Abstract
A series of new HIV-1 protease inhibitors with the hydroxyethylamine core and different phenyloxazolidinone P2 ligands were designed and synthesized. Variation of phenyl substitutions at the P2 and P2′ moieties significantly affected the inhibitors’ binding affinity and antiviral potency. In general, compounds with 2- and 4-substituted phenyloxazolidinones at P2 exhibited lower binding affinities than 3-substituted analogues. Crystal structure analyses of ligand-enzyme complexes revealed different binding modes for 2- and 3-substituted P2 moieties in the protease S2 binding pocket, which may explain the compounds’ different binding affinities. Several compounds with 3-substituted P2 moieties demonstrated pM binding affinity, low nM antiviral potency against patient-derived viruses from HIV-1 clades A, B and C, and most retained potency against drug-resistant viruses. Further optimization of these compounds using structure-based design may lead to the development of novel protease inhibitors with improved activity against drug-resistant strains of HIV-1.
Introduction
The global HIV-AIDSa epidemic causes an estimated 2 million deaths each year and remains one of the most serious healthcare challenges of our time.1 In the absence of curative or preventative treatments, suppressing HIV-1 replication in infected patients has become a critical goal in managing the disease. Among the anti-HIV drugs developed over the last two decades, inhibitors of HIV-1 reverse transcriptase and protease remain the most prominent in clinical use.2 These drugs are essential components of highly active antiretroviral therapy (HAART), which typically comprises two nucleoside reverse transcriptase inhibitors (NRTIs) and one protease inhibitor (PI).3,4 HAART is credited with significantly reducing AIDS-related mortality5,6 and is currently implemented throughout the world as the standard of care for HIV-AIDS treatment.
The efficacy of HIV-1 PIs has significantly improved since this drug class was first introduced in the mid-nineties.7 Structure-guided drug design efforts, both in academia and industry, have led to the development of highly potent second-generation PIs with improved potency, tolerability, and pharmacokinetic profiles. The introduction of low-dose ritonavir as a pharmacokinetic booster in most PI-based regimens has led to the sustained suppression of viral replication for prolonged periods, further improving clinical outcomes.8–10 Most importantly, the two most recently approved PIs, tipranavir (TPV)11 and darunavir (DRV),12–14 retain high antiviral potency against viral strains resistant to most other PIs. Despite these advances, PI therapies are associated with serious problems that significantly limit their effectiveness, including drug side effects, high dosing frequency due to poor pharmacokinetics, and the acquisition of many drug-resistant protease variants.15
Drug resistance occurs when mutations in the enzyme active site selectively impair drug binding without significantly affecting substrate recognition and processing. The high rate of HIV-1 replication coupled with the infidelity of its reverse transcriptase gives rise to viral quasi-species, which propagate under the selective pressure of drug therapy to confer resistance to these drugs.16 Alarmingly, even TPV- and DRV-based PI regimens have recently been reported to select multiple resistance mutations in the protease, leading to reduced virological response.17–19 Therefore, novel PIs must be developed that are less susceptible to drug resistance and maintain broad spectrum activity against multidrug-resistant (MDR) HIV-1 variants with improved pharmacokinetic profiles, providing effective treatments for HIV-AIDS.
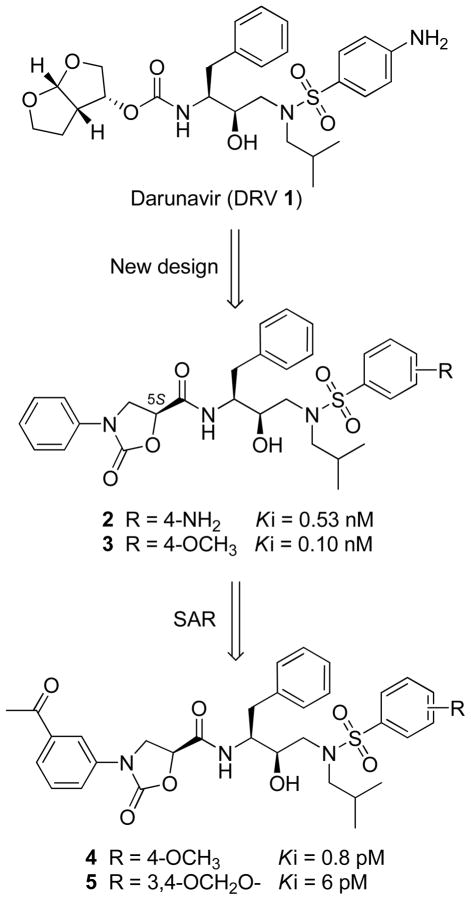
To reduce the probability of drug resistance in HIV-1 protease, we have been pursuing a structurebased strategy and recently discovered novel HIV-1 PIs which contain phenyloxazolidinones as P2/P2′ ligands.20,21 The initial inhibitor design (compounds 2 and 3, Figure 1) was based on the (R)-(hydroxyethylamino)sulfonamide isostere, the core scaffold of amprenavir (APV) and DRV, where the P2 tetrahydrofurynyl/bis-tetrahydrofurynyl moiety was replaced with a stereochemically defined N-phenyloxazolidinone-5-carboxamide. Exploration of the P1′ amine, P2 phenyloxazolidinone, and P2′ phenylsulfonamide moieties led to the identification of inhibitors 4 and 5 with exceptionally high binding affinities for wild-type protease (Ki = 0.8 pM and 6 pM, respectively).20 Although these binding affinities are comparable to those of lopinavir (LPV) (Ki = 5 pM) and DRV (Ki = 8 pM), compounds 4 and 5 exhibit lower antiviral potencies in cellular assays (EC50 = 30–35 nM)22 than LPV and DRV.
Figure 1.
Structures of darunavir (DRV) 1 and protease inhibitors 2–5.
The structural basis for these compounds’ high affinities was revealed by crystal structures of enzyme-inhibitor complexes from this series, involving a network of hydrogen bonds between the inhibitors’ oxazolidinone moiety and the invariant Asp29 residue of the protease.20,23 Based on these structures, we sought to enhance the antiviral potency of these compounds by exploring the structure-activity relationships (SAR) at the P2 phenyloxazolidinone and the P2′ phenylsulfonamide moieties. Herein, we report the structure-guided design, synthesis, and biological evaluation of new PIs incorporating a variety of P2 phenyloxazolidinone and P2′ phenylsulfonamide groups into the (R)-hydroxyethylamine isostere. The activities of these compounds were tested against wild-type HIV-1 protease and a MDR variant, and their antiviral potencies were determined in cellular assays against patient-derived wild-type viruses and two MDR HIV-1 variants. Several PIs showed low pM enzyme inhibitory potencies and high antiviral potencies against wild-type HIV-1 comparable to those of LPV. However, these PIs retained better potencies than LPV against drug-resistant viruses. We also report crystal structures of PIs with 2-substituted phenyloxazolidinones in complex with wild-type protease and compare them with the inhibitor 5-protease complex structure, revealing different binding modes for the 2- and 3-substituted P2 moieties in the S2 binding pocket of the enzyme.
Chemistry
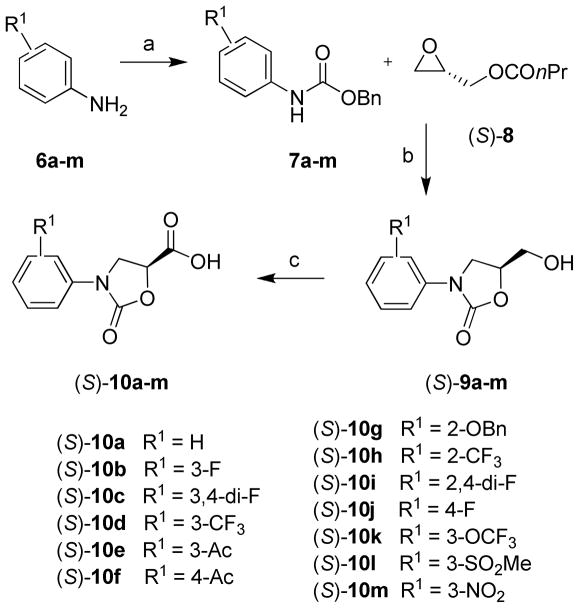
Synthesis of enantiomerically pure (S)-N-phenyloxazolidinone-5-carboxylic acids 10a–m is shown in Scheme 1. Reaction of Cbz-protected aniline derivatives 7a–m with (S)-glycidyl butyrate 8 promoted by n-BuLi provided enantiomerically pure alcohols, (S)-5-(hydroxymethyl)-3-phenyloxazolidine-2-ones, 9a–m. This one-pot, three-step cascade reaction involves the initial ring opening of chiral epoxide with N-lithium species followed by an intramolecular cyclization and finally an in situ ester hydrolysis.24 Oxidation of alcohols 9a–m by NaIO4 and catalytic RuCl3 provided N-phenyloxazolidinone-5-carboxylic acids 10a–m in good overall yields.
Scheme 1.
Synthesis of (S)-N-Phenyloxazolidinone-5-carboxylic acids 10a–ma
a Reagents and conditions: (a) NaHCO3, BnOCOCl, (CH3)2CO-H2O (2:1), 0 °C to rt, overnight; (b) n-BuLi, THF, −78 °C to rt, overnight; (c) NaIO4, RuCl3·H2O, CH3CN-CCl4-H2O (2:2:3), 0 °C to rt, 4–10 h.
