Abstract
Quantum mechanics/molecular mechanics calculations based on ab initio multiconfigurational second order perturbation theory are employed to construct a computer model of Bacteriorhodopsin that reproduces the observed static and transient electronic spectra, the dipole moment changes, and the energy stored in the photocycle intermediate K. The computed reaction coordinate indicates that the isomerization of the retinal chromophore occurs via a complex motion accounting for three distinct regimes: (i) production of the excited state intermediate I, (ii) evolution of I toward a conical intersection between the excited state and the ground state, and (iii) formation of K. We show that, during stage ii, a space-saving mechanism dominated by an asynchronous double bicycle-pedal deformation of the C10═C11─C12═C13─C14═N moiety of the chromophore dominates the isomerization. On this same stage a N─H/water hydrogen bond is weakened and initiates a breaking process that is completed during stage iii.
Keywords: photoisomerization, quantum mechanics/molecular mechanics (QM/MM), retinal proteins
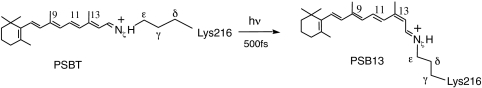
The light-activated proton pump bacteriorhodopsin (bR) is an Archaea receptor contained in the purple membrane of Halobacterium salinarium. As shown in Scheme 1, upon photoexcitation, the all-trans retinal chromophore (PSBT) bounded to Lys216 via a protonated Schiff base linkage is converted to the 13-cis isomer (PSB13). This event triggers a series of conformational changes that ultimately result in a proton translocation from the cytoplasmic to the extracellular domain (1).
Scheme 1.
The bR protein environment affects both the spectroscopic and photochemical properties of PSBT (2–4). First, the absorption maximum (568 nm) is red-shifted with respect to the one observed in solution (440 nm in methanol). Second, time-resolved spectroscopy reveals a dominant excited state lifetime component of 450 fs (5, 6) almost matching the 500-fs time scale for formation of the vibrationally hot primary photoproduct J (7, 8). In contrast, PSBT in solution features a biexponential decay dynamics with a dominant (ca. 10-fold longer) 2-ps component (9–11). Finally, although bR photoisomerizes stereoselectively (leading exclusively to PSB13) with high (ca. 67%) quantum yield (9, 12), irradiation of PSBT in solution leads to a mixture of different stereoisomers with a smaller (ca. 25%) total quantum yield (9, 13).
Low-temperature spectroscopic studies provided evidence for the existence of a tiny (≤ 1 kcal mol-1) energy barrier on the excited state (S1) potential energy surface of bR (14, 15). Such a barrier would explain the 450-fs short-lived quasistationary state observed by Ruhman et al. (5) that is assigned to the fluorescent state I (16–18) and precedes decay to the ground state (S0). Recent experiments by Léonard et al. (19) have also revealed that the photoinduced changes in the permanent dipole moment of PSBT are mirrored by the absorption of Trp86 up to the first stable (i.e., cryogenically trapped) photoproduct K (7). Relative to bR, K bears 11.7–15.9 kcal mol-1 stored photon energy (20).
The investigation of the photoisomerization mechanism of bR is of great interest because this receptor is the archetypal ion pump machine. Its comprehension may open previously undescribed perspectives for the use of bR in nanotechnology or for the design of unique molecular devices (21, 22). In recent years numerous theoretical works, involving various hybrid quantum mechanics/molecular mechanics (QM/MM) approaches, have investigated the tuning of the spectroscopic properties of the retinal chromophore (23–28), the mechanism, stereoselectivity, and dynamics of its photoisomerization (29–32), and the energy storage (33). However, these studies have not yet delivered a coherent mechanistic scenario. For instance, the counterion effects on the spectroscopy of PSBT, as well as the mechanism of the PSBT space-saving photoisomerization, are not resolved.
Our recent work on the retinal chromophore in vacuo (34, 35) shows that the CASPT2//CASSCF/6-31G* protocol yields balanced values for geometrical parameters, energy differences, spectroscopic properties, and change in dipole moment. Most important, it delivers an accurate description of the photochemically relevant potential energy surfaces (PESs). The same protocol has been successfully coupled with the AMBER molecular mechanics force field to investigate the photoisomerization of the visual pigment rhodopsin (Rh) (4, 36), returning energies that agree with the experiments. These studies support a two-state model for the retinal chromophore isomerization: the S1 path develops entirely along a charge transfer (i.e., hole-pair 1Bu-like) state and ends into a S1/S0 conical intersection (CI) where the reacting double bond features a ca. -90° twisted structure. Because of its geometrical and electronic structure, this point is consistent with that of a twisted intramolecular charge transfer (TICT) state (34, 37). The analysis of the S1 isomerization coordinate along the minimum energy path reveals that this is sequentially dominated by different, almost uncoupled, modes. First, a stretching mode leading to single-bond/double-bond order inversion occurs. Second, a one-bond-flip (38, 39) twisting of the reacting double bond is activated in vacuo, whereas in the visual pigment Rh we have documented (4, 36) a space-saving mechanism that includes the bicycle-pedal motion proposed by Warshel (40, 41). This mechanism has been recently demonstrated using trajectory computations in vacuo (42) and in Rh (43).
Below the CASPT2//CASSCF/AMBER protocol recently applied to Rh (4, 36, 43) is used to construct the S1 isomerization path of bR. Although structural (i.e., nondynamical), the data yield a mechanism of the initial stage of the photocycle [from the Franck–Condon (FC) point to the first isolable intermediate K] involving an asynchronous double bicyclo-pedal or folding-table (44) deformation. Indeed, it is shown that the ─C11═C12─ and ─C15═NH─ double bonds undergo a limited counterclockwise twisting, while the ─C13═C14─ bond is isomerizing in the clockwise direction. Remarkably, this photoinitiated motion induces a simultaneous hydrogen bond breaking.
