Abstract
IRGM, a human immunity related GTPase, confers autophagic defense against intracellular pathogens by an unknown mechanism. Here we report the unexpected mode of IRGM action. IRGM showed differential affinity for mitochondrial lipid cardiolipin, translocated to mitochondria, affected mitochondrial fission and induced autophagy. Mitochondrial fission was necessary for autophagic control of intracellular mycobacteria by IRGM. IRGM influenced mitochondrial membrane polarization and cell death. Overexpression of IRGMd but not IRGMb splice isoforms caused mitochondrial depolarization and autophagy-independent but Bax/Bak-dependent cell death. By acting on mitochondria IRGM confers autophagic protection or cell death, explaining IRGM action both in defense against tuberculosis and in damaging inflammation in Crohn's disease.
Introduction
Autophagy enables cells to sequester portions of their cytosol or damaged and surplus organelles into autophagosomes and degrade them in autolysosomes1. Autophagy promotes cell survival under starvation by digesting cell's own cytoplasm and resupplying essential anabolic functions2. Autophagy affects cell death 3, 4 through interactions with cell death pathways albeit its intrinsic pro-death action is debated5. Autophagy impacts aging, neurodegeneration, myodegeneration, and cancer1, and immunity6, 7. It contributes to control of intracellular pathogens7 including major human diseases tuberculosis8 and AIDS9, 10. Autophagy is an effector of Th1/Th2 polarization11, affects B and T cells12, 13, fuels endogenous antigen presentation6, assists pattern recognition receptors (PRR) by delivering cytosolic microbial products to endosomal Toll-like receptors14, suppresses IL-1β activation15, and acts as a PRR effector16. Autophagy affects central tolerance13 and chronic inflammatory illnesses such as Crohn's disease 15, 17, 18.
Autophagy is implicated8, 19-21 in the mechanism of control of intracellular pathogens by immunity related GTPases (IRG) 22. The mouse IRG family23 consist of 20 interferon-controlled complete IRG genes, Irgm1-Irgm3, Irgb1-Irg6, Irgb8-Irgb10, Irga1-Irga4, Irg6-Irga8 and Irgd. Their expression is driven by IRES (interferon-stimulated response sequences) and GAS (γ-activated sequences) elements. The abundant murine IRGs contrast with a derth of IRGs in humans23. The human genome encodes a single paralog of murine IRGs, IRGM23. IRGM traces back to prosimian IRGM9 with roots shared with the mouse Irgm124. IRGM is not under IFN-γ control, and is expressed from the human endogenous retrovirus element, ERV923. Nevertheless, IRGM is required for IFN-γ-induced autophagy and control of Mycobacterium tuberculosis in human macrophages20, whereas IRGM polymorphisms are a risk factor for tuberculosis25. IRGM has been identified26-28, along with another autophagy factor ATG16L129, as a risk locus for Crohn's disease. The roles of IRGM in autophagy, defense against mycobacteria, and inflammation in Crohn's disease require a definition of IRGM action. In this study, we report the surprising finding that IRGM translocates to mitochondria where it regulates autophagy in association with mitochondrial fission. We also show that a subset of IRGM splice variants can cause mitochondrial depolarization and cell death.
Results
IRGM localizes in mitochondria
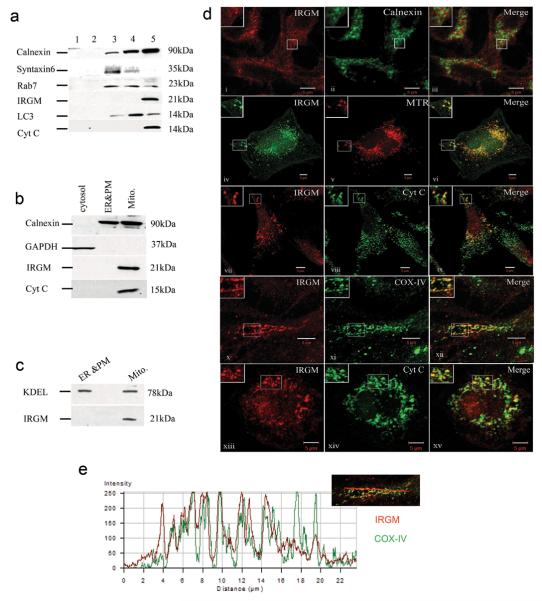
We investigated intracellular distribution of IRGM by sedimentation velocity separation of intracellular organelles. IRGM was enriched in U937 macrophage fractions containing the endoplasmic reticulum (ER) protein calnexin and mitochondrial protein cytochrome c (Fig. 1a). A mitochondrial proteomics kit placed IRGM in mitochondrial fractions (Fig. 1b), with calnexin in mitochondrial preparation likely originating from associated membranes whereas IRGM was absent from ER and plasma membrane fractions (Fig. 1b). IRGM showed similar distribution relative to another ER marker, KDEL (Fig. 1c). An identical pattern was obtained with HeLa cells (Suppl. Fig. S1a). Endogenous IRGM in HeLa was analyzed by microscopy with antibody against an epitope present in all IRGM splice isoforms20. There was no colocalization of IRGM with the ER marker calnexin (Fig. 1d, panels i-iii) or markers for trans-Golgi network and Golgi (syntaxin 6 and Golgi 58 kDa protein G58K), early endosomes, late endosomes, lysosomes and recycling endosomes (EEA1, Lamp2, CD63, CI-MPR, transferrin receptor), and autophagy organelles (p62 and LC3) (Suppl. Fig. S1b). A partial juxtaposition between IRGM and GFP-LC3 was noted (Suppl. Fig. S1b). IRGM colocalized with the mitochondrial dye MitoTracker Red (MTR) and with cytochrome c (Fig. 1d, panels iv-xii). IRGM colocalzied with mitochondrial inner membrane cytochrome oxidase complex IV (COX IV; Fig. 1d, panels x-xi, and e). IRGM also colocalized with mitochondria in primary human macrophages (Fig. 1d, panels xiii-xv).
Fig. 1. IRGM localizes to mitochondria.
a. Subcellular compartments (U937 cell extracts) were separated by sedimentation velocity on discontinuous sucrose gradient and probed for IRGM and indicated markers. b. Immunoblot analysis of mitochondria purified by Qproteome Mitochondria Isolation Kit (Qiagen). c. Analysis of KDEL vs IRGM distribution in fractions as in panel b. d. Intracellular localization of IRGM analyzed by confocal microscopy. Panels i-xi: Endogenous IRGM localization in HeLa cells relative to calnexin, Mitotracker Red (MTR), Cytochrome c, and COX IV. Panels xii-xiv: IRGM localization in primary human macrophages relative to mitochondria revealed with Cytochrome c antibody. e. analysis of IRGM (green) and COX IV (red) overlap along the line shown in the inset; the profiles correspond to panel ix.
