Abstract
Reactive oxygen species, generated endogenously and induced as a toxic response, produce several dozen oxidized or modified bases and/or single-strand breaks in mammalian and other genomes. These lesions are predominantly repaired via the conserved base excision repair (BER) pathway. BER is initiated with excision of oxidized or modified bases by DNA glycosylases leading to formation of abasic (AP) site or strand break at the lesion site. Structural analysis by experimental and modeling approaches shows the presence of a disordered segment commonly localized at the N- or C-terminus as a characteristic signature of mammalian DNA glycosylases which is absent in their bacterial prototypes. Recent studies on unstructured regions in DNA metabolizing proteins have indicated their essential role in interaction with other proteins and target DNA recognition. In this review, we have discussed the unique presence of disordered segments in human DNA glycosylases, and AP endonuclease involved in the processing of glycosylase products, and their critical role in regulating repair functions. These disordered segments also include sites for posttranslational modifications and nuclear localization signal. The teleological basis for their structural flexibility is discussed.
Keywords: Base excision repair, DNA glycosylases, End processing proteins, Disordered terminal segments, Single strand breaks, Reactive oxygen species, Repair complex, Protein–protein and protein–DNA interactions
Introduction
Damage to mammalian genomes, induced by a variety of reactive oxygen species (ROS) and reactive nitrogen species (RNS), that are generated either endogenously or by radiation and in response to other genotoxic and inflammatory agents, include a plethora of oxidatively damaged bases, abasic (AP) sites and DNA single-strand breaks (SSBs) that are often mutagenic and have etiological linkage to sporadic cancers and a variety of other pathophysiologies as well as aging [1, 2]. It is generally estimated that more than 104 base lesions and SSBs are induced daily in a mammalian cell genome [3]. These base lesions and SSBs are typically repaired via the evolutionarily conserved DNA base excision repair (BER) pathway. BER is initiated with excision of an oxidized, alkylated or aberrant base by a lesion-specific DNA glycosylase, generating an AP site. In the case of oxidized bases, the AP sites are further cleaved by the intrinsic AP lyase activity of DNA glycosylases, while in other cases, the AP sites are cleaved by an AP endonuclease (APE). The resulting DNA strand breaks have blocking groups either at the 3′ or 5′ end which need to be removed in order to generate a single-stranded gap that could then be filled in by a DNA polymerase followed by the sealing of the nick by a DNA ligase [4]. The SSBs with blocked ends are also generated directly by ROS that could include 3′-phosphate (3′-P), 3′-deoxyribosephosphate (3′-dRP) or 5′-deoxyriobosephosphate (5′-dRP) and oxidized sugar fragments such as 3′-phosphoglycolate [5, 6]. While 5′-dRP is processed by the dRP lyase activity of DNA polymerase β (Polβ) in mammalian cells, 3′-P and 3′-dRP are removed by polynucleotide kinase (PNK) and APE1, respectively [7]. Thus, the early step in repair of modified/oxidized bases and SSBs involves DNA glycosylases and nick-end processing enzymes.
Recent studies have suggested that BER is highly complex, involving a network of integrated pathways which are likely to be genome sequence-specific as well as cell cycle-specific [8, 9]. Several studies by us and others have shown that the early BER proteins form a complex to initiate repair and binarily interact with most if not all of the downstream proteins presumably for efficient co-ordination and sequential handover [7, 10–13].
The major challenge for the early BER proteins in mammalian cells is lesion recognition via efficient scanning of the gigabase size genome. Furthermore, a unique issue in BER, in contrast to nucleotide or mismatch excision repair pathways, is that the substrate lesion does not significantly distort the DNA helix and could retain near normal base pairing. These lesions do not block transcription or replication and are invariably bypassed. Thus, lesion recognition, particularly in highly condensed chromatin, poses a serious challenge which has not been extensively addressed. Interestingly, most early BER proteins have a stretch of disordered peptide segment invariably at one of the termini or which could serve sometimes as a linker bridging two domains, suggesting that such a common structural feature could be important for their functions. The focus of this review is to address the complexity of early BER activity in mammals and explore the common and unique structural features among these proteins that enable them to perform such an exigent function proficiently.
Basic BER mechanism
BER, first elucidated in E. coli and subsequently found to be universally conserved, is initiated with the recognition and excision of altered base lesions by about a dozen of distinct DNA glycosylases, each of which acts on a limited number of damaged bases [14, 15]. Uracil DNA glycosylase (Udg) was the first DNA glycosylase to be discovered in E. coli, which removes the U from DNA. U is generated due to deamination of cytosine and is mutagenic [14]. Subsequently, similar enzyme activities were discovered in mammalian cells and nuclear (UNG2)-specific and mitochondria (UNG1)-specific UDG variants were characterized [16]. Thymine-(T·G)–DNA glycosylase (TDG) is another mammalian DNA glycosylase that excises T and U paired with G [17]. DNA glycosylases specific for repair of alkylated bases have been characterized. The methylated bases generated by chemical reactions with endogenous 5-adenosylmethionine and exogenous alkylating agents, including many chemotherapeutic drugs and N-methylnitrosamine, are repaired either by direct reversal without DNA repair synthesis or via the BER pathway [18]. Ada in E. coli was the first repair protein discovered that carries out direct reversal of O 6-alkylguanine [18, 19]. Its mammalian ortholog O 6-methylguanine methyltransferase (MGMT) was subsequently cloned [20]. AlkB (E. coli) and its mammalian ortholog ABH are the other direct reversal enzymes for several alkylbase adducts [21]. N-methylpurine-DNA glycosylase (MPG, also named MAG) and its E. coli ortholog AlkA repair N-alkylpurines via the conventional BER pathway [22]. While DNA glycosylases in general recognize only abnormal bases in DNA, MutY, a mismatch-specific glycosylase discovered in E. coli and its mammalian homolog MYH excise normal base A from A·G and A·8-oxoG mispairs [23, 24]. The glycosylases for the repair of U, alkyl base adducts or base mispairs are monofunctional as they excise the base lesion, leaving an AP site without generating a strand break.
Oxidized base-specific DNA glycosylases in mammalian cells have been categorized in two families based on their tertiary structure and AP lyase reaction and named after their bacterial prototypes, Nth (endonuclease III) and Nei (endonuclease VIII) [25, 26]. OGG1 and NTH1 belonging to Nth family were the first to be discovered. Subsequently, we and others discovered NEIL1 and NEIL2 which belong to Nei family that also includes bacterial Fpg (formamidopyrimidine–DNA glycosylase) [1, 26–30]. NEIL3 was recently added to the list of oxidized base-specific glycosylases [31]; however, its biological activity is not well characterized. The Nth family glycosylases perform β-elimination at the AP site generating a 3′-phospho-αβ-unsaturated aldehyde, a dehydration product of deoxyribosephospate (3′-PUA also named as 3′-dRP) at the strand break. The members of Nei family catalyze βδ-elimination at the AP site to produce 3′-P [7, 32]. Thus, contrary to the APE-cleaved product of an AP site which contains 5′-dRP, and is removed by dRP lyase activity of Polβ, the oxidized base-specific DNA glycosylase-mediated strand breaks (BER intermediate) generate 3′ blocking groups, which need to be cleaned by specific end-processing enzymes in the subsequent BER step.