1. Results and Discussion
1.1. Chromophore Structure and Absorption Maximum.
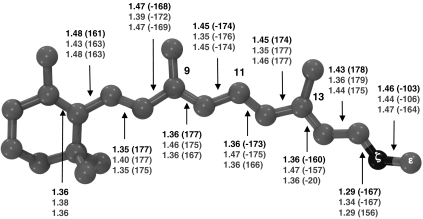
The optimized S0 bR structure (see Fig. 1) displays a -160° value of the PBST C12─C13─C14─C15 dihedral angle corresponding to a 20° clockwise twisted C13═C14 double bond. In contrast, the PSBT structure optimized in vacuo is planar (45). The bR C13═C14 pretwist is larger than the one observed for the reactive bond of Rh (4, 36) and bias the photoisomerization in the clockwise direction (as detailed by the computed path; see below). This suggests that the stereoselectivity observed in bR (see, e.g., ref. 31) is decided by the asymmetry of the protein cavity that tilts the S1 PES in order to favor a unidirectional C13═C14 photoisomerization.
Fig. 1.
Optimized structure for the first three states—bR, I, and K—of the proton–pump photocycle: The geometrical parameters (top, middle, and bottom values for bR, I, and K, respectively) are reported in Å and degrees (in parentheses).
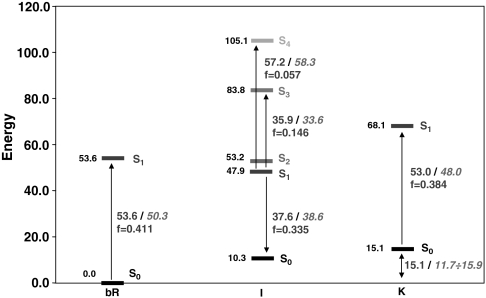
The computed 53.6 kcal mol-1 (534 nm) required for the vertical π → π∗ spectroscopically allowed (f = 0.411) S0 → S1 transition in bR compare well with the experimental value of 50.3 kcal mol-1 (568 nm); see Fig. 2. This value is blue-shifted (+6.9 kcal mol-1) as compared to that computed in vacuo (see SI Text for details). This effect agrees with previously reported values by Rajamani and Gao (∼4 kcal mol-1) (23), Vreven and Morokuma (2–6 kcal mol-1) (30), and Houjou et al. (∼7 kcal mol-1), (24, 25) and is due to electrostatic destabilization of the S1 state by the protein (2, 46). The blue-shift effect due to the complex counterion (Asp85, Asp212, Arg82) alone is much higher (+20.1 kcal mol-1 for the bare chromophore/counterion pair with respect to vacuo; see SI Text), revealing that the protein has a high shielding effect (ca. 65%). This conclusion is consistent with previous computational studies on Rh (4). A consequence of the above results is that the opsin-shift observed when going from solvent (440 nm) to bR (570 nm) (ca. -13 kcal mol-1) is mainly due to (i) the binding pocket structure that forces the β-ionone ring into a planar 6s-trans conformation (a skewed 6s-cis conformation is expected in solution), effectively increasing the length of the conjugated chain, and (ii) a dumping of the chromophore/counterion electrostatic interaction due to the protein environment and leading to a red-shifted absorption as compared to the unshielded counterion.
Fig. 2.
CASPT2 relative energies (in kcal mol-1) for the first three states—bR, I, and K—of the photocycle. Vertical electronic transitions (with their computed oscillator strength f) are also reported with the corresponding experimental values (in italics).
1.2. Excited State Intermediates and Transient Spectroscopy.
The PSBT displacement occurring after photoexcitation leads from the FC region to a shallow S1 minimum displaying bond order inversion and a negligible twisting of the reactive C13═C14 double bond (see Fig. 1). This structure is assigned to the fluorescent state I (5, 6). The corresponding computed and observed transition energies agree within 1–2 kcal mol-1 (see Fig. 2): (i) The ∼740 nm (38.6 kcal mol-1) fluorescence maximum observed by Schenkl et al. (16) is matched by the 37.6 kcal mol-1 (761 nm) energy gap computed for the S1 → S0 transition at this point. (ii) Hasson et al. (14) described a transient near-IR absorption peaking at 850 nm (i.e., 33.6 kcal mol-1). This band can be attributed to the significantly allowed S1 → S3 transition, which is computed to be 35.9 kcal mol-1 (797 nm). (iii) Finally, the transient absorption in the visible seen by Logunov et al. (47) and peaking at 490 nm (i.e., 58.3 kcal mol-1) can be assigned to the S1 → S4 transition, which shows an energy gap of 57.2 kcal mol-1 (500 nm) and a not negligible f. Notice that these properties are also consistent with the results of transient spectroscopic studies of artificial rhodopsins hosting a conformationally locked retinal analog (16–18).