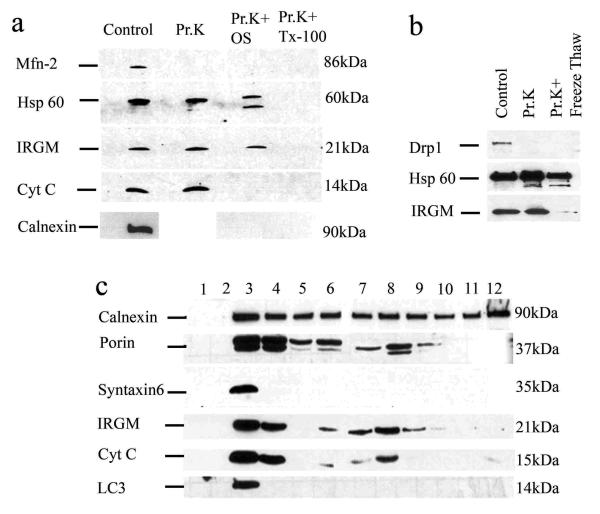
We used proteinase K-accessibility method for IRGM suborganellar localization in mitochondria (Fig. 2a). When mitochondrial preparations were digested with proteinase K, IRGM remained intact although Mfn2 (mitochondrial outer membrane tracer) was degraded (Fig. 2a, Pr.K). Upon osmotic shock to access intermembrane space, cytochrome c was degraded but IRGM remained undigested (Fig. 2a, Pr.K+OS). Calnexin was degraded when mitochondrial preparations were subjected to osmotic shock (Fig. 2a), indicating that calnexin and IRGM are not in the same compartment. Both inner membrane/matrix marker Hsp60 and IRGM were degraded when the membranes were solubilized with detergent (Fig. 2a, Pr.K+TX-100) or disrupted by freeze-thaw cycles (Fig. 2b). The IRGM protease-accessibility pattern indicated mitochondrial inner membrane or matrix localization. Finally, we subjected intracellular organelles to isopycnic separation and IRGM co-fractionated with mitochondria in two separate peaks (possibly reflecting two morphological or physiological states of mitochondria) (Fig. 2c).
Fig. 2. IRGM co-fractionates with mitochondria and localizes to their inner membrane or matrix.
a. Purified mitochondria were untreated or subjected to osmotic shock (OS), or total protein solubilized with TX-100, and preparations digested with Proteinase K (Pr.K) and analyzed by immunoblotting. b. Mitochondrial preparations as in e were subjected to membrane disruption by freeze-thaw cycles and accessibility of proteins to Proteinase K examined by digestion followed by immunobloting. c. Immunoblot analysis of membranous organelles from U937 cells separated by isopycnic sucrose density gradient centrifugation.
IRGM control of autophagy is associated with mitochondrial fission
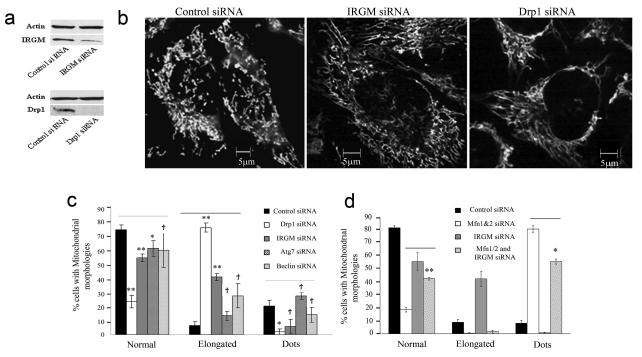
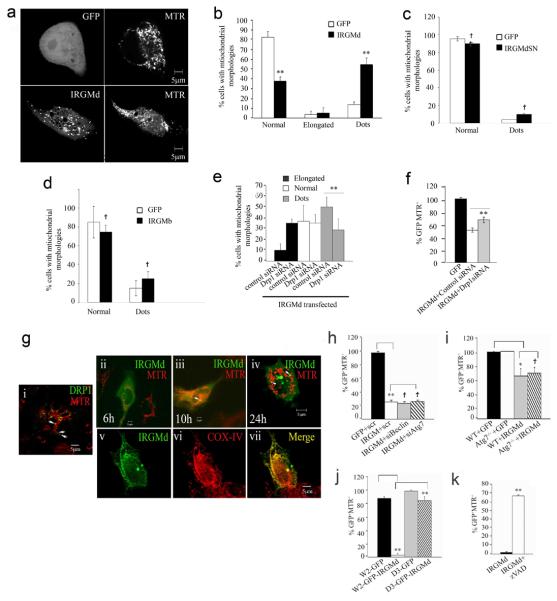
IRGs are remotely related to dynamins including the mitochondrial fission protein Drp130. Mitochondria are dynamic organelles undergoing fission and fusion, with fission or fission proteins promoting autophagy31-34. We tested whether IRGM affects mitochondrial dynamics. IRGM knockdown in HeLa (Fig. 3a) altered mitochondrial morphology (Fig. 3b,c) from reticulotubular plus punctiform to abnormally elongated mitochondria with few punctiform mitochondria (Suppl. Fig. S2b shows morphotypes), similarly to DRP1 knockdown. Thus, IRGM, like DRP1, supports mitochondrial fission.
Fig. 3. IRGM affects mitochondrial fission.
a. Knockdowns of IRGM and DRP1; cells were transfected with control siRNA or with siRNA to IRGM and DRP1 for 48 h and protein samples were analyzed by Western blotting with anti-IRGM and anti-Drp1 antibodies. Actin, loading control. b. HeLa cells treated with siRNA to either IRGM or DRP1 were labeled with MTR and analyzed by live microscopy. c. Cells were treated with either control siRNA or with DRP1, IRGM, ATG7 or BECN1 (Beclin 1) siRNAs, labeled with MTR and the % of cells with mitochondrial morphologies ranging from normal, punctiform (dots) and elongated were quantified. Definition of mito-morphologies and quantification criteria are given in Suppl. Fig. S2 legend. d. Cells were transfected with either control siRNA or with siRNA to MFN1&2, IRGM or IRGM and MFN1&2, labeled with MTR and analyzed for mitochondrial morphologies. Data, means ± SEM (n=3 panel c, n=4 panel d).*P<0.05, **P<0.01, †P>0.05 (t-test).
