Abstract
Background
Interaction between parental genomes is accompanied by global changes in gene expression which, eventually, contributes to growth vigor and the broader phenotypic diversity of allopolyploid species. In order to gain a better understanding of the effects of allopolyploidization on the regulation of diverged gene networks, we performed a genome-wide analysis of homoeolog-specific gene expression in re-synthesized allohexaploid wheat created by the hybridization of a tetraploid derivative of hexaploid wheat with the diploid ancestor of the wheat D genome Ae. tauschii.
Results
Affymetrix wheat genome arrays were used for both the discovery of divergent homoeolog-specific mutations and analysis of homoeolog-specific gene expression in re-synthesized allohexaploid wheat. More than 34,000 detectable parent-specific features (PSF) distributed across the wheat genome were used to assess AB genome (could not differentiate A and B genome contributions) and D genome parental expression in the allopolyploid transcriptome. In re-synthesized polyploid 81% of PSFs detected mid-parent levels of gene expression, and only 19% of PSFs showed the evidence of non-additive expression. Non-additive expression in both AB and D genomes was strongly biased toward up-regulation of parental type of gene expression with only 6% and 11% of genes, respectively, being down-regulated. Of all the non-additive gene expression, 84% can be explained by differences in the parental genotypes used to make the allopolyploid. Homoeolog-specific co-regulation of several functional gene categories was found, particularly genes involved in photosynthesis and protein biosynthesis in wheat.
Conclusions
Here, we have demonstrated that the establishment of interactions between the diverged regulatory networks in allopolyploids is accompanied by massive homoeolog-specific up- and down-regulation of gene expression. This study provides insights into interactions between homoeologous genomes and their role in growth vigor, development, and fertility of allopolyploid species.
Background
Genetic redundancy created by allopolyploidy is a source of new variation as well as the molecular basis for functional evolution [1-3]. The evidence for several rounds of recent and ancient polyploidization events found by analyzing the genomic sequence data suggests the importance of whole-genome duplication in the evolutionary success of angiosperms [4,5]. Analysis of natural and re-synthesized allopolyploids demonstrated that the combination of homoeologous genomes results in "genomic shock" accompanied by structural rearrangements [6,7], activation of transposons [8], expression changes [7,9,10] and epigenetic modifications [11-13]. Such changes are suggested to lead to the functional diversification of duplicated genes thereby promoting the establishment of new regulatory interactions and, ultimately, are responsible for phenotypic variability and the broader adaptability of allopolyploid species [5]. At a physiological level, allopolyploidy is often associated with plant vigor, adaptation to a broad range of environmental stress factors, resistance to pathogens, and increased fecundity and fertility [1,2,5].
While our knowledge of the molecular basis of phenotypic effects observed in allopolyploids is limited, cis- and trans-regulatory divergence between parental genomes and their interaction were shown to be important for transcription regulation in inter-species hybrids [14,15] and allopolyploids [16]. The role of cis-regulatory divergence between homoeologous genes in polyploids is demonstrated by transcriptional subfunctionalization and neofunctionalization of homoeologs in polyploid cotton [16], tissue- and development-dependent regulation of homoelogous genes in wheat [13,17] and Brassica [18]. Regulatory modifications can be facilitated by epigenetic mechanisms [12,13] that were also shown to be involved in regulation of circadian rhythms and growth vigor in Arabidopsis hybrids and allopolyploids [11]. Additionally, expression divergence between parental lines has correlated with non-additive gene expression in inter-species hybrids [15] and allopolyploids [9]. Recent studies in cotton using homoeolog-specific gene expression assays highlighted the importance of homoelogous genes in plant development and evolution of the allopolyploid transcriptome [10,19].
The genomic, genetic and cytogenetic resources developed for wheat and its wild ancestors make them an excellent system to study the mechanisms of allopolyploidy-induced processes. Evolutionary history of hexaploid wheat includes two polyploidizations events. The first event occurred about 0.5-0.36 million years ago (MYA) [20,21] involving hybridization of two diploid species T. urartu (A genome diploid ancestor, genome formula AuAu) and Ae. speltoides (closely related to the diploid ancestor of the wheat B genome, genome formula SS) resulting in origin of wild tetraploid wheat T. dicoccoides (wild emmer, genome formula BBAA) [22,23]. Hexaploid T. aestivum originated by the hybridization of cultivated tetraploid wheat T. turgidum (domesticated emmer, genome formula BBAA) with diploid goatgrass Ae. tauschii about 8,000 years ago [24]. In spite of significant reduction of genetic diversity during domestication bottleneck [25] polyploid wheat demonstrates broad range of phenotypic variability and ability to adapt to diverse environmental conditions [26] attributed to the plasticity of the allopolyploid genome. Analysis of gene expression in wheat allopolyploid showed that 16% of genes are expressed non-additively compared to the parental lines [27]. The cDNA-AFLP analysis found suppression of 11.4% and 3.3% of bands in the D and AB genomes, respectively, and induction of only 0.4% of bands in the AB genomes of the allopolyploid wheat [28]. When more distant diploid relatives of wheat genomes Ae. sharonensis (genome formula SlSl) and T. monococcum (genome formula AmAm) were used to synthesize allotetraploid wheat the alteration of gene expression was observed for 60 out 3072 (2%) AFLP bands [6]. Recently, Affymetix arrays were used to assess the expression in allopolyploid wheat [29] to show that only 7% of genes show non-additive type of gene expression. Although these studies demonstrate the impact of genome doubling on gene regulation in allopolyploid wheat, there are currently no large-scale studies focusing on genome-wide assessment of genome-specific expression in allopolyploid wheat.
Homoeologous gene expression in polyploids can be assessed using various SNP-based assays [16], expression and genotyping arrays [30,31] or next-generation sequencing technologies [32,33]. Recently, microarrays interrogating homoeolog-specific mutations were successfully used for estimating the relative contribution of homoeologous genes to the cotton transcriptome [10,19]. However, most of these assays require prior information about the homoeolog-specific mutations, which is currently limited for the wheat genome. The Affymetix microarrays have also been successfully used for the discovery of single feature polymorphisms (SFP) and genotyping crop species with large genomes for which, at the time when studies were performed, no complete genome sequences were available [34-38]. Here we applied Affymetrix expression array platform to discover homoeolog-specific mutations and use them to assess the relative expression of homoeologous gene copies in synthetic polyploid wheat created by the hybridization of Tetra-Cantach, the tetraploid derivative of hexaploid wheat, and the diploid ancestor of the wheat D genome Ae. tauschii [39]. Compared to extracted tetraploid parent the synthetic line showed normal growth vigor and fertility [39] and, therefore, is an excellent system to the study the role of homoeologous gene expression in physiological and morphological changes associated with polyploidization.
Methods
Plant material
Microarray hybridization experiments were conducted with the diploid ancestor of the wheat D genome Ae. tauschii (2n = 14, genome formula DD, accession TQ20), reconstituted tetraploid wheat Tetra-Cantach (2n = 28, genome formula BBAA) representing the extracted tetraploid component of the hexaploid wheat genome and synthetic hexaploid wheat (2n = 42, genome formula BBAADD) created by hybridization of Tetra-Cantach with TQ20 accession of Ae. tauschii [39]. This exact set of parental lines and the derived synthetic wheat have been previously characterized morphologically [39], cytogenetically and genetically [40].
Microarray hybridization
Affymetrix GeneChip Wheat Genome Arrays containing 61,178 probesets were hybridized with RNA isolated from the leaves of 4 week-old seedlings of Ae. tauschii, Tetra-Cantach and synthetic hexaploid wheat (henceforth referred to as AT, TC and SN). The plant leaves were collected at the four-leaf stage at the same time-point to avoid the effect of expression fluctuation during the day. Since at this stage wheat produces leaves with the rate of about one leaf every 4-5 days to total of 8-9 leaves before transition to tillering, our sampling point corresponds to the middle of the leaf development stage. This sampling approach should minimize the developmental effects on expression differences among lines. Additionally, this sampling time-point allows us to compare our results with the previously published studies with similar sampling time-points [27]. Four independent biological replicates for each line and 1:1 mixture of AT and TC RNA were used in the experiment. The fresh plant leaves were flash-frozen in liquid nitrogen and ground with mortar and pestle to obtain a fine powder. RNA was isolated from 100 mg of ground tissue using QIAGEN RNAesy Plant Mini Kit (Qiagen) according to the manufacturer recommended protocol. The intactness of RNA samples was assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies). RNA labeling and hybridization were performed according to standard Affymetrix protocols.
Data analyses
The Affymetrix arrays were used to obtain genome wide estimates of gene expression in a synthetic wheat line and its parents. Microarray intensity Cel files were background corrected and quantile normalized using the rma2 procedure implemented in the R package affy [41,42]. The comparison of expression between the synthetic wheat and its parents was performed using only perfect match (PM) probes (61,178 probesets × 11 probes = 672,958 array features). For all PM probes we fit the linear model Yijn = μ + pi + gj + εijn, where Yijn is the transcript abundance in replicate n (n = 1, ..., 4), μ is the overall mean of probe intensity, p is the effect of probe i (i = 1, ..., 11), g is the effect of genotype j, and ε is the error. The method of contrasts was used to compare the effect of genotype in AT vs. TC and SN vs. AT +TC comparisons. The false discovery rate was controlled using the Benjamini & Hochberg method maintaining FDR at level 0.05 [43].