Processing of blocked termini: common step in the repair of altered bases and SSBs
Single-strand interruption processing, the second step in the repair of oxidized bases and AP sites, is also required for repairing direct ROS-induced SSBs which invariably contain an unligatable blocking group, either 3′ or 5′ or both ends at the strand break [33]. Gap filling by a DNA polymerase and nick sealing by a ligase require 3′-OH and 5′-P at the break site. End processing is thus an obligatory step involving multiple essential enzymes, specific for the 5′ or 3′ blocking groups as well as their type.
5′ end processing at a strand break
APE cleavage of the AP site generates 3′-OH and 5′-dRP termini. DNA polymerase β in mammalian cells removes the unmodified 5′-dRP moiety via its lyase activity [34, 35]. However, Polβ cannot process 5′ blocking groups generated by the cleavage of oxidized AP site by APE, and such dirty ends are processed via the long-patch BER pathway which involves removal of displaced 2–8 nts along with the 5′ blocking group by FEN-1, as discussed later. A unique type of 5′ blocking groups are formed as intermediates during abortive DNA ligation, such as adenylate groups covalently linked to the 5′ P terminus at strand break, and such groups are processed by a protein named aprataxin [36, 37]. Aprataxin releases the adenylate moiety to form ligatable 5′-P.
3′ end processing at a strand break
The Nth family glycosylases with intrinsic β lyase activity generate 3′-dRP (3′-PUA) and 5′-P at the strand break as already mentioned. The 3′dRP is removed by the intrinsic phosphodiesterase activity of mammalian APE1 to generate the 3′-OH terminus. On the other hand, the βδ lyase activity of Nei family DNA glycosylases would generate the 3′-P. We had shown that 3′-P is a poor substrate of APE1 but is efficiently removed by PNK in mammalian cells [7]. In addition, the 3′ blocks at the ROS-induced SSBs, mainly 3′-phosphoglycolate and 3′-phosphoglycoladehyde, are processed by APE1 [38–40]. Tyrosylphosphodiesterase 1 (TDP1) is another 3′ end processing enzyme discovered in yeast and human cells, which removes the product of abortive topoisomerase 1 (Top1) reaction, namely Top1(Tyr)-linked 3′ termini to form 3′-P, a substrate for PNK [41–44].
While there are multiple APEs in prokaryotes and lower eukaryotes, APE1 is the predominant contributor of APE activity in mammalian cells. Its prototype in E. coli is exonuclease III (Xth). As AP sites and strand breaks are continuously generated in the genome, it is not surprising that multiple enzymatic processes involving APEs are evolved. We have recently reviewed the functions of multiple APEs in BER in Hegde et al. [1].
Furthermore, TREX1 and TREX2 DNA 3′ exonucleases are ubiquitous in mammalian tissues, whose primary function may be editing during replication by Polβ or Polα lacking constitutive 3′ exonuclease activity [45]. In addition, TREX1 has also been proposed to play a role in SSBR, and its deficiency has been linked to a severe brain disease [46].
XRCC1 and PARP are two other key proteins which play a role in early BER activity. While XRCC1 acts as a scaffold for recruiting BER proteins for excision or strand break repair, PARP acts a SSB sensor protein. We showed that XRCC1 physically interacts with NEILs suggesting its role in oxidized base repair [7, 47]. XRCC1 also interacts with end processing enzymes PNK and APE1, and other BER proteins, Polβ and LigIIIα [48]. Moreover, processing of 5′-OH and 3′-P termini at SSBs is reduced in XRCC1-deficient cells suggesting its role in end processing [49]. PARP present in mammalian cells and absent in E. coli is activated by SSBs and transfers ADP-ribose moiety from NAD to a variety of proteins including itself. PARP-1 and PARP-2, the two proteins of the PARP superfamily, have been shown to be important players in the repair of SSBs both as sensors and for recruiting other repair proteins to the strand break [50]. Thus, XRCC1 and PARP play an indirect but vital role in both base excision and end cleaning repair steps.
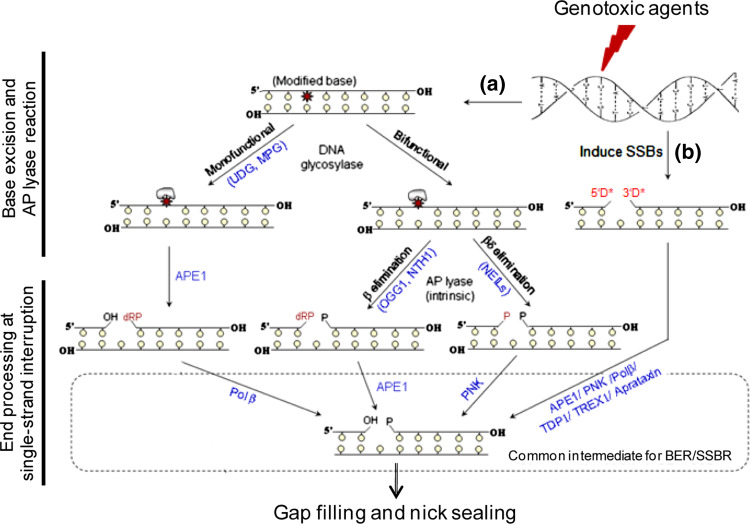
The early BER reactions for repair of modified bases and SSBs are schematically represented in Fig. 1.
Fig. 1.
Schematic representation of base excision (a) and single-strand break (b) repair steps in mammalian cells. Monofunctional DNA glycosylases (UDG, MPG) excise alkylated and modified bases from DNA to generate AP sites that are then cleaved by APE1. The 5′ blocking group at the break site is removed by Polβ to generate a single-nucleotide gap that is filled in by Polβ (and sealed by DNA ligase IIIα). Oxidized bases are excised by OGG1/NTH1 and NEILs which also cleave the DNA strand to generate 3′ blocking groups. DNA is directly cleaved by ROS/radiation and topoisomerases to generate 3′ or 5′ blocking groups (3′D* and 5′D*) which are removed by several end cleaning enzymes. Other details are described in the text
Complexities in BER and SSBR: multiple repair subpathways
Until recently, BER was believed to be the simplest among the DNA repair pathways involving four or five reaction steps. However, recent studies reveal that BER is much more complex, involving a network of distinct cell cycle-dependent as well as genome region-specific repair subpathways and could also involve non-BER proteins.
Short-patch versus long-patch repair pathways
In the simple mammalian BER model, excision of the lesion leaves a 1-nt gap at the damage site, which is subsequently filled by Polβ and the resulting nick sealed by DNA ligase IIIα (LigIIIα) to complete single-nucleotide repair (SN-BER), also called short-patch repair (SP-BER) [51].
