Abstract
Based on structural information derived from the F NMR data of labeled rhodopsins, rhodopsin crystal structure, and excited-state properties of model polyenes, we propose a molecular mechanism that accounts specifically for the causes of the well-known enhanced photoreactivity of rhodopsin (increased rates and quantum yield of isomerization). It involves the key features of close proximity of C-187 to H-12 and chromophore bond lengthening upon light absorption. The resultant “sudden punch” to H-12 triggers dual processes of decay of the Franck–Condon-excited rhodopsin, a productive directed photoisomerization and a nonproductive decay returning to the ground state as two separate molecular pathways [based on real-time fluorescence results of Chosrowjan, H., Mataga, N., Shibata, Y., Tachibanaki, S., Kandori, H., Shichida, Y., Okada, T. & Kouyama, T. (1998) J. Am. Chem. Soc. 120, 9706–9707]. The two processes are controlled by the local protein structure: an empty space provided by the intradiscal loop connecting transmembrane helices 4 and 5 and a protein wall composed of amino acid units in transmembrane 3. Suggestions, involving retinal analogs and rhodopsin mutants, to improve the unusually high photosensitivity of rhodopsin are proposed.
Keywords: photoisomerization, F NMR, protein perturbation
Rhodopsin is a heptahelical membrane protein responsible for scotopic vision. It is a photo-receptor protein activated by the 11-cis-retinal (1) (Structure 1) chromophore covalently bonded to Lys-296 through a protonated Schiff base (PSB) linkage. The photoisomerization of the chromophore initiates the vision process (1). The pigment is known to possess unusually high photochemical reactivity. For example, its quantum yield of isomerization is 0.65 (2, 3) (a recently refined value from the long accepted number of 0.67; ref. 4), more than two times higher than that of the same chromophore in solution (0.24, 0.22) (5, 6). The rate of isomerization is also much faster in protein (146 fsec) (7) than in solution (1–2 psec) (8). In this article, we propose a detailed molecular model accounting for the unusual protein assistance to the isomerization process.
Structure 1.
11-cis-Retinal (1) and its analogs (2–7).
Background Information
The Shape of the Binding Site of Rhodopsin from Analog Studies. Opsin, the apoprotein of rhodopsin, demonstrates some flexibility in being able to accept a number of retinal isomers (1, 9) and analogs (10) to form pigment analogs. An overlay of the atoms in all binding chromophores, in principle, will produce a 3D shape that reflects maximal flexibility of its binding site. Such a shape in two dimensions constructed from atoms of binding isomers of rhodopsin is in the literature (11). Additional relevant binding data from retinal analogs are: positive results with 5-, 10-, and 13-Me-substituted retinals (2–4) (12) and ring fused analogs connecting C-10 and 13-Me (5) (13) and negative results (or extreme low yield) with 12-substituted (6) (14) and 13-cis-retinals (11). The combined information paints a clear picture of the binding site: little space surrounding H-12 but a large space in the region between 5-Me and 13-Me, opposite to H-12 of the polyene chain.
However, there is clearly a preference in binding interaction with the 11-cis isomer as reflected in the fast rate of pigment formation, pigment stability, maximal spectral perturbation, and photosensitivity (15). And, the fact that the 9-cis isomer is the second best isomer in interacting with opsin is also a reflection of this selectivity. It is similar in shape to the 11-cis as demonstrated in molecular modeling studies of the PSBs (11, 16).
The 11-cis-retinyl chromophore of rhodopsin exhibits strong CD activity despite the absence of a single chiral carbon (17). The chirality originates from a defined, twisted conformation of the polyene chain, in particular large twists around the 6,7 and 12,13 single bonds. Recently the Columbia group, through the clever usage of conformationally rigid retinal analogs (7 the active form, the epimer does not bind with opsin), was able to define the helical sense of the twisted chromophore of rhodopsin (18).
The Structure of the Primary Photoproduct of Rhodopsin. The structure of the primary photoproduct of rhodopsin is still largely unknown. For many years bathorhodopsin (λmax = 535 nm) was considered the primary photoproduct of rhodopsin (1, 19). In the 1980s the Kyoto group (20, 21) identified an earlier intermediate in their time-resolved studies, named photorhodopsin (λmax = 570 nm). This early intermediate, however, is too unstable for trapping. Hence, little structural information is known other than that the 11,12-double bond must be highly twisted (on account of its large red-shift). Its existence was also demonstrated in rhodopsin analogs, e.g., that from 5d (22).
In contrast, much structural information on bathorhodopsin, the first stable intermediate, is available. Loppnow and Mathies (23) provided the most detailed information from vibrational spectroscopy (resonance Raman) and concluded that the chromophore has the all-trans-6-s-cis-retinyl structure. (For structure, see Scheme 1.) Later, 13C-NMR data of bathorhodopsin by Smith et al. (24) confirmed this structure.
Scheme 1.
(Upper) Primary photoproduct from rhodopsin, photorhodopsin, and the all-trans-6-cis structure of bathorhodopsin, the first stable intermediate. (Lower) The proposed HT-12 (giving a 12-s-cis-like structure) and ensuing BP-12,14 and BP-14,16 processes, leading to the same all-trans-6-cis structure.
