Abstract
microRNAs (miRNAs) are a class of ~22 nucleotide short non-coding RNAs that play key roles in fundamental cellular processes, including how cells respond to changes in environment or, broadly defined, stresses. Responding to stresses, cells either choose to restore or re-program their gene expression patterns. This decision is partly mediated by miRNA functions, in particular, by modulating the amount of miRNAs, the amount of mRNA targets, or the activity/mode of action of miRNA-protein complexes. In turn, these changes determine the specificity, timing and concentration of gene products expressed upon stresses. Dysregulation of these processes contributes to chronic diseases, including cancers.
Introduction
Stress, broadly defined, is the state when cells deviate from the status quo due to sudden environmental changes or frequent fluctuations in environmental factors. Such changes can damage existing macromolecules, including proteins, mRNAs, DNA and lipids, that need to be replenished; if the damage is not dealt with, metabolic imbalance can be incurred and redox potentials altered (Kultz, 2005). Depending on the severity and duration of stress encountered, cells either re-establish cellular homeostasis to the former state or adopt an altered state in the new environment. Such stress responses are mediated via multiple mechanisms (Kultz, 2005), such as induction of molecular chaperones (Buchberger et al., 2010; Richter et al., 2010), rapid clearance of damaged macromolecules (Kroemer et al., 2010), growth arrest, activation of certain gene expression programs (Spriggs et al., 2010) and, when cells can no longer cope with excessive damage, cell death. In this review, we will examine how microRNAs (miRNAs) are well positioned to help restore homeostasis upon sudden environmental changes, or, in the long run, enforce a new gene expression program so cells can tolerate the new environment. We will explore how stress alters the transcription, processing and turnover of miRNAs, the expression of mRNA targets and the activities of miRNA-protein complexes. To conclude, we will consider the possible roles of miRNA-mediated stress responses in the context of diseases and potential therapeutic avenues.
What are microRNAs?
As the name implies, miRNAs are short RNAs of ~22 nucleotides that modulate the stability and/or translational potential of their mRNA targets (Figure 1A) (Bushati and Cohen, 2007; Carthew and Sontheimer, 2009; Fabian et al., 2010). Over 60% of all mammalian mRNAs are predicted targets of miRNAs (Friedman et al., 2009) and miRNAs constitute a sizable class of regulators (e.g. ≥950 different miRNAs in humans according to the miRBase release 15 in April 2010; http://www.mirbase.org/), even outnumbering kinases and phosphatases, indicating their pervasive roles in regulation of cellular processes.
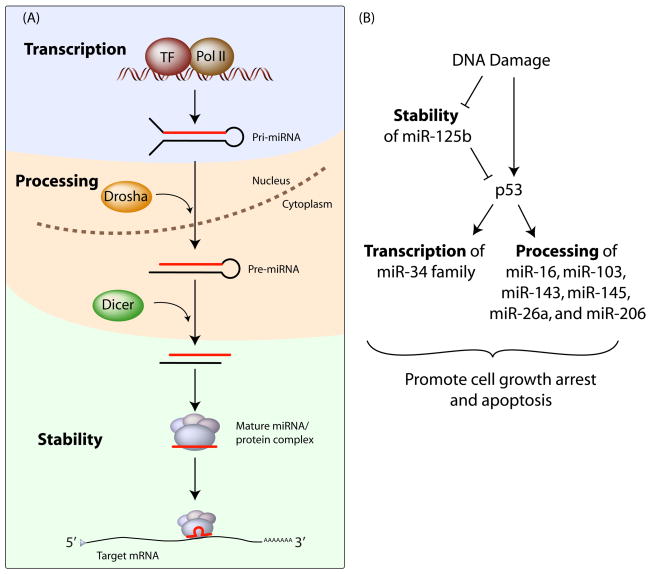
Figure 1. Biogenesis of mature miRNAs can be regulated at multiple levels.
(A) miRNAs are first transcribed as primary transcripts (pri-miRNA), which fold into hairpin structures and are subsequently processed – first by Drosha in the nucleus and then by Dicer in the cytoplasm. These processing steps result in a ~22 nucleotide duplex in which one strand, the mature miRNA, binds to the core protein Argonaute and associated protein factors to form the final effector complex. The activity of the effector complexes is determined by the half-lives of the miRNAs, the protein composition of the effector complexes and the post-translational modifications of the individual components. TF: Transcription Factor; Pol II: RNA polymerase II. (B) Upon DNA damage, transcription, processing and stability of specific miRNAs are modulated, resulting in a p53-mediated gene expression program that promotes cell growth arrest and apoptosis.
In particular, multiple lines of genetic evidence indicate that miRNAs play key roles in mediating stress responses (reviewed in (Leung and Sharp, 2007)). Perhaps somewhat surprisingly, systematic inactivation of individual miRNA in flies and worms has revealed that many miRNAs are dispensable for development or viability under standard laboratory conditions (Bushati and Cohen, 2007; Leaman et al., 2005; Miska et al., 2007). However, this is not the case when some of these animals are subjected to changing environments. For example, miR-7 knockout flies no longer develop their eyes properly when subjected to alternating temperatures (Li et al., 2009); miR-14 mutant flies are sensitive to high salinity (Xu et al., 2003); mice deficient in miR-208 cannot cope with cardiac overload (van Rooij et al., 2007); and inactivation of miR-8 renders zebrafish incapable of responding to osmotic stress (Flynt et al., 2009). Thus, miRNA mutants could appear apparently normal but exhibit phenotypic crisis in stress conditions.
microRNAs and stress
Emerging data suggest that stress conditions can alter the biogenesis of miRNAs, the expression of mRNA targets and the activities of miRNA-protein complexes. In the sections that follow, we first explore 8 specific examples in the literature regarding the multiple roles of miRNAs in stress responses. Then, we discuss how stress regulates miRNA activities and how cells mediate stress responses through miRNAs.