1.3. Photoisomerization, Photoproduct Formation, and Retinal Dipole Moment Changes.
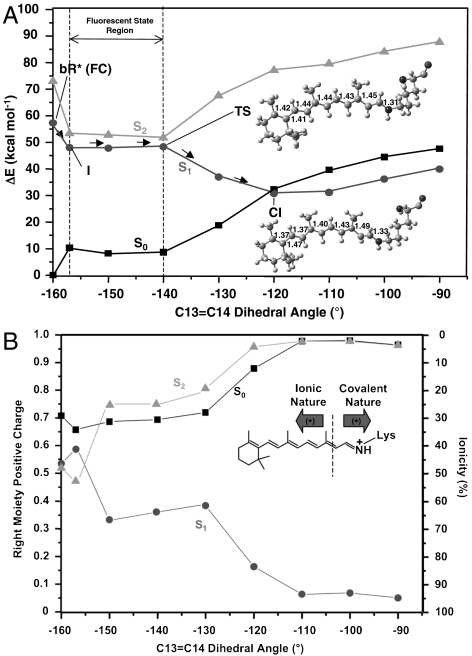
The < 1 kcal mol-1 barrier located along the flat region of the S1 path starting at I (see the difference between I and the transition state (TS) in Fig. 3) does not allow for a quantitative analysis. However, the existence of a barrier is in agreement with low-temperature experiments (14, 15) and with a quasistationary nature of I (5). The S2-S1 energy gap is initially decreasing [15.7 kcal mol-1, 5.3 kcal mol-1, and 3.3 kcal mol-1 for FC, I, and TS, respectively (see Fig. 3 and SI Text)], leading to mixed ionic/covalent S2 and S1 wave functions. Then, it rises again on the way to the twisted and peaked S1/S0 CI (which prompts radiationless decay to S0) and the path, after the TS, develops exclusively on a charge transfer S1 state [whereas S2 is a dark (π → π∗)2 covalent bound state. See charge analysis in Fig. 3B]. Consequently, the dark state shall not be involved in the photoisomerization and decay of bR, as found for Rh (4, 36) and for PSBT in vacuo. (35). This questions the S2 assignment from two-photon absorption experiments by Birge and Zhang (48) and supports the recent reassignment by Andersen and coworkers (49).
Fig. 3.
The photoisomerization channel mapped on S1. (A) CASPT2 corrected S0, S1, and S2 energy profiles (molecular parameters are reported in Å and degrees). (B) Corresponding evolution of the fractional positive (Mulliken) charge resident on the right chromophore moiety and ionic character (ionicity %). (Inset) See dotted line: Ionic/covalent states are characterized by a positive charge that stays preferentially on the left/right moiety.
The located CI features a -120° C13═C14 dihedral angle. This CI belongs to a S1/S0 crossing seam that develops along the isomerization coordinate as indicated by the fact that the S1 and S0 energy profiles remain parallel and energetically close up to a -90° twisting (less than 10 kcal mol-1 separation at the CASPT2 level and substantially degenerate at the CASSCF level; see Fig. 3). This situation parallels the one documented for Rh (50). Although in order to simulate/understand the reaction quantum yield, one should carry out trajectory calculations (that go beyond the scope of the present work), our results show that an extended low-lying S1/S0 intersection seam is energetically accessible upon relaxation from the Franck–Condon point. Therefore, trajectories that even slightly escape the computed path may access structurally different points of the seam contributing to the observed high photoisomerization efficiency.
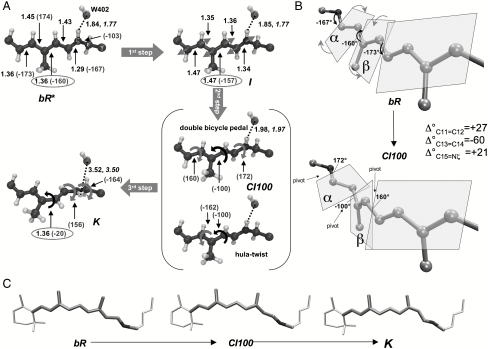
The S1 reaction coordinate displays three consecutive steps characterized by different structural changes (see Fig. 4A). The first (first step, bR∗ → I, < 50 fs) takes place immediately after light absorption and is characterized by the bond order inversion leading to the formation of the fluorescent state I (16, 51). The following step (second step, I → CI, ∼450 fs) occurs when the initial C13═C14 isomerization motion is accompanied by small rotations about the nearby bonds allowing the system to cross the barrier and reach the CI at 120° (in Fig. 4A the 100° twisted CI—CI100—is used to produce a clearer picture of the deformation needed to reach K). This step initiates the isomerization and is characterized by a concerted double bicycle-pedal mechanism (40) (illustrated in Fig. 4B) with a minor contribution of hula twist (52) involving tiny pyramidalizations at C12 and C13 [this pyramidalization was also reported by Hayashi et al. (31), but at a different location, C14, perhaps due to reduced QM moiety employed in the simulations]. As shown in Fig. 4C, this makes the isomerization facile as a limited change in the chromophore steric hindrance occurs from bR to K. The simultaneous three bond twist has been observed recently in crystals of all-cis-1,6-diphenyl-1,3,5-hexatriene (53) and also described in dynamics simulation of mechanically constrained protonated Schiff bases, named folding table (44).
Fig. 4.
Photoisomerization mechanism. Schematic representation of (A) the photoinduced three-steps (bR∗ → I → CI100 → K) motion [bonds in Å and angles in degrees (in brackets)], (B) the detailed double bicycle-pedal or folding-table motion occurring along the bR∗ → CI100 excited state stage (the variation in the twisting angle—Δ°—for C11═C12, C13═C14, and C15═Nζ is also reported), and (C) the space-saving character of the overall motion (the chromophore ends are at rest during the isomerization process). Structural parameters optimized for the mobile W402 setup are reported in italics when differing from those pertaining to the fixed W402 setup (see Computational Methods and SI Text).