In contrast to this simple model, recent discoveries document several BER subpathways involving at least a dozen more proteins. The second mode of BER, characterized by Matsumuto, Dogliotti and coworkers [34, 52, 53], involves a repair patch size of 2–8 nucleotides at the lesion site and was named long-patch repair (LP-BER). LP-BER was first observed during repair of an AP site analog lacking the aldehyde group. An upstream segment containing 2–8 nts including the 5′-blocking group is displaced as a single-strand flap during repair synthesis which is subsequently cleaved by flap endonuclease 1 (FEN-1). FEN-1’s normal function is to remove the 5′ RNA primers of Okazaki fragments during discontinuous replication of the lagging strand [54–56]. LP-BER is believed to utilize DNA replication proteins including DNA polymerase δ (Polδ), the sliding clamp PCNA, clamp loader replication factor-C (RF-C) and DNA ligase I (LigI) in addition to FEN-1 [57, 58]. However, Polβ has also been shown to participate in LP-BER via strand displacement, in collaboration with FEN-1 [59]. The choice of SN-BER versus LP-BER is a complex issue that is yet to be completely understood. Initial studies suggested that the nature of the 5′-phosphoribose terminus (normal vs oxidized) would be the deciding factor [60, 61]. However involvement of DNA replication proteins with LP-BER strongly suggests that LP-BER could be preferred in BER during DNA replication, irrespective of the 5′ terminal group.
DNA replication/transcription-specific BER subpathways
The mammalian genome at the replication fork or transcription bubble is relatively unfolded or nonchromatinized rendering it more prone to oxidative damage than nonreplicating chromatin. Replication of unrepaired oxidized base lesions which do not block replication could be mutagenic. Thus, there is an urgency to repair these lesions prior to replication (pre-replicative repair) in order to maintain genome integrity. Furthermore, incorporation of abnormal or oxidized bases (e.g., uracil or 8-oxoG) from the nucleotide pool into nascent DNA could be mutagenic as well, which also warrants urgent repair. Such repair was earlier described as post-replicative repair [62].
Recent studies by us and others have suggested that there are distinct BER subpathways for transcriptionally active versus inactive genomes as well as for quiescent versus replicating genome. Our initial studies with human NEIL1 and NEIL2 showed that both NEILs are active on bubble and fork-structured DNA substrates that mimic DNA replication or transcription intermediates [8]. At the same time, only NEIL1 is upregulated during the S-phase, based on which we had proposed that NEIL1 is preferentially involved in replication-associated repair (RA-BER) and NEIL2 in transcription-coupled BER (TC-BER) [8, 27, 28, 63]. Subsequently, our recent studies have shown NEIL1’s preferential association with DNA replication proteins including PCNA [11], Replication protein A (RPA; [13]), FEN-1 [10], and Werner’s helicase (WRN; [12]) that strongly support this hypothesis. Similarly, several other DNA glycosylases such as MYH and UNG (which are described later in this article) interact with replication proteins PCNA and RPA [64, 65]. Like NEIL1, UNG and MYH expression increases during the S-phase, and the UNG-PCNA-RPA complex co-localizes with replication foci suggesting preferential repair of nascent DNA [62, 66]. In contrast, our studies with NEIL2 which has cell cycle-independent expression suggested its association with transcription-associated BER. Although more studies are required to further characterize the genome region and cell-state-specific BER pathways, it is clear that additional complexities are associated with these distinct BER subpathways. For example, we have shown stable interaction between NEIL1 and 9-1-1 complex, a stress-activated sliding clamp implying a linkage between NEIL1-initiated BER and damage signaling pathways [67].
Involvement of non-BER proteins in the repair of base lesions/SSBs
While several non-BER proteins have been shown to be involved in BER, adding another dimension to the BER complexity, their precise in vivo role in repair has yet to be unraveled. We showed that NEIL2 interacts with YB-1, a Y-box binding protein, and it was suggested that YB-1 may be required for the fine-tuning of repair [68]. NTH1 was also shown to interact with and is stimulated by YB-1 [69]. The list of non-BER proteins interacting with BER proteins is still growing, which underscores the paradigm that the in vivo repair process is far more complex than in vitro repair demonstrated with minimal components.
Recent discoveries in BER
DNA glycosylases as hub proteins: binary interaction of DNA glycosylases with downstream proteins
Our initial characterization of NEIL1 and NEIL2 showed their similarity in in vitro repair of 5-OHU via SN-BER in a reconstituted system containing PNK, Polβ, LigIIIα and XRCC1. XRCC1 acts as a scaffold for recruiting BER proteins for excision or strand break repair. Both the NEIL immunoprecipitates from human cells contain these BER proteins [7, 47]. Furthermore, NEILs binarily interact in the absence of DNA with Polβ, LigIIIα and XRCC1, although not PNK. Direct interaction of NEILs with LigIIIα, the last enzyme in SN-BER, indicated that the repair is controlled or regulated by the initiating DNA glycosylase which acts as a hub protein. Similarly, as already discussed, NEIL1 binarily interacts with PCNA, RPA, FEN-1, and WRN in the absence of DNA, presumably for preferential BER during DNA replication. The stoichiometry of proteins in the repair complex and whether NEILs are present in distinct complexes or in a single complex are currently being investigated in our laboratories.
Repair complex versus sequential repair hand-off
The initial BER mechanism was proposed, based on co-crystal structure analysis of substrate-bound BER proteins, to involve ‘hand-off’ or ‘passing the baton’ process, wherein the repair product of each enzyme in the BER pathway is handed over to the next enzyme, primarily based on differential bending of DNA in each intermediate step [70, 71]. However, characterization of the BER interactome involving multiprotein interactions including stable complex of DNA glycosylase with DNA ligase, and the presence of multiprotein complexes has led to a new paradigm where complete repair occurs in the BER complex (BERosome). Although in vivo role of hand-off versus interactome modes of repair is not yet clear, we propose that preformed BER complexes predominantly repair endogenous base lesions, while repair via hand-off mechanism by sequential recruitment could occur with induced DNA damage. Further characterization of the dynamics of BERosomes is required to unravel the repair processes.
Common interaction interface of early BER proteins
For several years, our laboratory has focused on characterizing collaborations and mapping interactions among BER proteins. As already mentioned, NEIL1’s stable interaction with downstream repair proteins utilizes a common interaction interface located near its C-terminus (residues 289–349) [7, 10–13]. The segment is absent in NEIL1’s prototype Nei, and might have been acquired during evolution as a terminal addition [1]. We similarly identified the nonconserved N-terminal segment (65 residues) in APE1 which is involved in all its protein–protein interactions [72–74]. Although it is intriguing how NEIL1 or any other protein could simultaneously bind to so many proteins with high specificity via a small common interface, recent studies have indicated that it is not uncommon for the mammalian hub proteins to have such an interaction surface, which invariably have a disordered structure. The flexibility of the disordered domain may facilitate interaction with diverse partners [75, 76].
Disordered structure of the interaction interface
The C-terminal region of human NEIL1 (hNEIL1) spanning about 100 residues contains the common interaction interface whose disordered conformation was first suggested from the fact that NEIL1’s crystallization required deletion of 56 residues, while the proximal 44 residues did not form a defined structure [77]. We verified this conclusion using various protein structure prediction softwares.