I. Genome guardian p53 regulates miRNA biogenesis upon DNA damage
As numerous steps are required to produce mature miRNAs, this sequence of events offers various “tuning knobs” to precisely adjust the level of mature miRNAs (Figure 1A). One player that senses environmental stress and modulates the level of miRNAs is p53, a tumor suppressor that is commonly activated upon DNA damage.
p53 regulates the expression of specific miRNAs at two different levels – transcription and processing (Figure 1B). Upon DNA damage, p53 induces the transcription of the primary transcripts that encompass miR-34a, miR-34b and miR-34c from distinct genomic loci. In turn, these miRNAs together repress a number of shared target genes to promote growth arrest and apoptosis (reviewed in (Hermeking, 2007)). In addition, p53 enhances the processing of a restricted population of pri-miRNAs in cancer cells by associating with DDX5, a co-factor of Drosha (Suzuki et al., 2009). Overexpression of each of these miRNAs (miR-16, miR-103, miR-143, miR-145, miR-26a and miR-206) decreases the rate of cell proliferation. Interestingly, most p53 mutations found in cancers are located in a domain that is required for both the miRNA processing function and transcriptional activity (Junttila and Evan, 2009; Suzuki et al., 2009). Thus, a loss of p53 function in transcription and processing of specific miRNAs might contribute to tumor progression.
On the other hand, to ensure a robust DNA damage response, the expression level of p53 must be tightly regulated and this is partly mediated by the ubiquitin-mediated degradation of p53 (Junttila and Evan, 2009). To further keep its default expression level low, p53 is also regulated by a miRNA, miR-125b, in humans and zebrafish (Le et al., 2009). Such repression is relieved upon DNA damage by a decrease in miR-125b levels through an unknown mechanism. Therefore, during DNA damage, a relief in miR-125b-mediated repression of p53 is coupled with the activation of p53 through a cascade of kinases (Junttila and Evan, 2009), eventually resulting in upregulation of genes including miRNAs that promote cell cycle arrest and apoptosis (Figure 1B). In effect, the default repression of p53 by miR-125b raises the threshold required for p53 activation, providing another layer of safeguard before the DNA damage response can be triggered.
II. Stress-induced target expression overcomes miRNA-defined threshold
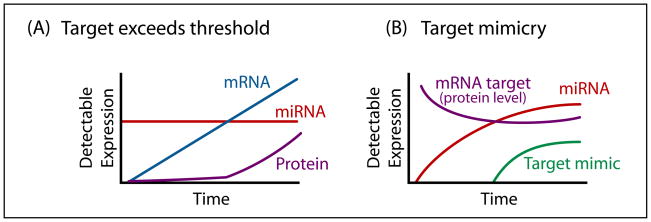
Apart from the cellular concentration of miRNAs, the level of target gene repression is also dependent on the concentration of mRNA target relative to the miRNA (e.g. (Doench et al., 2003)). This is illustrated in the case of two extracellular ligands MICA and MICB, which are induced by stresses like heat shock, oxidative stress, viral infection and DNA damage (Stern-Ginossar et al., 2008). These ligands are recognized by an immune activating receptor, NKG2D, which is expressed on Natural Killer cells and T cells to allow recognition and elimination of tumor and virus-infected cells. In normal cells, the translation of mRNAs encoding these ligands is inhibited by miR-20a, miR-93 or miR-106b. However, upon stress, while the levels of these miRNAs remain unchanged, the transcription of MICA and MICB mRNAs is substantially upregulated and significant protein levels are now detected. This result suggests that the increase in mRNA level presumably exceeds the quantity that can be “titrated” by the cellular miRNAs, resulting in the expression of MICA and MICB proteins (Stern-Ginossar et al., 2008). In effect, the relative levels of miRNAs and mRNA targets determine how much target protein is produced under normal conditions, thus providing a threshold. During stress, the high level of mRNAs encoding the ligands exceeds the threshold and contributes to an innate immune state (Figure 2A).
Figure 2. Stress-induced expression of mRNA targets or target mimics affects miRNA activity.
The protein output of an mRNA target is dependent on (A) the level of mRNA targets relative to the amount of miRNAs as well as (B) the level of mRNA targets relative to other mRNAs targeted by the same miRNA. (A) Translation of mRNA targets is generally suppressed by a constant level of miRNAs. However, when a particular mRNA target expression level exceeds the quantity of mRNA targets that can be titrated by the amount of miRNAs, translation of mRNA targets accelerates. (B) Translation of mRNA targets decreases along with the increase in miRNAs over time. Such decrease in translation halts as a result of the expression of target mimics, which compete with mRNA targets for the same miRNA.
III. Relief of miRNA repression by target mimicry
The level of miRNA-mediated repression depends not only on the ratio of a particular mRNA target relative to miRNA, but also on the amount of other mRNAs present in the transcriptome that are targeted by the same miRNA. Plant cells exploit this property to adapt to the amount of inorganic phosphate present in the environment (Franco-Zorrilla et al., 2007). Upon phosphate deprivation, plants remobilize phosphate from old leaves to new leaves by repressing a negative regulator of this process, PHO2, through transcriptional induction of miR-399. As expected, the level of PHO2 expression decreases as miR-399 accumulates and binds to the PHO2 mRNA; however, such reduction halts after 6 days, even though the level of miR-399 continues to rise. Franco-Zorrilla and colleagues discovered that the halt in miR-399-mediated repression of PHO2 is due to the induction, starting on day 6, of a conserved family of non-coding RNA species known as Mt4-TPS1 that harbors a miR-399 binding site. This binding site competes with PHO2 transcripts for the binding to miR-399. Therefore, overexpression of a target mimic is another means that cells can exploit to modulate the degree of miRNA-mediated repression during stress (Figure 2B).