Regiospecific Perturbation in F NMR of F-Rhodopsins. A F atom is only ≈25% larger than a H atom (25); therefore, the stereoselectivity exhibited by rhodopsin does not exclude possible formation of F-rhodopsin analogs. Indeed many have since been prepared (Table 1) (26). The slight variations of the absorption maxima are consistent with the expected role of the electronegative F atom (27).
Table 1. Vinyl F-rhodopsin analogs, pigment absorption maxima.
An extended effort (26, 28, 29) led to compilation of the 19F chemical shifts of 15 vinyl F-rhodopsin analogs. The corresponding PSBs were also prepared, and the F-shifts in solution were measured. The combined data yielded a set of parameters (fluorine opsin shift, FOS) that reflects the extent of local protein perturbation on the F-labels. They are summarized in Fig. 1, ranging from 3 to 19 ppm (26, 30).
Fig. 1.
FOSs of 11-cis (squares) and 9-cis (circles) vinyl fluoro-rhodopsins. FOS is the difference between the F chemical shift of a F-rhodopsin and its PSB. Unfilled marks are for F-rhodopsin, and filled marks are for F2-rhodopsins. The hatched area includes values for normal protein shifts (25). The original chemical shift data are available in the literature (26, 29, 30).
Factors affecting 19F chemical shifts include electron anisotropy (ring current effect), specific interactions (H-bonding), and weak electric field effects (31). Both ring current (32) and H-bonding effects (33) are small (≈1–2 ppm), which need not be of concern here. For electric field effect, De Dios and Oldfield (34) showed that weak electric field effects (α 1/r3) are the dominant factors, which obviated the need to invoke second-order van der Waals interactions (α 1/r6). Such a shift is a clear indication of effect of neighboring atoms on a 19F-label and is distance dependent.
The FOS values of the 9-cis chromophores showed a relatively narrow range of 4.4 to 7.9 ppm, which falls in the normal range of protein perturbation (9), reflecting the overall medium effect changing from solution to enclosed protein cavity environment. In the 11-cis series, the FOS values for 10F and 14F also were <8 ppm. However, those of 12F and 8F clearly exceed this range, mostly in the order of 12–13 ppm. These regiospecific perturbations can only suggest close proximity of protein residues to these labels, most likely in close van der Waals contact. The 8F is nearby the known hydrophobic pocket in opsin. Thus, the large FOS must be a reflection of its close interaction with a nonpolar amino acid unit (Trp-265, see below). It is not of immediate interest to the current discussion. But the specific amino acid residue responsible for the high FOS values of F-12 is germane to the current discussion.
X-Ray Crystal Structures of Rhodopsin. The amino acid residue responsible for the regiospecific perturbation at C-12 was readily identified through an examination of crystal structures of rhodopsin that have since become available, the first at 2.8-Å resolution (35) and the second at 2.6 Å (36). An examination of the latter crystal structure (Protein Data Bank ID code 1L9H) revealed that the chromophore in the protein cavity is closest to these amino acid units: Glu-113 (3.13 Å to N), Cys-187 (3.25 Å to C-12), and Trp-265 (3.49 Å to H-8). Glu-113 is the counter-anion to the iminium N, and Trp-265 is part of the hydrophobic pocket for the cyclohexenyl ring. For Cys-187, the actual distances between the F atom and the carbonyl oxygen should be much shorter than 3.25 Å inasmuch as the F atom extends one bond away from the carbon framework (C-F bond ≈1.41 Å) (37). Thus, it is safe to say that Cys-187 is the unit causing the observed large FOS at F-12 (30).‡
It should be noted that before these crystal structures there was a considerable amount of information about the primary and seven helical structure of rhodopsin (38, 39).
Models for Photoisomerization. Through analyses of Raman intensity of rhodopsin, Kim and Mathies (40) concluded that in twisting the 11,12-double bond the excess vibrational energy in the Franck-Condon (FC)-excited rhodopsin is concentrated in the chromophore. The transformation is believed to start with “the formal motion of the C12-H group” that “may dominate the development of this torsional distortion.” The final product involves a small twist of many bonds in addition to the >90° twist of the 11,12-bond (41). An earlier theoretical study suggested a similar process and structure for Batho (42). Thus, bathorhodopsin is similar in shape to the twisted rhodopsin.
Recently, the volume-conserving hula twist (HT) model for photoisomerization (43) has been shown to be generally applicable to simple organic polyene chromophores imbedded in rigid media (44). It involves simultaneous rotation of a pair of adjacent double and single bonds. When applied to rhodopsin isomerization, HT-12 (HT at center 12) was considered the primary photochemical process (45), an idea consistent with the photoreaction of a ring-fused analog (46) and a later modeling study (47). It was noted that the rigid protein structure would likely not allow completion of this motion. However, a nascent HT-12 structure (i.e., immediately beyond halfway of twisting) when followed by stepwise bicycle-pedal (BP) processes (48) (BP-12,14 and BP-14,16) could transfer the s-cis kink to the butyl tether giving the same all-trans-6-s-cis structure (45) (Scheme 1). On the other hand, Ishiguro et al. (49), based on molecular modeling studies, reaffirmed the earlier prediction that bathorhodopsin has the 12-s-cis structure.