In bR the concurrent twisting deformation of three adjacent double bonds C11═C12 (+27°), C13═C14 (-60°), and C15═Nζ (+21°) occurs in alternate directions (counterclockwise, clockwise, and counterclockwise, respectively). This parallels/extends the structural evolution during the isomerization of the PSB11 retinal chromophore in Rh where the C11═C12 reactive bond isomerizes in the counterclockwise direction while the adjacent C9═C10 bond twists in the clockwise direction. As in Rh, the mechanism can be described as asynchronous (the extent of twistings of the ─C11═C12─ and ─C15═Nζ─ bonds are smaller than the one experienced by the ─C13═C14─ bond). However, note that the combined +27° (C11═C12) plus +21° (C15═Nζ) angle variations almost fully compensate for the -60° twisting of C13═C14 at the CI. More remarkably, progressive lengthening of the Nζ─H/water-402 (W402) hydrogen bond occurs along the excited state path from bR to the CI (+0.14 and +0.2 Å for the fixed and mobile W402 set up, respectively; see Fig. 4A and SI Text), suggesting that photoexcitation prompts its breaking, and pointing to a W402 that stays always tightly bound to the complex counterion (see also Fig. 5). This process would be energetically demanding on S0. However, the chromophore N─H positive charge translocation occurring on S1 leads to a weakening of this hydrogen bond, which accounts for its lengthening. In fact (see SI Text for details), an instantaneous ∼3.0 kcal mol-1 destabilization accompanies S0 → S1 excitation that can be mainly due to hydrogen bond weakening. This effect is predicted to increase steadily as the system evolves along the S1 isomerization path. Indeed, positive charge translocation from the nitrogen head to the β-ionone ring progressively increases (till a net 100% transfer at the TICT point; see Fig. 3B).
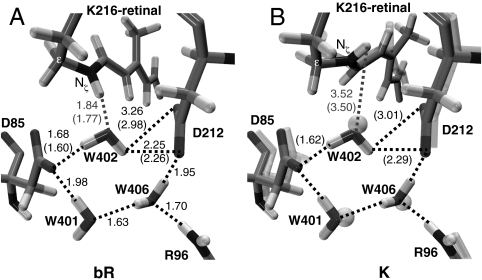
Fig. 5.
Structures of the optimized binding site (hydrogen bonds are highlighted and reported in Å; structural parameters pertaining the mobile W402 setup are reported in parentheses). (A) bR (modeled from the available X-ray crystallographic structure) (62). (B) The primary photoproduct [for comparison, the K structure from X-ray crystallography (56) is also shown in transparence]. bR and K crystal structures display a substantially unmodified binding pocket pointing to a W402 that is tightly bound to the complex counterion.
To the best of our knowledge, a double bicycle-pedal mode followed by hydrogen bond breaking has never been proposed or detected in dynamical studies of bR (31, 32). It is also different from the single bicycle-pedal mechanism recently found in Rh (4, 36) and described by the rotation of the plane hosting the ─C10─C11─ moiety. Indeed, in bR there are two such adjacent rotating units: the ═C14─C15═ unit (plane α) and the ═C12─C13═ unit (plane β). The direction of twisting during the isomerization is given in Fig. 4B. As anticipated above, besides bond order inversion and triple isomerization (i.e., about the ─C11═C12─, ─C13═C14─, and ─C15═N─ bonds), hydrogen bond weakening is activated on S1 and helps to release the electrostatic hindrances on retinal (as recently suggested by Schenkl et al.) (51).
The twisting of the reactive C13═C14 double bond initiated on S1 is finalized after decay to S0 through the CI, whereas the two adjacent bond revert to the original configuration (third step, CI → K). Indeed, S0 optimization leads to a stable PSB13 chromophore (even if still 20° twisted about the C13═C14 bond. See Figs. 1 and 4A). The aborted bicycle-pedal motion seems to be compensated by a 60 ° Nζ─Cε conformational change (see Fig. 4A) allowing minimum volume C13═C14 isomerization. Finally, during this third stage the breaking of the hydrogen bond between W402 and the Nζ─H retinal is completed. An animation of the simulated isomerization motion is included in SI Text.
The structural evolution described above seems to be consistent with the photoinduced changes in permanent dipole moment of the chromophore that have been experimentally monitored by Léonard et al. (19). In particular, the recorded transient response (i.e., modifications in UV-absorption spectrum) of the nearby Trp86 residue has been used as a probe of the ultrafast changes in the local PSBT charge distribution (i.e., the dipole moment variation—Δμ—with respect to the one recorded in ground state bR). The instantaneous rise (i.e., within the 50-fs time resolution of the experiments) of the Trp86 signal reveals the underlying retinal charge translocation upon photon absorption and the corresponding increase in μ (i.e., Δμ > 0). These experimental observations are consistent with our computations (see Table 1), because the latter show a significant increase in μ when going from bR to FC and I (Δμ ≈ 10) and even more (Δμ≥20) from I to the CI. Afterward, Δμ is shown to decrease with time constants that match S0 recovery and photoproduct K formation, although the initial bR value is not fully recovered (19), calling for a change in the local electrostatic environment that remains long after isomerization. This trend is consistent with the small value of Δμ calculated at K consistently with a charge distribution similar to that found in bR.
Table 1.
Changes of the difference dipole moment—Δμ (in debye)—along the photoisomerization channel
| Structure | FC | I | 150° | TS (140°) | CI | K |
| Δμ | 12.3 | 9.0 | 22.1 | 20.3 | 20.0 | 2.6 |
Δμ is calculated as the difference of the initial ground state bR dipole and the current transient dipole at each reported structure (this is the dipole computed on S1 for all the structures but K, where it is referred back to the ground state).