Similar phenomenon of target mimicry was also observed in animal cells (reviewed in (Ebert and Sharp, 2010)). Overexpression of artificially designed transcript decoys (termed “sponges”) has been shown to successfully ‘soak up’ specific miRNAs in mammalian cell culture and in animals, resulting in de-repression of endogenous targets (Ebert et al., 2007; Krol et al., 2010). As illustrated earlier, heat shock or viral infection induces the expression of MICA/MICB transcripts to such an extent that the existing level of miR-20a, miR-93 or miR-106b can no longer inhibit their translation (Stern-Ginossar et al., 2008), suggesting that these miRNAs are fully occupied. It will, therefore, be of interest to examine whether there is an increase in expression of other mRNAs that are targeted by the same miRNAs under these conditions.
IV. RNA-mediated association between miRNA-protein complexes and specific RNA-binding proteins determines miRNA activity during stress
Besides the relative concentration of miRNAs and mRNA targets, the functional outcome of a miRNA depends on the properties of other RNA-binding proteins that bind nearby to miRNA-protein complexes on the same mRNA target (reviewed in (Leung and Sharp, 2007)). For example, a cationic amino acid transporter (CAT-1) mRNA is normally translationally repressed by miR-122 but the repression is relieved during amino acid starvation (Bhattacharyya et al., 2006). Such relief of repression requires an AU-rich element binding protein (ARE-BP), HuR. Similarly, another ARE-BP, FXR1, mediates the switch for miR-369-3 from being a translational repressor to an activator for the expression of TNFα transcripts in quiescent cells upon serum starvation (Vasudevan et al., 2007). However, the molecular mechanisms by which these ARE-BPs alter the miRNA activity are unclear. Given that the local AU content around miRNA binding site is relatively high (Bartel, 2009) and that AU-rich elements are enriched near miRNA binding sites (Jacobsen et al., 2010), there might be more examples to be discovered for ARE-BPs in modulating miRNA activities.
Apart from these ARE-BPs, other RNA-binding proteins have been shown to modulate the activity of miRNA-protein complexes. Dnd1 inhibits miRNA access to target mRNAs (Kedde et al., 2007), whereas nhl-2/CGH-1 complexes enhances miRNA-mediated repression (Hammell et al., 2009). In some cases, an RNA-binding protein can have contrasting effects on miRNA activities depending on the sequence context. For example, binding of HuR adjacent to a binding site for the miRNA let-7 can relieve the repression of one mRNA target (CAT-1) (Bhattacharyya et al., 2006), but reduce the mRNA and translation of another (c-myc) (Kim et al., 2009). It remains to be determined whether this discrepancy is due to differences in secondary structure and, hence, local conformation in the miRNA recognition site, or a change in activities of these RNA binding proteins during different cellular states.
V. Some components of the miRNA-protein complex re-localize to stress granules upon multiple stress conditions
Another property of miRNA that changes upon stress is its subcellular location (Leung et al., 2006). The majority of miRNAs, mRNA targets and Argonaute proteins are found diffuse in the cytoplasm, with ~1% of Argonaute localized in cytoplasmic structures called P-bodies that are enriched with RNA degradation factors such as the decapping enzyme and other proteins related to the miRNA pathway, including GW182 and p54/rck/CGH-1. Upon multiple types of stresses, a subpopulation of miRNAs, mRNA targets, Argonaute and miRNA-protein complex components p54/rck/CGH-1, but not GW182, become enriched in another cytoplasmic structure called stress granules (SG) (Leung et al., 2006). The formation of these granules can be triggered by the phosphorylation of eIF2α, which is governed by five stress-sensing kinases (Anderson and Kedersha, 2008): (1) PKR responds to viral infection, heat and UV irradiation; (2) PERK senses ER stress; (3) GCN2 is activated by amino acid deprivation; (4) HRI senses oxidative stress and (5) Z-DNA kinases are involved in antiviral responses. Stress granules are enriched with many RNA regulators, such as FXR1, HuR and p54/rck/CGH-1 (Anderson and Kedersha, 2008), which, as described earlier, can modulate miRNA activity. It remains to be determined whether some of the changes in miRNA-mediated repression upon stress could be due to the local concentration of these RNA-binding proteins or other factors within these granules, and/or due to the exclusion of some components that are usually required for miRNA-mediated repression, such as GW182.