Prediction of disordered structure: various softwares
Contrary to the concept about spontaneous formation of secondary and tertiary structure in properly folded proteins that prevailed before the turn of this century, recent experimental evidence as well as genome-wide prediction of intrinsic disorder in eukaryotic proteomes have indicated that a large percentage of proteins have long disordered (unfolded) segments under physiological conditions. Further, these disordered regions are essential for their biological functions. Recent advancement in the softwares for accurate prediction of protein secondary structure and their predisposition to remain intrinsically disordered has furthered our understanding of the role of disordered structures in functional hierarchy. Commonly used disorder prediction tools include PONDR [78–80], PrDOS [81, 82], RONN [82], FoldIndex [83], GlobPlot [84], IUPred [85, 86], FoldUnFold [87], etc. in the public domain, which evaluate intrinsic disorder on per residue basis. Among these, PONDR is most widely used, an advanced version of which contains a reference collection set of VSL predictors (trained on variously characterized, short and long disordered regions). The PONDR developers point out that short and long disordered regions might have differences in their amino acid characteristics because predictors based on short regions of disorder fare poorly for long regions of disorder and vice versa [88–90]. The VSL predictors in PONDR take advantage of such differences to yield more accurate predictions. We used PONDR, PrDOS and RONN software, which generated similar disorder prediction in early BER proteins.
The commonly used approaches to characterize protein disorder are NMR and circular dichroism spectroscopy, and also small angle X-ray scattering, while the structural information thus generated is often based on the crystal structure of truncated proteins [91].
Signature sequences of intrinsic disorder
A major sequence characteristic of intrinsic disorder is the low content of bulky hydrophobic amino acid residues such as Val, Leu, Ile, Met, Phe, Trp and Tyr, which would normally form the core of a folded globular protein. In contrast, a high proportion of polar and charged residues such as Gln, Ser, Pro, Glu, Lys, and Gly, and sometimes also Ala, are characteristically present in disordered regions [78, 92]. The presence of such charged residue-rich sequences was first discovered in transcriptional regulatory proteins about three decades ago, which are often classified based on their amino acid composition, for example Glu-rich, Pro-rich and acidic activation domains [93]. Later, NMR spectroscopy and other biophysical studies confirmed intrinsically disordered nature of such sequences [91, 94].
While disordered regions have been variously described as intrinsically disordered, intrinsically unstructured, natively unfolded, natively disordered and highly flexible [91], we believe that the term ‘intrinsically disordered’ would be more appropriate than ‘unstructured’ because many disordered regions have been shown to contain partial or transient secondary and/or tertiary structural organization [95]. Dunker and Obradovic [96] proposed that intrinsically disordered regions may exist in two different structural forms: molten globule-like (collapsed) and random coil-like (extended), whereas Uversky and co-workers suggested existence of another extended form, the pre-molten globule, which is distinguishable from fully extended and molten globular conformations by the presence of an unstable secondary structure [97]. The recently proposed protein quartet hypothesis suggests that the protein functions in eukaryotes could depend on any of the three disordered forms along with the ordered form or on transitions between them [97].
Disorder predictions have been extensively utilized by protein crystallographers to design crystallization targets after deletion of disordered segments. One first application of the disorder predictor was in crystallization of Xeroderma pigmentosum group A (XPA) protein involved in DNA repair [98]. Extended disordered regions at the N- and C-termini of XPA with ordered central core as predicted was confirmed with partial proteolysis and NMR spectroscopy [99].