1.4. K and Energy Storage.
The results above indicate that the computed 13-cis photoproduct is K. The J intermediate is assigned to either a vibrationally hot state on S0 or S1 (e.g., I with activation of hydrogen-out-of-plane modes) (54) and cannot be characterized through static computations. This conclusion is consistent with the calculation of the energy difference between K and bR (15.1 kcal mol-1), which agrees with the experimental estimate of the energy storage in K (11.7–15.9 kcal mol-1) (20) and also with previous computational studies (16 kcal mol-1) (55). We have also compared the computed K geometrical parameters with those reported by Lanyi and Schobert (56) on the basis of X-ray diffraction data (see Fig. 5B). The two structures are in good agreement (the largest differences being located on the C13═C14 and C15═Nζ bonds, which correspond to the less-resolved regions in the experimental structure). Finally, the structure and energy analysis reported here for K (including the predicted Nζ─H/W402 hydrogen bond breaking) matches the results reported by Hayashi et al. (55) using a different QM/MM protocol. Thus, it is apparent that, despite a slightly overestimated simulated absorption maximum [the computed S0-S1 vertical energy gap is 53.0 kcal mol-1 (540 nm), i.e., 5 kcal mol-1 higher than the 48.0 kcal mol-1 (596 nm) experimental absorption maximum for K; see Fig. 1] (57), our data support the assignment of the optimized photoproduct to K.
The energy stored in K brings the proton pumping to the end. In the available literature its origin is explained with two different hypotheses (58). The first assigns it to the augmented steric strain of the PSB13 chromophore that is transferred to the protein in later steps (20). The second emphasizes the weakening of the interactions between the chromophore and the nearby polar groups (59). To disentangle these contributions, we have analyzed the energy difference between Rh and K (see SI Text for details). The van der Waals energy difference is only ∼0.7 kcal mol-1, revealing a negligible opsin/chromophore steric component. However, the intrinsic chromophore strain accounts for only 6.6 kcal mol-1. The weakening of the interactions between retinal and its complex counterion (most of which is due to the breaking of the Nζ─H/W402 hydrogen bond; see Fig. 5) contributes ca. twice that value (13.8 kcal mol-1). This value, which agrees with previous computations (33, 55) is, in part, compensated by the electrostatic effects due to the remaining protein environment leading to a total of 7.8 kcal mol-1 for the electrostatic component. It is thus concluded that, as also found for Rh (36), changes in both the intrinsic retinal strain and electrostatic interactions contribute almost equally to the energy stored at K, in line with the results of the pioneering work of Birge and Cooper (58) and more recent theoretical estimates (33, 55).
2. Conclusions
We have been able to construct a CASPT2//CASSCF/AMBER computational model that reproduces the observed spectral features of the initial part of the bR photocycle (i.e., bR → I → K) and the energy stored at K with a less than 5 kcal mol-1 error. It has been shown that bR blue-shifts the absorption maximum with respect to PSBT in vacuo, but not as much as the counterion alone would do: In fact, this is significantly quenched (> 60%) by the protein. The bR opsin-shift is due to an increased double-bond conjugation (in going from the skewed 6s-cis conformation in solvent to the fully planar 6s-trans in bR) and to an opsin-induced dumping of the counterion electrostatic effect. The bR photoisomerization mechanism involves three stages: (i) a backbone rearrangement leading to an inverted bond order intermediate I on S1 (first stage, ≤ 50 fs); (ii) the C13═C14 torsion accompanied by concurrent twistings of the adjacent bonds allowing the system to proceed along a space-saving isomerization coordinate reaching the deactivation region; a chromophore-NζH/W402 hydrogen bond breaking is also initiated during this stage (second stage, to ∼450 fs); (iii) the S0 relaxation leading to double-bond reconstitution and Nζ─Cε single bond twisting that result in an aborted bicycle-pedal progression and lead to the K state (third stage, till ∼3 ps). This final step is responsible for completing Nζ─H/W402 hydrogen bond breaking that is one of the main contributions to the energy storage.
3. Computational Methods
Consistently with previous works (4, 36, 50), the photoisomerization path is computed in terms of a relaxed scan (with the C13═C14 twisting angle fixed at specified values) carried out using our recently developed QM/MM potential [the GAUSSIAN98 (60) and TINKER (61) suite of programs are employed]. The retinal, Schiff base nitrogen and attached ε-carbon atom (see Scheme 1) constitute the QM part of the system, described at the CASSCF/6-31G* level (with an active space of 12 π-electrons in 12 π-orbitals), whereas AMBER is used for all MM atoms. All QM atoms and MM atoms comprising the Lys-216 side chain (CγH2, CδH2, and CβH2) are free to relax, whereas the protein framework [PDB ID code 1C3W (62) is used] is kept fixed: We assumed that this has no time to change (i.e., full thermal equilibration is prevented) during the subpicosecond time scale of the bR primary photoinduced event investigated here, a view that is supported by the similarity recognized between the bR and K crystal structures (see Fig. 5). W402 may be an exception: Although it belongs (and is tightly bound via hydrogen bonding) to the complex counterion (see Fig. 5), possibly limiting its motion, nevertheless it also directly/strongly interacts (via hydrogen bond) with the chromophore and can respond to its changes. Therefore, two sets of computations were employed to compute the path with a locked/unlocked W402, respectively, which eventually delivered the same mechanistic outcome (see Fig. 4A and SI Text). This validates the fixed W402 model, which the results reported in this work are generally referred to. The employed crystallographic structure is considered as a mean representation of the experimental position of the atoms (although at low temperature), and the computed path portrays a mean static view of the process. This approach to ultrafast photoinduced reactions has been already employed successfully to study the Rh primary event (4, 36, 43, 50). Multiconfigurational second-order perturbation theory computations [carried out with the CASPT2 (63) method using the MOLCAS5 (63) package] are then used to increase the accuracy of the energy profiles. Whenever possible, a single state approach is employed. Otherwise, when root flipping occurs, a state average procedure (by equally weighting the first five singlet roots) is used, together with a multistate approach (MS-CASPT2) (64) when required. Oscillator strengths (f) are computed using correlated (CASPT2) energies within the RASSI approach (65). See SI Text for further details on computations and protein setup.
