white light of the phyA mutation after 3 d FR irradiation, and then possible candidates were transferred to soil for seed harvesting. At the adult stage, genotyping for T-DNA insertion of AtGSTU17 involved use of the genomic DNA as templates isolated from the leaves of the possible double mutants. The primers used for genotyping of homozygous atgstu17 were the left border primer of T-DNA, LBa1: 5′-TGGTTCACGTAGTGGGCCATCG-3′, and two AtGSTU17-specific primers, CDS of GST30-F: 5′-GGAGGATCCAGAGAAAATGGCAAGCA-3′ and GST30-R: 5′-AAACCTAGTCCACCAACCAATTGGGC-3′. Respective gene expression levels and genotyping in the F3 generation further confirmed the putative double mutant atgstu17phyA.
Plasmid Constructs
To generate AtGSTU17-overexpressed transgenic lines, a 721-bp full-length cDNA was amplified by PCR with the primers GST30-F 5′-GGAGGATCCAGAGAAAATGGCAAGCA-3′ and GST30-R 5′-AAACCTAGTCCACCAACCAATTGGGC-3′ based on the sequence information deposited in the database of the National Center for Biotechnology Information. The resulting DNA fragment was cloned into the BamH1/Mfe1 site of the binary vector pCAMBIA1390-cMyc. To establish the transgenic lines of the AtGSTU17 promoter::GUS, a 1.7-kb BamH1/EcoR1 fragment containing the AtGSTU17 promoter region upstream of the 5′ untranslated region (UTR) transcriptional start site was amplified from Arabidopsis genomic DNA by PCR and cloned into the binary vector pCAMBIA1381Z.
Arabidopsis Transformation
The AtGSTU17-overexpressed or promoter::GUS construct was introduced into Agrobacterium tumefaciens (GV3101) by electroporation (Withers, 1995) and then transformed into wild-type Col and the phyA mutant by the floral-dip method (Clough and Bent, 1998). Transgenic seedlings were selected on 1% GM plates containing 25 μg/mL hygromycin. The resulting homozygous transgenic lines were used for phenotype examination.
RNA Gel-Blot Analyses
Total RNA from 3-d-old seedlings under different light conditions, including darkness, or 3-d-old dark-grown seedlings transferred to various light conditions was isolated as described (Hsieh et al., 1996). RNA samples were separated in a 1.2% denaturing agarose gel, transferred onto a positive-charged nylon membrane (Roche), and UV cross linked and then hybridized with probes. The riboprobe was synthesized by in vitro transcription according to Dig-labeling procedures (Roche). The gene-specific riboprobe was derived from a BamH1-digested construct of the 3′ UTR of AtGSTU17. All hybridization and washing conditions followed standard methods (Sambrook et al., 1989).
Measurement of Hypocotyl Length, Anthocyanin, Chlorophyll, and Flowering
The wild-type, atgstu17 mutants, phyA, double mutant atgstu17phyA, and AtGSTU17-overexpressing lines GSTU17OE and GSTU17OE-2phyA were grown for 3 d under cFR or other light conditions, then hypocotyl lengths and anthocyanin accumulation were determined as described (Hsieh et al., 2000), or were grown for another 2 d of white light after FR light treatment for measurement of chlorophyll content as described (Hsieh et al., 2000). For investigation of flowering time, the seedlings were grown to the adult stage under long-day (16-h-day, 8-h-dark) conditions. Flowering time was recorded by use of two different indexes: days from seedlings sown in the soil to appearance of the first inflorescence, and leaf number at bolting. More than 10 plants for each condition were recorded.
Assays of Hormone-Mediated Inhibition of Root Elongation
Seeds of the wild type, atgstu17 mutants, GSTU17OE, and phyA mutants were germinated on Murashige and Skoog medium for 7 d in white light, then transferred to medium plates containing 0.01 μm 2,4-D or 30 μm ABA for vertical growth of roots. The plates were incubated for 5 d under white light. The elongation lengths of new roots of seedlings were measured and recorded. For chlorophyll content and biomass of the wild type, atgstu17 mutants, and GSTU17OE under 0.5 μm 2,4-D treatment were also recorded. All these physiological responses to auxin and ABA were examined in at least two biological replicates.