VI. miRNAs up-regulated by NF-κB during inflammation down-regulate the pro-inflammatory signaling cascade
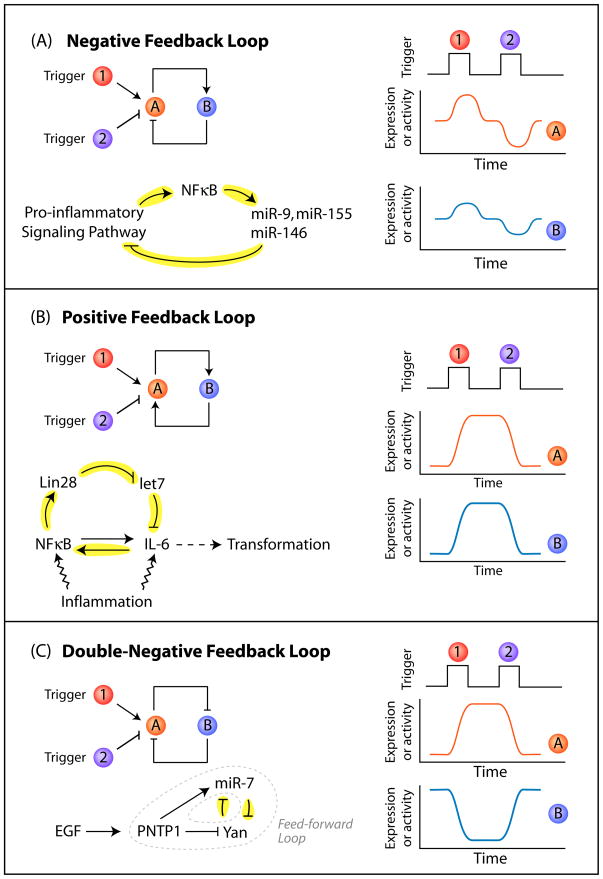
In some cases, a miRNA can act as a timer of the stress response. Timing becomes particularly important in the case of acute stress responses such as those during inflammation. In response to inflammation, NF-κB upregulates the transcription of miR-9, miR-155 and miR-146 along with other inflammatory-responsive genes through a signaling cascade in macrophages (O’Connell et al., 2010). Like other immediate early-response genes, the expression of these primary miRNA transcripts peaks within ~2 hours. However, the mature miRNAs take time to accumulate and peak at a much later time (e.g. miR-155 at ~24 hours) (O’Connell et al., 2007). Given that repression mediated by miRNAs would not be effective until the concentration of mature miRNAs reach a certain level in cells (Calabrese, 2008), this would create a delay between the emerging presence of mature miRNAs and the beginning of target repression. Interestingly, some of the mRNA targets of miR-9, miR-155 and miR-146 are themselves pro-inflammatory signaling molecules (e.g. NF-κB) that upregulate these miRNAs. Thus, these signaling molecules would only be expressed in a defined period of time before the increasing level of mature miRNAs switches off their expression. In effect, miRNAs activated by NF-κB reset the pro-inflammatory signaling pathway after activation (Figure 3A). Such temporal-controlled expression allows macrophages to elicit a strong inflammatory attack to pathogens while minimizing the duration to inflict unwanted damage to the host.
Figure 3. miRNAs are strategically located in (A) negative, (B) positive and (C) double-negative feedback loops to mediate stress responses.
(A) A negative feedback loop: A (red) activates B (blue), but B represses A. This circuitry results in the restoration of the original level of A and B after the triggering stimulus is removed. For example, inflammation triggers a signaling cascade that results in a NF-κB-dependent transcription of a set of miRNAs. These miRNAs in turn target the components of the pro-inflammatory pathway, hence resetting the activation status of the inflammation pathway. (B) A positive feedback loop: in this circuit, A activates B and B activates A. Therefore, there could be a stable state with both A and B on or both A and B off. Upon inflammation, an increase in IL-6 triggers activation of NF-κB and expression of LIN-28, which results in the reduction of let-7 expression. As let-7 normally represses the expression of IL-6, this reduction in let-7 results in an increase in expression of IL-6, thus further propagating the cycle of events. Unlike a negative feedback loop, this circuitry exhibits a persistent, self-perpetuating response long after the triggering stimulus is removed. (C) A double-negative feedback loop: A represses B and B represses A. Thus, there could be a steady state with A on and B off, or vice versa, but not both on or off. This is found in the case of miR-7 and Yan in the differentiation of Drosophila eyes. Similar to a positive feedback loop, this results in a self-perpetuating response long after the triggering stimulus is removed. Examples of feedback loops are highlighted in yellow on the left side of the panel and hypothetical timings of the trigger and resultant expression/activity of protein A and B are indicated on the right. A feed-forward loop is circled by dotted lines in panel C. Panel designs were adapted from Ferrell (2002) Current Opinion in Cell Biology 14:140–8.
VII. miRNAs enforce a new gene expression program upon inflammation
Even though the effect of miRNA-mediated repression is, in general, quantitatively modest (~2 fold) (Baek et al., 2008; Lim et al., 2005; Selbach et al., 2008), the signal can be amplified by a positive feedback loop during stress. This was recently demonstrated in a study by Iliopoulos and colleagues, in which they show that interleukin-6 (IL-6), commonly involved in inflammation, can mediate an epigenetic switch through let-7 to transform breast cells into stem cell-like cancer cells by means of a positive feedback loop (Iliopoulos et al., 2009). This feedback loop links the level of IL-6 with the expression of let-7 by indirectly activating LIN-28, a negative regulator of let-7 through the transcription factor NF-κB (Figure 3B). Remarkably, a single dose of IL-6 exposure for only 1 hour results in long-lasting cellular responses for many generations (>10 doublings) without requiring additional signal induction. In this case, the signal is amplified by the positive feedback loop and spreads to other cells through paracrine and autocrine signaling by the secreted IL6.
VIII. miRNAs ensure robust eye development in flies upon temperature fluctuations
miR-7 mutant flies are apparently normal in standard laboratory conditions but their eyes cannot develop properly when grown in an environment that constantly fluctuates in temperature (Li et al., 2009). This is partly because miR-7 normally stabilizes the developmental transition thanks to its strategic location within both a reciprocal negative feedback loop and a coherent feed-forward loop (Figure 3C). The reciprocal negative feedback loop involves miR-7 and the transcription factor Yan, where Yan represses the transcription of miR-7 in progenitor cells and miR-7 represses Yan post-transcriptionally in terminally differentiated cells. Since a negative feedback loop has the tendency to toggle between two states, the robustness of the switch from progenitor to terminally differentiated states can be instilled by coupling with an upstream feed-forward loop (Figure 3C). During development, the homeostasis of progenitor cells is broken by a transient induction of the epidermal growth factor (EGF) signaling cascade. EGF signaling upregulates the transcription factor PNTP1, which, in turn, induces the transcription of miR-7. In this case, both miR-7 post-transcriptionally and PNTP1 transcriptionally repress a common target, Yan. This resultant coherent feed-forward loop, in which PNTP1 regulates miR-7 and both negatively regulate Yan, creates stability in Yan repression against fluctuations in the level of PNTP1 and miR-7. As the signaling persists and photoreceptor cells terminally differentiate, miR-7 levels accumulate above a certain threshold. The accumulated high level of miR-7 and the maintenance of the feed-forward loop in differentiated cells therefore persistently repress Yan expression, ensuring that the change in cell fate will not be spontaneously reverted.