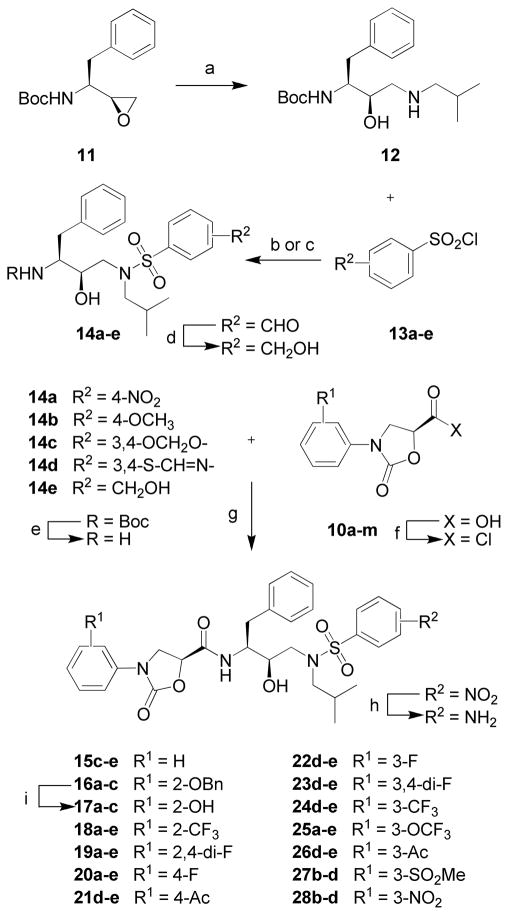
The synthesis of protease inhibitor series 15–28 incorporating various phenyloxazolidinone-based P2 ligands is illustrated in Scheme 2. The Boc-protected intermediates (R)-(hydroxyethylamino)sulfonamides 14a–e were prepared from the commercially available chiral epoxide 11, (1S,2S)-(1-oxiranyl-2-phenylethyl)carbamic acid tert-butyl ester, using previously developed methods.20,25 Briefly, ring opening of epoxide 11 with isobutylamine provided the amino alcohol 12. Reactions of 12 with sulfonyl chlorides 13a–d in the presence of a base, Na2CO3 in a CH2Cl2-H2O mixture for 13a–c and Et3N in dry CH2Cl2 for 13d, furnished the (R)-(hydroxyethylamino)sulfonamide derivatives 14a–d. The 4-(hydroxymethyl)benzenesulfonamide compound 14e was obtained in two steps, involving reaction of 12 with sulfonyl chloride 13e followed by reduction of the resulting 4-formylbenzenesulfonamide derivative with NaBH4 in MeOH. Deprotection of the Boc group in sulfonamides 14a–e followed by reacting the resulting amines with acid chlorides, obtained by activating the corresponding carboxylic acids 10a–m, afforded the target compound series 15–28 (Scheme 2). Inhibitors with the 4-aminobenzenesulfonamide at P2′ were obtained from the corresponding 4-nitrobenzenesulfonamide derivatives by reduction of the nitro group with SnCl2·2H2O. Removal of the benzyl protection in compounds 16a–c by Pd/C and HCO2NH4 in EtOAc provided inhibitors 17a–c.
Scheme 2.
Synthesis of Protease Inhibitors 15–28a
a Reagents and conditions: (a) iBuNH2, EtOH, 80 °C, 3 h; (b) aq. Na2CO3, CH2Cl2, 0 °C to rt, 6 h; (c) Et3N, CH2Cl2, 0 °C to rt, 6 h; (d) NaBH4, MeOH, 0 °C, 30 min; (e) TFA, CH2Cl2, rt, 1 h; (f) (COCl)2, rt, overnight; (g) Et3N, THF, 0 °C to rt, 6 h; (h) SnCl2·2H2O, EtOAc, 70 °C, 3 h; (i) Pd-C, HCO2NH4, EtOAc, rt, overnight.
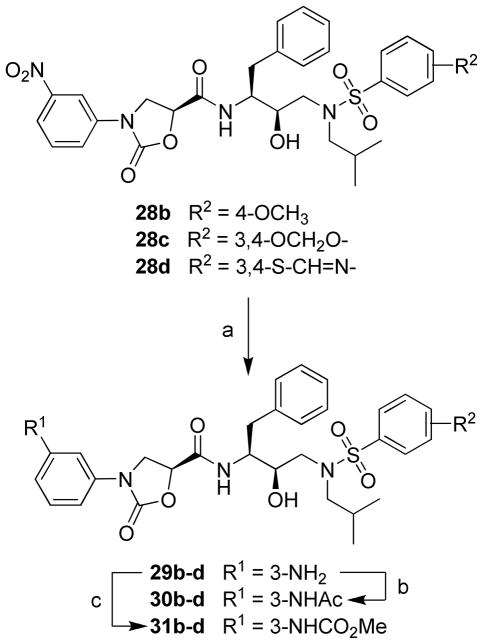
The protease inhibitors 29–31 were prepared from the corresponding 3-(nitrophenyl)oxazolidinone analogues 28b–d as shown in Scheme 3. Reduction of the nitro group in 28b–d using SnCl2·2H2O in EtOAc provided the corresponding inhibitors 29b–d with 3-(aminophenyl)oxazolidinones as P2 ligands. Further reactions of the 3-aminobenzene compounds 29b–d with acetic anhydride and methyl chloroformate in CH2Cl2 provided the 3-acetamidophenyl (30b–d) and methyl 3-phenylcarbamate (31b–d) analogues.
Scheme 3.
Synthesis of Protease Inhibitors 28–31a
a Reagents and conditions: (a) SnCl2.2H2O, EtOAc, 70 °C, 3 h; (b) Ac2O, CH2Cl2, rt, 2 h; (c) MeOCOCl, Na2CO3, CH2Cl2, 0 °C to rt, 2 h.
Results and Discussion
Analysis of crystal structures of inhibitors 4 and 5 in complex with wild-type HIV-1 protease revealed that the carbonyl oxygen of their oxazolidinone moieties form direct hydrogen bonds with the amide nitrogen and side-chain oxygen atoms of the conserved Asp29 residue in the enzyme S2 binding pocket. In addition, the nitrogen and oxygen atoms of the oxazolidinone ring form water-mediated hydrogen bonds with the carbonyl oxygen of Gly48 in the protease flap and the backbone amide nitrogen of Asp30, respectively.20,23 However, unlike DRV,14 inhibitors 4 and 5 do not directly interact with the backbone or side chain atoms of Asp30. Since the 2-position of the phenyl ring is observed close to Asp30, we reasoned that a substitution at this position might directly interact with the Asp30 residue of protease. Based on these observations, we designed a series of new HIV-1 protease inhibitors by incorporating a variety of 2-, 3- and 4-substituted (S)-N-phenyloxazolidinones into the (R)-(hydroxyethylamino)sulfonamide isostere. Variations at the P2 phenyloxazolidinone moiety were explored in combination with five different phenylsulfonamide groups as P2′ ligands.
Structure-Activity Relationships
The enzyme inhibitory potencies of all compounds were measured against wild-type HIV-1 protease (Q7K) and a drug-resistant variant (L10I, L63P, A71V, G73S, I84V, L90M) using fluorometric assays as previously described (Table 1).20,25 Selected compounds were further tested by Monogram Biosciences for antiviral activity in cellular assays against three patient-derived wild-type viruses from predominant HIV-1 clades (A, B, and C), and against a drug-resistant virus with protease mutations M46I, I54V, V82A and L90M. LPV and DRV were used as drug controls and a HIV-1 NL4-3-derived clade B laboratory strain (CNDO) and a MDR strain (MDRC4) were used as virus controls (Table 2).
Table 1.