Autophagic machinery is implicated upstream of mitochondrial fragmentation35. No increase in % of cells with elongated mitochondria following ATG7 or BECN1 (human Beclin 1) knockdowns was observed, in contrast to DRP1 or IRGM knockdowns (Fig 3c). A knockdown of fusion proteins MFN1 and MFN2 caused fragmentation of mitochondria whereas a concurrent knockdown of IRGM reversed these effects (Fig. 3d). Thus, IRGM effects on mitochondrial morphology are not indirect via inhibition of autophagy or mitofusins.
Mitochondrial fission has been linked to the generation of reactive oxygen species (ROS)36 whereas ROS generated by mitochondria induce autophagy through Atg4 37. We knocked down IRGM and examined ROS production in cells induced for autophagy by starvation using intracellular ROS indicator DFC (Suppl. Fig. S2b-d). The % of double positive cells (MTR+DCF+) increased in starved cells but was reduced upon IRGM knockdown (Suppl. Fig. S2b-d). Thus, IRGM supports generation of ROS under autophagy-inducing conditions.
Mitochondrial fission proteins are required for autophagic output
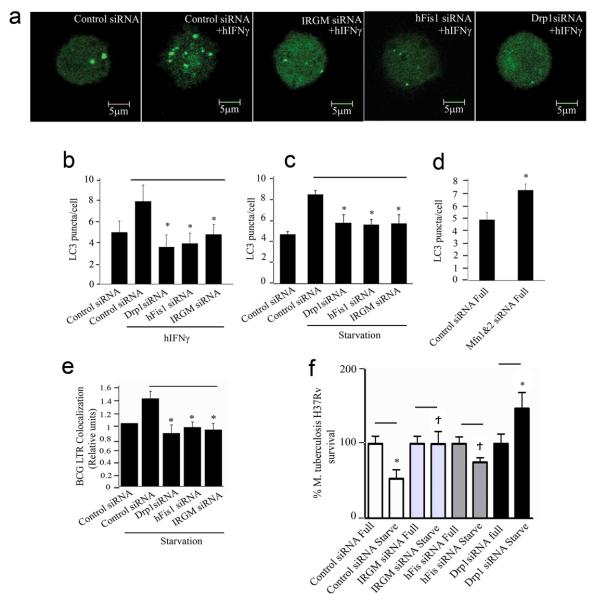
IRGM supports autophagy induction by diverse agonists (starvation, IFN-γ)20, suggesting a role in a core process, e.g. relationships between mitochondrial fission and autophagy31-34, 38. We compared effects of IRGM knockdowns on IFN-γ- and starvation-induced autophagy with those of mitochondrial fission regulators FIS139 and DRP130. LC3 puncta formation in response to IFN-γ or starvation was diminished in cells subjected to IRGM knockdown, similarly to FIS1 or DRP1 knockdowns (Fig. 4a-c). MFN1 and MFN2 knockdown increased LC3 puncta (Fig. 4d). Thus, fission proteins promote autophagy whereas mitofusins inhibit autophagy.
Fig. 4. Relationship between mitochondrial fission and autophagy and roles of IRGM, DRP1 and FIS1 in autophagic control of mycobacteria.
a and b. (a) U937 cells were transfected with GFP-LC3 and siRNA to FIS1, DRP1, IRGM or control and autophagy was induced with hIFN-γ (300 u/ml) for 24 h, and (b) LC3 puncta per cell quantified. c. As in (b) except that autophagy was induced by starvation (4 h). d. LC3 puncta per cell were quantified in cells transfected with either control or MFN1&2 siRNA. e. siRNA treated cells were infected with BCG, autophagy induced by starvation and phagosome maturation \ analyzed using LysoTracker (LTR), as a reporter of acidification 8, 20. f siRNA treated U937 were infected with virulent M. tuberculosis H37Rv, autophagy induced by starvation (4 h) or kept in full medium. CFU were counted to assess bacterial survival. Data, means ± SEM (n=6; two independent transfections, 6 independent infections; SD values are given in Supplementary Table S1). *P<0.05, **P<0.01, †P>0.05 (t-test).
If autophagic control of intracellular mycobacteria depends on IRGM via mitochondrial fission, it should also depend on DRP1 and FIS1. Cells that were knocked down for DRP1 or FIS1 failed to increase M. tuberculosis var. bovis BCG (BCG) phagosome maturation induced by starvation, similarly to IRGM silencing (Fig. 4e). A knockdown of DRP1 or FIS1 inhibited starvation-induced killing of BCG (Suppl. Fig. S3a) and virulent M. tuberculosis H37Rv (Fig. 4f).
Analysis of IRGM isoforms
IRGM has 4 different splice isoforms, IRGMa, IRGMb, IRGMc/e (referred herein as IRGMc) and IRGMd 23 differing chiefly within C-terminal tails, showing presence (IRGMb and IRGMd) or absence (IRGMa and IRGMc) of the putative G5 (SAK) motif (Fig. 5a). Endogenous expression of all IRGM isoform RNAs was detected in all cells tested (Suppl. Fig. S4a). Fluorescent protein fusions with IRGM isoforms were generated and tested at the RNA, protein, and cytoplasmic distribution levels (Suppl. Fig. S4b-f). Intracellular distribution of IRGM isoforms was quantified for: (i) diffuse cytosolic vs punctate (Suppl. Fig. S4g, inset); (ii) colocalization with mitochondrial markers (MTR and COX IV) and ER markers (calnexin and protein disulfide isomerase; PDI) (Suppl. Fig. S4g, main graph). The highest colocalization with mitochondria (COX IV) was seen with both endogenous IRGM and IRGMd (Suppl. FigS4g). A small subset of cells showed endogenous IRGM colocalization with PDI (Suppl. Fig. S5a, panels i-iii vs iv-vi; suggestive of a specialized subcompartment of ER). Time-lapse analyses following transfection with GFP-IRGMd showed transition to puncta associated with mitochondria (Movie 1). Thus, IRGMd was representative of a significant fraction of the endogenous IRGM detected in mitochondria.