Stress responses: restore or reprogram gene expression by miRNAs
miRNAs and their targets are often connected within a gene expression network: (1) multiple targets are linked together through common regulation by individual miRNAs; (2) transcription factors regulate the expression of miRNAs and (3) many transcription factors themselves are subject to extensive miRNA regulation (Shalgi et al., 2007). Depending on where a miRNA is embedded within the gene circuitry, it can either act as a restorer to homeostasis or as an enforcer of a new gene expression program during stress. If the miRNA is embedded in a negative feedback loop, it allows the cells to resume their original target expression patterns despite external triggers (Figure 3A). On the other hand, if the miRNA is embedded in a positive feedback loop (Figure 3B) or double-negative feedback loop (Figure 3C), it becomes part of a bistable switch to enforce a new gene expression pattern. Therefore, the involvement of miRNAs in negative or positive feedback loops commits cells either to stay on course or to change for good, respectively, in adapting to new environments.
Moreover, miRNAs are ideal for buffering transcript surges during stress. At the single-cell level, transcription in eukaryotic cells often occurs in a burst-like manner (reviewed in (Raj and van Oudenaarden, 2008)), causing the number of mRNAs per cell to fluctuate significantly over time. With this constant fluctuation, cells need to decide whether new transcripts should be committed for translation. Such decision-making becomes particularly essential in times of duress when transcriptional bursting can become more frequent (e.g. (Cai et al., 2008)). miRNAs can ensure a steady level of gene expression unless the stress signal is sustained long enough to increase the amount of transcripts over a certain threshold, as in the case of MICA/MICB described earlier. In addition, the induction of miRNA expression upon stress might partly explain the phenomenon of “stress hardening”, where the tolerance of a particular stress increases after pre-conditioning with low doses, or “cross-tolerance”, where preconditioning of one stress increases tolerance of another stress (Kultz, 2005). Due to the long half-lives of miRNAs, their sustained presence from the previous round of stress potentially allow certain miRNA-mediated gene regulation to be already in place to tackle future assaults.
Small effect of miRNAs writ large during stress?
A common conundrum in the miRNA field is how can such subtle effects (~2 fold) mediated by miRNAs be important for physiology. Yet, inactivation of the key processing enzyme Dicer clearly indicates that miRNAs are important for a wide array of biological processes (Bushati and Cohen, 2007) and apparently normal single miRNA mutants exhibit phenotypic crisis in stress conditions (Li et al., 2009; van Rooij et al., 2007; Xu et al., 2003). The manifestation of phenotypes upon subtle changes in gene dosage caused by the loss of particular miRNAs upon stress, but not in normal conditions, is reminiscent of the phenomenon of haploinsufficiency observed in yeast upon stress. Only 3% of heterozygous yeast mutants exhibit growth phenotypes in nutrient-rich media, but 66% of them display a growth or fitness defect in at least one condition of chemical or environmental stress (Hillenmeyer et al., 2008). Clinically, inactivation of one allele of certain genes in humans and mice can lead to cancers or other pathological conditions (Seidman and Seidman, 2002). Thus, in an analogous fashion, a 2-fold change in the mRNA target expression due to a loss of a particular miRNA could also be important for physiology in animals. The question is which mRNA targets are sensitive to such changes in concentration.
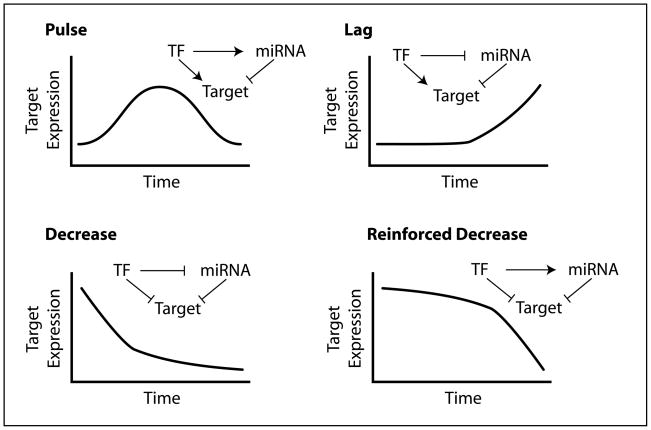
Based on observations in the literature, two particular classes of proteins are particularly sensitive to changes in concentration, where such changes can result in profound physiological effects during stress – transcription factors and signaling molecules. A change in the expression of transcription factors defines a new pattern of miRNA and mRNA expressions during stress. p53 defines a new gene circuitry with new miRNA and mRNA expression patterns upon DNA damage to promote growth arrest and apoptosis. On the other hand, overexpression of mRNA target mimics (Mt4-TPS1) or mRNA targets (MICA/MICB) results in an escape from miRNA suppression (Franco-Zorrilla et al., 2007; Stern-Ginossar et al., 2008). Since many transcription factors regulate the expression of both miRNAs and their mRNA targets (Tsang et al., 2007), we anticipate that the interplay amongst transcription factors, miRNAs and their mRNA targets can potentially generate different types of expression timing patterns (Figure 4). For example, a transcription factor can activate the transcription of both miRNAs and their mRNA targets. Initially, the level of miRNAs is not high enough to exert its suppressive effect and, therefore, an immediate increase in target mRNA expression results. As miRNAs accumulate over time, miRNAs exert their suppressive effect on the mRNA targets, causing a decrease in mRNA target expression. As a result, a pulse of mRNA target expression is observed. These different timing patterns including pulses, lags and varied forms of decreases are indeed commonly observed in gene expression responses of human cells to various stress states at mRNA levels (Murray et al., 2004).