The recently created databank of protein intrinsic disorder (http://www.disprot.org) suggests that proteins with long terminal extensions containing no or limited structure are common in eukaryotes and are involved in many key functions including cell cycle control, regulation, and signaling [99]. Such disordered tails were also shown to be more common in DNA binding proteins than in other proteins, particularly in the ones that are involved in target sequence binding that include early repair proteins and transcription factors [100, 101]. It is likely that their structural flexibility and plasticity provides major functional advantage.
Predictions of disorder using accurate bioinformatic tools are in fact helping design experiments to characterize their biological functions, and this is one of the fastest growing topics of protein studies, humorously dubbed as ‘the protein unfoldomics decade’. Although the number of proteins with experimentally determined disordered structure is still small, the behavior of the experimentally verified ones are highly consistent with the predictions [102].
Comparison of E. coli versus mammalian early BER proteins to identify disordered structures
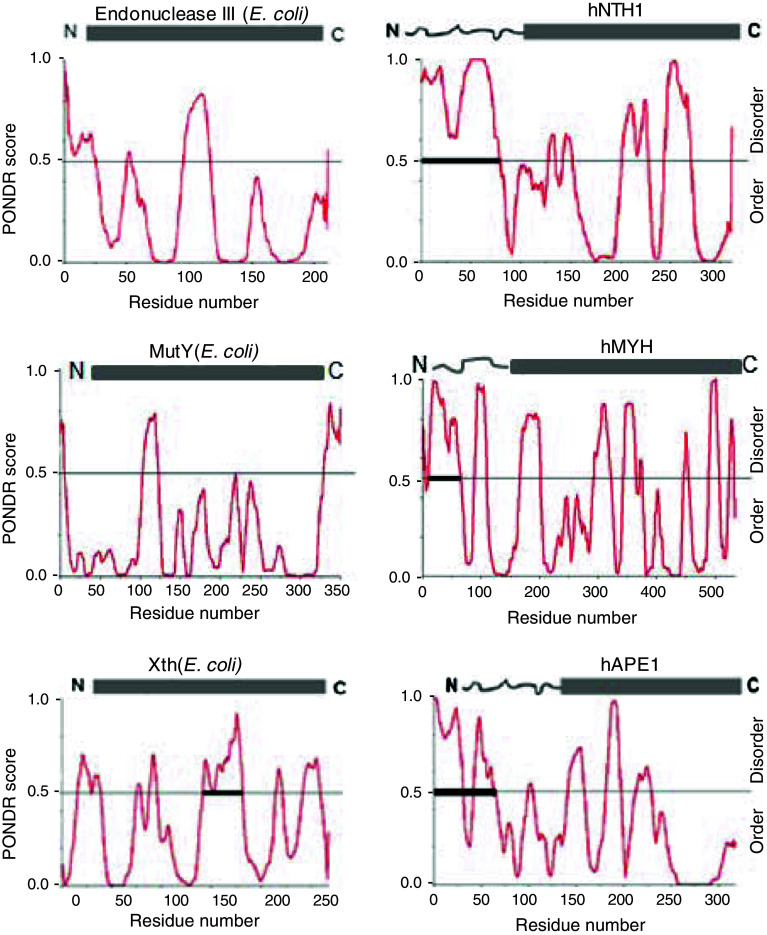
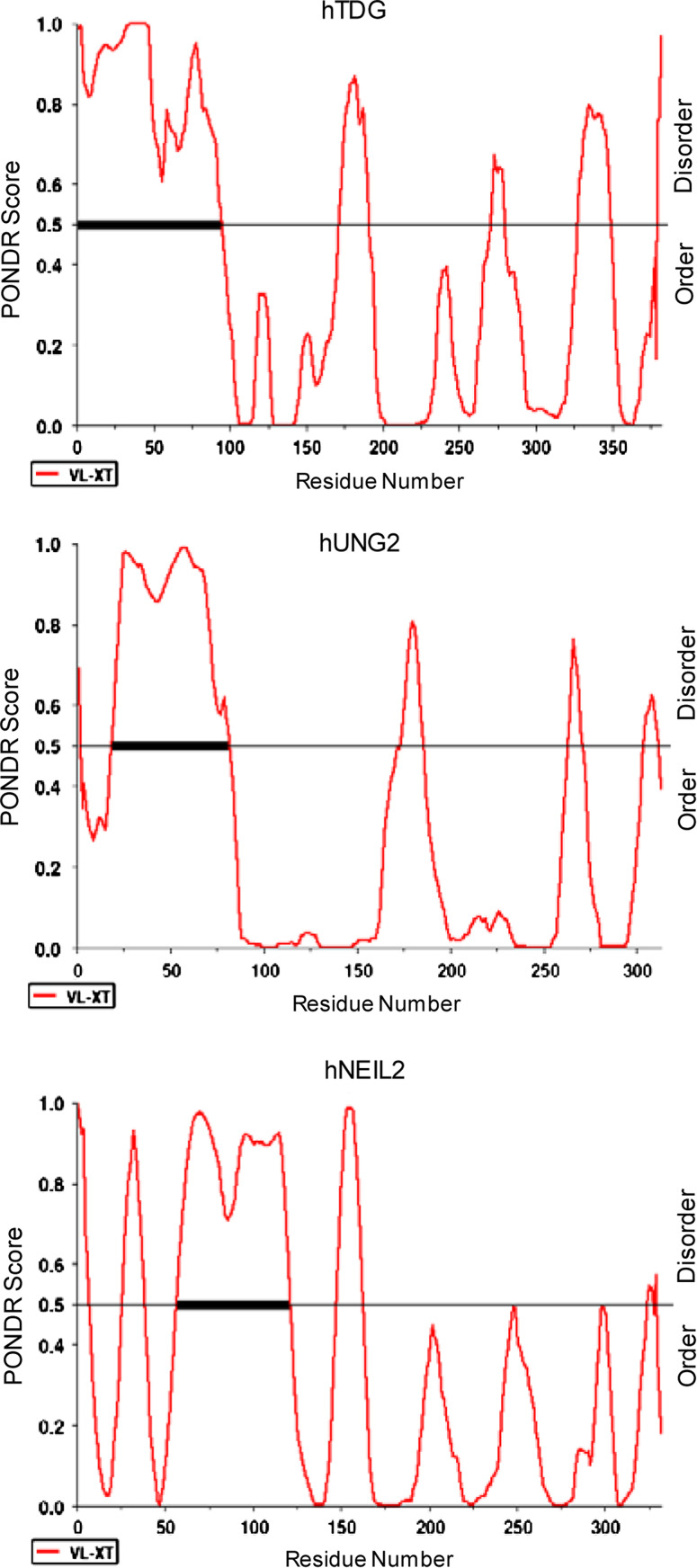
We used PONDR (prediction of natively disordered regions) and PrDOS (prediction of disordered structures) softwares to compare the secondary structure of human and bacterial early BER proteins and correlated them with their available structural information. As already mentioned, hNEIL1 contains an extended disordered region spanning about 100 residues in the C-terminus which is absent in E. coli Nei protein (Fig. 2). Comparison of predicted structures of human DNA glycosylases NTH1, MYH and their E. coli prototypes endonuclease III and MutY, respectively, indicates that both hNTH1 and hMYH have extended disordered tails at the N-terminus that are absent in the E. coli enzymes. Similarly, the N-terminal disordered region present in hAPE1 is absent in Xth, its E. coli prototype (Fig. 3). PONDR modeling showed that such disordered terminal sequences may also be present in other human DNA glycosylases including UNG2 and TDG (Fig. 4). Although the unfolded sequence generally exists at the N- or C-terminus, this could exist internally in some proteins, acting as a linker joining two domains. HNEIL2 is such a protein with an internal disordered segment near the N-terminus (residues 45–130) as indicated from the PONDR plot (Fig. 4). The average size of disordered extensions in early BER proteins ranges from 50 to 100 residues, with few exceptions, e.g., hOGG1 and human Polβ, which have short (~10 residues) disordered tails at both termini, as predicted by PrDOS (Fig. 5). The early BER proteins are generally small (30–50 kDa) and monomeric, whereas other DNA transaction proteins such as PCNA are multimeric and typically possess disordered linkers bridging different subunits.
Fig. 2.
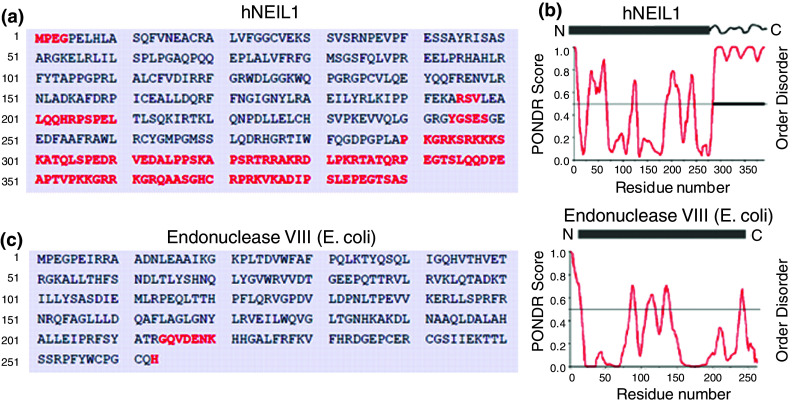
Secondary structure prediction of hNEIL1 and its E. coli prototype endonuclease VIII (Nei) by PrDOS (a,c) and PONDR (b,d) softwares. The protein sequences were obtained from NCBI database. Sequences in red in PrDOS and a score of 0.5 and above in the PONDR plot indicate disordered structures. The disordered C-terminal segment (wiggled line) of hNEIL1 is absent in Nei
Fig. 3.
PONDR plot of predicted secondary structures of hNTH1, hMYH, hAPE1 and their E. coli prototypes endonuclease III (Nth), MutY, Xth, respectively. Wiggled lines at the N-termini of human enzymes represent disordered segments
Fig. 4.
PONDR plot of disordered conformation at the N-terminus of hTDG and UNG2. HNEIL2 has an internal disordered region
Fig. 5.
PrDOS secondary structure prediction of hOGG1 and Polβ indicate short disordered segments (sequence in red) at both termini
Among early BER proteins, the disordered N-terminal sequence of hNTH1 has been extensively characterized. HNTH1 has a lower specific activity than E. coli Nth that lacks the N-terminal extension [103, 104], deletion of which increases hNTH1’s activity [105]. This suggests that this segment inhibits enzyme turnover in the absence of other BER proteins.