Fig. 5. IRGMd binds to cardiolipin and causes loss of mitochondrial membrane potential.
a. Splice variants of IRGM. G1-G5, GTPase motifs. The S47N mutation is indicated in blue. b. HeLa cells were transfected with IRGMb, IRGMd-WT (wild type IRGMd, unaltered) or IRGM-S47N mutant (SN) for 48 h, labeled with MTR and imaged by live microscopy. c. Fluorescence intensity analysis of green and red channels along a line drawn through two adjacent cells, one GFP-IRGMd positive and one GFP-IRGMd negative. d. Quantification of GFP+MTR+ cells. e,f. Analysis of GST-IRGMd binding to lipids by lipid protein binding dot blots (e) and cardiolipin bound agarose bead pull down assay (f; details in Methods). g. IRGMd, IRGMdS47 mutant, and IRGMb isoform concentration-dependent analysis of binding to lipids on strip blots (numbers identifying lipids correspond to the legend in panel e). GST control is in Suppl. Fig. S1d. Membranes in e-g were probed with anti-GST antibody.
IRGMd can induce mitochondrial depolarization
Cells transfected with GFP-IRGMd (but not with YFP-IRGMb) showed a progressive loss of mitochondrial capacity to stain with MTR, a Δψm (mitochondrial membrane potential) dependent dye. At 48 h posttransfection, cells expressing GFP-IRGMd were MTR− and rounded (Fig. 5b,c), preceded by mitochondrial clumping (Suppl. Fig. S5b). Knocking down endogenous IRGM affected Δψm, as measured by DiOC6(3): a trend was observed as transient protection (at 1.5 h, lost at 3 h) against mitochondrial depolarization caused by staurosporine (Fig. S5c). A mutant IRGMd (S47N; corresponding to inactive GTPases) did not cause MTR− and rounded phenotype (Fig. 5b,d). This was not due to decreased expression or stability of GFP-IRGMdS47N relative to the wild type (Fig. S4c,d). Cells transfected with YFP-IRGMb also did not show loss of MTR staining (Fig. 5b,d).
IRGMd binds to the mitochondrial lipid cardiolpin
IRGMd has no identifiable mitochondrial localization motifs (MITOPORT and PREDOTAR algorithms). IRGMa, IRGMb, IRGMc/e and IRGMd contain no obvious hydrophobic patch, amphipathic α-helices or cystein-motifs for targeting to mitochondria 40. We asked whether lipid binding preferences of IRGMd might explain its localization. Purified GST-IRGMd (Fig. S1c) was subjected to lipid dot-blot binding assays (Fig. 5e). IRGMd showed selective binding to the mitochondria-specific lipid cardiolipin (Fig. 5e), confirmed in assays with cardiolipin-agarose beads (Fig. 5f). Increasing GST-IRGMd concentration confirmed IRGMd preferential binding to cardiolipin (Fig. 5g, Suppl. Fig. S1d). No binding was observed to PtdIns(3,4,5)P3 and PtdIns(3,4)P2. reported to bind murine Irgm1 (LRG-47)41. Inspection of data in lipid blots by Tiwari et al.,41 revealed that Irgm1 (depicted in Fig. 4a) too had a capacity to bind cardiolipin. The IRGMdS47N mutant (GST-IRGMdS47N; Suppl. Fig. S1e) showed reduced binding to cardiolipin (Fig. 5g and Suppl. Fig. S1g). Instead, it displayed binding to phosphatidic acid when compared to GST-IRGMd (Fig. 5g) and GST alone (Suppl. Fig. S1d). Purified GST-IRGMb (Fig. S1f) did not bind cardiolipin (Fig. 5g and Suppl. Fig. S1g). Neither IRGMdS47N nor IRGMb caused MTR− mitochondrial phenotype (Fig. 5b,d), correlating binding to cardiolipin with effects on mitochondrial morphology.
IRGMd promotes mitochondrial fission and autophagy
Before mitochondria lost Δψm due to GFP-IRGMd transfection, cells showed increased MTR+ punctiform (dots) (Fig. 6a,b) and clumped mitochondria (Suppl. Fig. S5b). In contrast, cells transfected with cardiolipin non-binders (IRGMdS47N and IRGMb) showed no significant change in mitochondrial morphology (Fig. 6c and d), in keeping with absence of IRGMdS47N and IRGMb effects on Δψm (Fig. 4b,d). GFP-IRGMd expression increased LC3-II levels over the bafilomycin A1-sensitive fraction (Suppl. Fig. S5d), indicating that IRGMd expression promotes autophagy. These findings are complementary to endogenous IRGM knockdowns that inhibited autophagy.
Fig. 6. IRGMd translocates to mitochondria, induces mitochondrial fragmentation and causes loss of mitochondrial Δψm independent of autophagic but dependent on apoptotic machinery.
a. HeLa cells transfected with GFP or GFP-IRGMd for 24 h were stained with MTR and imaged. b-d. Quantification of mitochondrial morphologies in cells transfected with IRGMd, IRGMdS47N, or IRGMb fusions. e,f. IRGM acts in concert with DRP1. Cells were transfected with siRNAs (48 h) and GFP-IRGMd, and mitochondrial morphologies (e; 24 h) or MTR staining (f; % of GFP+ cells that were also MTR+ at 48 h) quantified. g. IRGMd translocates (ii-iv) from the cytosol to mitochondria and colocalizes with COX IV (v-vii). Cytosol-to-mitochondria translocation and mitochondrial localization was compared with the typical appearance of steady state Drp1 distribution (i). h. HeLa cells, co-transfected with GFP-IRGMd and control, BECN1 (Beclin 1) or ATG7 siRNA, were labeled with MTR 48 h post-transfection, imaged by live microscopy, and % of GFP+ cells that were MTR+ quantified. i. Atg7 wild type (WT) or Atg7−/− MEF transfected with GFP-IRGMd for 48 h were analyzed for mitochondrial staining as in g. j. Wild type W2 (Bax/Bak+/+) or mutant D3 (Bax/Bak−/−) BMK cells were transfected with GFP-IRGMd for 48 h and % of GFP+ cells that were MTR+ quantified. k. HeLa cells were transfected with GFP-IRGMd, treated with z-VAD, and after 48 h stained with MTR. Data, means ± SEM (n=3). †P≥0.05,*P<0.05, **P<0.01, (t-test).
IRGMd effects on mitochondrial fission requires DRP1
Knocking down DRP1 inhibited mitochondrial fission (Fig. 6e) and loss of Δψm (Fig. 6f) induced by GFP-IRGMd. DRP1 acts on mitochondrial outer membrane whereas IRGM, based on its localization to the inner membrane or matrix and binding to cardiolipin (localized to the inner mitochondrial membrane albeit present at outer membrane-inner membrane contact sites42) may function synergistically on the inner membrane.