Figure 4. Timing of mRNA target expression can be modulated by the interplay amongst transcription factors, miRNAs and mRNA targets.
Depending on whether a transcription factor (TF) activates or represses the level of miRNAs and/or mRNA targets, different timing of mRNA target expression results. The assumption is that it takes time for mature miRNAs to accumulate (deplete) upon transcriptional activation (repression) in suppressing (de-repressing) their mRNA targets. On the other hand, the change in the expression of mRNA targets is immediate.
The other ideal mRNA targets are signaling complexes. They are highly dynamic and non-stoichiometric in nature, thus making them particularly sensitive to changes in concentration of their components (reviewed in (Inui et al., 2010)). In fact, miRNAs play instrumental roles in mediating stress responses through components at all tiers of signaling pathways, from sensors to transducers to effectors. For example, in zebrafish, miR-8 represses the expression of a membrane-bound component NHERF1, which links extracellular sensing to actin cytoskeleton rearrangement, and, as a result, regulates Na+/H+ exchange activity upon osmotic stress (Flynt et al., 2009). Upon hypoxia, miR-210 is upregulated and targets iron-sulfur cluster assembly proteins ISCU1/2 that are critical for electron transport and mitochondrial oxidation/reduction, thereby shutting down the aerobic mitochondrial functions and switching the hypoxic cells to anaerobic metabolism (Chan et al., 2009). Thus, the subtle changes in the protein concentrations at these key nodes of signaling cascades allow miRNAs to transmit information in a timely manner and reconfigure cellular decisions.
Other potential modes of modulation of miRNA activities upon stress
Summarizing observations highlighted in previous sections, the functions of miRNAs during stress can be modulated by four factors: (a) the level of miRNAs, (b) the level of mRNA targets, (c) the activity or (d) the mode of action of miRNA-protein complexes (Figure 5). The changes in these factors are regulated by various signaling pathways. The level of miRNAs can be modulated at the transcription and/or processing level by stress-induced factors like p53 or NF-κB. In addition, the level of miRNAs is also determined by their half-lives. In most cases examined, miRNAs are relatively stable (e.g. half-life of 12 days in vivo (van Rooij et al., 2007)). However, increasing evidence suggests that the stabilities of subsets of miRNAs are subjected to regulation. For example, the decay of host miR-27 is triggered by herpesvirus infection (Cazalla et al., 2010), the turnover of miR-183/96/182 clusters, miR-204 and miR-201 is accelerated in dark-adapted retina (Krol et al., 2010) and the level of miR-125b decreases upon DNA damage (Le et al., 2009). Though the molecular mechanisms involved remain to be dissected, emerging data suggest that the specificity might lie at the 3′ end of the miRNAs (Ameres et al., 2010; Cazalla et al., 2010) and several enzymes/co-factors have been implicated (Kai and Pasquinelli, 2010). Therefore, the long half-lives of miRNAs and thus their suppressive activity could provide a stable memory of a cell state; or, when certain miRNAs are degraded, the ability to “forget” the cell state. Changes in the expression or stability of the cellular miRNAs upon stress could thus alter the homeostasis to a new cell state.
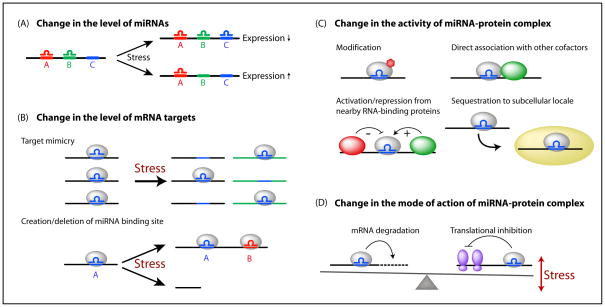
Figure 5. Different ways to modulate miRNA activities upon stress.
The miRNA function can be modulated at multiple levels by changing (A) the level of mature miRNAs, (B) the level of mRNA targets, (C) the activity of miRNA-protein complex and (D) the mode of action of miRNA-protein complex. (A) Shown is an mRNA target that has 3 binding sites for 3 different miRNAs. In normal condition, the target is repressed by miRNAs A and B. Upon stress, expression of mRNA target decreases if the level of miRNA C increases (hence 3 sites are bound by miRNAs); alternatively, the expression increases if the level of miRNA B decreases (only 1 site is bound). (B) The expression of mRNA targets (black) increases if the expression of mRNA target mimics (green) increases upon stress. These mimics compete with the same miRNAs as mRNA targets, thereby causing a relief in the repression of mRNA targets (“Target mimicry”). Alternatively, upon stress, cells could express different isoforms of the mRNA targets where miRNA binding sites could be created or deleted. (C) A change in the activity of miRNA-protein complex upon stress could be a result of post-translational modifications of components in the complex, direct association with other stress-specific co-factors, activation/repression from adjacent RNA-binding proteins bound on the same mRNA targets or sequestration to specific subcellular locales, such as stress granules or P-bodies. (D) Stress might alter the balance between the two major modes of action of miRNA-protein complexes: accelerating mRNA decay or inhibiting translation. Note that mRNA degradation is irreversible by nature and, hence, tipping towards this mode of action upon stress could potentially alter the composition of the transcriptome.