Inhibitory Activity of Compounds against Wild-Type HIV-1 Protease and an MDR Varianta
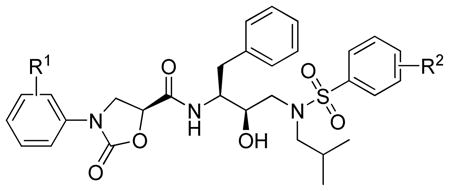 | ||||
|---|---|---|---|---|
| Compd. | R1 | R2 |
Ki (nM) |
|
| Wt | MDR | |||
| 15c | H | 3,4-OCH2O- | 0.206 | 5.50 |
| 15d | H | 3,4-S-C=N- | 0.033 | 1.21 |
| 15e | H | 4-CH2OH | 0.252 | NT |
| 17a | 2-OH | 4-NH2 | 7.36 | 40.46 |
| 17b | 2-OH | 4-OCH3 | 5.65 | 67.34 |
| 17c | 2-OH | 3,4-OCH2O- | 1.47 | NT |
| 18a | 2-CF3 | 4-NH2 | 0.136 | 5.71 |
| 18b | 2-CF3 | 4-OCH3 | 0.086 | 2.45 |
| 18c | 2-CF3 | 3,4-OCH2O- | 0.142 | 4.36 |
| 18d | 2-CF3 | 3,4-S-C=N- | 0.097 | 1.86 |
| 18e | 2-CF3 | 4-CH2OH | 0.235 | 4.57 |
| 19a | 2,4-di-F | 4-NH2 | 0.385 | NT |
| 19b | 2,4-di-F | 4-OCH3 | 0.063 | 10.54 |
| 19c | 2,4-di-F | 3,4-OCH2O- | 0.347 | NT |
| 19d | 2,4-di-F | 3,4-S-C=N- | 0.150 | 3.50 |
| 19e | 2,4-di-F | 4-CH2OH | 0.212 | NT |
| 20a | 4-F | 4-NH2 | 0.448 | NT |
| 20b | 4-F | 4-OCH3 | 0.128 | 9.57 |
| 20c | 4-F | 3,4-OCH2O- | 0.167 | 10.78 |
| 20d | 4-F | 3,4-S-C=N- | 0.133 | 3.0 |
| 20e | 4-F | 4-CH2OH | 0.207 | 9.56 |
| 21d | 4-Ac | 3,4-S-C=N- | 0.073 | 2.04 |
| 21e | 4-Ac | 4-CH2OH | 0.317 | NT |
| 22d | 3-F | 3,4-S-C=N- | 0.080 | 2.12 |
| 22e | 3-F | 4-CH2OH | 0.319 | NT |
| 23d | 3,4-di-F | 3,4-S-C=N- | 0.232 | 5.18 |
| 23e | 3,4-di-F | 4-CH2OH | 0.330 | NT |
| 24d | 3-CF3 | 3,4-S-C=N- | 0.016 | 2.96 |
| 24e | 3-CF3 | 4-CH2OH | 0.196 | 10.15 |
| 25a | 3-OCF3 | 4-NH2 | 0.225 | 9.95 |
| 25b | 3-OCF3 | 4-OCH3 | 0.130 | 16.3 |
| 25c | 3-OCF3 | 3,4-OCH2O- | 0.239 | 10.6 |
| 25d | 3-OCF3 | 3,4-S-C=N- | 0.026 | 3.38 |
| 25e | 3-OCF3 | 4-CH2OH | 0.286 | 7.49 |
| 26d | 3-Ac | 3,4-S-C=N- | 0.015 | 1.69 |
| 26e | 3-Ac | 4-CH2OH | 0.236 | NT |
| 27b | 3-SO2CH3 | 4-OCH3 | 0.049 | 1.74 |
| 27c | 3-SO2CH3 | 3,4-OCH2O- | 0.025 | 4.16 |
| 27d | 3-SO2CH3 | 3,4-S-C=N- | 0.003 | 2.45 |
| 28b | 3-NO2 | 4-OCH3 | 0.136 | 5.40 |
| 28c | 3-NO2 | 3,4-OCH2O- | 0.117 | 6.48 |
| 28d | 3-NO2 | 3,4-S-C=N- | 0.015 | 0.93 |
| 29b | 3-NH2 | 4-OCH3 | 0.020 | 2.68 |
| 29c | 3-NH2 | 3,4-OCH2O- | 0.113 | 6.55 |
| 29d | 3-NH2 | 3,4-S-C=N- | 0.008 | 1.84 |
| 30b | 3-NHAc | 4-OCH3 | 0.122 | 3.89 |
| 30c | 3-NHAc | 3,4-OCH2O- | 0.051 | 4.76 |
| 30d | 3-NHAc | 3,4-S-C=N- | 0.006 | 2.83 |
| 31b | 3-NHCO2CH3 | 4-OCH3 | 0.163 | 4.44 |
| 31c | 3-NHCO2CH3 | 3,4-OCH2O- | 0.289 | 3.68 |
| 31d | 3-NHCO2CH3 | 3,4-S-C=N- | 0.026 | 1.87 |
| LPV | 0.005 | 0.90 | ||
| DRV | 0.008 | 0.025 | ||
Wt: Q7K; MDR: L10I, L63P, A71V, G73S, I84V, L90M. NT = not tested
Table 2.
Antiviral Activity of Compounds against Wild-Type and MDR HIV-1 Strainsa
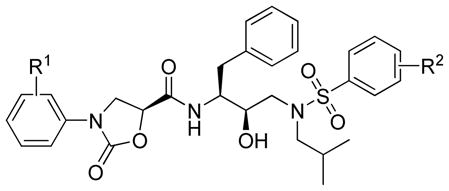 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compd. | R1 | R2 | EC50 (nM) (fold change) |
|||||
| WT | WT-A | WT-B | WT-C | MDR1 | MDR | |||
| 15d | H | 3,4-S-C=N- | 21.5 | 29.8 | 14.1 | 40.9 | 112.9 (5) | 123.1 (6) |
| 24d | 3-CF3 | 3,4-S-C=N- | 13.5 | 16.1 | 9.9 | 32.5 | 57.6 (4) | 53.4 (4) |
| 25d | 3-OCF3 | 3,4-S-C=N- | 22.1 | 32.5 | 12.7 | 57.8 | 104.5 (5) | 110.8 (5) |
| 26d | 3-Ac | 3,4-S-C=N- | 6.5 | 9.7 | 4.2 | 10.1 | 32.3 (5) | 34.7 (5) |
| 27b | 3-SO2CH3 | 4-OCH3 | 5.5 | 7.3 | 4.7 | 9.6 | 14.4 (3) | 18.6 (3) |
| 27c | 3-SO2CH3 | 3,4-OCH2O- | 4.0 | 7.8 | 3.5 | 7.6 | 13.6 (3) | 12.1 (3) |
| 27d | 3-SO2CH3 | 3,4-S-C=N- | 5.0 | 4.1 | 3.5 | 5.7 | 9.4 (2) | 10.5 (2) |
| 28d | 3-NO2 | 3,4-S-C=N- | 7.3 | 11.5 | 5.7 | 16.3 | 33.8 (3) | 37.7 (5) |
| 29b | 3-NH2 | 4-OCH3 | 4.0 | 8.5 | 3.0 | 9.4 | 19.0 (5) | 36.4 (9) |
| 29c | 3-NH2 | 3,4-OCH2O- | 7.1 | 6.1 | 2.8 | 12.3 | 27.9 (4) | 35.4 (5) |
| 29d | 3-NH2 | 3,4-S-C=N- | 1.7 | 2.6 | 1.3 | 2.5 | 11.9 (7) | 14.6 (9) |
| 30c | 3-NHAc | 3,4-OCH2O- | 10.9 | 9.0 | 6.3 | 21.1 | 41.9 (4) | 46.9 (4) |
| 30d | 3-NHAc | 3,4-S-C=N- | 5.8 | 10.1 | 7.7 | 9.5 | 25.4 (4) | 29.5 (5) |
| 31d | 3-NHCO2CH3 | 3,4-S-C=N- | 1.2 | 2.4 | 1.6 | 3.0 | 10.8 (9) | 13.0 (11) |
| LPV | 1.4 | 1.6 | 1.0 | 1.6 | 34.1 (24) | 39.7 (28) | ||
| DRV | 0.4 | 0.6 | 0.4 | 0.4 | 0.5 (1) | 1.0 (2) | ||
Antiviral assays were carried out by Monogram Biosciences. WT, wild-type HIV-1 control; WT-A, WT-B, WT-C, patient derived strains of wild-type HIV-1 from clades A, B, and C, respectively; MDR1, drug-resistant HIV-1 variant with protease mutations M46I, I54V, V82A and L90M; MDR, drugresistant HIV-1 control MDRC4; values in the parentheses represent change in EC50 values compared to the WT control.
The first series of inhibitors 15c–e, incorporating the unsubstituted phenyloxazolidinone moiety at the P2 position and various benzenesulfonamides at the P2′ position, displayed sub nM inhibitory potency against wild-type protease. The 1,3-benzodioxolane and 4-(hydroxymethyl)benzene analogues, 15c and 15e, are slightly less potent than inhibitors 2 and 3, but the benzothiazole compound 15d is more potent against both wild-type (Ki = 0.033 nM) and MDR (Ki = 1.21 nM) proteases. In cellular assays, 15d inhibited HIV-1 viral replication with EC50 values of 14–41 nM against wild-type viruses but lost about 6-fold potency against MDR viruses.
To test if 2-position substitutions at the P2 phenyloxazolidinone moiety could improve binding affinity to protease, we prepared inhibitors 17–19 with 2-hydroxy-, 2-trifluoromethyl-, and 2,4-difluoro-substitutions on the phenyl ring that were designed to interact with the Asp30 residue of the protease. Surprisingly, protease inhibitors containing the 2-hydroxyphenyl group at the P2 moiety 17a–c exhibited significantly lower inhibitory potency than the corresponding unsubstituted analogues (2, 3 and 15C), with Ki values of all three analogues in the nM range. However, inhibitors with the 2-trifluoromethylphenyl (18a–e) and 2,4-difluorophenyl (19a–e) groups were equipotent to inhibitors with unsubstituted phenyl rings. Similarly, compounds with the 4-fluorophenyl (20a–e) and 4-acetylphenyl (21d–e) groups on the P2 moiety did not have better potency than the corresponding unsubstituted phenyl compounds. In each series, inhibitors with the 4-methoxybenzene and benzothiazole moieties at P2′ position are more potent than compounds with other groups.