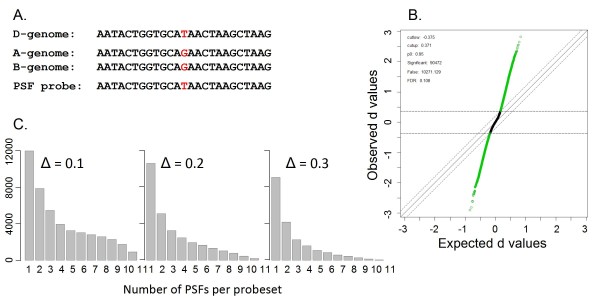
The Affymetrix data obtained for AT and TC RNA samples was used to identify oligonucleotide features hybridizing with sequences carrying mutations differentiating the AT (D genome) and TC (AB genomes) (Fig. 1A). The statistical procedure for identification of these mutations was similar to that used for identification of single-feature polymorphisms (SFPs) [37,44]. However, the term "SFP", which refers to an oligonucleotide feature differentially hybridizing with different genotypes due to intra-species polymorphisms, in its original meaning is not strictly applicable to our experiment. Most of the mutations differentiating AT and TC parents are fixed divergent mutations accumulated after the radiation of the diploid ancestors of the wheat A, B and D genomes. Therefore, the probes differentially hybridizing with AT and TC will be referred to as parent-specific features (PSF) or parent-specific probes. For all 672,958 PM probes on the array, we fit the same linear model Yijn = μ + pi + gj + εijn described above. Residuals containing the probe by genotype effect were tested for difference between genotypes by calculating the d-statistics [45] using the Significance Analysis of Microarrays (SAM) procedure implemented in the R package siggenes (Fig. 1B). The threshold Δ = 0.2 was selected to identify significantly different features with 0.1 false discovery rate (FDR). Parent-specific oligonucleotide features having higher hybridization intensity with Ae. tauschii had negative values of d-statistics (henceforth, AT-specific probes); PSFs showing higher hybridization intensity with Tetra-Cantach had positive values of d-statistics (henceforth, TC-specific probes). It is worth mentioning that the TC-specific probes cannot discriminate the level of expression of homoeologous genes in the A and B genomes of tetraploid TC. The intensity of TC-specific probes on the array is a product of hybridization of transcripts generated by either one of the genomes of tetraploid TC or by both of its genomes. The TC-specific probes can have greater hybridization intensity with targets in TC than with those in AT due to either 1) the perfect match of a TC-specific probe with both homoeologous sequences in the A and B genomes and presence of a mismatch in the D genome or 2) the perfect match of a probe with either of the two homoeologous sequences in the A and B genomes and presence of a mismatch in the D genome. Likewise, the AT-specific probes on the array hybridize more efficiently with AT due to the perfect match of a probe with AT transcripts and the presence of mismatch in either one or both TC genomes. The example of a AT-specific probe sequence perfectly matching the AT sequence and mismatching the A and B genome sequences in TC is depicted in the Fig. 1A.
Figure 1.
Identification of parent-specific features (PSFs) in diploid Ae. tauschii and tetraploid wheat Tetra-Cantach. A. Alignment of an AT-specific probe with the target sequences in the D-genome of Ae. tauschii and AB-genomes of Tetra-Cantach. Homoeolog-specific mutation discriminating both the A- and B-genomes from the D-genome is shown in red. B. The scatter plot of observed versus expected d-statistics obtained for each PM probe. Expected values were obtained by permutations of dataset. Threshold Δ = 0.2 was applied for identifying significantly different oligonucleotide features between Ae. tauschii and Tetra-Cantach. C. Frequency distribution of the number of PSFs per probeset under various values of threshold Δ.
The PSFs were experimentally verified by comparing the sequences of 25-mer PM probes with the cDNA sequences of Ae. tauschii (D genome ancestor) and the two lines of hexaploid wheat Triticum aestivum cv. Chinese Spring and T. aestivum cv. Jagger. The sequencing of full-length normalized cDNA libraries was performed using the pyrosequencing instrument GS LR70 (Roche) in the University of Oklahoma following the standard single read shotgun 454 sequencing protocol with LR70 kit (Roche). Plants were grown in greenhouse conditions under natural day light supplemented with artificial source of light. The total RNA was isolated from 4 week seedlings using the RNeasy Plant Mini Kit (QIAGEN). First-strand cDNA synthesis was performed according to SMART cDNA synthesis technology (Clontech Laboratories, Inc.) using modified 3' SMART CDS Primer II A (5'-AAGCAGTGGTATCAACGCAGAGTACTTTTGT(9)C T(10)VN-3') and SuperScript III reverse transcriptase (Invitrogen). Double-stranded cDNA was amplified by long-distance (LD) PCR using Advantage 2 PCR Enzyme System (Clontech Laboratories, Inc). Amplification was performed on thermal cycler (Applied Biosystem) with the following PCR parameters: 95°C for 1 min. followed by 16 cycles of 95°C - 15 sec., 65°C - 30 sec., 68°C - 6 min. The quality of double-stranded cDNA was checked on the 1.1% agarose/EtBr gel in 1× TAE buffer and purified with QIAquick PCR Purification Kit (QIAGEN).
The double-stranded cDNA was normalized using TRIMMER cDNA normalization Kit (EVROGEN), which is based on a unique DSN (duplex-specific nuclease) normalization technology and is specifically developed for normalization of cDNA enriched with full-length sequences. The program Lucy 1.19p was used for quality trimming of 454 sequence reads prior to further analyses. PM probe sequences were downloaded from the Affymetrix website [46] and compared with 454 sequence reads using the blastn program with e-value threshold set at 10-4. This e-value threshold was selected to allow for no more than one mismatch per 25 bp alignment.
In order to measure the expression of homoeologous genes in the synthetic polyploid we used an approach similar to that applied for assessing the allele-specific expression in a cross of two unrelated strains of Saccharomyces cerevisae [30]. Ronald et al (2005) used the observed to expected intensity of probe hybridization to assess the contribution of parental alleles to overall gene expression. The expected intensity of probe hybridization was modeled by taking into account the energy of RNA-DNA duplex formation during hybridization. This model considers two modes of probe hybridization, gene-specific binding and non-specific binding. The former refers to RNA-DNA duplex formation with complementary sequences and the latter refers to the duplex formation with many mismatches. The model ignores the duplexes with few mismatches assuming that probes are preselected against this type of binding [47]. However, in allopolyploid wheat, the presence of recently diverged homoeologous copies of genes makes this model's assumption invalid.
Therefore, to measure the expression of a homoeologous gene relative to total gene expression we used the following approach. All eleven PM probes within each probeset were classified into two categories based on difference in hybridization intensity between parental lines (using previously described d-statistics): those that showed detectable differences were called PSFs and those that did not show detectable differences were called non-PSFs. The averaged hybridization signal of non-PSFs (Ēt) was used to get an estimate of the total gene expression for each probeset. The contribution of AT or TC copies of a gene to total gene expression was assessed from the ratio of the PSF intensity Ep to the averaged non-PSF intensity Ēt (henceforth Ep/Ēt ratio). The ratio was calculated using the formula Ep/Ēt = log2Eiks - log2Ēims, where Eiks is the hybridization intensity of the parent-specific probe k in the probeset i (i = 1, 2, ..., 21,379) for species s (s = AT, TC or SN) and Ēims is the mean intensity of remaining m probes in the probeset i in species s. The linear model Yjn = μ + gj + εjin was fit to Ep/Ēt ratio data, where Yjn is the Ep/Ēt ratio in replicate n (n = 1, ..., 4), μ is the overall mean of Ep/Ēt ratio, g is the effect of genotype j (j = AT, TC or SN), and ε is the error. Contrasts were used to compare the intensity ratios in AT vs. SN, TC vs. SN, SN vs. (AT + TC)/2 and SN vs. (1/3AT +2/3TC) comparisons. The latter two contrasts were used to test the different ratios of parental gene expression in a synthetic polyploid. According to previous studies of allopolyploids, including wheat [9,27], 1 : 1 mid-parent model is the best fit to the expression values observed in allopolyploids. The false discovery rate (FDR) among significant features was estimated separately for each contrast using the Benjamini & Hochberg method maintaining FDR at level 0.05 [43]. The ability of AT- and TC-specific probes to correctly predict the expression divergence from the mid-parent value was verified experimentally by hybridizing Affymetrix microarray chips with the 1:1 mix of AT and TC RNA samples and fitting the model assuming the 1:1 ratio of AT and TC expression in the mix. The proportion of significant oligonucleotide features in this experiment to the proportion of significant oligonucleotide features in the contrast SN vs. (AT + TC)/2 corresponds to the experimentally determined false positive rate.