However, we must emphasize that the exact nature of the initial isomerization process has no bearing on the idea of enhancement mentioned in this article. Both Mathies' torsional relaxation process (41) and the HT concept involve initial motion of the C12-H moiety. For the sake of convenience, HT-12 is used to describe such a movement.
The Accelerated Rate of Photoisomerization in Rhodopsin. In an elegant application of femtosecond laser technology, Mathies and coworkers (50) showed in a time-resolved absorption study that the transition of the primary excited rhodopsin to the ground-state product is complete within 200 fsec. Subsequently, Shichida and coworkers (7) showed, by an amazing real-time spontaneous fluorescence method, that the FC-excited rhodopsin has a fast component (accounting for ≈70% of decay of FC rhodopsin) with a decay time of 146 fsec that leads directly to the primary product photorhodopsin. A second (≈30%) slower process (330 fsec) leads to vibrationally relaxed, electronically excited rhodopsin, which after 1–2 psec returns to rhodopsin with no detectable chemical transformation (7). Both processes are faster than the 0.5- to 3.1-psec range detected for deactivation of excited 11-cis PSB in methanol (8), a clear demonstration of protein assistance in the isomerization process. These dual accelerated decay processes of FC-excited rhodopsin was verified in a subsequent wavelength-dependent study by the same group (51).
A recent redetermination of quantum yield of photoisomerization of rhodopsin showed that the values depend on excitation wavelength (2, 3). Between 450 and 480 nm, the values are constant at 0.65, but decrease upon increase of excitation wavelength (reduced by 5% at 570 nm). But even for the reduced values, they are much higher than the corresponding value for photoisomerization of 11-cis-retinyl PSB in solution (ϕ = 0.22–24) (5, 6). Again, it reflects protein assistance. But, what is the specific nature of this protein assistance to the photochemical process?
Proposed New Concepts
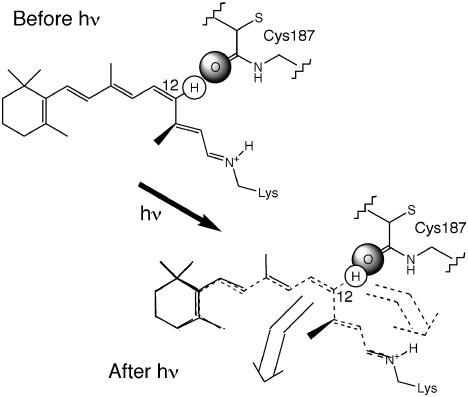
A Molecular Model of Protein Assistance to the Isomerization Process. Above, we summarized the F NMR and crystallographic evidence that led to the conclusion of close proximity of Cys-187 to H-12 of the retinyl chromophore. We now discuss a possible role of this amino acid unit in the accelerated isomerization process. In Fig. 2 is the partial crystal structure of rhodopsin (36) showing the 11-cis-retinyl chromophore with the added H-12 in close proximity to Cys-187. The estimated distance between H-12 and the O-atom of C O of Cys-187 is 2.23 Å, a remarkably short distance that allows one to conclude safely that the atoms must be in close van der Waals contact. Light absorption of the chromophore (in 1–10 fsec) results in promotion of an electron in a bonding molecular orbital to a more diffuse antibonding molecular orbital. The expected immediate consequence is bond lengthening of the chromophore in the excited state, within the common time scale for bond lengthening (≈100 fsec) (52). But from analysis of resonance Raman intensities, Mathies and Lugtenburg (41) concluded that in excited rhodopsin, the bond lengthening process is complete in 50 fsec. Such bond length changes for simple polyenes are well documented [e.g., for the Pariser-Pople-Parr/configuration interaction calculated results of hexatriene and octatetraene (53)].
O of Cys-187 is 2.23 Å, a remarkably short distance that allows one to conclude safely that the atoms must be in close van der Waals contact. Light absorption of the chromophore (in 1–10 fsec) results in promotion of an electron in a bonding molecular orbital to a more diffuse antibonding molecular orbital. The expected immediate consequence is bond lengthening of the chromophore in the excited state, within the common time scale for bond lengthening (≈100 fsec) (52). But from analysis of resonance Raman intensities, Mathies and Lugtenburg (41) concluded that in excited rhodopsin, the bond lengthening process is complete in 50 fsec. Such bond length changes for simple polyenes are well documented [e.g., for the Pariser-Pople-Parr/configuration interaction calculated results of hexatriene and octatetraene (53)].
Fig. 2.
Partial x-ray crystal structure of rhodopsin (36) showing the 11-cis retinyl chromophore and amino acids residues Lys-185, Ser-186, Cys-187, and Gly-188 of intradiscal loop 4,5 (O and N in dark gray, and S in light gray of larger circles). Cys-187 is closest to C-12. The estimated distance between the O atom of the carbonyl group of C-187, and the added H-12 is 2.23 Å.