Supplementary Material
Acknowledgments.
M.G. is grateful to Prof. Stefan Haacke for useful discussions. M.O. is grateful to the Center for Photochemical Sciences and the College of Arts and Sciences for start-up funding. This research has been supported by Ministero della Pubblica Istruzione, Università, Ricerca Scientifica e Tecnologica - Progetti di Rilevante Interesse Nazionale 2008 (“Tracking ultrafast photoinduced intra- and inter-molecular processes in natural and artificial photosensors,” project 2008JKBBK4).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007000107/-/DCSupplemental.
References
- 1.Oesterhelt D, Stoechenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 2.Cembran A, Bernardi F, Olivucci M, Garavelli M. Counterion controlled photoisomerization of retinal chromophore models: A computational investigation. J Am Chem Soc. 2004;126:16018–16037. doi: 10.1021/ja048782+. [DOI] [PubMed] [Google Scholar]
- 3.Coto PB, Strambi A, Ferré N, Olivucci M. The color of rhodopsins at the ab initio multiconfigurational perturbation theory resolution. Proc Natl Acad Sci USA. 2006;103:17154–17159. doi: 10.1073/pnas.0604048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomasello G, et al. Electrostatic control of the photoisomerization efficiency and optical properties in visual pigments: On the role of counterion quenching. J Am Chem Soc. 2009;131:5172–5186. doi: 10.1021/ja808424b. [DOI] [PubMed] [Google Scholar]
- 5.Ruhman S, Hou BX, Friedman N, Ottolenghi M, Sheves M. Following evolution of bacteriorhodopsin in its reactive excited state via stimulated emission pumping. J Am Chem Soc. 2002;124:8854–8858. doi: 10.1021/ja026426q. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt B, et al. Excited-state dynamics of bacteriorhodopsin probed by broadband femtosecond fluorescence spectroscopy. Biochim Biophys Acta. 2005;1706:165–173. doi: 10.1016/j.bbabio.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Dobler J, Zinth W, Kaiser W, Oesterhelt D. Excited-state reaction dynamics of bacteriorhodopsin studied by femtosecond spectroscopy. Chem Phys Lett. 1988;144:215–220. [Google Scholar]
- 8.Mathies RA, Cruz CHB, Pollard WT, Shank CV. Direct observation of the femtosecond excited-state cis-trans isomerization in bacteriorhodopsin. Science. 1988;240:777–779. doi: 10.1126/science.3363359. [DOI] [PubMed] [Google Scholar]
- 9.Hamm P, et al. Femtosecond spectroscopy of the photoisomerisation of the protonated Schiff base of all-trans retinal. Chem Phys Lett. 1996;263:613–621. [Google Scholar]
- 10.Logunov SL, Song L, El-Sayed MA. Excited-state dynamics of a protonated retinal Schiff base in solution. J Phys Chem. 1996;100:18586–18591. [Google Scholar]
- 11.Kandori H, Katsuta Y, Ito M, Sasabe H. Femtosecond fluorescence study of the rhodopsin chromophore in solution. J Am Chem Soc. 1995;117:2669–2670. [Google Scholar]
- 12.Kandori H, et al. Real-time detection of 60-fs isomerization in a rhodopsin analog containing eight-membered-ring retinal. J Am Chem Soc. 1996;118:1002–1005. [Google Scholar]
- 13.Freedman KA, Becker RS. Comparative investigation of the photoisomerization of the protonated and unprotonated normal-butylamine Schiff-bases of 9-cis-retinals, 11-cis-retinals, 13-cis-retinals, and all-trans-retinals. J Am Chem Soc. 1986;108:1245–1251. [Google Scholar]
- 14.Hasson KC, Gai F, Anfinrud PA. The photoisomerization of retinal in bacteriorhodopsin: Experimental evidence for a three-state model. Proc Natl Acad Sci USA. 1996;93:15124–15129. doi: 10.1073/pnas.93.26.15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro SL, et al. Picosecond and steady state, variable intensity and variable temperature emission spectroscopy of bacteriorhodopsin. Biophys J. 1978;23:383–393. doi: 10.1016/S0006-3495(78)85457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenkl F, et al. Ultrafast energy relaxation in bacteriorhodopsin studied by time-integrated fluorescence. Phys Chem Chem Phys. 2002;4:5020–5024. [Google Scholar]
- 17.Ye T, et al. On the nature of the primary light-induced events in bacteriorhodopsin: Ultrafast spectroscopy of native and C-13═C-14 locked pigments. J Phys Chem B. 1999;103:5122–5130. [Google Scholar]
- 18.Zhong Q, et al. Reexamining the primary light-induced events in bacteriorhodopsin using a synthetic C-13═C-14-locked chromophore. J Am Chem Soc. 1996;118:12828–12829. [Google Scholar]
- 19.Léonard J, et al. Functional electric field changes in photoactivated proteins revealed by ultrafast Stark spectroscopy of the Trp residues. Proc Natl Acad Sci USA. 2009;106:7718–7723. doi: 10.1073/pnas.0812877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birge RR, et al. A spectroscopic, photocalorimetric, and theoretical investigation of the quantum efficiency of the primary event in bacteriorhodopsin. J Am Chem Soc. 1989;111:4063–4074. [Google Scholar]