IRGMd translocation to mitochondria
The majority of the endogenous IRGM appeared associated with mitochondria, whereas the majority of overexpressed GFP-IRGMd was diffuse cytosolic early upon transfection. We tested whether newly made IRGMd transfers to mitochondria akin to Drp1 translocation. Drp1 is predominantly cytosolic and is recruited to mitochondrial outer membrane for fission43 and appears as dots44 (Fig. 6g sub-panel i). At 3 h - 6 h post-transfection, most of the GFP-IRGMd appeared diffuse cytosolic (Fig. 6g sub-panel ii). GFP-IRGMd translocated to mitochondria with time (Fig. 6g sub-panels iii-iv). As with Drp1, GFP-IRGMd changed from diffuse cytosolic to punctate distribution. The co-localization of GFP-IRGMd with MTR was less frequent than in the case of GFP-Drp1 (Fig. 6g, sub-panels i vs. iv). The punctate GFP-IRGMd shared only few points of colocalization as dots juxtaposed to or on MTR+ mitochondria (Fig. 6g, sub-panel iv) but displayed clear colocalization with CoxIV (Fig. 6g, sub-panels v-vii), indicating that many of the GFP-IRGMd+ MTR− puncta are mitochondria that have lost Δψm. Not all GFP-IRGMd puncta were mitochondrial in nature, as a small proportion of the GFP-IRGMd colocalized with peroxisomes (Suppl. Fig. S6a). This inidicates additional similarities between Drp1 and IRGMd versus intracellular organelles, since Drp145 and Fis146 show partial association with peroxisomes.
IRGMd-induced loss of Δψm requires apoptotic but not autophagic factors
IRGMd-dependent induction of autophagy and accessibility of fragmented mitochondria could lead to excessive autophagic mitochondrial elimination, a processes described as mitoptosis or programmed mitochondrial clearance47-49. However, the positive staining for mitochondrial Hsp60 in the MTR− cells expressing IRGMd (Suppl. Fig. S3d) ruled out en masse removal of mitochondria and suggested Δψm loss instead. The loss of Δψm in cells overexpressing IRGMd could be due to direct IRGMd action on mitochondria or due to processes downstream of IRGM-induced autophagy. This was addressed by knocking down ATG7 or BECN1 (Beclin 1) (Suppl. Fig. S3b,c) in GFP-IRGMd expressing cells. Blocking autophagy did not reverse the IRGMd-induced MTR− phenotype (Fig. 6h). Atg7−/− mouse embryonic fibroblasts (MEF) showed equal frequency of GFP-IRGMd MTR− phenotype as Atg7+ MEF (Fig. 6i). Thus, IRGMd induces loss of Δψm independently of autophagy.
Mitochondrial fission proteins have been functionally linked to Bax/Bak-dependent apoptosis43, 44, 47, 50, 51 whereas GFP-IRGMd induces cell rounding. Hence, we compared Bax/Bak wild type (W2) and Bax/Bak−/− (D3) isogenic cell lines52. IRGMd induced loss of mitochondrial MTR staining in W2 cells but not in the Bax/Bak−/− D3 cells (Fig. 6j). Thus, IRGMd-provoked loss of Δψm requires pro-apoptotic proteins Bax/Bak.
IRGMd expression causes cell death
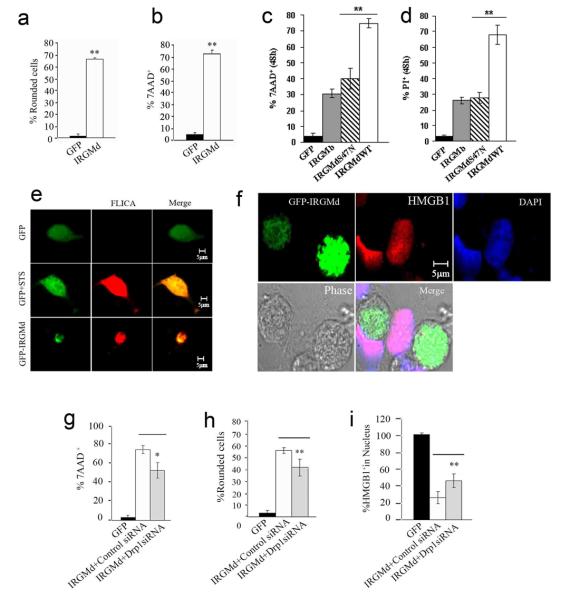
A connection with pro-apoptotic systems was further reflected in the sensitivity of the IRGMd-induced MTR− phenotype to the caspase inhibitor z-VAD (Fig. 6k). One of the earliest events associated with mitochondrial outer membrane permeabilization is that caspases gain access to a specific target within the electron transport chains in the mitochondrial inner membrane thus bringing about a loss of Δψm53. In addition to cell rounding (Figs. 5b, 7a), cell death was assessed with 7AAD staining. IRGMd transfected cells after 48 h were 7AAD-positive, confirming that IRGMd induced cell death (Fig. 7b). IRGMdS47N and IRGMb yielded fewer 7AAD-positive cells (Fig. 7c). IRGMd wild type induced higher cell death by PI-staining compared to IRGMdS47N (Fig. 7d). Endogenous IRGM also showed a role in cell survival in response to staurosporine-induced cell death (Fig. S6b).
Fig. 7. IRGMd induces cell death.
a. Fraction (%) of HeLa cells transfected with GFP or GFP-IRGMd that were rounded 48 h post-transfection. b. HeLa cells transfected with GFP or GFP-IRGMd for 48 h were stained with 7-AAD and % of GFP+ cells that were 7-AAD+ quantified. c. Comparison of 7-AAD staining in cells transfected with IRGMd vs. IRGMDS47N mutant and IRGMb (expressed as fluorescent protein fusions). d. Comparison of propidium iodide staining in cells transfected with IRGMd vs. IRGMDS47N mutant and IRGMb (expressed as fluorescent protein fusions). e. GFP-IRGMd transfected cells were stained for active caspases 3 and 7 using FLICA dye (Serotec). Staurosporine was used as a positive control. f. GFP-IRGMd transfected cells were immunostained for HMGB1 (absence of nuclear HMGB1 stain is a marker of HMGB1 release). g-i. Cells were transfected for 48 h with GFP or IRGMd plus control or DRP1 siRNA, and % of 7-AAD+ cells (g), rounded cells (h) and cells with HGMB1+ nuclei quantified. Data, means ± SEM (n=3). *P<0.05, **P<0.01 (t-test).