The lengthened chromophore after light absorption should lead to a displacement of atoms in the molecule but in a specific, defined manner because of the anchored nature of the chromophore (43). The hydrophobic pocket in the opsin binding site that recognizes the trimethylcyclohexenyl ring should hold the ring in place during the short period of light absorption. [Furthermore, time scale for rotational motions or bond twisting is 1 order of magnitude longer than that of bond lengthening (52).] Likewise, the ion pair nearby the PSB should keep the chain terminus in place. With the two ends rigidly “anchored,” the displacement of atoms caused by bond lengthening will have to be directed toward the “V-bend” of the 11-cis chromophore with the largest displacement at H-11 and H-12 (see Fig. 3 Lower). However, the protein including Cys-187 does not move during the excitation that happened elsewhere. Such a sudden displacement should produce a sudden large repulsive interaction between H-12 and Cys-187.§ Since two atoms cannot occupy the same space, the only possible reaction is the bending of the C12-H bond or a concerted twisting motion that is equivalent to, hence promoting, the HT-12 process. In fact, Mathies and Lugtenburg (41) estimated that it takes only 50 fsec to drive the 11-cis chromophore to twist to a local torsional angle of 90°.¶ Since this reaction is triggered by light, it may be viewed as a phototrigger for the accelerated rhodopsin isomerization.
Fig. 3.
(Upper) Before light absorption. A cartoon figure of the 11-cis-retinyl twisted at the 12,13 bond (following that of Nakanishi and coworkers, ref. 18). The carbonyl O of Cys-187 is in close contact with H-12. (Lower) Upon light absorption, expected lengthening of the π-bonds as a result of promoting a π-electron into a more diffuse antibonding orbital, the atoms are expected to relocate. In drawing the structure (dashed lines) we assumed an arbitrary 4% increase in bond lengths. Also, taking into consideration the anchored nature of the chromophore, we assumed that the six-ring and the charged N do not move during the short period of light excitation (hence overlapping rings and the iminium Ns before and after light absorption). Movement of atoms will have to be directed toward the 11,12 cis-bend, hence the largest displacement near H-11 and H-12. To avoid the resultant untenable steric strain near H-12 (overlapped H... O), the C12-H bond will have to bend inward (HT-right) or outward (HT-left), giving two potentially distinguishable processes.
The Role of Cys-187 in Rhodopsin Structure and Function. A search of the literature revealed limited discussion on the functional role of Cys-187 of rhodopsin. Much of the past interest seemed to center on the polar or aromatic amino acid residues that are responsible for the functions and color tuning of the pigment (54). However, Khorana and coworkers (55) demonstrated in a series of elegant mutation work that replacement of Cys-187 would disrupt the disulfide bridge (with Cys-110) and the tertiary structure of the protein. (This structural role of Cys-187 implies difficulties in determining effects of its replacement with amino acid units of varying sizes on photoreactivity.) Indeed, the mutants where Cys-187 was replaced by Ser, Thr, Tyr, or Glu were found not to bind 11-cis-retinal to give a pigment analog (55, 56). When replaced by Ala, the mutant (C187A) yielded a rhodopsin-like pigment for its absorption spectrum. But it has altered dark bleaching reactivities (and no quantum yield data reported) (57). We believe, whereas Cys-187 is obligatory for proper protein folding, it also plays the added role of accelerating the primary photo-process.
The Dual Decay Pathways of FC-Excited Rhodopsin. Another piece of important information regarding rhodopsin photochemistry is the presence of one, and only one, extra ultrafast decay process (330 fsec) of the FC-excited rhodopsin in addition to the faster isomerization process (146 psec). The slower vibrational relaxation process led to the relaxed excited rhodopsin (7), which harmlessly returned to the ground state (8). These two processes suspiciously suggest that they might correspond to the two rotational modes of the anchored chromophore prompted by the immovable Cys-187. This scenario coincides with the two directions of reaction depicted in Fig. 3. Therefore, it became imperative to examine the protein crystal structure, looking for possible asymmetric protein environment surrounding the reaction center.
Examination of the protein structure (36) indeed revealed exactly the structural features needed for the dual decay processes. Fig. 4 reproduces a portion of the protein structure that contains the retinyl chromophore (viewing from the top of the C11—C12 bond) all seven transmembrane (TM) helices and intradiscal loops, in particular that connecting TMs 4 and 5. The large difference of the protein structure on two sides of the 11,12 bond is most evident: on the left is the protein wall, specifically TM 3 (aqua), and to the right is the open space surrounded by the 4,5-loop.
Fig. 4.
Partial crystal structure of rhodopsin (36) showing the centered chromophore (in green) and its orientation with respect to the seven helices: TM 1 (purple), TM 2 (blue), TM 3 (aqua), TM 4 (dark green), TM 5 (yellow), TM 6 (orange), and TM 7 (red). The left side faces the helix 3 in aqua color. The right side of the 11,12-bond faces the open region bound by the ribbon of the 4,5-intradiscal loop, i.e., not completely shielded by the helical bundle.
Fig. 5 is a close-up of the protein structure in space-filling models showing the retinyl chromophore and nearby amino acid residues (within 3.8 Å). The 11,12-cis-bend is in the center of Fig. 5, projecting outward and the remaining portions of the chromophore projecting inward. To the left of the 11,12 bond is the backbone of TM helix 3. Specifically Ala-117 is 3.53 Å away from C-12. It is the “front man” of the protein wall needed for the nonreactive channel of deactivation.