One of the limitations of the approach used in our study is that the Ep/Ēt ratio does not discriminate the expression of the A genome homoeolog from that of the B genome. The hybridization intensity signal of TC-specific probes is composed of two components: one is contributed by hybridization with the A genome transcripts and another is contributed by the hybridization with the B genome transcripts. Another limitation of the wheat Affymetrix microarray platform for the analysis of homoeologous gene expression is that each identified PSF is efficient for measuring the expression of gene copies from only one of the parental lines, AT or TC, depending on which one of them have sequences with a perfect match to the PSF. The absence of a probe that matches the sequences of another parent precludes the estimation of expression of both parental gene variants in the polyploid. Therefore, we used AT-specific probes for assessing only AT-type expression (or D genome expression) and TC-specific probes for assessing only TC-type expression (combined expression of both A and B genomes) in the synthetic allopolyploid wheat.
Distribution of parent-specific features across the wheat genome
The blasn program was used to compare the sequences of ESTs used for the Affymetrix probe design with the sequences of ESTs mapped to the wheat deletion bin map [48]. The best blast hits passing the e-value threshold set at 10-10 and having the alignment length more than 100 bp were selected. The bin map locations were extracted from the GrainGenes database [49]. Only those loci that mapped to the deletion bins in synthenic positions on the same homoeologous chromosome were retained for further analysis. Bins were grouped according to the distances of their midpoints from the centromere into five equal intervals along chromosome arms. The number of mapped ESTs matching the Affymetrix probesets was calculated in each of five intervals.
Quantitative RT-PCR
We performed a two-step RT-PCR in which RNA was first converted into cDNA and then amplified with homoeolog-specific primers. The reaction was performed on the BioRad iQ iCycler real time PCR system. The transcript levels were determined in SN and 1:1 mixture of AT and TC RNA using the SYBR Green detection system. The primers for amplification were designed to harbor homoeolog-specific mutations at their 3'-ends. The mutations differentiating homoeologous sequences were identified by comparing EST sequences generated by 454 sequencing of transcriptomes of hexaploid wheat and its diploid ancestors. The ability of the primers to specifically amplify targets in the parental genomes was tested by PCR with AT and TC cDNAs. The primer amplification efficiency (E) was tested on series of 2-fold diluted cDNAs.
For accurate relative quantification of expression levels, we calculated the theoretical value R0 using the formula R0 = RCt (1 + E)-Ct, where R0 is the starting fluorescence, RCt is the fluorescence at the threshold cycle Ct and E is the amplification efficiency [50]. This method of quantification takes into account the effect of amplification efficiency on the accumulation of PCR products. All R0 values were normalized relative to the R0 obtained for the actin gene followed by log-transformation. The log-transformed R0 values obtained for SN were compared with R0 values obtained for 1:1 mixture of AT and TC RNAs. The difference in expression levels was tested using the one-tailed t-test statistics assuming equal variance.
Functional annotation
Affymetrix probesets containing parent-specific probes were classified into functional categories using the TIGR Gene Ontology assignments [51]. The contigs of wheat genes used for the array design were downloaded from the Affymetrix website and compared with the sequences of TIGR tentative contigs using the blastn program. The blast hits were filtered using the e-value threshold 10-10 and an alignment length above 80 bp. The tentative contigs showing the best blast hits were then selected to extract the TIGR GO assignments. The gene annotations were mapped to high-level broader parent terms, referred to as GO Slim terms, using the GO Slimmer tool [52]. Additional functional annotations were performed using the MapMan software [53]. MapMan's Scavenger module was used to assign the genes on the Affymetrix chip to non-redundant functional categories. The Wilcoxon rank-sum test was used to identify functional bins that demonstrate the coordinated change in the expression of multiple genes that belong to the same pathway.
Results
Discovery and validation of probes differentially hybridizing with the AT and TC
We used the PM probes on the wheat array for the discovery of probes hybridizing to sequences harboring mutations that differentiate TC (AB genome) and AT (D genome) (Fig. 1A). Background corrected, normalized and log2 transformed expression values of PM probes were extracted from each of 61,127 probesets on the Affymetrix wheat array. The data was analyzed using the linear model described in the Materials and Methods. A total of 90,472 PSFs distributed among 28,679 probesets were discovered at 0.1 FDR (Fig. 1B). Most of the probesets (63%) contained two or more PSFs (Fig. 1C).
For further analysis, we selected only 21,379 probesets with no more than 4 PSFs per probeset. This cutoff value is based on the expected number of nucleotide substitutions in a probeset given the nucleotide substitution rate and the divergence time of diploid ancestors of polyploid wheat [20]. The total length of each probeset is 275 bp (11 probes × 25 bp = 275 bp), mostly represented by coding sequences or non-coding 3'-UTRs. If we assume that the neutral mutation rate in non-coding wheat sequences is 5.5 × 10-9 per site per year (the mutation rate in coding regions is significantly lower) and set the divergence time of the diploid ancestors of the wheat genomes to 2.7 million years [20], we would expect no more than 4 mutations within the 275 bp sequence. This procedure should eliminate probesets that contain excessive number of parent-specific features due to their hybridization with the transcripts encoded by the diverged copies of multigene families. Also, by excluding probesets with more than 4 PSFs we eliminate probes that were falsely detected as PSFs if a gene was silenced or expressed at the low level in one parent, but was expressed at high level in another parent. Under this scenario, all probes in a probeset would appear as PSFs. The resulting dataset consisted of 40,281 PSFs including 18,293 AT-specific features and 21,988 TC-specific features (Additional File 1).
This dataset was used for experimental validation of discovered parent-specific features by comparing oligonucleotide feature sequences with the Expressed Sequence Tags (ESTs) generated using the GS LR70 pyrosequencing platform (also referred to as 454). A total of 270,230 raw reads were generated with an average length of 216.3 nucleotides (Table 1). After quality filtering to remove adaptors, polyA tails, and low quality sequences, we obtained 224,926 reads with the average length 211 bp totaling to 47,699,354 bases (Table 1). A total of 10.1, 20.6 and 17 Mb of cDNA sequence data was obtained for the diploid ancestor of the wheat D genome Ae. tauschii and the two lines of hexaploid wheat T. aestivum cv. Chinese Spring and T. aestivum cv. Jagger, respectively. The sequencing data have been submitted to the NCBI Short Read Archive (accession number SRA012746).
Table 1.
454 transcriptome sequencing of Ae. tauschii and wheat cultivars Chinese Spring and Jagger
| Summary of 454 run | AT | CS | JG | Total |
|---|---|---|---|---|
| No. raw reads | 61,664 | 113,594 | 94,972 | 270,230 |
| Total length of raw reads, bp | 12,931,816 | 24,692,552 | 20,878,927 | 58,503,295 |
| No. trimmed reads | 48,126 | 97,148 | 79,652 | 224,926 |
| Total length of trimmed reads, bp | 10,112,016 | 20,553,679 | 17,033,659 | 47,699,354 |
| Average length of trimmed reads, bp | 210 | 211 | 214 | 212 |
| Median length of trimmed reads, bp | 211 | 214 | 216 | 214 |
The sequences of 40,281 PSFs were compared with the 454 sequence reads using the blastn program applying e-value threshold 10-4. Eighty one PSFs producing significant 25 bp alignments with the cDNAs of Ae. tauschii and both wheat lines were classified into the two groups (Additional File 2). The first group included 39 PSFs that are expected to be TC-specific. These PSFs had one mismatch with a Ae. tauschii cDNA sequence and at least one mismatch and one perfect match with cDNA sequences of both wheat lines. The second group included 42 PSFs (expected to be AT-specific) that have a perfect match with Ae. tauschii cDNA sequences and at least one mismatch and one perfect match with cDNA sequences of both wheat lines. For all 81 features we compared the value of d-statistics (see Material and Methods) and their assignments to the first and second groups (Additional File 2). In the case of perfect concordance between the PSFs identified on the basis of d-statistics and DNA sequence data, all probes in the first group should have positive d values and the probes in the second group should have negative d values. In the first group, 33 out of 39 probes had positive d values leaving 6 with negative d values; in the second group, 34 out of the total 42 probes had negative d values while 8 probes had positive d values. Thus, 10% FDR applied in SAM results in 20% (8/(33+8) × 100% ≈ 20%) of falsely-discovered TC-specific features and 15.0% (6/(34+6) × 100% = 15%) of falsely-discovered AT-specific features.