Since compounds with the 2-substituted phenyl group at the P2 moiety were generally less potent than the corresponding unsubstituted analogues, we explored various 3-substituted phenyl groups at the P2 oxazolidinone moiety. Compounds 22–23 with the 3-fluorophenyl and 3,4-difluorophenyl groups at the P2 moiety showed inhibitory potency in the sub nM range, with analogues 22d and 23d containing benzothiazole moieties at P2′ being the most potent. Introducing a 3-trifluoromethylphenyl group at the P2 markedly improved potency, as previously observed;20 compound 24d with the benzothiazole group at P2′ exhibited a Ki of 16 pM. Replacing the 3-trifluoromethylphenyl group with a 3-trifluoromethoxylphenyl (compounds 25a–e) failed to improve potency; only the P2′ benzothiazole inhibitor 25d showed inhibitory potency in the pM range (Ki = 0.026 nM).
Incorporating the 3-acetylphenyloxazolidinone as the P2 ligand in combination with the 4-methoxyphenyl and 1,3-benzodioxolane groups at the P2′ position has previously provided highly potent protease inhibitors such as 4 and 5.20 However, these compounds showed lower antiviral potency in cellular assays than LPV and DRV, presumably due to their higher lipophilicity (cLogP > 5 for both compounds). Combining this high affinity P2 ligand with the more polar benzothiazole group at P2′ position provided the analogue 26d with similarly high inhibitory potency against wild-type (Ki = 15 pM) and MDR (Ki = 1.69 nM) proteases. Compared to inhibitors 4 and 5, compound 26d exhibited better antiviral potency against wild-type viruses (EC50 = 4.2–10.1 nM), though it lost 5-fold potency against MDR viruses.
Based on these encouraging results, we explored various 3-substituted phenyloxazolidinones that could mimic the key interactions of the 3-acetylphenyloxazolidinone moiety of 4 and 5. Replacing the 3-acetyl group on the phenyl ring with an isosteric 3-methylsulfonyl moiety greatly improved the cellular antiviral potency of resulting analogues 27b–d. Thus, compounds with the P2′ 4-methoxyphenyl (27b, Ki = 49 pM) and 1,3-benzodioxolane (27c, Ki = 25 pM) groups showed high antiviral potency against wild-type and MDR viruses (EC50 = 4.7–18.6 nM and 3.5–13.6 nM, respectively). Analogue 27d with the P2′ benzothiazole group showed the best binding affinity (Ki = 3 pM), high cellular antiviral potency against wild-type viruses (EC50 = 3.5–5.7 nM), and retained significant potency against MDR viruses (EC50 = 9.4–10.5 nM). Interestingly, 27c and 27d retain antiviral potencies against MDR viruses and have relatively flat potency profiles against both wild-type and MDR viruses, while LPV, which is more potent against wild-type viruses (EC50 = 1.4–1.6 nM), displayed significantly lower potency against MDR viruses (EC50 = 34.1–39.7 nM).
To further manipulate the 3-position of the phenyl ring using simple transformations, we next introduced a nitro group at this position. Among the compounds with the 3-nitrophenyl group, 28b–d, the benzothiazole analogue 28d exhibited potent inhibitory activity (Ki = 15 pM) and good antiviral potency in cellular assays (EC50 = 5.7–37.7 nM) against the viruses tested. Reducing the nitro group provided the 3-aminophenyl analogues 29b–d with better inhibitory potencies than those of the parent nitrophenyl compounds, and all three potently inhibited HIV-1 replication. The analogues 29b and 29d (Ki = 20 pM and 8 pM, respectively), with the 4-methoxyphenyl and benzothiazole groups at P2′, showed low nM antiviral potency; 29d (EC50 = 1.7 nM) was equipotent to LPV against wild-type viruses but retained better potency against the MDR variants (EC50 = 11.9–14.6 nM).
Further transformation of the 3-aminophenyl analogues 29b–d to the 3-acetamidophenyl and methyl 3-phenylcarbamate derivatives did not significantly improve the inhibitory or antiviral potency of resulting compounds 30b–d and 31b–d. However, the P2′ benzothiazole analogues 30d and 31d showed highly potent activity (Ki = 6 pM and 26 pM, respectively) in both enzymatic and cellular assays. The antiviral potency of the carbamate analogue 31d (EC50 = 1.2–3.0 nM) is comparable to that of LPV against wild-type viruses, but 3-fold better against the MDR HIV-1variants.
These findings indicate that the antiviral activity in this series can be significantly improved by changing substitutions at the P2 phenyloxazolidinone and P2′ phenylsulfonamide moieties, and these changes can also improve the resistance profile of inhibitors. Analogues 27d, 29d, and 31d all have the same benzothiazole moiety at P2′, but variations at the phenyl ring of the P2 moiety resulted in different activity profiles against wild-type and MDR viruses. Compound 27d is slightly less potent than LPV against wild-type viruses but it lost only 2-fold potency against the two MDR viruses tested, whereas LPV lost 24- to 28-fold potency against the same viruses. On the other hand, compounds 29d and 31d inhibited wild-type viruses with high potency comparable to that of LPV, but lost relatively more potency against MDR viruses, though they retained better potency than LPV. Although the new PIs contain the same (R)-(hydroxyethylamino)sulfonamide core as DRV, and exhibit similar binding affinities to protease, their antiviral potencies against both wild-type and MDR viruses are lower than DRV. Further optimization of these PIs may lead to compounds with improved antiviral potencies and better resistance profiles.
Crystal Structures of Protease Complexes
To understand why the observed protease inhibitory activities were significantly different for compounds with 2- and 3-substituted P2 phenyloxazolidinone moieties, we determined crystal structures of 17c and 18d in complex with the wild-type HIV-1 protease and compared them with the structure of the inhibitor 5-protease complex.23 The three complexes differ mainly at the P2 phenyloxazolidinone moieties where they have different substituent groups at the 2-(17c and 18d) and 3-position (5) on the phenyl ring; 18d also has a different P2′ moiety (benzothiazole). These differences at the P2 moiety resulted in the phenyl rings adopting different orientations in the three ligand-enzyme complexes; the phenyl ring in 17c and 18d is perpendicular to the oxazolidinone ring, whereas both rings in 5 are almost in the same plane (Figure 3). As a result, all three structures show variations in the backbone near one of the flap regions where the inhibitor’s phenyloxazolidinone moiety interacts with the protease. Other than these differences, the three structures superpose well onto each other with a maximum RMSD of 0.264 for all Cα atoms.
Figure 3.
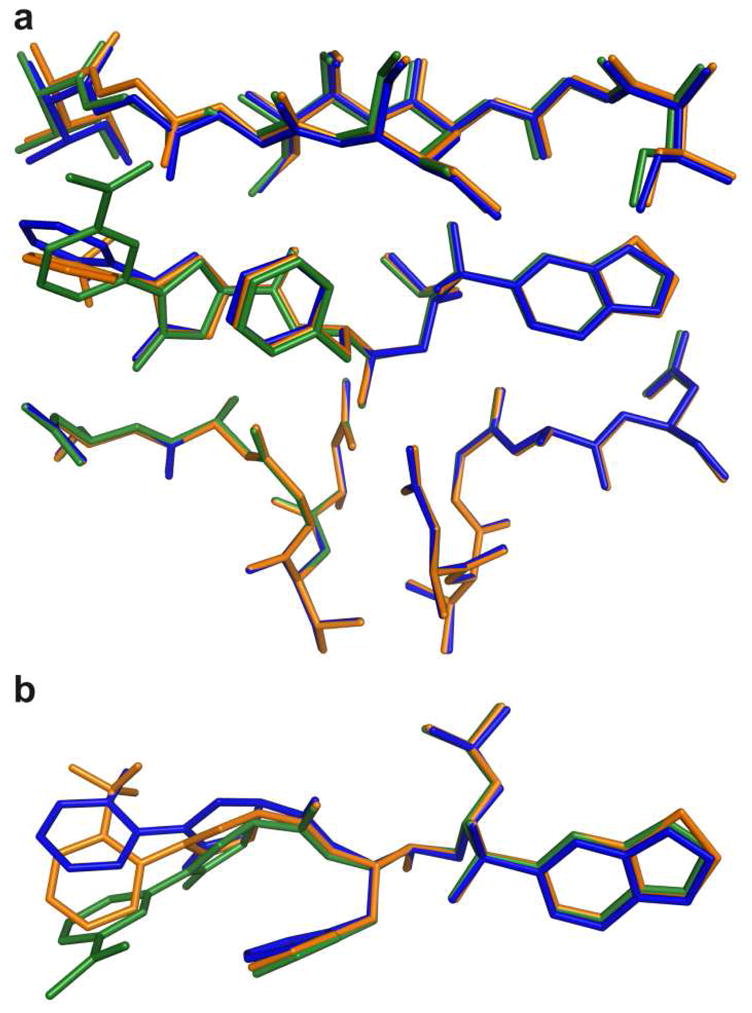
Superposition of protease-inhibitor complexes of 5 (green), 17c (blue) and 18d (orange); inhibitors in the protease active site (A), inhibitors only (B).
