Abstract
The balance between protein acetylation and deacetylation controls several physiological and pathological cellular processes, and the enzymes involved in the maintenance of this equilibrium—acetyltransferases (HATs) and deacetylases (HDACs)—have been widely studied. Presently, the evidences obtained in this field suggest that the dynamic acetylation equilibrium is mostly maintained through the physical and functional interplay between HAT and HDAC activities. This model overcomes the classical vision in which the epigenetic marks of acetylation have only an activating function whereas deacetylation marks have a repressing activity. Given the existence of several players involved in the preservation of this equilibrium, the identification of these complex networks of interacting proteins will likely foster our understanding of how cells regulate intracellular processes and respond to the extracellular environment and will offer the rationale for new therapeutic approaches based on epigenetic drugs in human diseases.
1. Introduction
Lysine acetylation is a reversible and highly regulated posttranslational modification discovered on histones in 1968 [1], but the enzymes responsible for acetyl group addition to or removal from target proteins, known as histone acetyltransferases (HATs) and deacetylases (HDACs), respectively, had not been identified until 1995 [2]. In the past decade, the knowledge about this modification has grown exponentially with targets rapidly expanding from histones to transcription factors and other proteins such as metabolic enzymes and signaling regulators in the cytoplasm. Thus, lysine acetylation has emerged as a major posttranslational protein modification rivaling phosphorylation.
Numerous protein properties are regulated through lysine acetylation, including DNA-protein interaction, subcellular localization, transcriptional activity, stability, and involvement in signaling pathways [3–5]. Besides, the dynamic state of posttranslational protein acetylation is intimately linked to aging and to several major diseases such as cancer, retroviral pathogenesis, neurodegenerative disorders, and cardiovascular diseases [6–8].
At the chromatin level, it has been widely demonstrated that the balance between acetylation and deacetylation of histone and nonhistone proteins plays a pivotal role in the regulation of gene expression. The general model of transcription is based on the interaction among RNA Pol II, general transcription factors, coactivators, corepressors, and sequence-specific DNA-binding proteins (DBPs) [9–11], which confer tissue and signal-dependent specificity. Coactivator and corepressor complexes contain a variety of chromatin-modifying enzymes, including HATs and HDACs.
HATs are classified into two groups, HAT A and HAT B, depending on the mechanism of catalysis and on cellular localization. The members of the HAT A family are found in the nucleus, where they transfer the acetyl group from Acetyl-CoA to an ε-NH2 group of histone N-tails after the assembly into nucleosomes. The HAT A family can be further divided into three subclasses depending on the homology with yeast proteins. Conversely, the members of the HAT B family act in the cytoplasm and transfer the acetyl group from Acetyl-CoA to an ε-NH2 group of free histones prior to their deposition on the DNA (Table 1).
Table 1.
Mammalian Members of HAT Family.
| Mammalian HATs | |||||
| Class | Subclass | Homology to yeast | Mammalian members | Mechanism of catalysis | Cell localization |
| A | GNAT-family | Gcn5 | GCN5L | Transfer of acetyl group from acetyl-CoA to ε-NH2 group of histone N-tails after the assembly into nucleosomes | Nucleus |
| PCAF | |||||
| MYST-family | Esa1; Sas2; Sas3 | Tip60 | |||
| HBOI | |||||
| MORF | |||||
| MOZ | |||||
| CLOCK | |||||
| NCOAT | |||||
| MOF | |||||
| Others | HATI; Elp3; Hpa2; NutI | p300/CBP | |||
| TFIIIC complex | |||||
| ACTR/SRC-I | |||||
| ATF-2 | |||||
| B | Hat1 | HAT1 | Transfer of acetyl group from acetyl-CoA to ε-NH2 group of free histones prior to their deposition on DNA | Cytoplasm | |
HDACs can be grouped into four classes in relation to their phylogenetic conservation [12]. Class I, class II, and class IV, which are related to the yeast Rpd3, Had1, and Hos3 proteins, respectively, encompass the classical family of zinc-dependent HDACs, while class III consists of the NAD+-dependent yeast Sir2 homologues, which comprise the sirtuin family [13, 14] (Table 2).
Table 2.