GST Activity Assay
GST activity of AtGSTU17 as purified recombinant proteins His-6-GSTU17 was determined as described by Chen et al. (2007).
Analysis of GSH
About 200 mg of seedlings was ground in liquid nitrogen. Subsequently, 2 mL of 1 mm EDTA and 6% (v/v) metaphosphoric acid, pH 2.8, were added and mixed, then centrifuged at 15,000g for 20 min; the supernatant used for determination of total GSH and GSSG by a spectrophotometric assay was as described by Griffith (1980).
Histochemical GUS Staining of AtGSTU17 Promoter Activity
Transgenic seedlings harboring the pAtGSTU17::GUS construct were selected on 1% Murashige and Skoog medium containing 25 mg/L hygromycin. The resulting transgenic plants were selected to homozygous lines and underwent histochemical GUS staining as described (Chen et al., 2007).
Microarray Experiment
Seedlings of the wild-type and atgstu17 mutants were grown in the dark for 2 d, then transferred to FR for 6 h; or germinated on Murashige and Skoog medium for 7 d in white light, then transferred to medium plates containing 100 μm ABA for another 5 d under white light. Total RNA was extracted from treated plant materials as described (Hsieh et al., 1996). cDNA and cRNA synthesis and hybridization to 22 K Affymetrix Gene Chips (ATH1) were performed according to Affymetrix instructions. Genes with 2-fold expression difference between the wild-type and atgstu17 mutants were selected for further analysis and validated by RT-PCR.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtGSTU17 (At1g10370), phyA (At1g09570), phyB (At2g18790), CRY1 (At4g08920), CRY2 (At1g04400), and FIN219 (At2g46370).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AtGSTU17 alignment and enzymatic activity assays.
Supplemental Figure S2. Phenotypic examination of AtGSTU17 T-DNA inserted lines in Arabidopsis Col.
Supplemental Figure S3. Induction of AtGSTU17 transcripts by different phytohormones.
Supplemental Figure S4. Characterization of the phyA mutant.
Supplemental Figure S5. Measurement of GSH levels in AtGSTU17 mutant lines and the overexpression line under auxin and ABA treatments.
Supplemental Table S1. A list of selected genes misregulated in atgstu17 under FR light.
Supplemental Table S2. A list of selected genes affected in atgstu17 in the presence of ABA.
Supplementary Material
Acknowledgments
We thank the ABRC (The Ohio State University, Columbus, OH) for providing AtGSTU17 T-DNA inserted mutant seeds and Ramanarayan Krishnamurthy (Institute of Plant Biology, National Taiwan University, Taipei, Taiwan) for reading and critically commenting on the manuscript. We also thank Min-Yan Kuo at Academia Sinica, Taiwan, for assistance in microarray experiments.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V. (1998) Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Baginsky S, Mulo P, Summer H, Aro EM, Link G. (2001) Chloroplast transcription at different light intensities: glutathione-mediated phosphorylation of the major RNA polymerase involved in redox-regulated organellar gene expression. Plant Physiol 127: 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH. (1996) Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP. (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Donald G, Reid DM, Yeung EC, Stasolla C. (2005) Alterations of the glutathione redox state improve apical meristem structure and somatic embryo quality in white spruce (Picea glauca). J Exp Bot 56: 2355–2364 [DOI] [PubMed] [Google Scholar]
- Chen IC, Huang IC, Liu MJ, Wang ZG, Chung SS, Hsieh HL. (2007) Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol 143: 1189–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- DeRidder BP, Goldsbrough PB. (2006) Organ-specific expression of glutathione S-transferases and the efficacy of herbicide safeners in Arabidopsis. Plant Physiol 140: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Cole DJ, Edwards R. (2000) Characterisation of a zeta class glutathione transferase from Arabidopsis thaliana with a putative role in tyrosine catabolism. Arch Biochem Biophys 384: 407–412 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Davis BG, Edwards R. (2002a) Functional divergence in the glutathione transferase superfamily in plants: identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem 277: 30859–30869 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. (2002b) Plant glutathione transferases. Genome Biol 3: reviews3004.1–reviews3004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Del Buono D, Fordham M, Skipsey M, Brazier M, Dixon DP, Cummings I. (2005) Differential induction of glutathione transferases and glucosyltransferases in wheat, maize and Arabidopsis thaliana by herbicide safeners. Z Naturforsch C 60: 307–316 [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. (2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Espunya MC, Díaz M, Moreno-Romero J, Martínez MC. (2006) Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ 29: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Gong H, Jiao Y, Hu WW, Pua EC. (2005) Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro. Plant Mol Biol 57: 53–66 [DOI] [PubMed] [Google Scholar]