The conformational changes at the P2 phenyloxazolidinone moiety resulted in different hydrogen bonding patterns and van der Waals (vdW) interactions in all three complexes. The nitrogen atom of the oxazolidinone moiety forms a water-mediated hydrogen bond with the oxygen atom of the Gly48 in 5 and a water-mediated hydrogen bond with the nitrogen atom of Asp30 in 17c, whereas no water-mediated hydrogen bond is associated with this nitrogen atom in the crystal structure of 18d. The acetyl group on the phenyl ring in 5 does not form any hydrogen bond with the protease but forms vdW contacts with the residues Gly48 and Gly49 as well as the side chain of Phe53 in the flap region. The hydroxyl group on the phenyl ring in 17c forms a direct hydrogen bond with the side chain of Asp30 and a water-mediated hydrogen bond with the main chain nitrogen of Asp30. Although 17c forms more hydrogen bonds than 5, the binding affinity of 17c is significantly lower than that of 5, emphasizing that the binding affinity of an inhibitor is a product of both enthalpic interactions, including hydrophilic and hydrophobic contributions, as well as the entropy of binding.26,27
HIV-1 protease is known to be highly plastic and can adopt different conformations when binding to various ligands.23 In these three complexes, however, the conformational changes in the ligands are more pronounced than those in the protease. These conformational differences at P2 moieties alter the compounds’ interactions with the protease in the S2 binding pocket, which likely account for their significantly different binding affinities.
Conclusion
We have evaluated the structure-activity profiles of a series of HIV-1 protease inhibitors containing phenyloxazolidinone-based P2 ligands to improve the cellular antiviral potency. Based on the structures of ligand-enzyme complexes, a number of (S)-N-phenyloxazolidinone-5-carboxylic acids were designed, synthesized and incorporated into the (R)-hydroxyethylamine isostere as P2 ligands in combination with several phenylsulfonamides as P2′ ligands. In nearly all cases, compounds with the 2-substituted phenyloxazolidinone moieties, designed to improve binding in the S2 region of the enzyme, exhibited lower binding affinities than the 3-substituted analogues. Comparison of the crystal structures of PIs with 2- and 3-substituted P2 phenyloxazolidinone moieties revealed different orientations of the P2 moieties in the S2 binding pocket of the protease, possibly explaining the reduced potencies of the 2-substituted analogues.
Our SAR studies show that P2 moieties with polar substituents at the 3-position of the phenyl ring significantly enhance antiviral potency in resulting compounds. The polarity and size of the P2′ moiety also strongly influenced the enzymatic and antiviral potencies; compounds with large polar groups at P2′ have better potencies than those with smaller groups. In each series, compounds with the benzothiazole moiety at P2′ were the most potent, exhibiting the highest enzymatic and antiviral potencies. These efforts identified three compounds, 27d, 29d, and 31d, with the benzothiazole moiety at P2′ and different phenyloxazolidinone moieties at P2, which demonstrated excellent antiviral potencies against wild-type viruses and better potencies against MDR viruses than LPV. Since variations at the P2 and P2′ moieties significantly affected the antiviral activities of compounds against wild-type and MDR viruses, further optimization of these groups may lead to PIs with improved antiviral potencies against MDR HIV-1 variants.
Experimental Section
Chemistry General
Proton (1H NMR) and carbon nuclear magnetic resonance (13C NMR) spectra were recorded with a Varian Mercury 400 MHz NMR spectrometer operating at 400 MHz for 1H and 100 MHz for 13C NMR. Chemical shifts are reported in ppm (δ scale) relative to the solvent signal, and coupling constant (J) values are reported in hertz. Data are represented as follows: chemical shift, multiplicity (s) singlet, d) doublet, t) triplet, q) quartet, m) multiplet, br) broad, dd) doublet of doublet), coupling constant in Hz, and integration. High-resolution mass spectra (HRMS) were recorded on a Waters Q-TOF Premier mass spectrometer by direct infusion of solutions of each compound using electrospray ionization (ESI) in the positive mode. cLogP values were calculated using ChemBioDraw (CambridgeSoft) version 11. All reactions were performed in oven-dried round bottomed or modified Schlenk flasks fitted with rubber septa under dry N2 atmosphere, unless otherwise noted. Flash and column chromatography was performed using silica gel (230–400 mesh, Merck KGA). Analytical thin-layer chromatography (TLC) was performed using silica gel (60 F-254) coated aluminum plates (Merck KGA), and spots were visualized by exposure to ultraviolet light (UV) and/or exposure to an acidic solution of p-anisaldehyde (anisaldehyde) followed by brief heating. Dichloromethane was dried over P2O5 and distilled, tetrahydrofuran (THF) was distilled from sodium/benzophenone. All other chemicals, reagents, and solvents were purchased from commercial sources and used as received.
The purity of all final compounds was determined by analytical reversed-phase high performance liquid chromatography (HPLC) using two different systems and was found to be ≥ 95%. Analytical HPLC was performed on a Waters Separation Module 2695 system equipped with an auto sampler and a Waters 996 photodiode array detector. Purity of the final compounds was determined using two different chromatographic systems. First system: column, Waters XTerra RP-C18 (3.5 μm, 4.6 mm × 150 mm); mobile phase A, 10 mM ammonium acetate in water; mobile phase B, acetonitrile. Using a flow rate of 1.0 mL/min, gradient elution was performed from 50% B to 100% B over 10 min. Second system: column, Alltech Prevail RP-C18 (3.0 μm, 4.6 mm × 150 mm); mobile phase A, 10 mM ammonium acetate in water; mobile phase B, acetonitrile. Gradient elution was performed from 50% B to 100% B over 10 min at a flow rate of 1.0 mL/min. HPLC retention times and purity data for each target compound is provided in the Supporting Information.
Typical Procedures for the Synthesis of Protease Inhibitors
(S)-5-(Hydroxymethyl)-3-(3-nitrophenyl)oxazolidin-2-one (9m)
A solution of the Cbz-protected aniline derivative 6m (9.45 g, 34.7 mmol) in dry THF (150 mL) was cooled to −78 °C under dry nitrogen atmosphere. A solution of n-BuLi (1.6 M in hexanes; 25 mL, 40 mmol) was slowly added keeping the reaction temperature below −70 °C. After stirring the reaction mixture at −78 °C for 45 min, a solution of (S)-glycidyl butyrate 7 (5 g, 34.7 mmol) in dry THF was slowly added. The resulting mixture was stirred at −78 °C for 2 h and then slowly warmed to room temperature and stirred overnight. Reaction was quenched by the addition of saturated aqueous NH4Cl solution (50 mL). EtOAc (200 mL) and water (50 mL) were added and layers separated, the aqueous layer was further extracted with EtOAc (3 × 150 mL). Combined organic extract was washed with saturated aqueous NaCl solution (100 mL), dried (Na2SO4), filtered, and evaporated under reduced pressure to yield a pale yellow solid. This solid was triturated with a mixture of chloroform and hexanes (1:3) (50 mL) and filtered to provide pure alcohol 9m (5.2 g, 63%) as a pale yellow solid. 1H NMR (400 MHz, CDCl3) δ 8.27 (t, J = 2.0 Hz, 1H), 8.13 (m, 1H), 7.99 (m, 1H), 7.55 (t, J = 8.4 Hz, 1H), 4.82 (m, 1H), 4.15–4.04 (m, 3H), 3.80 (dd, J = 8.4, 6.0 Hz, 1H), 2.18 (br s, 1H); MS (ES+) m/z 239.1 [M + H]+.
(S)-3-(3-Nitrophenyl)-2-oxooxazolidine-5-carboxylic acid (10m)
To an ice-cooled solution of NaIO4 (35 mmol) in water (75 mL) was added a solution of the alcohol 9m (2.4 g, 10 mmol) in a mixture of CH3CN and CCl4 (1:1) (100 mL). Solid RuCl3.H2O (0.105 g, 0.5 mmol) was added and the reaction mixture was stirred at 0 °C for 30 min, warmed to room temperature and stirred for 6 h. Reaction was quenched by adding CH2Cl2 (100 mL), layers were separated, and the aqueous layer was further extracted with CH2Cl2 (2 × 100 mL); combined organic extract was dried (Na2SO4), filtered, and evaporated. The residue was purified by column chromatography on silica gel, eluting with 30% CH3CN in CH2Cl2 + 1% HCO2H, to provide the acid 10m (2.1 g, 83%) as a pale yellow solid. 1H NMR (400 MHz, CD3OD) δ 8.59 (t, J = 2.4 Hz, 1H), 8.01 (ddd, J = 8.0, 2.4, 1.2 Hz, 1H), 7.91 (ddd, J = 8.0, 2.4, 1.2 Hz, 1H), 7.63 (t, J = 8.0 Hz, 1H), 5.21 (dd, J = 9.6, 5.2 Hz, 1H), 4.47 (t, J = 9.2 Hz, 1H), 4.24 (dd, J = 9.6, 5.6 Hz, 1H); MS (ES−) m/z 251.1 [M − H]−.