Mammalian Members of HDAC Family.
| Mammalian HDACs | ||||
| Class | Homology to yeast | Mammalian members | Mechanism of catalysis | Cell localization |
| I | Rpd3 | HDAC1 | Zn2+ ion dependent | Ubiquitous |
| HDAC2 | ||||
| HDAC3 | ||||
| HDAC8 | ||||
| II | Hda1 | HDAC4 | Zn2+ ion dependent | |
| HDAC5 | ||||
| HDAC7 | Shuttle between | |||
| HDAC9 | nucleus and cytoplasm | |||
| HDAC6 | ||||
| HDAC10 | ||||
| III | Sir2 | SIRT1 | NAD+ dependent | Nucleus |
| SIRT2 | Cytoplasm | |||
| SIRT3 | Mitochondria | |||
| SIRT4 | Mitochondria | |||
| SIRT5 | Mitochondria | |||
| SIRT6 | Nucleus | |||
| SIRT7 | Nucleus | |||
| IV | HOS3 | HDAC11 | Zn2+ ion dependent | Nucleus |
In eukaryotes, HATs and HDACs are involved in several aspects of cellular homeostasis. For example, in yeast, the HAT Gcn5 is required for the regulation of various cellular processes such as cell response to stress, meiosis, and DNA replication [15–17]. In mammals, the HAT p300/CBP plays a pivotal role in cell growth, myotube differentiation, and apoptosis [18–20]. Additionally, PCAF, an HAT enzyme originally identified as a p300/CBP-binding protein, is known to play a key role in regulating myofilament contractile activity, the myogenic program, and adipocyte proliferation [21, 22]. The G1-S phase progression in the cell cycle is mediated by class I HDACs; homologous recombination involves members of the sirtuin family; members of HDACs are found in complexes with transcriptional repressors in multipotent neural progenitor; HDACs play a role in the prevention of cytoxicity arising from protein aggregation in neural cells [23–29].
In the last years, various studies showed that HATs and HDACs are both targeted to the transcribed regions of active genes marked by phosphorylated RNA Pol II. These data give further complexity to the general model of gene expression, suggesting that the dynamic cycle of acetylation and deacetylation by the transient binding of HATs and HDACs may poise primed genes for future activation [19, 30, 31].
The present knowledge in this field suggests that the balance between acetyltransferases and deacetylases provides a major contribution to the regulation of cellular functions. Given the key role of this equilibrium in cell physiology and considering that it is lost in various pathological conditions, targeting acetyltransferases and/or deacetylases might represent an effective therapeutic approach for human diseases.
2. Histone Targets of HATs and HDACs: Epigenetic Regulation
Histone modifications, together with factors responsible for adding, interpreting, and removing epigenetic marks, regulate specific responses of the eukaryote genome, and this represents the basis of the “histone code hypothesis.” Indeed, epigenetic marks are sites of recognition for specific readers and effectors. In the case of acetylation marks, certain modified lysines represent specific binding surfaces for bromodomain-containing proteins, which are part of large complexes controlling chromatin architecture. Singular or combinatorial histone modifications impact on chromatin organization and structure. Well-studied examples of this mechanism are the contribution of H4K16 acetylation to the regulation of chromatin structure and the interaction between nonhistone proteins and chromatin fibers [32, 33]. Bromodomain-containing proteins represent a large class of chromatin-associated factors with at least 75 members expressed in humans [34]. Some of them have been identified as part of chromatin-remodeling complexes [35]. Indeed, acetylation of histones H3K4 and H3K14 plays a central role in the recruitment of SWI/SNF chromatin-remodeling complexes and of the general transcription factor TFIID during transcription initiation [36].
Recent studies have proposed various regulatory mechanisms for histone acetylation and deacetylation on gene activity. Several evidences have shown that HAT or HDAC enzymes are stepwise recruited to a specific locus by various types of transcription factors. For example, in vivo experiments have revealed the existence of different kinetics for the accumulation of different components of SWI/SNF remodeling and SAGA-containing HAT complexes at a condensed chromatin locus [37]. In yeast, studies conducted on various genes indicated that multiple chromatin regulators are recruited in a temporal order [38, 39] and that the recruitment of HATs or HDACs depends on the kind of factors involved in the transcriptional program [40].
Other works suggest that these two enzymatic activities are both present simultaneously on the regulatory regions of target genes, and the transcription activation or repression depends on the activation of different pathways and/or the type of enzymes which stabilize these interactions. In mammals, the association of HATs and HDACs in the same complex has been demonstrated to support transcriptional competence during myogenesis and p53-dependent transcription. Based on these data, we have previously proposed an experimental model in which a deacetylase activity is recruited by the C/H3 region of p300 antagonizing p300 functionality [19].