Fig. 5.
A close-up crystal structure of rhodopsin (36) in space-filling models showing all amino acid residues within 3.8 Å of the retinyl chromophore (green). N atoms are in blue, O in red, and S in yellow. The 11-cis linkage is centered in the middle with the remaining portions of the retinyl chromophore projecting inward. On the left is the TM helix 3 with Ala-117 nearby C-12. The empty space on the right is encircled by amino acids 187–191. Cys-187, Ile-189, and Tyr-192 are within 3.8 Å, thus visible here. The units in between (Gly-188 and Asp 190) are outside 4.0 Å, thus not visible here. Their unique locations encircles the empty space on the right side of C-12.
To the right in Fig. 5 is an open space. In fact, a closer inspection revealed that the empty space is surrounded by Gly-188, Ile-189, Asp-190, and Tyr-191 of the intradiscal loop connecting TM 4 and TM 5. Ile-189 and Tyr-191 are within 3.8 Å of the chromophore, hence they are shown in Fig. 5. The connecting amino acid units (Gly-188 and Asp-190) are, however, outside 4.0 Å, thus not appearing in Fig. 5. The clear implication is that these amino acid units encircle an empty space for the isomerization reaction. In other words, the surprising revelation to us is that the location of the chromophore is such that it is not completely encapsulated by the “binding cavity” (the helical bundle). Instead, one side of the cis-bend of the 11-cis-retinyl chromophore faces an empty space surrounded by the largest yet rigid (caused by the Cys-187, Cys-110 disulfide bond) 4,5 intradiscal loop.
Interestingly, Cys-110 and Cys-187 (and Cys-316) are conserved in all vertebrate visual pigments and seven helix hormone receptors, as noted by Khorana and colleagues (55). It is reasonable to conclude that these units along with amino acids 188–191 are essential for the photoisomerization. Unfortunately, functional roles of amino acid units in the connecting loops have not been examined in detail (54), although these amino acid units are known to be highly conserved in different visual pigments (58). It marks a new direction of research for protein chemists.
So, it seems that in addition to the many trumpeted advantages of the 11-cis configuration of the retinyl chromophore (15), we must now add the following. The protruded cis-bend “feels” the pressure of Cys-187 at its top and on one side it “overlooks” the wide open “pasture” provided by the 4,5-loop, making possible the rapid, directed isomerization process.
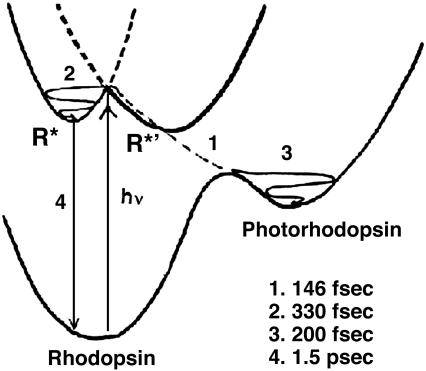
The rhodopsin photochemistry can now be summarized by Fig. 6 that contains double excited-state potential wells to describe the directed, dual relaxation processes of the FC-excited rhodopsin. This added feature makes Fig. 6 different from the ideas originally suggested by Kandori and coworkers (7, 51) and described in more detail by Mathies and Lugtenburg (41).
Fig. 6.
Double excited-state potential wells for excited rhodopsin. The FC-excited rhodopsin can either undergo diabatic photoisomerization directly to photorhodopsin (process 1) (≈70% of the absorbed quanta) or relaxation to the vibrationally relaxed excited rhodopsin (R*) (process 2) (≈30% of the absorbed light quanta) (7). Processes 3 and 4 are product appearance and decay of R*. R*′ is probably a conical intersecting point (41), thus an undetectable species. (See text for origin of the double potential wells from the unique structure of rhodopsin.) The time scales for the four processes are those of Kandori and coworkers (7, 8) and Mathies and coworkers (50).
Implications of the Current Model for the Rapid, Directed Photoisomerization. Pursuing this specific model for the accelerated isomerization process, we now examine possible ways to enhance the reactivity of the native rhodopsin. We already discussed that replacement of Cys-187 with a bulkier amino acid residue for increased light induced repulsive interaction is an untenable approach (likely to destroy the 3D structure). An obvious alternative is to increase the steric bulk at the H-12 position. Since H-12 and the carbonyl oxygen of Cys-187 are in close contact, any increase of the size of the substituent replacing H-12 would exacerbate the situation. This is probably the cause of the low pigment yields for 12-chloro- and 12-methyl-rhodopsin (14). Would these crowded rhodopsin analogs have a higher quantum yield of isomerization than rhodopsin? Such numbers for these pigment analogs were not reported. However, it should be pointed out that there is a possibility that the quantum yield might be reduced rather than increased, because the increased steric crowding should make the 12-X substituent “tilted” to the side of the carbonyl oxygen in forming the pigment analog. Clearly, only one of the two possible sides will lead to an increased quantum yield of isomerization. The other side is the dead-end channel, leading back to rhodopsin.