GFP control transfectants remained negative for active caspase stain (fluorochrome inhibitor of caspases; FLICA) (Fig. 7e, top row). Staurosporine treatment activated caspases 3 and 7 (Fig. 7e, middle row). GFP-IRGMd expressing cells stained positive for FLICA (Fig. 7e, bottom row). The IRGMd-induced cell death was accompanied by release from the nucleus of HMGB1 (Fig. 7f), a nuclear protein released primarily during necrosis but also released from some apoptotic cells54-56. Cells were rescued from IRGMd-induced cell death in the absence of DRP1, showing diminished 7-AAD staining (Fig. 7g) and rounding (Fig. 7h). Nuclear HMGB1 was partially recovered upon depletion of DRP1 in IRGMd overexpressing cells (Fig. 7i). Thus, IRGMd, in addition to inducing mitochondrial fission and autophagy, can cause mitochondrial depolarization and cell death in cooperation with other mitochondrial fission factors.
We examined the remaining IRGM isoforms, IRGMa and IRGMc (Fig. 8). Overexpression of IRGMa induced potent loss of MTR staining, with substantial conversion to MTR− phenotype by 24 h of transfection (Fig. 8a). IRGMc resembled IRGMd showing only mild effects at 24 h and strong Δψm effects at 48 h post-transfection (Fig. 8a,b). IRGMa caused a major rounding of cells at 24 h (Fig. 8c), whereas IRGMc and IRGMd cought up with this effect at 48 h post-transfection (Fig. 8d). IRGMa caused at 24 h, and IRGMc and IRGMd at 48 h, appreciable increase in % of 7-AAD+ cells (Fig. 8e,f). Equal HMGB1 release was found in IRGMa-, IRGMc-, and IRGMd-overexpressing cells (Fig. 8g,h). Thus, the majority of IRGM isoforms, with the exception of IRGMb, when overexpressed can cause cell death with different kinetics but similar end points.
Fig. 8. Comparison of IRGMa, b and c effects.
HeLa cells were transfected with GFP, IRGMa, IRGMc or IRGMd for 24 h (a) or 48 h (b), stained with Mitotracker Red, and % of GFP+ that were also MTR+ cells quantified. c and d. % of IRGMa, IRGMc, IRGMd or GFP transfected cells that were rounded 24 h and 48 h after transfection. e and f. Cells were transfected with IRGMa, c or d isoforms for 24 h (e) or 48 h (f), labeled with 7-AAD and % of GFP+ cells that were 7-AAD+ determined. g and h. Cells transfected with indicated plasmids for 24 h (g) or 48 h (h) were processed by immunofluorescence to assess retention of HGMB1 in the nucleus. Data, means ± SEM (n=3 a-f; n=4 g,h). *P<0.05, **P<0.01 (t-test).
Discussion
IRGM, the sole sensu stricto human immunity related GTPase and a genetic risk factor in Crohn's disease26, 27 and tuberculosis25, localizes to mitochondria and in addition to autophagy20 affects mitochondrial fission, mitochondrial Δψm, and cell death. The mitochondrial localization and function of IRGM, while unexpected given the prior history of IRG studies, explains its mode of action. The presence of IRGM on mitochondria is in keeping with recognition of mitochondria as core regulators of autophagy32, 37, 48, 49. The effects of mitochondrial fission in regulating innate immunity defenses are not limited to IRGM, since other previously characterized mitochondrial fission regulators, DRP1 and FIS1, as shown here, play a role in autophagic control of mycobacteria. These, previously unappreciated functions of mitochondrial fission factors indicate a general nature of the relationship between mitochondrial division and autophagy in its role of a cell-autonomous defense against microbes. An emerging model posits that IRGM immune function is via effects on mitochondria, a mechanism compatible with the absence of IRGM from mycobacterial phagosomes (Suppl. Fig. S6c). The effects of IRGM on mitochondrial morphology, autophagy, and cell death, suggest that IRGM may have roles in cellular homeostasis beyond innate immunity.
IRGM displays specific affinity for mitochondrial lipid cardiolipin, providing in part the molecular basis for IRGM partitioning to mitochondria: (i) Cardiolpin is a key mitochondrial lipid known to affect, aside from its other roles, import into the mitochondria57. (ii) An IRGM isoform that does not bind to cardiolipin shows diffuse cytosolic distribution and no mitochondrial phenotypes. (iii) The IRGM isoform IRGMd that binds to cardiolipin causes mitochondrial and cellular phenotypes. (iv) An IRGMd mutant that no longer binds cardiolipin has no mitochondrial or cellular phenotypes. Furthermore, dependence of IRGM on cardiolipin may link the physiological state of mitochondria with IRGM localization and action. Cardiolipin, abundant in the mitochondrial membranes, affects diverse processes (Suppl. Fig. S7a) from mitochondrial protein import57 to apoptosis42, 58. Cardiolipin binds to electron transport chain components and regulates their function and cytochrome c membrane association42, 58. Cardiolipin is a fusogenic lipid and affects oligomerization of GTPases controlling mitochondrial inner membrane fusion59. The IRGM effects on mitochondria reported here parallel many of the previously identified functions of cardiolipin42, 58, 59.
The large number of murine IRG genes contrasts with the dearth of IRG genes in humans (Suppl. Fig. S7b). Why the evolution led to IRG family reduction in primate evolution, leaving only IRGM as the sole somatic cell member of the IRG family in humans (Suppl. Fig. S7b) is puzzling. IRGM, with its truncated domain organization, may have augmented its mitochondrial, autophagic and cell death functions relative to more primitive species (e.g. murine IRGs that display full length N- and C-terminal domains; Fig. 5a). The low baseline IRGM expression suggests that it is tightly regulated, as enhanced expression of IRGM isoforms a, c and d can be detrimental to the cell. Thus, IRGM may act as a double-edged sword, protecting against invading microbes via autophagy when expressed in moderation but causing cell death and inflammation when its isoforms IRGMa, c or d are overexpressed. In contrast, IRGMb overexpression is not overtly harmful to the cells, indicating that there may be a diversification of functions among IRGM isoforms. Whereas human IRGM expression is uncoupled from IFN-γ (unlike murine IRG genes), IFN-γ can nevertheless promote mitochondrial morphology changes (Suppl. Fig. S7c). Hence, IFN-γ and IRGM actions remain functionally linked since IRGM is required for IFN-γ-induced autophagy in human cells20.