The hypothesis that acetylating and deacetylating enzymes bind simultaneously to regulatory loci also arises from observations on the effects produced by deacetylase inhibitors, which cause general and local histone hyperacetylation in yeast and mammalian cells [41–43]. In human pancreatic and breast cancer cells, the expression of the TGF β type II receptor gene (TβRII) is mediated by modulation of the components present in multiprotein complexes that bind to its promoter. Both the p300 and PCAF acetyltransferases and the HDAC1 deacetylase are potential components of these complexes, and treatment of cells with HDAC inhibitors leads to the recruitment of PCAF and p300, resulting in the activation of the TβRII promoter and in the decrease of the amount of HDAC1 associated to the complexes [41, 44]. Besides, a very recent genomewide analysis carried out in yeast showed that the acetyltransferase Gcn5 colocalizes with one or more HDACs both in ORFs (open reading frames) and IGRs (intergenic regions). Moreover, Gcn5 binds significantly to ORF regions that are hyperacetylated on histones H3K9 and H3K14, which are Gcn5 substrates. In these loci, Gcn5 collaborates antagonistically with the class II histone deacetylase Clr3 to modulate acetylation levels and transcriptional elongation. These data suggest the existence of a functional link between HATs and HDACs in regulating the balance of histone acetylation [45]. Due to the similarity between yeast HATs and HDACs and mammalian complexes, these results are likely to be relevant also in mammals.
Taken together, these data support an epigenetic model in which the activity of HATs and HDACs and the position of acetylated or deacetylated histones within genes play a major role in gene regulation.
3. Nonhistone Targets of HATs and HDACs: The Acetylome
The cellular and physiological functions of lysine acetylation are not limited to the regulation of gene expression. Lysine acetylation assumes a wider significance in many physiological processes, as it also targets nonhistone proteins. Following the identification of additional localizations of HATs and HDACs in other cell compartments, a search for new targets has begun with the aim of determining potential novel biologic functions of these enzymes.
A proteomic analysis of lysine acetylation has identified 388 acetylation sites in 195 proteins derived from HeLa cells and mouse liver mitochondria proving a potential link between acetylation and mitochondrial function. Among nonhistone proteins, the authors found RNA splicing factors (HnRNPA1), chaperones (Hsp70, Hsp27, and Hsp90), structural proteins (actin and tropomyosin), signaling proteins (phospholipase Cβ1 and annessin V) and also proteins involved in energy metabolism, and longevity-related mitochondrial proteins [4]. These new targets for the activity of HATs and HDACs comprise the so-called “Acetylome”.
Additionally, in a very recent work, high-resolution mass spectrometry was used to identify new lysine acetylation sites and evaluate acetylome changes after the inhibition of HDACs [4]. In this study, Choudhary and colleagues used the SILAC (stable-isotope labeling by amino acid in cell culture) technology coupled with an LTQ Orbitrap mass spectrometer. By labeling cellular proteomes with isotopes of different molecular weight, SILAC allows simultaneous quantification of specific acetylated peptides of mixed proteomes prepared under different experimental conditions with a reported false-discovery rate of only 0.1 to 0.3%. This strategy revealed that the acetylation pattern is conserved in cells derived from different tissue types and that acetylation preferentially targets large macromolecular complexes involved in several major nuclear processes, such as cell cycle-associated chromatin remodeling (SWI-SNF and methyltransferases complexes), protein turnover (the BRE1A and BRE1B ubiquitin ligases, the USP14 and Ubch37 deubiquitylases), and DNA damage and repair (phosphoinositide 3 kinase-related protein kinases [PIKKs]). In addition, since HDACs are common targets in cancer and neurodegenerative diseases, the authors characterized acetylome changes in response to HDACs inhibition at a global level. Using two different inhibitors (suberoylanilide hydroxamic acid [SAHA] and MS-275), they showed that the increase in acetylation was not equal in all histone sites and nonhistone proteins. This observation suggests that the activity of these inhibitors is highly specific to particular HDAC members; thus, a global understanding of these processes could reveal an unexpected clinical specificity of HDACs inhibitors.
Several evidences suggest that the acetylation balance is also very important for cell viability. Indeed, it has been shown that this balance (I) controls the stability of various proteins such as p53 [46], β-catenin [47], and SMAD7 [48], thereby modulating the signaling pathways in which these proteins are involved [49, 50]; (II) plays a key role in DNA replication, recombination, and repair by regulating the stability of WRN, a multifunctional protein responsible for these processes [51]; (III) regulates proteins involved in nucleocytoplasmic shuttling, such as Importin α, or in translocation to the nucleus [52]; (IV) suppresses toxic protein aggregation through the interaction between HDACs (HDAC4, HDAC6, SIRT2) and the members of a subclass of the DNAJB family or members of heat shock proteins (HSP90) which are known to counteract protein misfolding and aggregation associated with cytoxicity and what is mentioned in [27, 53–55].