The latter line of reasoning suggests a different approach to enhance quantum efficiency of photoisomerization, i.e., to redirect the known ≈30% wasted energy of vertical decay back to the isomerization channel. The solution will have to “move closer” the protein wall on the left side. Ala-117 is the closest amino acid residue in the protein wall. One possibility is to replace this unit with a bulkier one. As it turns out several Ala-117 mutants are in the literature: A117F (59), A117G, A117V, A117I, A117M, and A117W (60), and A117Y (54). The common theme was the similarity of absorption of these mutants compared to that of rhodopsin. A modeling study based on earlier 2D crystal structure suggested that the side chain of A117 mutant is directed away from the chromophore (60). This conclusion is not in disagreement with the current 3D crystal structure that shows that there is a small gap between TM 3 and the chromophore (the closest distance between Ala-117 and C-12 is 3.53 Å). Quantum yield data for the primary photoisomerization of these mutants also were not available. Replacement of other amino acid unit(s) on the “wall” should also be considered. Hence, the issue of possible redistributing the two decay pathways of FC-excited rhodopsin remains an open one.
Such a reshaped wall not only could reapportion the ≈70:30 distribution of productive quanta from the FC-excited rhodopsin but also it might skew the starting chromophore so that the reaction site (X-12) leans into the isomerization channel even before light excitation. It is our hope that the protein chemists will soon be able to provide data showing whether such a mutant will indeed have a higher efficiency of isomerization. A related question will be: whether, e.g., A117G will have a lower quantum yield of photoisomerization.
A combination of retinal analog and protein mutation work will be interesting. At the same time, we caution that an improved quantum yield of isomerization of a mutant does not guarantee a better visual pigment. It only improves the first step of the very complex visual transduction process (39). Nevertheless, here is a thrilling possibility that a manmade visual pigment can partly be better than the native system.
It is of interest to note that the limited quantum yields of photoisomerization reported for different native visual pigments are not identical. Thus, the Kyoto group reported that the quantum yield of chicken rhodopsin is ≈5% larger than that of bovine rhodopsin, whereas that of the chicken iodopsin is ≈6% smaller (61). [The latter cone pigment is known to be more “exposed to the molecular surface than that of rhodopsin” (61, 62).] Unfortunately, crystal structure for the latter two pigments are not available for examination whether the varied reactivities are based on the structural variation of the individual pigment.
Concluding Remarks. A molecular model for the enhanced reactivity of rhodopsin has been proposed based on simple premises of close proximity of Cys-187 to H-12 and displacement of H-12 as a result of π-bond lengthening through light absorption. The conception of this model is not caused by any new experimental results on our part. In fact, in reviewing the literature, it became apparent that much suggestive information has been available for some time. However, the association of bond lengthening in an excited chromophore with rhodopsin photochemical reactivity occurred to us only recently. It was probably prompted by the simultaneous preparation of two papers. In one (30), we attempted to interpret the accumulated F NMR chemical shifts on F-rhodopsins based on recently available crystal structures. And, in the other (63), one of us attempted to understand the accumulated photochemical results of polyenes with the known concepts derived from ultrafast kinetic studies available in recent literature. This sudden realization allowed us to connect groundstate structural properties (closeness of Cys-187) with excitedstate reactivities and to apply our understanding in volume-controlled photoisomerization reactivity in model systems (64).
The above statement, we hope, is a sufficient explanation for the emphasis on our own data in presenting the concepts described in this article. Very likely, we did not give full credit to many research groups that contributed much valuable information on rhodopsin in manners that we can only marvel at and in many ways that we benefited from.
Acknowledgments
R.S.H.L. thanks Professors R. A. Mathies and H. Kandori for the preprints and reprints on fast kinetic studies of rhodopsin and G. S. Hammond for the mechanistic trains of thought instilled in him since his graduate student days at the California Institute of Technology. D. Chang assisted in producing figures of partial rhodopsin crystal structure. The unexpected termination of the long-term support of the U.S. Public Health Services to our retinoid program (Grant DK-17806, 1973–2000) gave R.S.H.L. the additional time to digest accumulated experimental and literature information. The recent support from National Science Foundation Grant CHE-0132250 provided the funding necessary to sustain the isomerization research program at Hawaii.
Abbreviations: BP, bicycle pedal; FC, Franck-Condon; FOS, fluorine opsin shift; HT, hula twist; PSB, protonated Schiff base; TM, transmembrane.
Footnotes
Dr. T. Mirzadegan (Hoffmann–La Roche) first brought to our attention the unique location of Cys-187 (personal communication).
In other words, light absorption causes H-12 to “bang” itself into the protein wall (Cys-187) at lightning speed, a Hawaiian punch (?).
Deuterium isotope effects (at H-11 and H-12) on the early excited-state processes of rhodopsin are in the literature (65). But the effect is relatively small and appears to be rather complex, varying at different time delays.