Role of disordered domain in function of early BER proteins
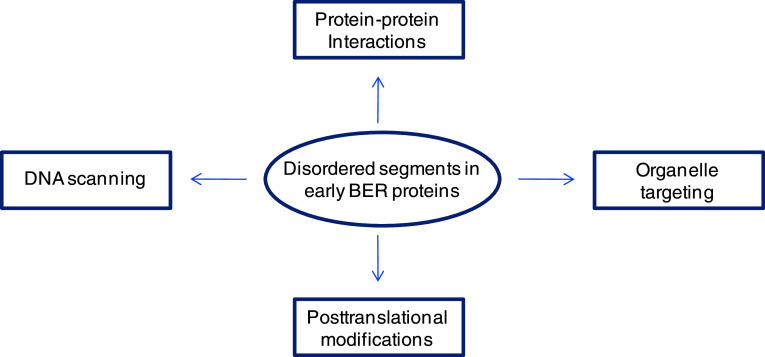
The presence of disordered extensions in proteins involved in transcriptional regulation, signal transduction, cell cycle control, DNA damage sensing and repair suggest their involvement in diverse functions. Furthermore, such disordered regions may regulate formation of large multiprotein complexes [106, 107]. An exhaustive discussion of this topic is beyond the scope of this review, focusing on the early BER proteins, in which these disordered segments include sites of posttranslational modifications, subcellular targeting, DNA scanning as well as common interface for protein–protein interactions, as already discussed (Fig. 6).
Fig. 6.
Schematic of multiple regulatory functions of disordered regions in early BER enzymes
Posttranslational modifications in disordered segments
Posttranslational modifications of proteins such as phosphorylation, acetylation, ubiquitylation, ADP-ribosylation, sumoylation and methylation play a critical role in diverse cellular processes including DNA repair [108]. The modification sites are invariably localized in disordered regions, e.g., in the N-terminal segment in hAPE1 [74, 109], N- and C-terminus of p53 [110], and C-terminal region in hNEIL1 (Bhakat et al., unpublished). Our laboratory identified and characterized acetylation of hAPE1 at Lys6 or Lys7, and this modification plays an important role in APE1’s transcriptional regulatory functions [74]. Recently, APE1 was also shown to be ubiquitynated at N-terminal Lys residues, which regulates its degradation as well as cellular functions [109]. Such covalent modifications may have multiple physiological effects on these proteins, including stability, interaction with DNA or other proteins, organelle targeting, and enzymatic activity [111]. We also showed that hNEIL2 is acetylated at Lys49 and Lys153 both in vitro and in cells [112]. Acetylation of Lys49 located in the disordered region (Fig. 4) inactivated NEIL2’s base excision and AP lyase activity while acetylation of Lys150 had no effect on the activity. We have proposed that acetylation of Lys49 could act as a regulatory switch for NEIL2’s activity [112]. TDG is acetylated in the N-terminal segment, Lys70, 94, 95 and 98 which is within the disordered segment of 100 residues [113]. PONDR modeling of TDG sequence indicates that the N-terminal 100 residues are in disordered conformation (Fig. 3). Strong acetylation sites in TDG were identified. Acetylation of TDG by CBP/p300 does not affect its binding to G·T or G·U base mispairs but indirectly deregulate TDG-coupled repair by releasing CBP/p300 from DNA bound complex leading to reduced interaction with APE and in turn suppressing APE-dependent repair [113]. Thus, TDG acetylation could contribute to genomic instability and cancer susceptibility. We had earlier characterized acetylation of hOGG1 at Lys338 and Lys341 within its short disordered C-terminus, which increases its DNA glycosylase activity by reducing affinity for the product AP site [114].
The flexibility of disordered region appears to be a prerequisite for these modifications, presumably because the amino acid side chains in the flexible sequence are accessible for modifying enzymes, like kinases, phosphatases, acetyltransferases and deacetylases, methylases, and ubiquitin ligases, etc. [91].
Subcellular localization
Organelle localization signals such as nuclear localization signal (NLS) or mitochondrial transport signal (MTS) are contained in short segments (generally <20 residues) that mediate transport to the target organelle. Multiple types of mammalian NLS sequences have been identified, the major ones belonging to the classical type consisting of seven basic residues and the bipartite NLS with two strings of basic residues separated by a short intervening sequence [115]. Recent studies showed that almost all NLS sequences with overall basic nature are disordered [99, 116].
We mapped the NLS of hAPE1 to the disordered N-terminal 20 residues, the deletion of which markedly diminishes its translocation to the nucleus [117]. Our preliminary studies of GFP-fusion polypeptide of truncated NEIL1 suggest the presence of putative NLS at the disordered C-terminal region (unpublished observation). Similarly, the disordered N-terminal tails in hNTH1, UNG2 and TDG contain putative NLS and MTS [118–120]. Taken together, these studies show that subcellular distribution of many human repair proteins is mediated through signals localized in their disordered regions.
DNA scanning
Burg et al. have shown that target DNA search by proteins could be achieved via facilitated diffusion comprising four mechanisms, namely, one-dimensional (1D) sliding, hopping, 3D search and intersegmental transfer. An efficient search mechanism involves combination of these different modes [121].
Recent studies have shown that the most efficient and rapid scanning of the DNA for the target site involves 80% hopping and intersegmental transfer and 20% sliding by the DNA binding proteins, which invariably contain a disordered terminal extension, or a disordered linker for multidomain or multisubunit proteins [101]. Bioinformatics analysis has suggested that nearly 70% of DNA binding proteins have such disordered tails compared to about 25% for non-DNA binding proteins [100, 101]. In addition, the disordered segments are about seven residues longer on an average for DNA binding proteins compared to all proteins with disordered tails [100]. Another unique characteristic of the disordered tails in DNA binding proteins is clustering of positively charged residues in the distal region, which turned out to be important for the scanning. Mutating such residues in HOXD9 and NK-2 markedly decreased scanning efficiency [101]. Similar results were obtained when the N-terminal segment in these proteins were deleted, suggesting that the initial scanning is mediated by a non-specific, mostly electrostatic, transient DNA binding via the basic disordered segment which is followed by target DNA sequence binding by the active site.
In light of the above studies, we examined hNEIL1’s C-terminus, which possesses most characteristics required for DNA scanning as described above, including the clustered basic residues. Our recent biochemical studies using C-terminal deletion NEIL1 mutants showed that the C-terminus is important for NEIL1’s substrate scanning and efficiency of damage recognition, via its non-specific DNA binding (Hegde et al., unpublished). Although limited studies are available on the role of disordered regions of other early BER proteins in such activity, we expect that all of them have similar functions.
Intrinsic disorder and hub proteins: dynamic repair complexes mediated by disorder/disorder interactions
As mentioned earlier, the ‘hub’ proteins like NEIL1 with several partners (usually >10) form a network of complexes [122]. Recent studies have indicated that ‘hub’ protein complexes are widely present in higher eukaryotes, whose formation mostly involves interaction with disordered regions [123]. The crucial role of intrinsic disorder in hub proteins was reviewed earlier [91, 124]. Bioinformatics analysis of known protein interactions suggested that such interactions among disordered structures are significantly preferred among human proteins [123]. Disorder-mediated interactions could either involve disorder–disorder or disorder–order types. Both modes of interactions are prevalent in BER proteins, for example NEIL1–FEN-1 interaction involves disorder–disorder interfaces [10], while NEIL1–Polβ could involve disorder–order type of contacts [7]. Specific recognition and binding of hub proteins is achieved as a result of the flexibility of domain itself, because it facilitates conformational rearrangements and induced-fit with specific partners [99, 107]. Although it was inconceivable a few years ago that such specificity could be obtained via disorder–disorder interactions, these are quite common, particularly for hub proteins. Disorder-mediated interactions in fact confer advantages over order-mediated interactions because of the rapid and easy interconversion among diverse conformers, allowing formation of dynamic complexes [107]. The dynamics of such complexes could be further regulated by posttranslational modifications of one or more partners, as already discussed. Furthermore, disorder-mediated interactions have kinetic benefits, because the larger capture radius of disordered states facilitates faster on-rates for binding [125, 126]. Finally, disorder-mediated interactions have steric advantages by providing a large surface area for binding interface for wrapping around partners resulting in stronger specificity [127, 128].