When overexpressed, IRGMd promotes mitochondrial fission (observed before Δψm loss), followed by a loss of Δψm and cell death. This is reflected in the sequence of events: (i) First, presence of diffuse cytosolic GFP-IRGMd; (ii) Second, formation of GFP-IRGMd puncta associated with mitochondria; (iii) Third, most of the mitochondria associated with GFP-IRGMd puncta become depolarized and only a few GFP-IRGM+ mitochondria remain MTR+; (iv) Fourth, once mitochondria that have recruited GFP-IRGMd lose Δψm and MTR-staining, they can be revealed as COXIV-positive GFP-IRGM-positive depolarized mitochondria. During these events, IRGM interactions with cardiolipin may affect cardiolipin or its remodeling products and alter fusion potential of mitochondrial membranes (thus promoting fission), electron transport chain and mitochondrial polarization, and unbalance apoptosis suppression42, 58, 59.
Our studies show that IRGM partakes in autophagy as a cell survival process and in cell death processes in addition to its role in control of intracellular pathogens. When IRGMd expression enhanced cell death, this was independent of autophagy and instead required apoptotic machinery (caspases, Bax and Bak). IRGM may contribute to cell death in a manner proposed for other mitochondrial fission proteins participating in apoptosis43, 44, 47, 50, 51. Mitochondrial fission is the only universally conserved process that accompanies programmed cell death including the species (e.g. nematodes, flies) where apoptosis proceeds in the absence of release of cytochrome c from mitochondria60. Our findings furthermore suggest a hierarchy of events, with autophagy being downstream of IRGM during cell's initial responses, followed by cell death processes that can outrun autophagy upon increased expression of the majority of IRGM isoforms.
The IRGM functions in autophagy, Δψm, and cell death suggest that there are two faces (protective and inflammatory) of IRGM as a cell-autonomous autophagic effector against M. tuberculosis20, a tuberculosis risk locus in human populations25, and a Crohn's disease risk locus17, 27, 28. We also found that IRGM induced cell death was associated with the release of HMGB1, a major pro-inflammatory alarmin/DAMP, normally present in the nucleus of live cells but released from necrotic and some apoptotic cells54-56. This points to a potential but hitherto unappreciated role in Crohn's disease of HMGB154-56. Our findings pose broader questions regarding the relationship between mitochondria, autophagy, and apoptosis in innate immunity. Mitochondria are central players in many cellular functions including oxidative metabolism, autophagy, and programmed cell death, and are believed to have originated as symbiont α-proteobacteria of the pre-eukaryotic cell. We propose that mitochondrial fission, controlled by Drp1 in all eukaryotic cells and, as shown here, affected by IRGM in human cells, may have been a primordial signal of microbial presence in the cytosol, and that it has become evolutionarily hardwired into induction of autophagy as a mechanism for elimination of intracellular pathogens, or, when autophagic elimination fails, elimination of infected cells by cell death, either way limiting the spread of infection.
Online Methods
Cell culture, transfections, DNA constructs, siRNA knockdowns and fluorogenic probes
Human U937, THP-1 cells and HeLa cells were from ATCC. Atg7−/− and Atg7 wild type MEF were from Juntendo University, Japan. Baby mouse kidney (BMK) wild type (W2) and Bax/Bak−/− (D3) cell lines were previously described 52. U937 cells were transfected by nucleoporation using Nucleofector Reagent Kit C (Amaxa biosystems), HeLa and BMK cells were transfected with Kit V. IRGM splice variants IRGMa, IRGMb, IRGMc/e, IRGMd and IRGMd-S47N were cloned by DNA synthesis and insertion into SacI and SalI restriction sites of pCFP-C1 (IRGMa), pYFP-C1 (IRGMb) and pEGFP-C1 (IRGMc, IRGMd and IRGMd S47N). GFP-Drp1 was from A. van der Bliek. For knockdowns, cells were transfected with siGENOME SMARTpool reagent (Dharmacon) for human mRNAs encoding IRGM (CCACAACCCUGGAGAACUA; CCAAAGAUGUGCCUCCUAU; GCAAUGGGAUGUCCACCUU; AGAAGGAGCGGGUAUGUGA), DRP1 (GAAAGAAGCAGCUGAUAUG; GGAGCCAGCUAGAUAUUAA; CAAAGGCAGUAAUGCAUUU; CGUAAAAGGUUGCCUGUUA), FIS1 (CGAGAAGGCCUUAAAGUAC; CCAAGAGCACGCAGUUUGA; ACUACCGGCUCAAGGAAUA; CGGACAAGGUACAAUGAUG), MFN1 (GAAGAGCUCUGUUAUCAAU; GCACAGAUGUCACUACAGA; GAUACUAGCUACUGUGAAA; CUGGAUAGCUGGAUUGAUA), MFN2 (ACUAUAAGCUGCGAAUUAA; GAUCAGGCGCCUCUCUGUA; GGUUACCUAUCCAAAGUGA; GGUUACCUAUCCAAAGUGA; CAACUAUGACCUAAACUGU), ATG7 (CCAAAGUUCUUGAUCAAUA; GAUCAAAGGUUUUCACUAA; GAAGAUAACAAUUGGUGUA; CAACAUCCCUGGUUACAAG), and BECN1 (human Beclin 1; GGAUGACAGUGAACAGUUA; UAAGAUGGGUCUGAAAUUU; GCCAACAGCUUCACUCUGA; UUGAAAACCAGAUGCGUUA). Non-targeting siRNA pool was used as a control. Cells were transfected with 1.5 μg of siRNA by nucleoporation as described above. Propidium iodide (PI), 5,6-carboxy-2′,7′dichlorodihydrofluorescein (DCF), MTR, 7-amino actinomycin D (7-AAD), and DiOC6(3) were from Molecular Probes.