References
- 1.Wald, G. (1968) Science 162, 232–239. [Google Scholar]
- 2.Kim, J. E., Tauber, M. J. & Mathies, R. A. (2001) Biochemistry 40, 13774–13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, J. E., Tauber, M. J. & Mathies, R. A. (2003) Biophys. J. 84, 2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dartnall, H. (1968) Vision Res. 8, 339–358. [DOI] [PubMed] [Google Scholar]
- 5.Becker, R. (1990) Photochem. Photobiol. 48, 369–399. [DOI] [PubMed] [Google Scholar]
- 6.Koyama, Y., Kubo, K., Komori, M., Yasuda, H. & Mukai, Y. (1991) Photochem. Photobiol. 54, 433–443. [DOI] [PubMed] [Google Scholar]
- 7.Chosrowjan, H., Mataga, N., Shibata, Y., Tachibanaki, S., Kandori, H., Shichida, Y., Okada, T. & Kouyama, T. (1998) J. Am. Chem. Soc. 120, 9706–9707. [Google Scholar]
- 8.Kandori, H., Katsuta, Y., Ito, M. & Sasabe, H. (1995) J. Am. Chem. Soc. 117, 2669–2670. [Google Scholar]
- 9.Liu, R. S. H., Matsumoto, H., Kini, A., Asato, A. E., Denny, M., Kropf, A. & DeGrip, W. J. (1984) Tetrahedron 40, 473–482. [Google Scholar]
- 10.Nakanishi, K. & Crouch, R. (1995) Isr. J. Chem. 35, 253–272. [Google Scholar]
- 11.Liu, R. S. H. & Mirzadegan, T. (1988) J. Am. Chem. Soc. 110, 8617–8623. [Google Scholar]
- 12.Liu, R. S. H. & Asato, A. E. (1990) in Chemistry and Biology of Synthetic Retinoids, eds. Dawson, M. & Okamura, W. H. (CRC, Boca Raton, FL) pp. 51–75.
- 13.Balogh-Nair, V. & Nakanishi, K. (1990) in Chemistry and Biology of Synthetic Retinoids, eds. Dawson, M. & Okamura, W. H. (CRC, Boca Raton, FL) pp. 147–176.
- 14.Liu, R. S. H., Asato, A. E., Denny, M. & Mead, D. (1984) J. Am. Chem. Soc. 106, 8298–8300. [Google Scholar]
- 15.Nakanishi, K. (1991) Pure Appl. Chem. 63, 161–170. [Google Scholar]
- 16.Hirano, T., Lim, I. T., Kim, D. M., Zheng, X.-G., Yoshihara, K., Oyama, Y., Imai, H., Shichida, Y. & Ishiguro, M. (2002) Photochem. Photobiol. 76, 606–615. [DOI] [PubMed] [Google Scholar]
- 17.Lou, J., Tan, Q., Karnaukhova, E., Berova, N., Nakanishi, K. & Crouch, R. (2000) Methods Enzymol. 315, 219–237. [DOI] [PubMed] [Google Scholar]
- 18.Fujimoto, Y., Fishkin, N., Pescitelli, G., Decatur, J., Berova, N. & Nakanishi, K. (2002) J. Am. Chem. Soc. 124, 7294–7302. [DOI] [PubMed] [Google Scholar]
- 19.Yoshizawa, T. & Kandori, H. (1991) Prog. Ret. Res. 11, 33–55. [Google Scholar]
- 20.Shichida, Y., Matuoka, Y. & Yoshizawa, T. (1984) Photobiochem. Photobiophys. 7, 221–228. [Google Scholar]
- 21.Kandori, Y., Shichida, Y. & Yoshizawa, T. (1989) Biophys. J. 56, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizukami, T., Kandori, H., Shichida, Y., Chen, A.-H., Derguini, F., Caldwell, C. G., Bigge, C. F., Nakanishi, K. & Yoshizawa, T. (1993) Proc. Natl. Acad. Sci. USA, 90, 4072–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loppnow, R. G. & Mathies, R. A. (1988) Biophysics 54, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, S. O., Courtin, J., deGroot, H., Gebhard, R. & Lugtenburg, J. (1991) Biochemistry 30, 7409–7415. [DOI] [PubMed] [Google Scholar]
- 25.Gerig, J. T. (1978) Biol. Magn. Reson. 1, 139–203. [Google Scholar]
- 26.Colmenares, L. U., Niemczura, W. P., Asato, A. E. & Liu, R. S. H. (1996) J. Phys. Chem. 100, 9175–9180. [Google Scholar]
- 27.Colmenares, L. U., Asato, A. E. & Liu, R. S. H. (2001) Spectrum 14, 8–12. [Google Scholar]
- 28.Liu, R. S. H., Matsumoto, H., Asato, A. E., Denny, M., Shichida, Y., Yoshizawa, T. & Dahlquist, F. W. (1981) J. Am. Chem. Soc. 103, 7195–7201. [Google Scholar]
- 29.Colmenares, L. U., Zou, X.-L., Liu, J., Asato, A. E., Liu, R. S. H., de Lera, A. & Alvarez, R. (1999) J. Am. Chem. Soc. 121, 5803–5804. [Google Scholar]
- 30.Colmenares, L. U. & Liu, R. S. H. (2003) J. Photosci. 10, 81–87. [Google Scholar]
- 31.Emsley, J. W. & Phillips, L. (1971) Prog. Nucl. Magn. Reson. Spectrosc. 10, 85–756. [Google Scholar]
- 32.Perkins, S. J. (1982) Biol. Magn. Reson. 4, 193–336. [Google Scholar]
- 33.Doddress, D., Wenkert, E. & Demarco, P. (1969) J. Mol. Spectrosc. 32, 162–165. [Google Scholar]
- 34.De Dios, A. C. & Oldfield, E. (1994) J. Am. Chem. Soc. 116, 7453–7454. [Google Scholar]
- 35.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 36.Okada, T., Fujiyoshi, Y., Silow, M., Navarro, J., Landau, E. M. & Shichida, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 5982–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pauling, L. (1960) The Nature of the Chemical Bond (Cornell Univ. Press, Ithaca, NY).