Our recent studies using size fractionation chromatography of human cell nuclear extract suggest that the BER proteins indeed exist in large, stable complexes, presumably in the absence of DNA. Characterization of such complexes, the dynamics of their formation and regulation as well as their stoichiometry are warranted.
Evolutionary advantages of protein disorder
Disordered regions in proteins generally show higher rates of mutation, presumably because changes in their protein sequence may not affect protein stability and function as severely as that in ordered regions [107, 129, 130]. Although such an analysis of mutation distribution and mutation tolerance is yet to be carried out for BER proteins, a similar situation is likely to exist. The unique presence of disordered segments in eukaryotic proteins but not in the prokaryotic counterparts, with the highest degree of disorder in mammals, suggests its evolutionary development [107]. The disordered regions also enable alternative splicing in eukaryotic proteins without the risk of perturbing structured regions [131]. In addition, the disorder provides advantage of limiting molecular size as complexity increases, by providing common interface for multiprotein binding and sites of modifications. To achieve a similar goal, folded proteins need to be considerably larger, and thus disorder may help higher organisms to limit protein size and to reduce intracellular crowding [132].
Disordered extensions in early BER proteins as potential targets of cancer therapy
The BER proteins have been explored as targets for cancer therapy which generally involved inactivation of key BER reactions such as ligation [133] or damage sensing proteins such as PARP [134]. DNA glycosylases turned out to be poor targets for such therapy, because of the nonessentiality of individual glycosylases due to back up functions of other glycosylases [1]. Although PARP inhibitors have been proven highly successful in recent therapeutic studies, inhibition of a key BER reaction poses the challenge of accurate targeting and dosage regulation to prevent their undesired effect on normal cells [134]. Based on the emerging evidence for a disorder-mediated repair regulatory switch in early BER hub proteins that controls the repair pathway, we propose that the disordered regions of early BER proteins could be targeted for cancer therapy which would disrupt repair regulation.
Conclusions and perspectives
A combination of experimental studies and structural predictions has revealed a critical role of disordered segments in many mammalian early BER enzyme functions including both protein–protein and protein–DNA interactions. Although few disordered regions have been experimentally characterized so far, we predict that such disordered segments are ubiquitous and essential for efficient repair. Future studies should address the role of disordered sequences in other mammalian repair pathways and their evolutionary significance in complex repair regulation.
Acknowledgments
The research in the authors’ laboratory is supported by USPHS grants, R01 CA81063, R01 CA53791, P01 CA92586 and P30 ES06676 (S.M.) and R01 CA 102271, R21 ES017353 (T.K.H). Because of the limited focus of the article on protein disorder in early BER proteins, many appropriate references could not be included, for which the authors apologize. We thank Mitra lab members for various stimulating discussions during preparation of this review.
Abbreviations
- BER
Base excision repair
- SSBR
Single-strand break repair
- AP
Abasic
- APE
AP endonuclease
- ROS
Reactive oxygen species
- RNS
Reactive nitrogen species
- SSB
Single-strand break
- PONDR
Prediction of naturally disordered regions in proteins
References
- 1.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Mitra S, Hazra TK, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- 5.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 6.Demple B, DeMott MS. Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene. 2002;21:8926–8934. doi: 10.1038/sj.onc.1206178. [DOI] [PubMed] [Google Scholar]
- 7.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. Ap endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases neil1 and neil2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 9.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. Hung2 is the major repair enzyme for removal of uracil from u:A matches, u:G mismatches, and u in single-stranded DNA, with hsmug1 as a broad specificity backup. J Biol Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 10.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase neil1 and flap endonuclease 1. J Biol Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase neil1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 12.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. The human werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase neil1. J Biol Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 13.Theriot CA, Hegde ML, Hazra TK, Mitra S. Rpa physically interacts with the human DNA glycosylase neil1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair (Amst) 2010;9:643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindahl T. An n-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 17.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitra S. Mgmt: a personal perspective. DNA Repair (Amst) 2007;6:1064–1070. doi: 10.1016/j.dnarep.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demple B, Sedgwick B, Robins P, Totty N, Waterfield MD, Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci USA. 1985;82:2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tano K, Shiota S, Collier J, Foote RS, Mitra S. Isolation and structural characterization of a cdna clone encoding the human DNA repair protein for o6-alkylguanine. Proc Natl Acad Sci USA. 1990;87:686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Mitra S, Kaina B. Regulation of repair of alkylation damage in mammalian genomes. Prog Nucleic Acid Res Mol Biol. 1993;44:109–142. doi: 10.1016/S0079-6603(08)60218-4. [DOI] [PubMed] [Google Scholar]
- 23.Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nghiem Y, Cabrera M, Cupples CG, Miller JH. The muty gene: a mutator locus in Escherichia coli that generates g.C…t.A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazra TK, Izumi T, Kow YW, Mitra S. The discovery of a new family of mammalian enzymes for repair of oxidatively damaged DNA, and its physiological implications. Carcinogenesis. 2003;24:155–157. doi: 10.1093/carcin/24.2.155. [DOI] [PubMed] [Google Scholar]
- 26.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 27.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 29.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease viii. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/S1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 30.Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease iii homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of neil3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the fpg family of DNA glycosylases. DNA Repair (Amst) 2003;2:839–862. doi: 10.1016/S1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 33.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 34.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 35.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 36.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 37.Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem. 2007;282:9469–9474. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 38.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. Xrcc1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/S0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 40.Parsons JL, Dianova II, Dianov GL. Ape1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase i covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 42.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 43.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type i topoisomerases. Proc Natl Acad Sci USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a tyr-DNA phosphodiesterase that repairs topoisomerase i complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 45.Mazur DJ, Perrino FW. Structure and expression of the trex1 and trex2 3′ → 5′ exonuclease genes. J Biol Chem. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 46.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3′–5′ DNA exonuclease trex1 cause aicardi-goutieres syndrome at the ags1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 47.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. Neil2-initiated, ape-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidal AE, Boiteux S, Hickson ID, Radicella JP. Xrcc1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(adp-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 51.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the xrcc1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumoto Y. Molecular mechanism of pcna-dependent base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:129–138. doi: 10.1016/S0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- 53.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for dnase iv (fen1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maga G, Villani G, Tillement V, Stucki M, Locatelli GA, Frouin I, Spadari S, Hubscher U. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein a, proliferating cell nuclear antigen, and flap endonuclease-1. Proc Natl Acad Sci USA. 2001;98:14298–14303. doi: 10.1073/pnas.251193198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh P, Zheng L, Chavez V, Qiu J, Shen B. Concerted action of exonuclease and gap-dependent endonuclease activities of fen-1 contributes to the resolution of triplet repeat sequences (ctg)n- and (gaa)n-derived secondary structures formed during maturation of okazaki fragments. J Biol Chem. 2007;282:3465–3477. doi: 10.1074/jbc.M606582200. [DOI] [PubMed] [Google Scholar]