The level and species of miRNAs expressed during stress in turn determine the specificity of mRNA targets. Friedman and colleagues estimated that 70% of mRNA targets contain multiple binding sites for different miRNAs and on average ~4 different conserved sites per target (Friedman et al., 2009). Such binding site architecture implies that expression of the mRNA target is likely to be under combinatorial control of multiple miRNAs (Krek et al., 2005). The repression effect of individual binding sites is additive and can be cooperative when sites are nearby (Doench et al., 2003; Krek et al., 2005). Potentially, some of these sites could be considered as “stress” control elements that would only affect the target expression under stress conditions when specific miRNAs are expressed (Figure 5A).
Apart from changes in the absolute amounts of miRNAs and mRNA targets, the availability and nature of a particular miRNA binding site can also be altered by RNA editing, alternative splicing or the use of alternative polyadenylation sites – processes that are known to be regulated by changes in cellular status or environment (Biamonti and Caceres, 2009; Farajollahi and Maas, 2010; Neilson and Sandberg, 2010) (Figure 5B). In addition, it has become increasingly evident that miRNA binding sites, although commonly discovered and examined in the 3′ untranslated regions, can also be present in the coding sequences (Chi et al., 2009; Hafner et al., 2010; Zisoulis et al., 2010). In particular, these miRNA binding sites can be located near sites containing rare codons (Hafner et al., 2010). Therefore, miRNA accessibility to those binding sites might be sensitive to the level of tRNA charging, which is commonly altered during stress (e.g. (Zaborske et al., 2009)).
The activity of miRNA-protein complexes is another determinant of miRNA function that is regulated by signaling pathways in response to environmental cues (Figure 5C). The activity of the complexes can be modulated by its direct association with a stress-sensing chaperone Hsp90 (Johnston et al., 2010; Pare et al., 2009), or by post-translational modifications of individual components (Heo and Kim, 2009). For example, the core miRNA binding protein Argonaute has been shown to be modified by different post-translational modifications, including phosphorylation by p38 MAP kinase pathway (Zeng et al., 2008) and proline hydroxylation (Qi et al., 2008), whose enzymes are upregulated upon hypoxia. Alternatively, the miRNA processing complex or the miRNA-protein complex itself can contain components that sense the environment. For example, Drosha cofactor DGCR8 is a heme-binding protein and heme binding stimulates Drosha-DGCR8 activity in vitro, suggesting a potential connection between miRNA processing and a heme-regulated, oxygen sensing signaling cascade in vivo (Faller et al., 2007).
Finally, miRNAs have two major modes of action: accelerating decay and/or inhibiting translation of their mRNA targets (Bushati and Cohen, 2007; Carthew and Sontheimer, 2009; Fabian et al., 2010). Yet, the consequences for mRNA targets are very different: mRNA degradation results in an irreversible removal from the transcriptome whereas the level of mRNA targets can remain constant upon inhibition of translation. Depending on which mode of action is taken, miRNAs can effectively mold the composition of the transcriptome upon stress. It has recently been shown in worms that the repression of lin-14 by miRNA lin-4 at mRNA level is alleviated upon nutrient deprivation while its suppressive effect at protein level remains unchanged (Holtz and Pasquinelli, 2009). Therefore, it will be of interest to determine whether other stresses or miRNA/mRNA target pairs favor one mode of miRNA action over the other (Figure 5D).
Diseases and potential therapeutics
Organisms face constantly changing environments in the wild, including fluctuations in temperature and limited availability of food and water; on the other hand, modern humans in the developed world face the other extreme, such as excess nutrients, new dietary components and a lack of physical activity. In either case, homeostasis is achieved through biological processes associated with stress responses. When the system cannot cope with these challenges, homeostasis breaks down, which can result in diseases such as cancers, diabetes, neurodegenerative disorders, cardiovascular diseases, viral infections and many others. In some of these cases, homeostasis can be mediated via miRNAs. For example, herpes simplex virus 1 expresses its own set of miRNAs that downregulate viral immediate early proteins to lie dormant in the trigeminal nerve of the face (Umbach et al., 2008). However, upon excessive sunlight, fever or other stress, such miRNA-mediated homeostasis is broken down and results in cold sores. In this section, we will further describe how dysregulation of miRNA expression and activity can contribute to stress-related chronic diseases, using cancer as a model for discussion.
As tumor cells grow, they must be highly adaptive to a dynamic microenvironment, avoiding host immune system detection, proliferating, surviving and, in some cases, metastasizing (Hanahan and Weinberg, 2000; Kroemer and Pouyssegur, 2008; Luo et al., 2009). To do so, cancer cells usually subvert normal physiological processes, such as proliferation, apoptosis, metabolic pathways and stress responses, many of which are regulated by miRNAs. One obvious way to take away such controls in cancer cells is to reduce the total amount of miRNAs and/or mutate key components of the miRNA pathways, including Dicer and Argonaute family members. Indeed, such reductions and mutations are commonly observed in tumors (Esquela-Kerscher and Slack, 2006; Kumar et al., 2007; Lu et al., 2005). Take the positive feedback loop involving let-7 and IL-6 as an example (Figure 3B): a reduction of let-7 level, which is commonly observed in tumors (Esquela-Kerscher and Slack, 2006; Kumar et al., 2008; Lu et al., 2005), lowers the threshold of extracellular IL-6 that is required to trigger cancer cell transformation (Iliopoulos et al., 2009). On the other hand, certain miRNAs are commonly upregulated in cancer cells (Calin and Croce, 2006). For example, metastatic tumor cells can potentially avoid immune recognition by upregulating miRNAs (including miR-373 and miR-502c) that repress the stress-induced ligands MICA/MICB, which serve as extracellular signals for elimination by Natural Killer cells or T lympocytes (Huang et al., 2008; Stern-Ginossar et al., 2008).