(5S)-N-[(1S,2R)-3-[(6-Benzothiazolylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-3-(3-nitrophenyl)-2-oxooxazolidine-5-carboxamide (28d)
Excess oxalyl chloride (6 mL) was added to the acid 10m (0.253 g, 1.0 mmol) and the resulting mixture was stirred at room temperature overnight. Oxalyl chloride was removed by distillation under reduced pressure and the residue was dried under high vacuum for 30 min. The resulting acid chloride was dissolved in dry THF (10 mL) and was used in the coupling reaction.
To a solution of the N-[(1S,2R)-3-[(6-benzothiazolylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-carbamic acid-1,1-dimethylethyl ester25 14d (0.534 g, 1 mmol) in CH2Cl2 (10 mL) was added trifluoroacetic acid (4 mL) and the resulting mixture was stirred at room temperature for 1 h. The reaction mixture was concentrated under reduced pressure; the residue was dissolved in toluene (10 mL) and evaporated at reduced pressure to dryness. The resulting de-protected amine was dissolved in dry THF (10 mL) and the solution was cooled 0 °C. Et3N (0.3 mL, 2.15 mmol) was added and after 15 min the above acid chloride solution was slowly added; the resulting reaction mixture was stirred at 0 °C for 15 min, allowed to warm to room temperature, and stirred until reaction was complete (monitored by TLC). Water (10 mL) and EtOAc (50 mL) were added and layers were separated, the organic portion was washed with saturated aqueous NaCl solution (30 mL), dried (Na2SO4), filtered, and evaporated. The residue was purified by flash chromatography on silica gel, eluting with a mixture of EtOAc-hexanes (4:2), to afford the target compound 28d (0.56 g, 84%) as a white solid. 1H NMR (400 MHz, CDCl3) δ 9.23 (s, 1H), 8.50 (d, J = 1.6 Hz, 1H), 8.28 (d, J = 8.4 Hz, 1H), 8.22 (t, J = 2.0 Hz, 1H), 8.05 (m, 1H), 7.93–7.90 (m, 2H), 7.60 (t, J = 8.4 Hz, 1H), 7.13 (d, J = 7.2 Hz, 2H), 7.02 (t, J = 7.2 Hz, 2H), 6.85 (t, J = 7.2 Hz, 1H), 6.79 (d, J = 10.0 Hz, 1H), 4.84 (dd, J = 9.6, 5.2 Hz, 1H), 4.29 (m, 1H), 4.09 (t, J = 9.2 Hz, 1H), 3.99 (m, 1H), 3.41 (dd, J = 9.2, 6.0 Hz, 1H), 3.29 (dd, J = 15.6, 9.6 Hz, 1H), 3.14–3.05 (m, 3H), 2.93 (dd, J = 13.2, 6.4 Hz, 1H), 2.78 (dd, J = 13.6, 10.4 Hz, 1H), 1.89 (m, 1H), 0.96 (d, J = 6.8 Hz, 3H), 0.91 (d, J = 6.4 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 168.36, 158.38, 155.90, 152.82, 148.86, 138.69, 137.44, 135.68, 134.69, 130.33, 129.61 (2C), 128.62 (2C), 126.67, 125.05, 124.75, 123.92, 122.59, 119.35, 113.01, 72.58, 69.98, 59.12, 53.91, 53.45, 48.10, 35.53, 27.57, 20.38, 20.13; HRMS (ESI) m/z: calcd for C31H34N5O8S [M + H]+ 668.1849; found 668.1852.
(5S)-3-(3-Aminophenyl)-N-[(1S,2R)-3-[(6-benzothiazolylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-2-oxooxazolidine-5-carboxamide (29d)
A mixture of the above nitro compound 28d (0.335 g, 0.5 mmol) and SnCl2.2H2O (0.565 g, 2.5 mmol) in EtOAc (20 mL) was heated at 80 °C for 3 h. Reaction mixture was allowed to cool to ambient temperature and treated with saturated aqueous NaHCO3 solution (15 mL). It was diluted with EtOAc (50 mL) and layers were separated, aqueous layer was further extracted with EtOAc (2 × 50 mL); combined organic extract was washed with saturated aqueous NaCl solution (2 × 40 mL), dried (Na2SO4), filtered, and evaporated. The residue was purified by flash chromatography on silica gel, eluting with a mixture of 1% MeOH in EtOAc, to provide the target compound 29d (0.29 g, 91%) as a white solid. 1H NMR (400 MHz, CDCl3) δ 9.21 (s, 1H), 8.49 (d, J = 2.0 Hz, 1H), 8.27 (d, J = 8.8 Hz, 1H), 7.92 (dd, J = 8.8, 1.6 Hz, 1H), 7.17–7.12 (m, 3H), 7.09–7.04 (m, 3H), 6.96 (t, J = 7.6 Hz, 1H), 6.90 (d, J = 9.6 Hz, 1H), 6.55 (dd, J = 8.0, 1.6 Hz, 1H), 6.51 (dd, J = 8.0, 2.0 Hz, 1H), 4.73 (dd, J = 10.0, 6.4 Hz, 1H), 4.21 (m, 1H), 4.0 (t, J = 9.6 Hz, 2H), 3.85 (br s, 1H), 3.75 (br s, 1H), 3.32 (dd, J = 9.6, 6.4 Hz, 1H), 3.25 (dd, J = 15.2, 9.2 Hz, 1H), 3.14 (dd, J = 14.0, 4.8 Hz, 1H), 3.09–3.03 (m, 2H), 2.93 (dd, J = 13.6, 6.8 Hz, 1H), 2.73 (dd, J = 13.6, 10.8 Hz, 1H), 1.89 (m, 1H), 0.95 (d, J = 6.4 Hz, 3H), 0.91 (d, J = 6.4 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 169.07, 158.34, 155.87, 153.14, 147.55, 138.46, 137.47, 135.69, 134.66, 130.10, 129.53 (2C), 128.70 (2C), 126.85, 125.09, 124.72, 122.60, 111.81, 108.37, 105.48, 72.49, 68.87, 59.11, 53.99, 53.74, 48.47, 35.84, 27.55, 20.38, 20.12; HRMS (ESI) m/z: calcd for C31H36N5O6S2 [M + H]+ 638.2107; found 638.2098.
(5S)-3-[3-(Acetylamino)phenyl]-N-[(1S,2R)-3-[(6-benzothiazolylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-2-oxooxazolidine-5-carboxamide (30d)
To a solution of compound 29d (0.1 g, 0.157 mmol) in dry CH2Cl2 was added Ac2O (30 μL), and the resulting solution was stirred at room temperature for 2 h. The solvents were evaporated under reduced pressure and the residue was purified by flash chromatography, eluting with 1% MeOH in EtOAc, to afford the title compound 30d (0.1 g, 94%) as a white solid. 1H NMR (400 MHz, CDCl3) δ 9.20 (s, 1H), 8.49 (d, J = 2.0 Hz, 1H), 8.23 (d, J = 8.8 Hz, 1H), 7.95 (s, 1H), 7.91 (dd, J = 8.8, 1.6 Hz, 1H), 7.72 (s, 1H), 7.48 (d, J = 7.2 Hz, 1H), 7.35–7.26 (m, 2H), 7.14 (d, J = 7.2 Hz, 2H), 7.02 (t, J = 7.6 Hz, 2H), 6.90 (t, J = 7.6 Hz, 1H), 6.84 (d, J = 8.0 Hz, 1H), 4.76 (dd, J = 9.6, 6.0 Hz, 1H), 4.22 (m, 1H), 4.11 (m, 2H), 4.0 (t, J = 10.0 Hz, 1H), 3.31–3.15 (m, 4H), 3.05 (dd, J = 13.2, 7.6 Hz, 1H), 2.96 (dd, J = 13.6, 7.2 Hz, 1H), 2.83 (dd, J = 14.0, 10.8 Hz, 1H), 2.19 (s, 3H), 1.92 (m, 1H), 0.92 (d, J = 6.8 Hz, 3H), 0.90 (d, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 168.97, 158.34, 155.83, 153.43, 139.14, 137.98, 137.81, 135.84, 134.63, 129.87, 129.61 (2C), 128.58 (2C), 126.75, 125.08, 124.66, 122.56, 116.25, 113.81, 110.08, 72.63, 70.02, 58.96, 53.89, 53.80, 48.48, 36.02, 27.44, 24.89, 20.36, 20.11; HRMS (ESI) m/z: calcd for C33H38N5O7S2 [M + H]+ 680.2213; found 680.2225.