Distribution of PSFs across the wheat genome
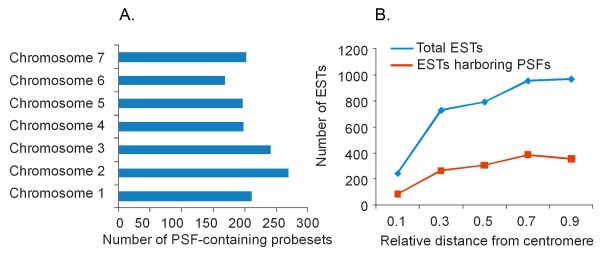
The distribution of the discovered 40,281 PSFs across the wheat genome was inferred by comparing the EST sequences used for Affymetrix array design with the sequences of deletion-bin mapped ESTs [49]. A total of 1,906 PSF-containing probesets showed similarity with the mapped wheat ESTs. We excluded those ESTs that were mapped to more than one chromosomal location in the wheat genome and presumably belong to multigene families. The remaining 1,401 ESTs were distributed similarly among all seven homoeologous groups of chromosomes (Additional File 3, Fig. 2A). The distribution of ESTs showing similarity to PSF-containing probesets along the chromosomal arms mirrors the distribution of all single-copy ESTs, with higher number of parent-specific features mapped to the distal 30% of the chromosome arms (Fig. 2B).
Figure 2.
Distribution of parent-specific features across the wheat chromosomes. A. Distribution of PSF-containing probesets among the 7 homoeologous groups of chromosomes. B. Correlation between the distribution of ESTs harboring PSFs and total ESTs along the wheat chromosomes. The distance from the centromere is given in fractions of chromosome arm length where 0 represents centromere and 1.0 represents telomere.
Using parent-specific features for assessing the homoeolog-specific gene expression
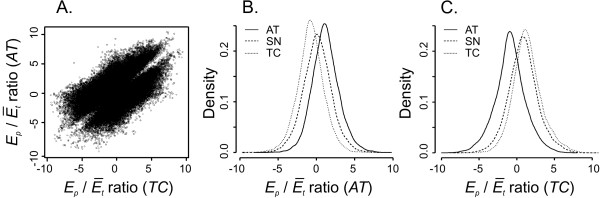
Given the ability of the Affymetrix microarray to discriminate between AT- and TC- specific gene expression, we used this platform for measuring the expression of parental genes in the D and AB genomes of SN. The Ep/Ēt intensity ratios of PSFs having a perfect match with either AT or TC target sequences are expected to be higher than the intensity ratios of probes hybridizing with mismatching target sequences (Fig. 3B,C, and 4). This is evident from the well-defined two-cluster pattern produced by plotting the Ep/Ēt intensity ratios estimated for 40,281 probes in AT (y-axis) and TC (x-axis) lines (Fig. 3A). As expected, the AT-specific probes had greater Ep/Ēt intensity ratio in AT than in TC and TC-specific probes had greater Ep/Ēt ratio in TC than in AT (Fig. 3A and 4).
Figure 3.
Comparison of Ep/Ēt ratios in AT, TC and SN obtained for 40,281 probes. A. A scatterplot showing the comparison of Ep/Ēt ratios calculated for AT (y-axis) and TC (x-axis) lines. B. Density distribution of Ep/Ēt ratios in AT, TC and SN lines calculated for probes having perfect match with the AT sequences (AT-specific). C. Density distribution of Ep/Ēt ratio in AT, TC and SN lines for probes preferentially hybridizing with the TC sequences (TC-specific).
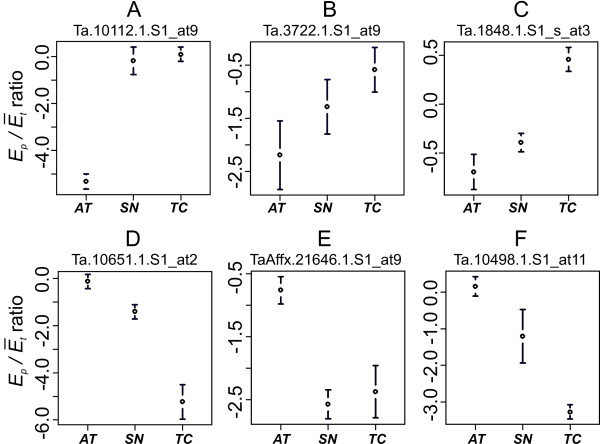
Figure 4.
Ep/Ēt ratio estimates in AT, SN and TC for selected PSFs. The graphs are showing the means and 95% confidence intervals of Ep/Ēt ratio estimates. The top three probes hybridize preferentially with TC (TC-specific); the bottom three probes hybridize preferentially with AT (AT-specific). The names of PSF probes are indicated on the top of each plot.
The significant fraction of Ep/Ēt intensity ratio values in SN for AT-specific probes falls between the intensity ratios obtained for AT and TC lines (Fig. 3B), consistent with previous observations that the gene expression in polyploids tends to stabilize at mid-parent level [9,27]. The mean of Ep/Ēt intensity ratios in SN for TC-specific probes was shifted toward TC Ep/Ēt intensity ratios (Fig. 3C) suggesting higher level of gene expression in the A and B genomes combined.
The analysis of contrasts was used to test the two possible ratios of parental gene expression in the synthetic polyploid assuming 1:1 and 1:2 (henceforth, 1:1 and 1:2 models, respectively) in silico ratio of AT :TC gene expression in the SN transcriptome. The former model corresponds to cases when parental genomes make equal contribution to the SN transcriptiome, whereas the latter model assumes that the tetraploid parent (TC) contributes two parts and diploid parent (AT) contributes only one part to total gene expression. Based on analysis of contrasts, we classified genes into several expression categories: 1) expression is not significantly different from the assumptions of both 1:1 and 1:2 models (mid-parent expression); 2) expression is significantly different from the assumptions of both models (non-additive expression); 3) expression is not significantly different from the assumptions of 1:1 model but different from 1:2 model; and 4) expression is not significantly different from the assumptions of 1:2 model but different from 1:1 model (Table 2). Both models were fit to the probeset expression data obtained for all genes on the Affymetrix chip and to Ep/Ēt ratio data calculated for PSFs (Additional File 4).
Table 2.
Comparison of the fit of two models to probeset and Ep/Ēt ratio expression data
| Expression ratio models | 1AT : 2TC model | ||||
|---|---|---|---|---|---|
| Probeset expression data | Ep/Ēt ratio data | ||||
| Significant | Not significant | Significant | Not significant | ||
| 1AT : 1TC model | Significant | 24,070 | 8,486 | 6,430 | 4,863 |
| Not significant | 2,946 | 25,676 | 1,879 | 21,497 | |
The models assume 1AT : 1TC and 1AT : 2TC expression ratio of parental genes in the synthetic allopolyploid. False discovery rate was controlled at level 0.05.
The analysis of probesets showed that nearly 61% of all genes in SN are expressed at mid-parent level corresponding to either 1:1 or 1:2 parental expression ratios. The expression of 13.9% of genes fit the expectations of the 1:2 model but was different from the assumptions of the 1:1 model, whereas 4.8% of genes were expressed at levels that fit the expectations of the 1:1 model but was different from that of the 1:2 model. A total of 39% of genes were expressed at the levels suggesting non-additive mode of expression.
The Ep/Ēt ratio dataset was used to assess relative contribution of parent-specific gene expression in allopolyploid wheat transcription. Only 20,643 out of 21,379 previously selected probesets were included into the final analysis (Additional File 4). A total of 736 probesets with more than one PSF per probeset were removed from the analysis because they contained PSFs failing to predict the expression change in SN in the same direction relative to the mid-parent value. For example, a probeset that contains two AT-specific probes, one predicting the up-regulation of gene expression in SN and another down-regulation of gene expression in SN, would be removed from dataset. The analysis of Ep/Ēt ratio data showed that 81% of homoeologous copies of genes are expressed at the levels fitting the expectations of 1:1 or 1:2 models, and 19% are expressed non-additively. The examples of PSFs detecting the mid-parent levels of homoeologous gene expression are shown in Fig. 4B and 4F. The AT-specific probes detected a higher number of genes with mid-parent level of expression in SN than the TC-specific probes (Table 3). The differences in the proportion of additively and non-additively expressed genes between the entire probeset data and the Ep/Ēt ratio data (Table 2) are mostly explained by the filtering step included in the selection of 20,643 PSF-containing probesets for calculation of Ep/Ēt ratio. When only these 20,643 probesets were included into the analysis of contrasts, the proportion of genes fitting 1:1 and 1:2 models increased from 61% to 73%.
Table 3.
Parent-specific gene expression in allopolyploid wheat
| Gene regulation | AT- type expression | TC- type expression |
|---|---|---|
| Down-regulated | 548 (705) | 317 (406) |
| Up-regulated | 1,297 (1,601) | 2,988 (3,718) |
| Mid-parent | 11,683 (13,840) | 12,388 (14,399) |
The table shows the number of homoeologous genes interrogated by PSFs (provided in the brackets) in the AB and D genomes of SN.