As observed for histone acetylation/deacetylation, the dynamic balance in the acetylation of nonhistone proteins seems to be maintained by a physical and functional interplay between HAT and HDAC activities. Indeed, we reported that deacetylase inhibitors (DIs) could enhance the autoacetylation activity of p300 immunoprecipitated from nuclear extracts, but not that of the same purified recombinant enzyme [19], indicating that the presence of HDAC in p300 multiprotein complexes could also affect nonhistone targets. Subsequently, several studies have identified a similar mechanism for other members of the HAT and HDAC families. An example is the interaction between p300 and Sirt2, for which a model has been proposed stating that p300 indirectly increases the transcriptional activity of p53 through acetylation and subsequent attenuation of the deacetylase function of Sirt2. The existence of this interaction network suggests that the transcriptional activation mediated by the p300 coactivator is not regulated solely through epigenetic modification of histones and transcription factors. Indeed, the direct interplay between the opposing enzymatic activities of HATs and HDACs also seems to play a nodal role in this model, and their ability to regulate each other's activity appears involved in the control of common targets. In support of this hypothesis, it has been demonstrated that acetylation of Sirt2 by p300 attenuates α-tubulin deacetylation by Sirt2 [56]. Consistent with this idea, a recent study has shown that p300 can inactivate HDAC6 affecting its ability to interact with other signaling modulators [57]. One of these is Hsp90, whose interaction with HDAC6 is functional to the regulation of chaperone-dependent activation of the glucocorticoid receptor [58]. Taken together, these data imply a new function for p300 and other members of the HAT family, which is opposed to their well-characterized positive regulatory effect, suggesting that they can also play a negative regulatory role on target proteins.
4. Acetylation Balance at the Crossroad of Cell Proliferation and Differentiation
The maintenance of an undifferentiated state requires that chromatin architecture sustains the silencing of target genes involved in lineage progression. This implies an acetylation balance strongly shifted towards deacetylation. The opposite occurs during lineage progression, when these genes need to be activated, and thus the balance must be weighted towards acetylation. Any modification of this equilibrium will interfere with the proper execution of the proliferating/differentiating program and may contribute to the development of a pathologic condition. Hence, HATs and HDACs play a pivotal role in the differentiation/proliferation balance of several cells and tissues.
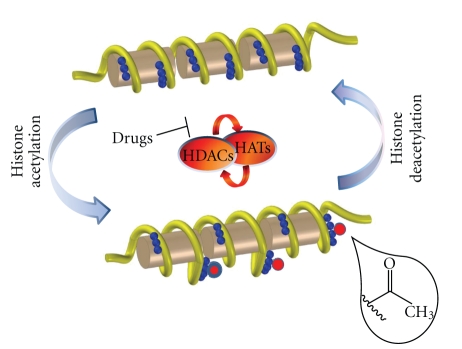
As described above, several studies conducted to decipher histone acetylation and deacetylation dynamics suggested that the simultaneous presence of HATs and HDACs and their physical interaction play a key role in the regulation of the acetylation balance [59–61] (Figure 1). Increasing evidences indicate that the interaction between HATs and HDACs occurs in a dynamic fashion depending on the physiological conditions of the cell. Thus, acetylation homeostasis has to be considered intimately linked to cell homeostasis, and global changes in epigenetic modulators are important in the genetic reprogramming during cell proliferation or differentiation. Several examples of the role of writers and eraser of acetylation marks in these processes have been unveiled. Gcn5 and HDAC1 form a complex in mammalian cells, and their dynamic interaction is influenced by physiological processes such as cell differentiation. Indeed, treatment with TPA (phorbol ester tetradecanoyl phorbol acetate), which is known to induce differentiation, causes the replacement of Gcn5 with PCAF [59].
Figure 1.
Physical and Functional interaction between HATs and HDACs regulates gene expression; HATs and HDACs enzymes are simultaneously present on the regulatory regions of target genes, and their opposing activities play a pivotal role for transcriptional competence. The use of selective HDAC inhibitors allows to restore the acetylation balance lost in several pathological conditions.