- 56.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between pcna and DNA ligase i is critical for joining of okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–922. doi: 10.1016/S0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 58.Prasad R, Dianov GL, Bohr VA, Wilson SH. Fen1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J Biol Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 60.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 61.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 62.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R, Krokan HE. A sequence in the n-terminal region of human uracil-DNA glycosylase with homology to xpa interacts with the c-terminal part of the 34-kDa subunit of replication protein a. J Biol Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 65.Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the muty repair protein (hmyh) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 66.Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, Otterlei M, Horning O, Jensen ON, Krokan HE, Slupphaug G. Cell cycle-specific ung2 phosphorylations regulate protein turnover, activity and association with rpa. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu AL. The human checkpoint sensor rad9-rad1-hus1 interacts with and stimulates neil1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, Wilson SH, Hazra TK. Stimulation of neil2-mediated oxidized base excision repair via yb-1 interaction during oxidative stress. J Biol Chem. 2007;282:28474–28484. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marenstein DR, Ocampo MT, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Stimulation of human endonuclease iii by y box-binding protein 1 (DNA-binding protein b). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem. 2001;276:21242–21249. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- 70.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 71.Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/S0959-440X(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 72.Izumi T, Mitra S. Deletion analysis of human ap-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis. 1998;19:525–527. doi: 10.1093/carcin/19.3.525. [DOI] [PubMed] [Google Scholar]
- 73.Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/S0300-483X(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 74.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human ap-endonuclease (ape1/ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 77.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease viii-like 1 (neil1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 79.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 80.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61(Suppl 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 81.Ishida T, Kinoshita K. Prdos: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang ZR, Thomson R, McNeil P, Esnouf RM. Ronn: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 83.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Foldindex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 84.Linding R, Russell RB, Neduva V, Gibson TJ. Globplot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 86.Dosztanyi Z, Csizmok V, Tompa P, Simon I. Iupred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 87.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Foldunfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- 88.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/S0219720005000886. [DOI] [PubMed] [Google Scholar]
- 90.Romero Obradovic, Dunker K. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform Ser Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 91.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 92.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 93.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 94.O’Hare P, Williams G. Structural studies of the acidic transactivation domain of the vmw65 protein of herpes simplex virus using 1 h nmr. Biochemistry. 1992;31:4150–4156. doi: 10.1021/bi00131a035. [DOI] [PubMed] [Google Scholar]
- 95.Golovanov AP, Chuang TH, DerMardirossian C, Barsukov I, Hawkins D, Badii R, Bokoch GM, Lian LY, Roberts GC. Structure-activity relationships in flexible protein domains: regulation of rho gtpases by rhogdi and d4 gdi. J Mol Biol. 2001;305:121–135. doi: 10.1006/jmbi.2000.4262. [DOI] [PubMed] [Google Scholar]
- 96.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 97.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iakoucheva LM, Kimzey AL, Masselon CD, Bruce JE, Garner EC, Brown CJ, Dunker AK, Smith RD, Ackerman EJ. Identification of intrinsic order and disorder in the DNA repair protein xpa. Protein Sci. 2001;10:560–571. doi: 10.1110/ps.29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toth-Petroczy A, Simon I, Fuxreiter M, Levy Y. Disordered tails of homeodomains facilitate DNA recognition by providing a trade-off between folding and specific binding. J Am Chem Soc. 2009;131:15084–15085. doi: 10.1021/ja9052784. [DOI] [PubMed] [Google Scholar]
- 101.Vuzman D, Azia A, Levy Y. Searching DNA via a “Monkey bar” mechanism: the significance of disordered tails. J Mol Biol. 2010;396:674–684. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 102.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 103.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease iii crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human nth1, a homolog of Escherichia coli endonuclease iii. Direct identification of lys-212 as the active nucleophilic residue. J Biol Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 105.Liu X, Roy R. Truncation of amino-terminal tail stimulates activity of human endonuclease III (hnth1) J Mol Biol. 2002;321:265–276. doi: 10.1016/S0022-2836(02)00623-X. [DOI] [PubMed] [Google Scholar]
- 106.Stein A, Pache RA, Bernado P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 107.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 108.Krueger KE, Srivastava S. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics. 2006;5:1799–1810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 109.Busso CS, Iwakuma T, Izumi T. Ubiquitination of mammalian ap endonuclease (ape1) regulated by the p53-mdm2 signaling pathway. Oncogene. 2009;28:1616–1625. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 111.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 112.Bhakat KK, Hazra TK, Mitra S. Acetylation of the human DNA glycosylase neil2 and inhibition of its activity. Nucleic Acids Res. 2004;32:3033–3039. doi: 10.1093/nar/gkh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of cbp/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/S1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 114.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-W. [DOI] [PubMed] [Google Scholar]
- 116.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jackson EB, Theriot CA, Chattopadhyay R, Mitra S, Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (ape1/ref1) Nucleic Acids Res. 2005;33:3303–3312. doi: 10.1093/nar/gki641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarker AH, Ikeda S, Nakano H, Terato H, Ide H, Imai K, Akiyama K, Tsutsui K, Bo Z, Kubo K, Yamamoto K, Yasui A, Yoshida MC, Seki S. Cloning and characterization of a mouse homologue (mnthl1) of Escherichia coli endonuclease iii. J Mol Biol. 1998;282:761–774. doi: 10.1006/jmbi.1998.2042. [DOI] [PubMed] [Google Scholar]
- 119.Ikeda S, Kohmoto T, Tabata R, Seki Y. Differential intracellular localization of the human and mouse endonuclease iii homologs and analysis of the sorting signals. DNA Repair (Amst) 2002;1:847–854. doi: 10.1016/S1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- 120.Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nuclear and mitochondrial splice forms of human uracil–DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 122.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shimizu K, Toh H. Interaction between intrinsically disordered proteins frequently occurs in a human protein–protein interaction network. J Mol Biol. 2009;392:1253–1265. doi: 10.1016/j.jmb.2009.07.088. [DOI] [PubMed] [Google Scholar]
- 124.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 125.Pontius BW. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem Sci. 1993;18:181–186. doi: 10.1016/0968-0004(93)90111-Y. [DOI] [PubMed] [Google Scholar]
- 126.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27kip1 cyclin-dependent-kinase inhibitor bound to the cyclin a-cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 128.Kiss R, Bozoky Z, Kovacs D, Rona G, Friedrich P, Dvortsak P, Weisemann R, Tompa P, Perczel A. Calcium-induced tripartite binding of intrinsically disordered calpastatin to its cognate enzyme, calpain. FEBS Lett. 2008;582:2149–2154. doi: 10.1016/j.febslet.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 129.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 130.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 131.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 133.Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, MacKerell AD, Jr, Tomkinson AE. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res. 2008;68:3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Drew Y, Plummer R. Parp inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist Updat. 2009;12:153–156. doi: 10.1016/j.drup.2009.10.001. [DOI] [PubMed] [Google Scholar]