The remaining 6,430 PSFs detected the non-additive mode of gene expression in allopolyploid wheat. For AT-specific probes, a decrease in the Ep/Ēt ratio in SN relative to the mid-parent value indicates the decrease of the AT-parent contribution to total expression (Fig. 3E), whereas an increase in the Ep/Ēt intensity ratio in SN relative to the mid-parent value would suggest increase of the AT-type part in total gene expression (Fig. 3D). Following the same reasoning, the decrease in the Ep/Ēt ratio in SN relative to the mid-parent value for TC-specific probes is evidence of down-regulation of TC-type expression (Fig. 3C), while an increase of SN Ep/Ēt ratio relative to the mid-parent value suggests an increase in TC-type expression (Fig. 3A). The number of non-additively expressed genes detected by TC-specific probes compared to that detected by AT-specific probes was almost 2 times higher, which points to differential patterns of homoeologous gene activation and repression in the D and AB genomes after polyploidization. The proportion of up- and down-regulated probes among non-additively expressed genes was 82.7% and 17.3%, respectively. Out of 2,306 AT-specific probes, 705 (30.6%) showed the decrease of AT-type expression in SN, whereas 1,601 (69.4%) showed the increase of AT-type expression in SN transcriptome. Among the TC-specific probes 4,124, showed statistically significant departure of Ep/Ēt intensity ratios from the expected mid-parent values with 3,718 probes (90.2%) showing the increase of TC parent contribution to SN expression whereas 406 probes (9.8%) demonstrated the decrease of TC proportion in total gene expression. The number of genes with increased TC-type expression was significantly higher than the number of genes with decreased TC-type expression (χ2-test, P < 1 × 10-10). Out of all up-regulated homoeologous genes, 69.9% showed increased proportion of AB genome parental expression and 30.1% showed increased contribution from the D genome parent. The opposite trend was discovered for down-regulated genes out of which 63.5% were down-regulated in the D genome and 36.5% were down-regulated in the AB genomes.
We have experimentally estimated the rate of false positives among the probes capable of detecting the bias from mid-parent gene expression by measuring the expression in the mix of AT and TC RNA. Four Affymerix microarrays were hybridized with the mix of parental RNA and the Ep/Ēt ratios of 34,669 probes were compared between the mix and in silico mid-parent values. We fit the linear model that correctly assumes that all genes in the mix are expressed at half the level of gene expression in the parental genomes. The significant probes in this case will correspond to false positives misidentified as cases of parent-specific bias in gene expression. Out of all 34,669 probes 3,804 (~11%) showed intensity ratios that deviate from the expected mid-parent value at FDR 0.05.
We tested the effect of expression divergence between parental genomes on gene expression in the synthetic polyploid wheat. The analysis of Affymetrix data at the probeset level was used to assess the expression divergence between AT and TC lines. Approximately 57% (34,809/61,178) of genes showed significant difference in the expression level at FDR 0.05. Expression divergence between the parental transcriptomes explains the significant proportion of non-additive expression variation in the synthetic polyploid wheat. Out of 6,430 probes whose Ep/Ēt ratio indicates departure from the mid-parent value in SN, 83.6% were found in the genes that are also differentially expressed between the AT and TC parents. Among 1986 PSFs that were located in the genes expressed at higher levels in AT than in TC, 1,414 and 28 probes were found in the genes that are up- and down-regulated in the D genome of SN, respectively, and 188 and 356 probes were found in the genes that are up- and down-regulated in the AB genome of SN, respectively. Among 3,389 PSFs found in the genes expressed at higher levels in TC than in AT, 32 and 653 PSFs were found in the genes that are up- and down-regulated in the D genome of SN, while 2,689 and 15 probes were found in the genes that are up- and down-regulated in the AB genomes of SN, respectively.
We have validated the expression of several parental copies of a random set of genes in SN by quantitative RT-PCR assay. A total of 6 genes in the wheat A, B and D genomes representing 4 homoeologous sets of loci were selected and the expression of homoeologous copies of these genes was measured in SN and a 1:1 mixture of AT and TC RNA. Although, previous studies indicate that due to differences in discriminatory power between the RT-PCR and microarray methods, the confirmation rate can vary [54], we were able to confirm expression change for all these genes. In all cases the direction of expression change predicted by microarray hybridization and quantitative RT-PCR were consistent (Additional File 5).
Functional annotation of homoeologous genes
To better understand how polyploidy impacts the regulation of homoeologous genes involved in various biological processes, we used the GO annotations. In order to map gene annotations to high-level, broader parent terms we used the GO Slimmer tool [52]. The results of this analysis are presented in Table 4 show that the majority of genes subjected to regulatory modifications also belong to most abundant functional categories including genes involved in metabolism, biosynthesis, organism development, cellular organization, translation, transport and stress response. We have tried to identify statistically over- and under-represented functional categories using the Fisher exact test. The p-values of the test statistic were corrected for multiple comparisons maintaining FDR at level 0.05. Only one functional category was significantly overrepresented for up-regulated genes (GO:0006091) including D genome homoeologous genes involved in the generation of precursor metabolites and energy. The functional category GO:0007049 was over-represented for down-regulated AB genome homoeologs involved in cell cycle regulation (Table 4). Among down-regulated genes in the D genome of SN the genes that are involved in catabolic processes and carbohydrate metabolism were under-represented. Lower level of up-regulation than expected was observed for the D genome homoeologs that belong to translation (GO:0006412) and cell communication (GO:0007154) functional categories. Under-representation of down-regulated AB genome homoeologous genes was detected only for amino acid metabolism functional category (GO:0006519). The AB genome homoeologs involved in carbohydrate metabolic process (GO:0005975) and cell communication (GO:0007154) were also under-represented among up-regulated genes.
Table 4.
GO annotation of up- and down-regulated homoeologous copies of genes in the synthetic polyploid wheat
| GO term | Biological process | D genome (AT) | AB genome (TC) | ||
|---|---|---|---|---|---|
| suppression | activation | suppression | activation | ||
| GO:0009987 | cellular process | 264 | 512 | 145 | 1231 |
| GO:0008152 | metabolic process | 214 | 454 | 137 | 1039 |
| GO:0009058 | biosynthetic process | 127 | 170 | 53 | 494 |
| GO:0019538 | protein metabolic process | 115 | 199 | 78 | 545 |
| GO:0016043 | cellular component organization | 72 | 123 | 39 | 290 |
| GO:0006412 | translation | 64 | 54** | 29 | 232 |
| GO:0006810 | transport | 61 | 108 | 42 | 287 |
| GO:0006139 | nucleic acid metabolic process | 55 | 101 | 21 | 255 |
| GO:0006950 | response to stress | 53 | 65 | 24 | 201 |
| GO:0007275 | Multi-cellular organism development | 51 | 101 | 31 | 229 |
| GO:0008150 | Biological process | 45** | 118 | 45 | 295 |
| GO:0006464 | protein modification process | 42 | 95 | 30 | 205 |
| GO:0000003 | reproduction | 38 | 69 | 33 | 154 |
| GO:0009628 | response to abiotic stimulus | 30 | 32 | 15 | 88 |
| GO:0009790 | embryonic development | 29 | 52 | 24 | 121 |
| GO:0009791 | post-embryonic development | 27 | 41 | 19 | 102 |
| GO:0040007 | growth | 23 | 50 | 21 | 111 |
| GO:0007049 | cell cycle | 22 | 49 | 26* | 93 |
| GO:0006091 | precursor metabolites and energy | 21 | 62* | 4 | 97 |
| GO:0006350 | transcription | 19 | 56 | 10 | 106 |
| GO:0006519 | cellular amino acid metabolic process | 18 | 17 | 0** | 72 |
| GO:0009653 | anatomical structure morphogenesis | 17 | 42 | 10 | 78 |
| GO:0007165 | signal transduction | 17 | 55 | 22 | 139 |
| GO:0009056 | catabolic process | 14** | 98 | 21 | 172 |
| GO:0009607 | response to biotic stimulus | 13 | 19 | 6 | 36 |
| GO:0009719 | response to endogenous stimulus | 13 | 24 | 2 | 55 |
| GO:0008219 | cell death | 11 | 19 | 7 | 39 |
| GO:0030154 | cell differentiation | 11 | 35 | 5 | 71 |
| GO:0016265 | death | 11 | 19 | 7 | 41 |
| GO:0019748 | secondary metabolic process | 10 | 7 | 0 | 12 |
| GO:0005975 | carbohydrate metabolic process | 9** | 80 | 14 | 94** |
| GO:0006259 | DNA metabolic process | 9 | 17 | 3 | 49 |
| GO:0006629 | lipid metabolic process | 9 | 24 | 5 | 39 |
| GO:0007154 | cell communication | 8 | 16** | 5 | 47** |
| GO:0019725 | cellular homeostasis | 7 | 6 | 4 | 34 |
| GO:0009605 | response to external stimulus | 6 | 10 | 2 | 31 |
| GO:0007267 | cell-cell signaling | 5 | 4 | 1 | 13 |
| GO:0015979 | photosynthesis | 5 | 10 | 3 | 15 |
| GO:0016049 | cell growth | 3 | 10 | 3 | 14 |
| GO:0009991 | response to extracellular stimulus | 3 | 5 | 2 | 17 |
| GO:0007610 | behavior | 2 | 9 | 2 | 34 |
| GO:0009908 | flower development | 2 | 3 | 0 | 8 |
| GO:0009838 | abscission | 1 | 0 | 0 | 1 |
| GO:0040029 | regulation of gene expression, epigenetic | 1 | 8 | 0 | 11 |
* GO categories showing statistically significant overrepresentation of genes (Fisher exact test, FDR ≤ 0.05)
** GO categories showing statistically significant underrepresentation of genes (Fisher exact test, FDR ≤ 0.05)
We have also used the Wilcoxon rank-sum test implemented in the MapMan software to identify the sets of co-regulated homoeologous genes assigned to the same functional pathways or genetic networks whose expression deviates from the response of all other genes [55]. The Benjamini and Hochberg correction was used to control for the false discovery rate [43]. The MapMan software performs functional classification of genes using the redundancy-reduced ontology [55]. Contrary to GO annotation, MapMan assigns a gene to the most appropriate functional category (BIN) rather than sorting it into multiple BINs. We decided to test if this reduction of redundancy will improve our ability to identify functional categories of genes that are co-regulated in SN. Parent-specific co-regulation of large sets of genes in SN was observed for only a few functional categories (Table 5). The coordinated up-regulation of genes involved in protein biosynthesis pathways was observed in the A and B genomes of SN. The genes involved in photosynthetic pathways, protease inhibitor/seed storage/lipid transfer and protein folding processes were also co-activated in the A and B genomes of SN. Contrary to this observation, the D genome in SN experienced massive down-regulation of genes involved in protein synthesis pathways (Table 5).