The interplay between HDACs and HATs is also linked to adipocyte differentiation. Downregulation of HDAC1 activity results in preferential histone hyperacetylation at the promoter regions of adipocyte marker genes. Specifically, HDAC1 directly interacts with PPARγ, the master adipogenic factor, and represses its transcriptional activity. Thus, the downregulation of HDAC1 promotes PPARγ activity by relieving it from repression. A very similar mechanism occurs during osteoblast differentiation suggesting that the modulation of HDAC expression and activity may be a general way of regulating cell differentiation [62].
Additionally, a recent work showed that neuronal outgrowth is driven by intrinsic and extrinsic factors ultimately affecting the balance between HAT and HDAC activities. Indeed, the addition of TSA leads to hyperacetylation of specific proneuronal outgrowth gene promoters. This suggests the presence of a positive feedback loop initiated by the relative increase in acetyltransferase activity through HDAC inhibition. This leads to histone hyperacetylation and activation of the CBP, p300, and PCAF promoters. p300/CBP and PCAF in turn promote p53 acetylation which plays a key role in neuronal outgrowth [63].
The genetic reprogramming driving neuronal and ologodendrocyte lineage progression depends on the interplay between pluripotency-associated factors and epigenetic modulators. Thus, the acetylation balance plays a pivotal role in this process together with the histone trimethylation pattern. Several works showed that adult multipotent neural progenitor cells differentiated predominantly into neurons in the presence of the HDAC inhibitor valproic acid (VPA). VPA treatment also actively suppressed glial differentiation, even in conditions favoring lineage-specific differentiation [64, 65]. Moreover, the progressive restriction of cell lineage during differentiation from multipotent neural stem cells to oligodendrocyte progenitors (OPCs) is characterized by the progressive decrease of genes such as Sox2 (pluripotency-associated factor) and chromatin modifications on astrocytic and neuronal genes that are initiated by the activity of HDACs and are antagonized by Brca1 and Brm [66, 67]. The alteration of the HAT/HDAC balance can revert committed progenitors to multipotent cells displaying Sox2 expression [68].
A very recent study proposed a critical role in the differentiation of neural precursor cells for MRG15, a chromodomain-containing nuclear protein. The authors found that Mrg15-deficient neuronal precursor cells exhibit differentiation defects in addition to growth defects, suggesting the presence of a common pathway for HAT/HDAC activity modulation [69]. Interestingly, MRG15 associates in complexes both with the HAT Tip60 and with mSin3 and HDACs [70–75].
Besides, HDACs and HATs are also implicated in the regulation of E2F-responsive genes that control cell cycle progression. These genes are repressed by the coordinated activity of HDAC and the retinoblastoma protein, whose association requires the recruitment of HAT-TRRAP (an ATM-related protein) [76].
The simultaneous presence of acetyltransferases and deacetylases on regulatory regions of certain genes might explain the rapid changes occurring in promoter acetylation that drive the regulation of genes whose expression fluctuates rapidly (e.g., p21). This hypothesis arises from our recent work in which we showed the contribution of HDAC-HAT interaction to MyoD- and p53-dependent transcription [19]. The myogenic program is mediated by the MRF family of transcription factors—MyoD, Myf-5, myogenin, and MFR-4—which act sequentially to regulate the expression of genes involved in the early phase of determination and in the late differentiation phase. MyoD, the best studied MRF, is regulated by a dynamic flow of acetylation and deacetylation that influences its DNA binding ability [77–80].
How does this HAT-HDAC flow work? We have proposed a model in which transcriptional competence is conferred by the physical interaction between the MyoD transcription factor and HATs or HDACs [19]. In precommitted myoblasts, MyoD is expressed but inactive, because it is complexed with HDACs [78, 81]. The replacement of HDAC1 by PCAF helps MyoD, to drive differentiation by conferring transcriptional competence [19, 79]. Indeed, PCAF acetylates MyoD and the acetyl marks mediate the recruitment of a bromodomain protein such as p300/CBP. Then, PCAF and p300/CBP coordinately acetylate lysine residues in the N-terminal tails of nucleosomal histones [82–85]. We have recently completed the picture by identifying the two signals that influence the composition of the muscle-specific transcriptome: p38MAPK, which is required for the recruitment of SWI-SNF, and Akt, which is involved in HDAC displacement and HAT recruitment and function [80].