One other recently established hallmark of cancer is the presence of stress phenotypes (Luo et al., 2009). These stress phenotypes, including those resulting from DNA damage/replication stress, proteotoxic stress, mitotic stress, metabolic stress and oxidative stress, may not be responsible for initiating tumorigenesis. Instead, cancer cells develop tolerance to these stresses, and, in turn, become dependent on stress response pathways to maintain their growth and survival. Dependency on these stress response pathways regulated by miRNAs thus provides new avenues for therapeutic intervention by modulating miRNA levels or activities.
One advantage of using miRNAs as therapeutic targets is that they often regulate multiple mRNA targets that belong to the same signaling pathway or protein complexes at the same time (Tsang et al., 2010). The key is to identify which miRNAs and which targets are involved in each particular disease. High-throughput biochemical techniques have been developed to identify endogenous mRNA targets (Chi et al., 2009; Hafner et al., 2010; Zisoulis et al., 2010). The identification of certain miRNAs that act as enforcers of a new gene expression program might allow the use of a relatively small amount of a miRNA mimic to trigger an amplification cycle within a specific positive feedback loop. There are ongoing efforts to deliver miRNA mimics or perfectly complementary “antagomir” inhibitors to increase or decrease, respectively, the levels of specific miRNAs in vivo. An antagomir against miR-122 has been demonstrated in animals to lower HCV replication levels and cholesterol levels (Elmen et al., 2008; Krutzfeldt et al., 2005; Lanford et al., 2010). In cases where a specific miRNA-mRNA target interaction should be modulated, short oligos termed ‘target protectors’ have been successfully applied in mammalian cells and zebrafish (Choi et al., 2007). The idea of a target protector is that a single-strand oligonucleotide could specifically interfere with the interaction of a miRNA with a single target while leaving the regulation of other targets of the same miRNA unaffected.
Alternatively, multiple signaling molecules in stress responses modulate the transcription, processing and stability of miRNAs. These provide further avenues for modulating the miRNA pathway. For example, a small molecule has been identified from a pilot screen for positive modulators of miRNA processing (Shan et al., 2008). This raises the possibility of restoring the global miRNA level in cancer cells to a level similar to their normal counterparts. As our understanding of these fundamental processes deepens, therapeutic possibilities will continue to rise.
Conclusions and perspectives
Our current understanding of miRNA pathways is mostly based on experiments performed under standardized laboratory “normal” conditions. However, by doing so, we might be overlooking the “normal” function of miRNAs in stressful conditions that cells commonly face. For example, endothelial cells are constantly under exposure to shear stress, epidermal cells to varying light and temperature levels, and transitional epithelial cells to fluctuations in pH and osmotic pressures. When these cells fail to adapt, pathological conditions arise. Yet, miRNA functions in stress responses are not just restricted to pathological conditions; instead, these stress-mediated functions are actually parts of the natural physiological processes taking place in multicellular organisms every day.
Currently, there are only a handful of observations in the literature, which are likely to be the first of many examples to be discovered, concerning the roles of miRNAs in stress responses and how stress modulates miRNA activities. Multiple studies have shown that miRNA mutant animals appear normal and viable until subjected to stress. miRNAs can behave as temporal agents to control a process over time or to set a threshold in determining when target gene expression becomes sufficiently strong to signal a transition. miRNA functions can be modulated at multiple, distinct steps by various signaling pathways. Depending on where miRNAs are embedded in the gene regulatory network, they can either function to restore homeostasis or to enforce a new program, allowing cells to adapt to the new environment until conditions change yet again. Together, this emerging evidence clearly indicates that miRNAs play indispensable roles in stress responses. Yet, in many of these cases, the molecular mechanisms remain unclear and should be investigated further to elucidate these fundamental roles of miRNAs in controlling mRNA regulation during stress.
Acknowledgments
We thank M. Lindstrom for figure illustrations, A. Ravi, M. Ebert and J. Wilusz for insightful comments. This work was supported by RO1-CA133404 from NIH, PO1-CA42063 from NCI to PAS and partially by Cancer Center Support (core) grant P30-CA14051 from NCI. AKLL was a special fellow of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the Endoplasmic Reticulum: Brothers in Arms. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.10.001. [THIS ISSUE] [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM. Dicer deletion and short RNA expression analysis in mouse embryonic stem cells. Department of Biology, Cambridge: Massachusetts Institute of Technology; 2008. [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz J. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010 doi: 10.1016/j.cub.2010.08.052. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Thatcher EJ, Burkewitz K, Li N, Liu Y, Patton JG. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J Cell Biol. 2009;185:115–127. doi: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz J, Pasquinelli AE. Uncoupling of lin-14 mRNA and protein repression by nutrient deprivation in Caenorhabditis elegans. RNA. 2009;15:400–405. doi: 10.1261/rna.1258309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010 doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, Evan GI. p53--a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.09.023. [THIS ISSUE] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, Sandberg R. Heterogeneity in mammalian RNA 3′ end formation. Exp Cell Res. 2010;316:1357–1364. doi: 10.1016/j.yexcr.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20:3273–3284. doi: 10.1091/mbc.E09-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. Life on the verge of death: the heat shock response revisited. 2010. [THIS ISSUE] [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. Transcription factor haploinsufficiency: when half a loaf is not enough. J Clin Invest. 2002;109:451–455. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol. 2008;26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.09.028. [THIS ISSUE] [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell. 2010;38:140–153. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem. 2009;284:25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]