Table 5.
Homoeolog-specific co-regulation of various functional groups of genes in allopolyploid wheat
| MapMan BIN | MapMan functional classes | Genome | Expression | P value* |
|---|---|---|---|---|
| 29 | Protein biosynthesis | AB | up | 6.7 × 10-3 |
| 1.3.1 | Photosynthesis: calvin cycle, rubisco large subunit | AB | up | 8.8 × 10-3 |
| 1.1 | Photosynthesis: light reaction | AB | up | 0.01 |
| 26.21 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | AB | up | 0.02 |
| 29.6 | Protein folding | AB | up | 0.05 |
| 29.2 | Protein synthesis | D | down | 2.1 × 10-5 |
| 29.2.1.2 | Ribosomal protein synthesis | D | down | 2.1 × 10-5 |
| 29.2.1 | Protein synthesis, ribosomal proteins | D | down | 3.2 × 10-5 |
| 29.2.1.2.2 | Ribosomal protein synthesis 60S subunit | D | down | 0.02 |
* P value is subjected to Benjamini and Hochberg (1995) correction for multiple test comparison
Discussion
Most genes in the synthetic allopolyploid wheat were expressed at mid-parent level fitting the expectations of either 1:1 or 1:2 parental expression ratios, which indicates that polyploidization did not have detectable impact on their regulation. A significant amount of functional redundancy retained after polyploidization found in our study was also consistently found in other artificially created polyploids [7,9,27]. This phenomenon apparently underlies the evolutionary flexibility of polyploid wheat [26], its ability to withstand large scale chromosomal deletions or even loss of entire chromosomes [56,57], and to accumulate high density of EMS-induced mutations [58].
The analyses of probeset data resulted in the higher proportion of non-additively expressed genes (39%) in synthetic wheat than that previously detected using long-oligonucleotide microarray platforms in wheat [27,29], Arabidopsis [9] or Brassica [7]. The differences in non-additive gene expression between our study and previously reported estimates can potentially be attributed to technological capacities of short- and long-oligonucleotide microarray platforms and differences between genotypes used to create allopolyploids. The usage of a short oligonucleotide array system sensitive to the presence of mutations discriminating one homoeolog from another may have increased the rate of non-additive expression discovery. In both previous studies [27,29] natural tetraploid wheat accessions were used to create allohexaploid synthetics. Apparently, differences accumulated during the co-evolution of AB and D genomes of hexaploid wheat contribute to higher proportion of non-additive expression in reconstituted TetraCantach/Ae. tauschii allopolyploid observed in our study.
We used an Affymetrix microarray system for the simultaneous detection of divergent nucleotide mutations differentiating the wheat AB genomes from the D genome and to measure the relative contribution of parents to total expression in the synthetic polyploid wheat. According to the results of quantitative PCR and microarray hybridization with mid-parent mixture of parental RNA samples, this approach can correctly predict significant departure from the expected mid-parent level of expression with 24% false positive rate. The Ep/Ēt ratio data was more consistent with the observations made in previous studies; the majority of the parental genes in synthetic polyploid wheat (81%) were expressed at the mid-parent levels corresponding to 1:1 or 1:2 expression ratios between the parental genomes. Non-additive contribution from the D or AB genome homoeologs was observed for the remaining 19% of genes.
One of the interesting aspects of homoeologous gene expression in allopolyploid species is transcriptional dominance of one of the parental genomes [9,10,19,59]. The bias toward TC- type parental expression in allopolyploid wheat suggests the transcriptional dominance of TC. Our study demonstrated that out of all genes showing the increase of parental expression in SN about 70% were of TC-type. In contrast, the D genome homoeologs comprised about 63% of all genes showing decreased contribution of parental expression to SN transcriptome. Similar proportion of significantly biased genes (80%) was discovered by studying homoelog-specific gene expression in cotton [10]. Several mechanisms have been proposed to account for parent-specific expression bias in allopolyploids including the incompatibility of regulatory elements [28], removal of gene imprinting [17,60,61], chromatin modification [9] and activation of transposable elements adjacent to the genes [8]. Apparently, all of these mechanisms play some role during the adjustment of two diverged parental regulatory systems. Regulation of homoeologous genes in the polyploid wheat suggests that divergent mutations in cis- and trans-acting regulatory elements or epigenetic modifications can result in spatial and/or temporal partitioning of gene expression [17,60]. Likewise, compensatory co-evolution of cis- and trans-acting factors in different lineages of Drosophila can lead to dysregulation of about 30% of genes in the hybrids [14]. The role of interaction between the diverged regulatory elements in modulating the expression of homoeologous genes in polyploids is confirmed by our data, which shows that 84% of non-additive changes in gene expression in SN can be explained by the transcription divergence between Ae. tauschii and Tetra-Cantach. This result is also consistent with the data reported for Arabidopsis and cotton [9,10]. The proportion of genes for which we observed the expression divergence between the parental lines (~57%) was similar to those reported for Arabidopsis (47%) and cotton (42-53%) [9,59], and higher than 32% divergence reported for Brassica [7]. An even higher level of expression divergence (78%) was reported for Ae. tauschii and T. turgidum [27] using a long oligonucleotide array system.
Interestingly, transcription divergence has been shown to correlate with the divergence of regulatory elements [62], which, in turn, depends on the evolutionary history of diverged lineages and time passed since their divergence. Recent studies in Drosophila demonstrated that the divergence of cis-trans elements occurs rapidly, resulting in about 40% of genes showing regulatory changes in ~2.5 million years of divergence [14,31]. In genus Gossypium (L.), the divergence time between the diploid species (5-10 MYA) was positively correlated with the expression divergence (42% - 53%) [59]. The diploid ancestors of polyploid wheat genomes diverged about 2.7 MYA followed by an initial polyploidization event resulting in the origin of T. dicoccoides (AB genome) that occurred about 0.5 MYA [20,21]. Therefore, when compared to other inbreeding allopolyploid species, the level of expression divergence reported for tetraploid wheat and the diploid ancestor of the wheat D genome shows an acceleration of regulation divergence after the first polyploidization event. The acceleration of expression divergence is probably driven by functional redundancy created by the first round of whole genome duplication and facilitated by epigenetic modifications and regulatory mutations. This process may play important role in gene expression evolution and in the generation of evolutionary novelties and functional variation upon which selection can act.
We hypothesized that compensatory co-evolution of cis-trans elements in the parental genomes may lead to the divergence of whole networks of genes that can be regulated independently in allopolyploids. The level of regulatory incompatibility between the networks in allopolyploids probably vary for different gene ontology categories and most likely are defined by the strength of natural selection. Those cis- and trans- acting elements that experienced strong purifying selection might remain compatible even after a long period of divergent evolution. Experimental studies demonstrated that transcription factors may co-evolve with the subset of targets that they regulate [14,63]. The parent-specific bias in gene regulation documented in allopolyploids can partially be explained by this process [9,19,59]. Additionally, parent-specific regulation of groups of genes could be important for maintaining correct gene dosage. It was hypothesized that stoichiometric ratios in macromolecular complexes or balances of gene products involved in multiple steps in regulatory cascades are targeted by positive selection and important for optimal organism fitness [64,65]. According to the gene balance hypothesis, if the gene product's concentration does not follow stoichiometry of the complex or cascade, then fitness is lowered [65]. One intriguing possibility is that the restoration of fecundity and growth vigor of the extracted tetraploid TetraCantach after hybridization with Ae. tauschii is partially the result of gene dosage restoration through adjustment of gene expression.