- 38.Ridge, K. D. & Abdulaev, N. G. (2000) Methods Enzymol. 315, 59–70. [DOI] [PubMed] [Google Scholar]
- 39.Pepe, I. M. (1999) J. Photochem. Photobiol. 48, 1–10. [DOI] [PubMed] [Google Scholar]
- 40.Kim, J. E. & Mathies, R. (2002) J. Phys. Chem. A 106, 8508–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathies, R. A. & Lugtenburg, J. (2000) in Handbook of Biological Physics, eds. Stavenga, D. G., deGrip, W. J. & Pugh, E. N., Jr. (Elsevier, Amsterdam), Vol. 3, pp. 55–90. [Google Scholar]
- 42.Warshel, A. & Barboy, N. (1982) J. Am. Chem. Soc. 104, 1469–1476. [Google Scholar]
- 43.Liu, R. S. H. & Asato, A. E. (1985) Proc. Natl. Acad. Sci. USA 82, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, R. S. H. & Hammond, G. S. (2000) Proc. Natl. Acad. Sci. USA 97, 11153–11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imamoto, Y., Kataoka, M. & Liu, R. S. H. (2002) Photochem. Photobiol. 76, 584–589. [DOI] [PubMed] [Google Scholar]
- 46.Asato, A. E., Denny, M. & Liu, R. S. H. (1986) J. Am. Chem. Soc. 108, 5032–5033. [Google Scholar]
- 47.Ishiguro, M. (2000) J. Am. Chem. Soc. 122, 444–451. [Google Scholar]
- 48.Warshel, A. (1976) Nature 260, 679–683. [DOI] [PubMed] [Google Scholar]
- 49.Ishiguro, M., Hirano, T. & Oyama, Y. (2003) ChemBioChem 4, 228–231. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Q., Schoenlein, R. W., Peteanu, L. A., Mathies, R. A. & Shank, C. V. (1994) Science 266, 422–424. [DOI] [PubMed] [Google Scholar]
- 51.Kandori, H., Furutani, Y., Nishimura, S., Shichida, Y., Chosrowjan, H., Shibata, Y. & Mataga, N. (2001) Chem. Phys. Lett. 334, 271–276. [Google Scholar]
- 52.Zewail, A. H. (2000) Angew. Chem. Int. Ed. 39, 2586–2631. [DOI] [PubMed] [Google Scholar]
- 53.Zerbetto, F., Zgierski, M. Z., Negri, F. & Orlandi, G. (1988) J. Chem. Phys. 89, 3681–3688. [Google Scholar]
- 54.Lin, S. W., Han, M. & Sakmar, T. P. (2000) Methods Enzymol. 315, 116–130. [DOI] [PubMed] [Google Scholar]
- 55.Karnik, S. S., Sakmar, T. P., Chen, H.-B. & Khorana, H. G. (1988) Proc. Natl. Acad. Sci. USA, 85, 8459–8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anukanth, A. & Khorana, H. G. (1994) J. Biol. Chem. 269, 19738–19744. [PubMed] [Google Scholar]
- 57.Davidson, F. F., Loewen, P. C. & Khorana, H. G. (1994) Proc. Natl. Acad. Sci. USA 91, 4029–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okano, T., Kojima, D., Fukada, Y., Shichida, Y. & Yoshizawa, T. (1992) Proc. Natl. Acad. Sci. USA 89, 5932–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakayama, T. A. & Khorana, H. G. (1991) J. Biol. Chem., 266, 4269–4275. [PubMed] [Google Scholar]
- 60.Shieh, T., Han, M., Sakmar, T. P. & Smith, S. O. (1997) J. Mol. Biol. 272, 373–384. [DOI] [PubMed] [Google Scholar]
- 61.Okano, T., Fukada, Y., Shichida, Y. & Yoshizawa, T. (1992) Photochem. Photobiol. 56, 995–1001. [DOI] [PubMed] [Google Scholar]
- 62.Matsumoto, H., Tokunaga, F. & Yoshizawa, T. (1975) Biochim. Biophys. Acta 404, 300–308. [DOI] [PubMed] [Google Scholar]
- 63.Liu, R. S. H. & Hammond, G. S. (2003) Photochem. Photobiol. Sci. 2, 835–844. [DOI] [PubMed] [Google Scholar]
- 64.Liu, R. S. H., Asato, A. E. & Denny, M. (1983) J. Am. Chem. Soc. 105, 4829–4830. [Google Scholar]
- 65.Kakitani, T., Akiyama, R., Hatano, Y., Imamoto, Y., Shichida, Y., Verdegan, P. & Lugtenburg, J. (1998) J. Phys. Chem. B 102, 1334–1339. [Google Scholar]