- Gonneau J, Mornet R, Laloue M. (1998) A Nicotiana plumbaginifolia protein labeled with an azido cytokinin agonist is a glutathione S-transferase. Physiol Plant 103: 114–124 [Google Scholar]
- Griffith OW. (1980) Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 106: 207–212 [DOI] [PubMed] [Google Scholar]
- Gullner G, Tóbiás I, Fodor J, Kömives T. (1999) Elevation of glutathione level and activation of glutathione-related enzymes affect virus infection in tobacco. Free Radic Res (Suppl) 31: S155–S161 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. (1998) SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Hsieh HL, Tong CG, Thomas C, Roux SJ. (1996) Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol Biol 30: 135–147 [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol 148: 1583–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampranis SC, Damianova R, Atallah M, Toby G, Kondi G, Tsichlis PN, Makris AM. (2000) A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. J Biol Chem 275: 29207–29216 [DOI] [PubMed] [Google Scholar]
- Kilili KG, Atanassova N, Vardanyan A, Clatot N, Al-Sabarna K, Kanellopoulos PN, Makris AM, Kampranis SC. (2004) Differential roles of tau class glutathione S-transferases in oxidative stress. J Biol Chem 279: 24540–24551 [DOI] [PubMed] [Google Scholar]
- Kirik V, Kölle K, Wohlfarth T, Miséra S, Bäumlein H. (1998) Ectopic expression of a novel MYB gene modifies the architecture of the Arabidopsis inflorescence. Plant J 13: 729–742 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Mugford ST, Kopriva S. (2010) Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep 29: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Loyall L, Uchida K, Braun S, Furuya M, Frohnmeyer H. (2000) Glutathione and a UV light-induced glutathione S-transferase are involved in signaling to chalcone synthase in cell cultures. Plant Cell 12: 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA. (1996) The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 127–158 [DOI] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V. (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Moons A. (2003) Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Lett 553: 427–432 [DOI] [PubMed] [Google Scholar]
- Moons A. (2005) Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam Horm 72: 155–202 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Goodman CD, Silady RA, Walbot V. (2000) AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol 123: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Tasaka Y, Mino M, Tanaka Y, Iwabuchi M. (2001) Association of glutathione with flowering in Arabidopsis thaliana. Plant Cell Physiol 42: 524–530 [DOI] [PubMed] [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ. (2007) Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50: 429–438 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Smith AP, Nourizadeh SD, Peer WA, Xu J, Bandyopadhyay A, Murphy AS, Goldsbrough PB. (2003) Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant J 36: 433–442 [DOI] [PubMed] [Google Scholar]
- Sung DY, Lee D, Harris H, Raab A, Feldmann J, Meharg A, Kumabe B, Komives EA, Schroeder JI. (2007) Identification of an arsenic tolerant double mutant with a thiol-mediated component and increased arsenic tolerance in phyA mutants. Plant J 49: 1064–1075 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Noji M, Yamaguchi-Shinozaki K, Shinozaki K. (1996) A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J 10: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al. (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F. (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Wiśniewska J, Xu J, Seifertová D, Brewer PB, Růžička K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Withers HL. (1995) Direct plasmid transfer between bacterial species and electrocuring. Methods Mol Biol 47: 47–54 [DOI] [PubMed] [Google Scholar]
- Xu P, Xiang Y, Zhu H, Xu H, Zhang Z, Zhang C, Zhang L, Ma Z. (2009) Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol 149: 760–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettl R, Schell J, Palme K. (1994) Photoaffinity labeling of Arabidopsis thaliana plasma membrane vesicles by 5-azido-[7-3H]indole-3-acetic acid: identification of a glutathione S-transferase. Proc Natl Acad Sci USA 91: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao G, Qu LJ, Gu H. (2009) Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genomics 36: 99–107 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XR, Wang YZ, Smith JF, Chen R. (2008) Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol 178: 532–543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.