Subcellular fractionation, immunoblot analysis, and antibodies
The following panel of independent organelle purification and subcellualr compartment separation methods were used: (i) A previously applied method to study IRG subcellular distribution 61. (ii) Proteome-grade purification protocol using Qproteome Mitochondria Isolation kit (QIAGEN). (iii) Isopycnic separation by sucrose density equilibrium centrifugation 62. Cells were homogenized in 210 mM manitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES-NaOH, pH 7.5 (MB). The postnuclear supernatant was centrifuged at 13,000 g for 10 min. The pellet fraction was resuspended in MB and layered on top of a pre-formed sucrose gradient consisting of 1.2 M sucrose, on top of 1.6 M sucrose. The sample was centrifuged at 28,000 rpm in a Beckman SW 40 rotor at 4 °C. Fractions were collected from the top of the gradient and washed in MB for further analysis. IRGM is a very low abundance protein (when its expression is slightly elevated this can lead to cell death). Hence, when crude extracts are examined by immunoblotting additional bands in the 35-50 kDa range show up on films. These additional bands correspond to cytosolic cross-reactive proteins, and once membranous fractions are separated, they are no longer the most prominent bands on blots; these cross-reactive bands are not susceptible to IRGM siRNA knockdowns. Subcellular fractions were analyzed by Western blotting using the previously reported antibodies against IRGM (Singh et al., 2006), used interchangeably for immunoblotting of subcellular organelles with Abcam (ab69494) IRGM antibody (side-by-side comparisons are shown in Suppl. Fig. S7d). Peptide competition characterization is shown in Suppl. Fig. S7e,f, and endogenous IRGM vs. GFP-IRGMd distribution in Suppl. Fig. S7g. Subcellular fractions were analyzed in parallel with antibodies to calnexin (Santa Cruz Biotechnology), cytochrome c (BD biosciences), LC3 (Sigma), syntaxin 6 (Transduction labs) and Rab7 (L. Huber); staining was revealed with Super Signal West Dura chemiluminescent substrate (Pierce).
Sub-compartment localization by accessibility to proteinase K
Purified mitochondria were resuspended in reaction buffer (250 mM sucrose, 10mM Tris pH7.4) and were incubated with or without Proteinase K (40 μg). For osmotic shock, mitochondria were incubated with 20 mM HEPES for 30 min on ice to remove the outer mitochondrial membrane. 1% TX-100 was used to solubilize proteins. Following incubation at 37°C for 30 min, PMSF was added to the final concentration of 2 mM to stop the reaction. The samples were boiled in SDS sample buffer and probed for mitochondrial proteins with anti-cytochrome c, anti-IRGM, anti-Mfn-2 (Abcam) and anti-Hsp60 (Abcam).
IRGM protein purification and IRGM-lipid binding
GST-IRGM fusions were expressed in E. coli and purified by affinity chromatography (Suppl. Fig. S1). Nitrocellulose filters spotted with a panel lipids (Echelon) were blocked with 1% nonfat-dry milk in PBS for 1 1h at room temperature. Filters were incubated with 90 nM of GST (Abcam) or GST-IRGM for 1 h in blocking buffer, followed by three washes with PBS, 0.1% Tween-20. Filters were incubated with goat anti-GST (HRP) (Abcam) (1/2,000) for 1 h. After three washes, GST was detected using ECL kit (Pierce). Cardiolipin bound agarose beads and control beads (Echelon) were incubated with 5 μg of GST (Abcam) or GST-IRGMd overnight at 4°C, followed by five washes in 10 mM HEPES, 150 mM NaCl, 0.25% Igepal. To elute proteins, equal volume of 2X Laemmli buffer was added to beads, samples boiled for 2 min, and proteins analyzed by Western blotting using antibodies to IRGM (Abcam).
Confocal microscopy
HeLa, BMK and MEF were transfected with GFP-IRGMd for 48 h followed by staining with 200 nM MTR (Invitrogen) for 10 min. Cover slips were mounted in a 5% CO2 humidified chamber, with temperature maintained at 37°C, and imaged using a Zeiss LSM 5 Live microscope (laser wavelength, 488 nm and 543 nm). Fixed cells were imaged using a Zeiss LSM 510 Meta confocal microscope. For immunofluorescence analysis of endogenous IRGM, antibody described by Singh et al. 20 was used (peptide competition characterization and co-analysis in GFP-IRGMd transfected cells are shown in Suppl. Fig. S7c,d).
Mycobacterial phagosome maturation and mycobacterial survival
LysoTracker Red staining, morphomterics, and microbiological analyses of bacterial viability were carried out as previously described 8, 20, 63.
Autophagic and cell death assays
FLICA (Serotec) was used to stain intracellular active caspase 3 and caspase 7. LC3-GFP puncta assay, LC3-I-to-LC3-II conversion in the presence of baffilomycin A1, 7-AAD staining, and HGMB1 staining were carried as previously described 54, 64.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI069345, RC1AI086845, and AI42999, a grant from Crohn's & Colitis Foundation of America, and a grant from Bill and Melinda Gates Foundation.
Footnotes
Author contributions
SS, WO, IV, JN, MD, ER, MP, SM carried out planning, experimental work and data analysis, EW and MK contributed MEFs and plans for use, VD carried out project planning, experimental work, and integration.
Competing financial interest statement
Authors declare no competing financial interests.
Refererences
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 3.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 7.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 9.Kyei GB, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchet FP, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris J, et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Miller BC, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 13.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 14.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 15.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 16.Delgado M, et al. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 18.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 21.Feng CG, et al. The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat Immunol. 2008;9:1279–1287. doi: 10.1038/ni.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard J. The IRG proteins: a function in search of a mechanism. Immunobiology. 2008;213:367–375. doi: 10.1016/j.imbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Bekpen C, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bekpen C, et al. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Intemann CD, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craddock N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes L.C. & Scorrano. L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 32.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. Embo J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dagda RK, et al. Loss of pink1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009 doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benard G, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 37.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat Immunol. 2009;10:907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schug ZT, Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim Biophys Acta. 2009;1788:2022–2031. doi: 10.1016/j.bbamem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 45.Koch A, et al. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 46.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnoult D, et al. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 48.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Estaquier J, Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14:1086–1094. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- 52.Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E. Bax and Bak independently promote cytochrome C release from mitochondria. J Biol Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- 53.Ricci JE, et al. Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell. 2004;117:773–786. doi: 10.1016/j.cell.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 55.Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 56.Kazama H, et al. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gebert N, et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19:2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 59.DeVay RM, et al. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oberst A, Bender C, Green DR. Living with death: the evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ. 2008;15:1139–1146. doi: 10.1038/cdd.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaiser F, Kaufmann SH, Zerrahn J. IIGP, a member of the IFN inducible and microbial defense mediating 47 kDa GTPase family, interacts with the microtubule binding protein hook3. J Cell Sci. 2004;117:1747–1756. doi: 10.1242/jcs.01039. [DOI] [PubMed] [Google Scholar]
- 62.Antonsson B, Montessuit S, Sanchez B, Martinou JC. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J Biol Chem. 2001;276:11615–11623. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 63.Ponpuak M, Delgado MA, Elmaoued RA, Deretic V. Monitoring autophagy during Mycobacterium tuberculosis infection. Methods Enzymol. 2009;452:345–361. doi: 10.1016/S0076-6879(08)03621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.