Remarkably, using the non-redundant functional classification implemented in MapMan software we were able to identify several pathways that showed coordinated changes in gene expression that was restricted to only one of the parental genomes. We observed the coordinated up-regulation of genes involved in photosynthetic and protein biosynthesis and folding pathways in the A and B genomes of allopolyploid wheat. This process was accompanied by the coordinated down-regulation of protein biosynthesis pathway in the D genome. Functional classification using GO categories showed an over-representation of genes involved in the generation of precursor metabolites and energy in the D genome of SN.
Our ability to identify coordinated regulation probably depends on the relative contribution of cis- and trans- divergence to gene regulation in allopolyploids. Since mutations in transcription factors have large pleiotropic effects [66], any processes that are regulated by few diverged trans- factors that co-evolved with a large set of cis-targets should be readily detectable. The prevalence of cis-type regulation would localize regulatory effects to the level of a single gene. This type of regulation seems to be a major contributor to gene regulation in allopolyploids [16] and intra- and inter-species hybrids [15,31] and may explain the low number of co-regulated homoeologous genes found in our study. More rigorous testing including various functional classes of genes combined with the analysis of cis-regulatory divergence needs to be performed to estimate the relative impact of cis- and trans-effects on gene regulation in allopolyploids.
Interaction between the diverged ancestral genomes in newly formed allopolyploids plays an important role in evolution by generating new functional variation and contributing to increased vigor and fertility, broader adaptability and phenotypic variability of polyploid species [1,3,67,68]. In yeast, novel cis-trans interactions established in a hybrid were shown to be involved in environmental response [69]. The plasticity of the wheat genome, arising from genetic and functional variation contributed by allopolyploidy, was considered to be a key factor defining its adaptability to diverse environments [26,70]. One of the mechanisms proposed to explain the growth vigor of Arabidopsis allopolyploids and hybrids includes the regulation of photosynthesis and sugar metabolism through the epigenetic modification of circadian clock genes [11]. Similarly, up-regulation of the AB genome homoeologous genes involved in protein biosynthesis and photosynthesis may contribute to restoration of growth vigor and fertility in allopolyploid wheat compared to that of tetraploid wheat Tetra-Cantach. Kerber (1964) proposed that the existence of genetic changes in the AABB component of the wheat genome accumulated since the origin of hexaploid wheat result in reduced growth vigor and fertility of reconstituted tetraploids which can be compensated by hybridization with Ae. tauschii. Our results further suggest that the interaction between the D and AB genomes is critical in modulating homoeolog-specific expression which is apparently responsible for physiological and morphological characteristics of allopolyploid wheat.
Conclusions
In summary, the approach was developed that utilizes the Affymetrix array system for measuring homoeolog-specific gene expression in allopolyploid wheat. The expression data showed the dominance of the AB genome in the transcriptome of the re-synthesized allopolyploid. The expression divergence between the transcriptomes of parental lines explained most of the observed non-additive expression in allopolyploid wheat. Evidence of homoeolog-specific coordinated up- and down-regulation of several functional gene categories, including those involved in protein synthesis and photosynthesis processes, suggests the co-evolution of cis- and trans-regulators. Additionally, the co-evolution of cis- and trans-elements may lead to divergence and incompatibility of regulatory networks in allopolyploids. The up-regulation of protein synthesis and photosynthetic pathways in the AB genome of synthetic polyploid wheat that result in an increase in vigor and restoration of fertility provide good targets for studying the molecular basis of these described phenomena in allopolyploids. While this causal relationship still requires experimental verification, it suggests the existence of established regulatory interactions between homoeologous genomes that lead to increased growth vigor, fertility and biomass in allopolyploid wheat.
List of abbreviations
PSF: parent-specific features; SFP: single-feature polymorphism; MYA: million years ago; PM: perfect match; MM: mismatch
Authors' contributions
AA prepared samples for Affymetrix hybridization and 454 sequencing, performed validation of Affymetrix results by 454 sequencing, and helped to draft the manuscript; HL performed analysis of 454 sequencing data; RM performed experimental validation of Affymetrix results; EA conceived the experiment, performed the statistical analysis and drafted the manuscript. All authors read and approved the final version of the manuscript.
Supplementary Material
List of 40,281 parent-specific oligonucleotide features. File contains the list of 40,281 Affymetrix oligonucleotide features differentially hybridizing with AT and TC parental transcripts. The linear model described in Methods was fit to PM probe intensity data. Residuals containing the probe by genotype effect were tested for difference between genotypes by calculating the d-statistics [45] using the Significance Analysis of Microarrays (SAM) procedure implemented in the R package siggenes. The threshold Δ = 0.2 was selected to identify significantly different features at 0.1 false discovery rate.
Validation of parent-specific features by comparing with 454 sequence data. Validation of parent-specific features by comparing with 454 sequence data. The table provides the list of PSFs showing significant blast hits with sequence reads obtained for cDNA libraries of Ae. tauschii, T. aestivum cv. Chinese Spring and T. aestivum cv. Jagger; PM - Affymetrix probe perfectly matches the 454 sequence read; MM - Affymetrix probe have a mismatch with the 454 sequence read; d-value is used in SAM for identification of PSFs.
Distribution of PSFs across the wheat genome. List of Affymetrix probesets with parent-specific features (PSF), their locations on the deletion bin map and a list of deletion bin mapped ESTs showing similarity to transcripts interrogated by Affymetrix probesets. The map locations were calculated by averaging the deletion bin midpoints of homoeologous chromosomes and assigning them to one of the five intervals (0-0.2, 0.2-0.4, 0.4-0.6, 0.6-0.8, 0.8-1.0).
Parent-specific gene expression in allopolyploid wheat. The method of contrasts was used to compare Ep/Ēt intensity ratios between allopolyploid wheat and its parents. Two possible ratios of parental gene expression in the synthetic polyploid were tested assuming 1:1 (AT + TC)/2) and 1:2 (1AT+2TC)/3) in silico ratio of AT:TC gene expression in the SN transcriptome. The FDR was maintained at 0.05.
Validation of Affymetrix microarray hybridization results by quantitative RT-PCR. Expression levels were converted to theoretical value R0 using the formula R0 = R(Ct) (1 + E)(-Ct), where R0 is the starting fluorescence, R(Ct) is the fluorescence at the threshold cycle Ct and E is the amplification efficiency. The R0 values were normalized to R0 of actin gene followed by log-transformation. The expression levels in SN and 1:1 mixture of AT and TC RNA were compared using the t-test.
Contributor Information
Alina R Akhunova, Email: akhunova@ksu.edu.
Rustam T Matniyazov, Email: rmat@ksu.edu.
Hanquan Liang, Email: hliang@ksu.edu.
Eduard D Akhunov, Email: eakhunov@ksu.edu.
Acknowledgements
We would like to thank Jan Dvorak for kindly providing us with the seeds of Ae. tauschii, Tetra-Cantach and synthetic polyploid wheat, Gary Gadbury for helpful discussions on the statistical analyses of microarray data and Cyrille Saintenac, Miranda Gray and four anonymous reviewers for valuable comments on the earlier versions of the manuscript. Affymetrix microarray hybridizations and validation of Affymetrix data by quantitative PCR were performed in the UC Davis Microarray Core Facility and KSU Integrated Genomics Facility. This work was supported by the funds provided by the KSU Agricultural Experimental Station.