(5S)-N-[(1S,2R)-3-[(6-benzothiazolylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]amino]carbonyl]-2-oxo-3-oxazolidinyl]phenyl]-carbamic acid methyl ester (31d)
To an ice-cooled solution of compound 29d (0.1 g, 0.157 mmol) in CH2Cl2 (5 mL) was added an aqueous solution of Na2CO3 (0.026 g, 0.25 mmol) in water (2 mL) followed by the slow addition of methylchloroformate (12.3 μL, 0.157 mmol). After 15 min, the reaction mixture was warmed to ambient temperature and stirred till no starting material was detected by TLC (2 h). Reaction mixture was diluted with CH2Cl2 and layers were separated, organic extract was washed with saturated aqueous NaCl solution (5 mL), dried (Na2SO4), filtered, and evaporated. The residue was purified by flash chromatography on silica gel, eluting with a mixture of 2% MeOH in EtOAc, to afford the target compound 31d (0.095 g, 87%) as a white solid. 1H NMR (400 MHz, CDCl3) δ 9.21 (s, 1H), 8.49 (d, J = 1.6 Hz, 1H), 8.25 (d, J = 8.8 Hz, 1H), 7.91 (dd, J = 8.8, 2.0 Hz, 1H), 7.76 (t, J = 1.6 Hz, 1H), 7.34–7.26 (m, 2H), 7.13 (d, J = 7.2 Hz, 2H), 7.06–6.89 (m, 6H), 4.76 (dd, J = 10.0, 6.0 Hz, 1H), 4.23 (m, 1H), 4.03 (t, J = 10.0 Hz, 2H), 3.82 (br s, 1H), 3.79 (s, 3H), 3.34 (dd, J = 9.2, 6.0 Hz, 1H), 3.25 (dd, J = 15.6, 9.2 Hz, 1H), 3.19–3.10 (m, 2H), 3.05 (dd, J = 13.6, 8.0 Hz, 1H), 2.94 (dd, J = 13.6, 7.2 Hz, 1H), 2.79 (dd, J = 13.6, 10.8 Hz, 1H), 1.90 (m, 1H), 0.94 (d, J = 6.8 Hz, 3H), 0.90 (d, J = 6.4 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 168.94, 158.31, 155.85, 154.23, 153.21, 139.12, 138.18, 137.57, 135.78, 134.63, 129.94, 129.57 (2C), 128.65 (2C), 126.82, 125.08, 124.69, 122.58, 114.88, 113.11, 108.87, 72.60, 69.94, 59.04, 53.89, 53.75, 52.73, 48.45, 35.88, 27.51, 20.36, 20.12; HRMS (ESI) m/z: calcd for C33H38N5O8S2 [M + H]+ 696.2162; found 696.2158.
HIV-1 Protease Inhibition Assays
The HIV-1 protease inhibitory potencies of all compounds were determined by a fluorescence resonance energy transfer (FRET) method.20,25,28 Protease substrate, (Arg-Glu(EDANS)-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln Lys(DABCYL)-Arg) was purchased from Molecular Probes. The energy transfer donor (EDANS) and acceptor (DABCYL) dyes were labeled at two ends of the peptide, respectively, to perform FRET. Fluorescence measurements were carried out on a fluorescence spectrophotometer (Photon Technology International) at 30 °C. Excitation and emission wavelengths were set at 340 and 490 nm, respectively. Each reaction was recorded for about 10 min. The expression, isolation, and purification of wild-type and mutant HIV-1 protease used for binding experiments were carried out as previously described.29 Wild-type HIV-1 protease (Q7K) and its MDR variant (L10I, L63P, A71V, G73S, I84V, L90M) were desalted through PD-10 columns (Amersham Biosciences). Sodium acetate (20 mM, pH 5) was used as the elution buffer. Apparent protease concentrations were around 50 nM as estimated by UV spectrophotometry at 280 nm. All inhibitors were dissolved in dimethylsulfoxide (DMSO) and diluted to appropriate concentrations. Protease (2 μL) and inhibitor (2 μL) or DMSO were mixed and incubated for 20–30 min at room temperature before initializing the substrate cleavage reaction. For all experiments, 150 μL of a 1 μM substrate were used in substrate buffer [0.1 M sodium acetate, 1 M sodium chloride, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 2% DMSO, and 1 mg/mL bovine serum albumin (BSA) with an adjusted pH of 4.7]. Inhibitor binding dissociation constant (Ki) values were obtained by nonlinear regression fitting (GraFit 5, Erithacus software) to the plot of initial velocity as a function of inhibitor concentrations based on the Morrison equation.30 The initial velocities were derived from the linear range of reaction curves.
Antiviral Assays
Drug susceptibility assays were carried out by Monogram Biosciences against three patient-derived strains of wild-type HIV-1 from clades A, B, and C, and against two multidrug-resistant HIV-1 variants. Assays were carried out according to protocols detailed by Petropoulos et. al.31 and at the website http://www.monogrambio.com. Genbank accession numbers for the PR/RT regions of the six HIV-1 strains used in antiviral assays are as follows: WT-Control: HQ179654; WT-A: HQ179655; WT-B: HQ179656; WT-C: HQ179657; MDR: HQ179658; MDR1: HQ179659.
Protein Crystallography
The expression, isolation, and purification of wild-type and mutant HIV-1 proteases used for crystallization and binding experiments were carried out as previously described.29 Co-crystals of the inhibitors with the wild-type protease were grown by hanging drop vapor diffusion method. The protease solution of concentration 1.2–1.8 mg/ml was equilibrated with 3-fold molar excess of inhibitors for at least one hour prior to crystallization. The reservoir solution consisted of 126 mM phosphate buffer at pH 6.2, 63 mM sodium citrate and ammonium sulfate in a range of 24–29%.
The crystals used for data collection were mounted in Mitegen Micromounts and flash frozen over a nitrogen stream. Intensity data were collected at −80 °C on an in-house Rigaku X-ray generator equipped with an R-axis IV image plate. 180 frames were collected per crystal with an angular separation of 1° and no overlap between frames. The data processing of the frames was carried out using the programs DENZO and ScalePack,32,33 respectively; data collection statistics are listed in Table 3.
Table 3.
Crystallographic Data Collection and Refinement Statistics for Complexes of 17C and 18d with the Wild-Type HIV-1 Proteasea
| 17c | 18d | |
|---|---|---|
| Resolution (Å) | 1.95 | 1.85 |
| Space group | P212121 | P212121 |
| a (Å) | 50.67 | 50.79 |
| b (Å) | 57.86 | 58.25 |
| c (Å) | 61.91 | 62.01 |
| Z | 4 | 4 |
| Rmerge (%) | 10.2 | 8.7 |
| Completeness (%) | 99.7 | 98.4 |
| Total no. of reflections | 91018 | 100852 |
| No. of unique reflections | 13809 | 16073 |
| Rfree (%) | 20.1 | 22.3 |
| Rfactor (%) | 16.5 | 18.0 |
| RMSDa in: | 0.008 | 0.009 |
| Bond lengths (Å) | ||
| RMS Angle (°) | 1.340 | 1.317 |
| Temperature (°C) | −80 | −80 |
| PDB code | 3MXD | 3MXE |
RMSD, root mean square deviation; PDB, protein data bank
The crystal structures were solved and refined using the programs within the CCP4 interface.34 Structure solution was obtained with the molecular replacement package AMoRe,35 with 1F7A36 as the starting model. Upon obtaining solution, the molecular replacement phases were further improved using ARP/wARP37 to build solvent molecules into the unaccounted regions of electron density. Model building was performed using the interactive graphics program Coot.38 Conjugate gradient refinement using Refmac539 was performed by incorporating Schomaker and Trueblood tensor formulation of TLS (translation, libration, screw-rotation) parameters.40–42 The working R (Rfactor) and its cross validation (Rfree) were monitored throughout the refinement. The refinement statistics are also shown in Table 3.
Supplementary Material
Figure 2.
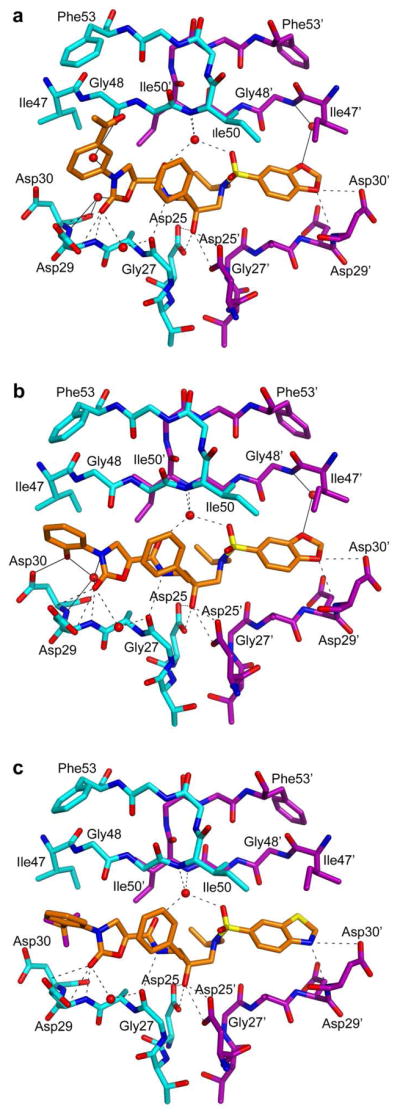
Crystal structures of inhibitors 5 (A), 17c (B), and 18d (C) in complex with HIV-1 protease. Dashed lines represent hydrogen bonds common to all three inhibitors and solid lines represent hydrogen bonds specific to each inhibitor.
Acknowledgments
We thank Kaneka USA for generous gifts of chiral epoxides, Ellen A. Nalivaika for providing the wild-type and MDR HIV-1 proteases, the AIDS Research and Reference Reagent Program (NIAD, NIH) for reference protease inhibitors, Monogram Biosciences for antiviral testing, and members of Rana and Schiffer laboratories for helpful discussions. This work was supported in part by a grant from the National Institutes of General Medical Sciences of the NIH (PO1-GM066524).