- 21.Dugave C, Demange L. Cis-trans isomerization of organic molecules and biomolecules: Implications and applications. Chem Rev. 2003;103:2475–2532. doi: 10.1021/cr0104375. [DOI] [PubMed] [Google Scholar]
- 22.Hampp N. Bacteriorhodopsin as a photochromic retinal protein for optical memories. Chem Rev. 2000;100:1755–1776. doi: 10.1021/cr980072x. [DOI] [PubMed] [Google Scholar]
- 23.Rajamani R, Gao J. Combined QM/MM study of the opsin shift in bacteriorhodopsin. J Comput Chem. 2002;23:96–105. doi: 10.1002/jcc.1159. [DOI] [PubMed] [Google Scholar]
- 24.Houjou H, Inoue Y, Sakurai M. Study of the opsin shift of bacteriorhodopsin: Insight from QM/MM calculations with electronic polarization effects of the protein environment. J Phys Chem B. 2001;105:867–879. [Google Scholar]
- 25.Houjou H, Inoue Y, Sakurai M. Physical origin of the opsin shift of bacteriorhodopsin. Comprehensive analysis based on medium effect theory of absorption spectra. J Am Chem Soc. 1998;120:4459–4470. [Google Scholar]
- 26.Hayashi S, Ohmine I. Proton transfer in bacteriorhodopsin: Structure, excitation, IR spectra, and potential energy surface analyses by an ab initio QM/MM method. J Phys Chem B. 2000;104:10678–10691. [Google Scholar]
- 27.Fujimoto K, Hasegawa JY, Hayashi S, Kato S, Nakatsuji H. Mechanism of color tuning in retinal protein: SAC-CI and QM/MM study. Chem Phys Lett. 2005;414:239–242. [Google Scholar]
- 28.Fujimoto K, Hayashi S, Hasegawa JY, Nakatsuji H. Theoretical studies on the color-tuning mechanism in retinal proteins. J Chem Theor Comput. 2007;3:605–618. doi: 10.1021/ct6002687. [DOI] [PubMed] [Google Scholar]
- 29.Vreven T, Morokuma K. The ONIOM (our own N-layered integrated molecular orbital plus molecular mechanics) method for the first singlet excited (S-1) state photoisomerization path of a retinal protonated Schiff base. J Chem Phys. 2000;113:2969–2975. [Google Scholar]
- 30.Vreven T, Morokuma K. Investigation of the S0 → S1 excitation in bacteriorhodopsin with the ONIOM(MO∶MM) hybrid method. Theor Chem Acc. 2003;109:125–132. [Google Scholar]
- 31.Hayashi S, Tajkhorshid E, Schulten K. Molecular dynamics simulation of bacteriorhodopsin’s photoisomerization using ab initio forces for the excited chromophore. Biophys J. 2003;85:1440–1449. doi: 10.1016/S0006-3495(03)74576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warshel A, Chu ZT. Nature of the surface crossing process in bacteriorhodopsin: Computer simulations of the quantum dynamics of the primary photochemical event. J Phys Chem B. 2001;105:9857–9871. [Google Scholar]
- 33.Hayashi S, Tajkhorshid E, Kandori H, Schulten K. Role of hydrogen-bond network in energy storage of bacteriorhodopsin’s light-driven proton pump revealed by ab initio normal-mode analysis. J Am Chem Soc. 2004;126:10516–10517. doi: 10.1021/ja047506s. [DOI] [PubMed] [Google Scholar]
- 34.González-Luque R, et al. Computational evidence in favor of a two-state, two-mode model of the retinal chromophore photoisomerization. Proc Natl Acad Sci USA. 2000;97:9379–9384. doi: 10.1073/pnas.97.17.9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cembran A, González-Luque R, Serrano-Andrés L, Merchán M, Garavelli M. About the intrinsic photochemical properties of the 11-cis retinal chromophore: Cmputational clues for a trap state and a lever effect in Rhodopsin catalysis. Theor Chem Acc. 2007;118:173–183. [Google Scholar]
- 36.Andruniów T, Ferré N, Olivucci M. Structure, initial excited-state relaxation, and energy storage of rhodopsin resolved at the multiconfigurational perturbation theory level. Proc Natl Acad Sci USA. 2004;101:17908–17913. doi: 10.1073/pnas.0407997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garavelli M, et al. Photoisomerization path for a realistic retinal chromophore model: The nonatetraeniminium cation. J Am Chem Soc. 1998;120:1285–1288. [Google Scholar]
- 38.Turro NJ. Modern Molecular Photochemistry. Menlo Park, CA: Benjamin-Cummings; 1991. [Google Scholar]
- 39.Gilbert A, Baggott J. Essentials of Molecular Photochemistry. Oxford UK: Blackwell Science; 1991. [Google Scholar]
- 40.Warshel A. Bicycle-pedal model for the first step in the vision process. Nature. 1976;260:679–683. doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- 41.Warshel A. Energetics of enzyme catalysis. Proc Natl Acad Sci USA. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schapiro I, Weingart O, Buss V. Bicycle-pedal isomerization in a rhodopsin chromophore model. J Am Chem Soc. 2009;131:16–17. doi: 10.1021/ja805586z. [DOI] [PubMed] [Google Scholar]
- 43.Polli D, et al. Conical intersection dynamics of the primary photoisomerization event in vision. Nature. 2010;467:440–443. doi: 10.1038/nature09346. [DOI] [PubMed] [Google Scholar]