References
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–46. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee HS, Comai L, Madlung A, Doerge RW, Colot V, Martienssen RA. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/S0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–8. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Bell CD, Kim S, Soltis PS. Origin and early evolution of angiosperms. Ann N Y Acad Sci. 2008;133:3–25. doi: 10.1196/annals.1438.005. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Yoo SY, Pires JC, Doerge RW, Chen ZJ, Osborn TC. Analysis of gene expression in resynthesized Brassica napus Allopolyploids using arabidopsis 70 mer oligo microarrays. PLoS One. 2009;4:e4760. doi: 10.1371/journal.pone.0004760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33:102–6. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Chen ZJ. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics. 2006;173:965–74. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L, Udall J, Nettleton D, Wendel J. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 2008;6:16. doi: 10.1186/1741-7007-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–31. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ha M, Lackey E, Wang J, Chen ZJ. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics. 2008;178:1845–58. doi: 10.1534/genetics.107.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley A, Koebner RM. Variation for homoeologous gene silencing in hexaploid wheat. Plant J. 2008;56:297–302. doi: 10.1111/j.1365-313X.2008.03586.x. [DOI] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–22. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell. 2007;19:2391–402. doi: 10.1105/tpc.107.052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM, Wendel JF. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (gossypium) Genetics. 2009;182:503–17. doi: 10.1534/genetics.109.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Ishihara A, Yanagita RC, Endo TR, Iwamura H. Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. Proc Natl Acad Sci USA. 2005;102:16490–5. doi: 10.1073/pnas.0505156102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Buchwaldt L, Rimmer SR, Brkic M, Bekkaoui D, Hegedus D. Differential expression of duplicated peroxidase genes in the allotetraploid Brassica napus. Plant Physiol Biochem. 2009;47:653–6. doi: 10.1016/j.plaphy.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Hovav R, Udall JA, Chaudhary B, Rapp R, Flagel L, Wendel JF. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci USA. 2008;105:6191–5. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Akhunov ED. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics. 2005;171:323–32. doi: 10.1534/genetics.105.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P. Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA. 2008;105:9691–6. doi: 10.1073/pnas.0803981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Zhang HB. Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc Natl Acad Sci USA. 1990;87:9640–9644. doi: 10.1073/pnas.87.24.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorák J, Terlizzi P, Zhang HB, Resta P. The evolution of polyploid wheats: identification of the A genome donor species. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- Willcox G. In: The Origins of Agriculture and Crop Domestication. Damania AB, Valkoun J, Willcox G, Qualset CO, editor. ICARDA, IPGRI, FAO and UC/GRCP, ICARDA, Aleppo, Syria; 1997. Archaeobotanical evidence for the beginnings of agriculture in Southwest Asia; pp. 25–38. [Google Scholar]
- Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, Hochu I, Poirier S, Santoni S, Glémin S, David J. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol. 2007;24:1506–1517. doi: 10.1093/molbev/msm077. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumphrey M, Bai J, Laudencia-Chingcuanco D, Anderson O, Gill BS. Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics. 2009;181:1147–57. doi: 10.1534/genetics.108.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Friebe BR, Gill BS, Zhou JM. Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol Biol. 2003;52:401–14. doi: 10.1023/A:1023965400532. [DOI] [PubMed] [Google Scholar]
- Chague V, Just J, Mestiri I, Balzergue S, Tanguy A, Huneau C, Huteau V, Belcram H, Coriton O, Jahier J, Chalhoub B. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytologist. 2010;187:1181–1194. doi: 10.1111/j.1469-8137.2010.03339.x. [DOI] [PubMed] [Google Scholar]
- Ronald J, Akey JM, Whittle J, Smith EN, Yvert G, Kruglyak L. Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 2005;15:284–91. doi: 10.1101/gr.2850605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics. 2009;183:547–61. doi: 10.1534/genetics.109.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Long Y, Meng J, Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J. 2009;7:334–46. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks TW, Jordan MC, Somers DJ. Single-Feature Polymorphism Mapping in Bread Wheat. The Plant Genome. 2009;2:167–178. doi: 10.3835/plantgenome2009.02.0009. [DOI] [Google Scholar]
- Cui X, Xu J, Asghar R, Condamine P, Svensson JT, Wanamaker S, Stein N, Roose M, Close TJ. Detecting single-feature polymorphisms using oligonucleotide arrays and robustified projection pursuit. Bioinformatics. 2005;21:3852–3858. doi: 10.1093/bioinformatics/bti640. [DOI] [PubMed] [Google Scholar]
- Gore M, Bradbury P, Hogers R, Kirst M, Verstege E, van Oeveren J, Peleman J, Buckler E, van Eijk M. Evaluation of Target Preparation Methods for Single Feature Polymorphism Detection in Large Complex Plant Genomes. The Plant Genome. 2007;S2:S–135-S-148. [Google Scholar]
- Rostoks N, Borevitz JO, Hedley PE, Russell J, Mudie S, Morris J, Cardle L, Marshall DF, Waugh R. Single-feature polymorphism discovery in the barley transcriptome. Genome Biol. 2005;6:R54. doi: 10.1186/gb-2005-6-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo AN, Bradbury PJ, Ma H, Hu S, Bowden RL, Buckler ES, Bai G. Discovery and mapping of single feature polymorphisms in wheat using Affymetrix arrays. BMC Genomics. 2009;10:251. doi: 10.1186/1471-2164-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ER. Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science. 1964;143:53–255. doi: 10.1126/science.143.3603.253. [DOI] [PubMed] [Google Scholar]
- Liu B, Vega JM, Feldman M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome. 1998;41:535–542. doi: 10.1139/gen-41-4-535. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc, Ser B. 1995;57:289–300. [Google Scholar]
- Borevitz J. Genotyping and mapping with high-density oligonucleotide arrays. Methods Mol Biol. 2006;323:137–45. doi: 10.1385/1-59745-003-0:137. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix. http://www.affymetrix.com/index.affx
- Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21:818–21. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorák J, Linkiewicz AM, Ratnasiri A, Dubcovsky J, Bermudez-Kandianis CE, Greene RA, Kantety R, La Rota CM, Munkvold JD, Sorrells SF, Sorrells ME, Dilbirligi M, Sidhu D, Erayman M, Randhawa HS, Sandhu D, Bondareva SN, Gill KS, Mahmoud AA, Ma XF, Miftahudin, Gustafson JP, Conley EJ, Nduati V, Gonzalez-Hernandez JL, Anderson JA, Peng JH, Lapitan NL, Hossain KG, Kalavacharla V, Kianian SF, Pathan MS, Zhang DS, Nguyen HT, Choi DW, Fenton RD, Close TJ, McGuire PE, Qualset CO, Gill BS. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics. 2004;168:701–12. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrainGenes 2.0. http://wheat.pw.usda.gov/GG2/index.shtml
- Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIGR Gene Indexes. ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/Triticum_aestivum
- AmiGO. http://amigo.geneontology.org/cgi-bin/amigo/go.cgi
- MapMan. http://mapman.gabipd.org
- Poole R, Barker G, Wilson ID, Coghill JA, Edwards KJ. Measuring global gene expression in polyploidy; a cautionary note from allohexaploid wheat. Funct Integr Genomics. 2007;7:207–219. doi: 10.1007/s10142-007-0046-7. [DOI] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environment. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Endo TR, Gill BS. The deletion stocks of common wheat. J Hered. 1996;87:295–307. [Google Scholar]
- Sears ER. Cytogenetic Studies with Polyploid Species of Wheat. II. Additional Chromosomal Aberrations in Triticum Vulgare. Genetics. 1944;29:232–246. doi: 10.1093/genetics/29.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp RA, Udall JA, Wendel JF. Genomic expression dominance in allopolyploids. BMC Biol. 2009;7:18. doi: 10.1186/1741-7007-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N, Tahira C, Kassai K, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K, Ogihara Y, Murai K. Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell. 2007;19:1723–37. doi: 10.1105/tpc.107.051813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N. On the relation between promoter divergence and gene expression evolution. Mol Syst Biol. 2008;4:159. doi: 10.1038/msb4100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol. 2002;22:8612–25. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10:575–9. doi: 10.1016/S0959-437X(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/S0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–61. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Feldman M, Levy AA. Allopolyploidy--a shaping force in the evolution of wheat genomes. Cytogenet Genome Res. 2005;109:250–258. doi: 10.1159/000082407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of 40,281 parent-specific oligonucleotide features. File contains the list of 40,281 Affymetrix oligonucleotide features differentially hybridizing with AT and TC parental transcripts. The linear model described in Methods was fit to PM probe intensity data. Residuals containing the probe by genotype effect were tested for difference between genotypes by calculating the d-statistics [45] using the Significance Analysis of Microarrays (SAM) procedure implemented in the R package siggenes. The threshold Δ = 0.2 was selected to identify significantly different features at 0.1 false discovery rate.
Validation of parent-specific features by comparing with 454 sequence data. Validation of parent-specific features by comparing with 454 sequence data. The table provides the list of PSFs showing significant blast hits with sequence reads obtained for cDNA libraries of Ae. tauschii, T. aestivum cv. Chinese Spring and T. aestivum cv. Jagger; PM - Affymetrix probe perfectly matches the 454 sequence read; MM - Affymetrix probe have a mismatch with the 454 sequence read; d-value is used in SAM for identification of PSFs.
Distribution of PSFs across the wheat genome. List of Affymetrix probesets with parent-specific features (PSF), their locations on the deletion bin map and a list of deletion bin mapped ESTs showing similarity to transcripts interrogated by Affymetrix probesets. The map locations were calculated by averaging the deletion bin midpoints of homoeologous chromosomes and assigning them to one of the five intervals (0-0.2, 0.2-0.4, 0.4-0.6, 0.6-0.8, 0.8-1.0).
Parent-specific gene expression in allopolyploid wheat. The method of contrasts was used to compare Ep/Ēt intensity ratios between allopolyploid wheat and its parents. Two possible ratios of parental gene expression in the synthetic polyploid were tested assuming 1:1 (AT + TC)/2) and 1:2 (1AT+2TC)/3) in silico ratio of AT:TC gene expression in the SN transcriptome. The FDR was maintained at 0.05.
Validation of Affymetrix microarray hybridization results by quantitative RT-PCR. Expression levels were converted to theoretical value R0 using the formula R0 = R(Ct) (1 + E)(-Ct), where R0 is the starting fluorescence, R(Ct) is the fluorescence at the threshold cycle Ct and E is the amplification efficiency. The R0 values were normalized to R0 of actin gene followed by log-transformation. The expression levels in SN and 1:1 mixture of AT and TC RNA were compared using the t-test.