Footnotes
Abbreviations: HIV-1, human immunodeficiency virus type 1; AIDS, acquired immune deficiency syndrome; HAART, highly active antiretroviral therapy; MDR, multidrug-resistant; PI, protease inhibitor; RTV, ritonavir; DRV, darunavir; TPV, tipranavir; APV, amprenavir; LPV, lopinavir; THF, tetrahydrofuran; SAR, structure-activity relationships; vdW, van der Waals.
PDB codes for HIV-1 protease complexes with 17c and 18d are 3MXD and 3MXE, respectively.
Supporting Information Available: Characterization data for additional target compounds and HPLC purity data for all compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Celia A. Schiffer, Email: Celia.Schiffer@umassmed.edu.
Tariq M. Rana, Email: trana@sanfordburnham.org.
References
- 1.The Joint United Nations Program on HIV/AIDS (UNAIDS) 2008 Report on the Global AIDS Epidemic. UNAIDS; Geneva, Switzerland: 2008. [Google Scholar]
- 2.Hammer SM, Eron JJ, Jr, Reiss P, Schooley RT, Thompson MA, Walmsley S, Cahn P, Fischl MA, Gatell JM, Hirsch MS, Jacobsen DM, Montaner JSG, Richman DD, Yeni PG, Volberding PA. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the international AIDS society-USA panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 3.Gulick RM, Mellors JW, Havlir D, Eron JJ, Meibohm A, Condra JH, Valentine FT, McMahon D, Gonzalez C, Jonas L, Emini EA, Chodakewitz JA, Isaacs R, Richman DD. 3-Year suppression of HIV viremia with indinavir, zidovudine, and lamivudine. Ann Intern Med. 2000;133:35–39. doi: 10.7326/0003-4819-133-1-200007040-00007. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JA, DeMasi R, Quinn J, Moxham C, Rousseau F. Overview of the effectiveness of triple combination therapy in antiretroviral-naive HIV-1 infected adults. AIDS. 2001;15:1369–1377. doi: 10.1097/00002030-200107270-00006. [DOI] [PubMed] [Google Scholar]
- 5.Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. The, H. I. V. O. S. I. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 6.Hogg RS, Heath KV, Yip B, Craib KJP, O’Shaughnessy MV, Schechter MT, Montaner JSG. Improved survival among HIV-infected individuals following initiation of antiretroviral therapy. JAMA. 1998;279:450–454. doi: 10.1001/jama.279.6.450. [DOI] [PubMed] [Google Scholar]
- 7.Mehellou Y, De Clercq E. Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go? J Med Chem. 2010;53:521–538. doi: 10.1021/jm900492g. [DOI] [PubMed] [Google Scholar]
- 8.Kempf DJ, Marsh KC, Kumar G, Rodrigues AD, Denissen JF, McDonald E, Kukulka MJ, Hsu A, Granneman GR, Baroldi PA, Sun E, Pizzuti D, Plattner JJ, Norbeck DW, Leonard JM. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob Agents Chemother. 1997;41:654–660. doi: 10.1128/aac.41.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeldin RK, Petruschke RA. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J Antimicrob Chemother. 2004;53:4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- 10.Youle M. Overview of boosted protease inhibitors in treatment-experienced HIV-infected patients. J Antimicrob Chemother. 2007;60:1195–1205. doi: 10.1093/jac/dkm364. [DOI] [PubMed] [Google Scholar]
- 11.Turner SR, Strohbach JW, Tommasi RA, Aristoff PA, Johnson PD, Skulnick HI, Dolak LA, Seest EP, Tomich PK, Bohanon MJ, Horng MM, Lynn JC, Chong KT, Hinshaw RR, Watenpaugh KD, Janakiraman MN, Thaisrivongs S. Tipranavir (PNU-140690): A potent, orally bioavailable nonpeptidic HIV protease inhibitor of the 5,6-dihydro-4-hydroxy-2-pyrone sulfonamide class. J Med Chem. 1998;41:3467–3476. doi: 10.1021/jm9802158. [DOI] [PubMed] [Google Scholar]
- 12.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother. 2003;47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Meyer S, Azijn H, Surleraux D, Jochmans D, Tahri A, Pauwels R, Wigerinck P, de Bethune MP. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob Agents Chemother. 2005;49:2314–2321. doi: 10.1128/AAC.49.6.2314-2321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surleraux DL, Tahri A, Verschueren WG, Pille GM, de Kock HA, Jonckers TH, Peeters A, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PB. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J Med Chem. 2005;48:1813–1822. doi: 10.1021/jm049560p. [DOI] [PubMed] [Google Scholar]
- 15.Esté JA, Cihlar T. Current status and challenges of antiretroviral research and therapy. Antivir Res. 2010;85:25–33. doi: 10.1016/j.antiviral.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Clavel F, Hance AJ. HIV drug resistance. N Engl J of Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 17.Baxter JD, Schapiro JM, Boucher CAB, Kohlbrenner VM, Hall DB, Scherer JR, Mayers DL. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J Virol. 2006;80:10794–10801. doi: 10.1128/JVI.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Meyer S, Vangeneugden T, Van Baelen B, De Paepe E, Van Marck H, Picchio G, Lefebvre E, De Béthune MP. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res Hum Retroviruses. 2008;24:379–388. doi: 10.1089/aid.2007.0173. [DOI] [PubMed] [Google Scholar]
- 19.Shafer RW, Schaprio JM. HIV-1 drug resistance mutations: An updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 20.Ali A, Reddy GSKK, Cao H, Anjum SG, Nalam MNL, Schiffer CA, Rana TM. Discovery of HIV-1 protease inhibitors with picomolar affinities incorporating N-aryl-oxazolidinone-5-carboxamides as novel P2 ligands. J Med Chem. 2006;49:7342–7356. doi: 10.1021/jm060666p. [DOI] [PubMed] [Google Scholar]
- 21.Reddy GSKK, Ali A, Nalam MNL, Anjum SG, Cao H, Nathans RS, Schiffer CA, Rana TM. Design and synthesis of HIV-1 protease inhibitors incorporating oxazolidinones as P2/P2′ ligands in pseudosymmetric dipeptide isosteres. J Med Chem. 2007;50:4316–4328. doi: 10.1021/jm070284z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali A, Reddy GSKK, Cao H, Anjum SG, Nalam MNL, Schiffer CA, Rana TM. Design, synthesis, and biological evaluation of HIV-1 protease inhibitors incorporating phenyloxazolidines as novel P2 ligands. American Chemical Society; 2007. p. MEDI-094. [Google Scholar]
- 23.Nalam MNL, Ali A, Altman MD, Reddy GSKK, Chellappan S, Kairys V, Ozen A, Cao H, Gilson MK, Tidor B, Rana TM, Schiffer CA. Evaluating the substrate-envelope hypothesis: Structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J Virol. 2010;84:5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brickner SJ, Hutchinson DK, Barbachyn MR, Manninen PR, Ulanowicz DA, Garmon SA, Grega KC, Hendges SK, Toops DS, Ford CW, Zurenko GE. Synthesis and antibacterial activity of U-100592 and U-100766, two oxazolidinone antibacterial agents for the potential treatment of multidrug-resistant gram-positive bacterial infections. J Med Chem. 1996;39:673–679. doi: 10.1021/jm9509556. [DOI] [PubMed] [Google Scholar]
- 25.Altman MD, Ali A, Reddy GSKK, Nalam MNL, Anjum SG, Cao H, Chellappan S, Kairys V, Fernandes MX, Gilson MK, Schiffer CA, Rana TM, Tidor B. HIV-1 Protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J Am Chem Soc. 2008;130:6099–6113. doi: 10.1021/ja076558p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discovery Today. 2008;13:869–874. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010 doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matayoshi ED, Wang GT, Krafft GA, Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 29.King NM, Melnick L, Prabu-Jeyabalan M, Nalivaika EA, Yang SS, Gao Y, Nie X, Zepp C, Heefner DL, Schiffer CA. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 2002;11:418–429. doi: 10.1110/ps.25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco WR, Hakala MT. Evaluation of methods for estimating the dissociation constant of tight binding enzyme inhibitors. J Biol Chem. 1979;254:12104–12109. [PubMed] [Google Scholar]
- 31.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minor W. XdisplayF. Purdue University; West Lafayette, Indiana: 1993. [Google Scholar]
- 33.Otwinowski Z, Minor W, Charles W, Carter . Methods in Enzymology. Vol. 276. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project. Number 4. The CCP4suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 35.Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 36.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J Mol Biol. 2000;301:1207–1220. doi: 10.1006/jmbi.2000.4018. [DOI] [PubMed] [Google Scholar]
- 37.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr. 2002;D58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Schomaker V, Trueblood KN. On the rigid-body motion of molecules in crystals. Acta Crystallogr. 1968;B24:63–76. [Google Scholar]
- 41.Kuriyan J, Weis WI. Rigid protein motion as a model for crystallographic temperature factors. Proc Natl Acad Sci USA. 1991;88:2773–2777. doi: 10.1073/pnas.88.7.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tickle IJ, Moss DS. IUCr99 Computing School. IUCr; London: 1999. Modeling rigid-body thermal motion in macromolecular crystal structure refinement. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.