- 44.Szymczak JJ, Barbatti M, Lischka H. Is the photoinduced isomerization in retinal protonated Shiff bases a single- or double-torsional process? J Phys Chem A. 2009;113:11907–11918. doi: 10.1021/jp903329j. [DOI] [PubMed] [Google Scholar]
- 45.Cembran A, et al. Structure, spectroscopy, and spectral tuning of the gas-phase retinal chromophore: The beta-ionone “handle” and alkyl group effect. J Phys Chem A. 2005;109:6597–6605. doi: 10.1021/jp052068c. [DOI] [PubMed] [Google Scholar]
- 46.Cembran A, Bernardi F, Olivucci M, Garavelli M. The retinal chromophore/chloride ion pair: Structure of the photo isomerization path and interplay of charge transfer and covalent states. Proc Natl Acad Sci USA. 2005;102:6255–6260. doi: 10.1073/pnas.0408723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logunov SL, Volkov VV, Braun M, El-Sayed MA. The relaxation dynamics of the excited electronic states of retinal in bacteriorhodopsin by two-pump-probe femtosecond studies. Proc Natl Acad Sci USA. 2001;98:8475–8479. doi: 10.1073/pnas.141220198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birge RR, Zhang C-F. Two-photon double-resonance spectroscopy of bacteriorhodopsin. Assignment of the electronic and dipolar properties of the low-lying 1Ag-like and 1Bu*(-)-like π,π* states. J Chem Phys. 1990;92:7178–7195. [Google Scholar]
- 49.Nielsen IB, Lammich L, Andersen LH. S1 and S2 excited states of gas-phase Schiff-base retinal chromophores. Phys Rev Lett. 2006;96:018304. doi: 10.1103/PhysRevLett.96.018304. [DOI] [PubMed] [Google Scholar]
- 50.Migani A, et al. Structure of the intersection space associated with Z/E photoisomerization of retinal in rhodopsin proteins. Faraday Discuss. 2004;127:179–191. doi: 10.1039/b315217k. [DOI] [PubMed] [Google Scholar]
- 51.Schenkl F, et al. Insights into excited-state and isomerization dynamics of bacteriorhodopsin from ultrafast transient UV absorption. Proc Natl Acad Sci USA. 2006;103:4101–4106. doi: 10.1073/pnas.0506303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu RSH, Asato AE. The primary process of vision and the structure of bathorhodopsin: Amechanism for photoisomerization of polyenes. Proc Natl Acad Sci USA. 1985;82:259–263. doi: 10.1073/pnas.82.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saltiel JPD, et al. Photoisomerization of all-cis-1,6-diphenyl-1,3,5-hexatriene in the solid state and in solution: Asimultaneous three-bond twist process. Angew Chem Int Edit. 2009;48:8082–8085. doi: 10.1002/anie.200902724. [DOI] [PubMed] [Google Scholar]
- 54.Terentis AC, Ujj L, Abramczyk H, Atkinson GH. Primary events in the bacteriorhodopsin photocycle: Torsional vibrational dephasing in the first excited electronic state. Chem Phys. 2005;313:51–62. [Google Scholar]
- 55.Hayashi S, Tajkhorshid E, Schulten K. Structural changes during the formation of early intermediates in the bacteriorhodopsin photocycle. Biophys J. 2002;83:1281–1297. doi: 10.1016/S0006-3495(02)73900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
56.Lanyi JK, Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and
 intermediates of the photocycle. J Mol Biol. 2003;328:439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
intermediates of the photocycle. J Mol Biol. 2003;328:439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar] - 57.Logunov SL, El-Sayed MA, Song L. Photoisomerization quantum yield and apparent energy content of the K intermediate in the photocycles of bacteriorhodopsin, its mutants D85N, R82Q, and D212N, and deionized blue bacteriorhodopsin. J Phys Chem. 1996;100:2391–2398. [Google Scholar]
- 58.Birge RR, Cooper TM. Energy storage in the primary step of the photocycle of bacteriorhodopsin. Biophys J. 1983;42:61–69. doi: 10.1016/S0006-3495(83)84369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulten K, Tavan P. A mechanism for the light-driven proton pump of Halobacterium halobium. Nature. 1978;272:85–86. doi: 10.1038/272085a0. [DOI] [PubMed] [Google Scholar]
- 60.Frisch MJ, et al. Gaussian98-Revision A.6. Pittsburgh, PA: Gaussian Inc.,; 1998. [Google Scholar]
- 61.Ponder JW, Richards FM. An efficient Newton-like method for molecular mechanics energy minimization of large molecules. J Comput Chem. 1987;8:1016–1024. [Google Scholar]
- 62.Luecke H, Schobert B, Richter H-T, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 angstrom resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 63.Andersson K, et al. MOLCAS 5.0. Lund: Lund University; 1999. [Google Scholar]
- 64.Malmqvist PA, Roos BO, Serrano-Andres L. The multi-state CASPT2 method. Chem Phys Lett. 1998;288:299–306. [Google Scholar]
- 65.Malmqvist P-Å, Roos BO. The CASSCF state interaction method. Chem Phys Lett. 1989;155:189–194. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.