The equilibrium between HATs and HDACs is also a nodal point in cell proliferation processes. It is well known that p300/CBP are involved in cell cycle control by regulating the transition from the G1 to the S phase. Indeed, cells lacking p300 activity display proliferation defects [86, 87]. The HAT activity of p300 is regulated by an intricate network of interactions between sumoylation and deacetylation epigenetic marks. A domain named CRD1 (Cell Cycle Regulatory Domain 1) consisting of two tandem SUMO modification sites has been found in p300/CBP proteins. The addition of SUMO to this domain is necessary for HDAC6 recruitment, thus promoting the transcriptional repressor activity of p300 [81]. This interaction mechanism might explain how a single enzymatic activity can both activate and repress transcription.
As described above, the reciprocal interplay between HATs and HDACs regulates various physiological cell processes; thus identification of the actors involved in the preservation of their equilibrium is highly desirable.
5. Epigenetic Drugs: A Matter of Acetylation Balance
The epigenetic etiology of many human diseases has led to the development of “epigenetic” therapies. As discussed above, the acetylation balance of chromatin regulates cell determination and cell fate suggesting that epigenetic drugs could prove useful for the treatment of muscle diseases, neurodegenerative disorders, and cancer. In the last decade, we provided several evidences in vitro and in vivo indicating that DIs could be a valid tool for pharmacological interventions in muscle dystrophies [19, 42, 88–90].
Deregulation of the equilibrium between HATs and HDACs has also been detected in several cancer types. The first evidences on how this balance is compromised in cancer cells derive from studies on the pathogenesis of leukemias [7].
In acute myeloid leukemia (AML) cells, various HDAC inhibitors have been used—including valproic acid (VPA), benzamide derivative (MS275), and suberoylanide hydroxamic acid (SAHA)—showing an anticancer action mediated by the expression of the tumor death ligand TRAIL and p21 [91]. In acute promyelocytic leukaemia (APL), which is characterized by chromosomal rearrangements leading to fusion proteins that involve the retinoic acid receptor (RAR), the fusion proteins maintain the ability to bind genes responsive to retinoic acid (RA), while exerting a modified biological function. In normal cells, the physiological concentration of RA induces the displacement of HDACs from RARs leading to their replacement with HATs at RA-regulated genes. In APL cells, the physiological concentration of RA is not sufficient to achieve this effect. In this case, the simultaneous treatment with RA and HDAC inhibitors is efficient in restoring the correct activation of RA target genes [92].
At present, a promising strategy to reverse aberrant epigenetic changes associated with cancer is based on the use of HDAC inhibitors. Indeed, it has been demonstrated that HDAC inhibition induces proliferation arrest, differentiation, and apoptosis of cancer cells in culture and in animal models [93, 94].
The involvement of several enzymatic activities in cell transformation has stimulated the development of combinatory therapies. Cancer is characterized by the loss of cell cycle check points, and recent studies have identified an important cross talk between proteins involved in the cell cycle regulatory apparatus (Cdks) and proteins regulating histone acetylation. This observation suggests that the combined therapy with agents targeting both the acetylation balance and the Cdks might prove effective [95].
The treatment with HDAC inhibitors is beneficial also in B-cell lymphomas, in which the pathogenesis is caused by the deregulation of the BCL6 proto-oncogene. BCL6 is negatively regulated by p300 acetylation, which disrupts its ability to recruit HDACs, and it has been shown that pharmacological inhibition of HDAC activity causes the accumulation of the inactive acetylated form of BCL6 leading to cell cycle arrest and apoptosis of B-cell lymphoma cells [96].
DIs are a potential arm also in neurodegenerative disorders. Indeed, recent works have revealed that inhibition of HDACs ameliorates the cognitive and motor deficits characteristic of Huntington's, Parkinson's, and Alzheimer's diseases (HD, PD, and AD). A common theme in these neurodegenerative disorders is the concept that intraneuronal aggregates such as plaques interfere with transcription and cause deficits in plasticity and cognition [97]. Therefore, if these aggregates interact with HAT/HDAC complexes, it might be possible to use epigenetic drugs for countering degeneration. For example, in PD α-synuclein mutated proteins aggregate in the nucleus and inhibit HAT-mediated acetyltransferase activity, thereby promoting neurotoxicity. In this case, HDAC inhibition is able to rescue α-synuclein-induced toxicity in vivo or in vitro [98]. In HD, nuclear translocation of mutated huntingtin proteins enhances ubiquitination and degradation of CBP through proteasome activity [99]. Neurodegeneration-coupled HAT activity loss is a molecular event that also characterizes AD; in this condition, the Presenilin1-dependent epsilon-cleavage product N-Cad/CTF2 binds to CBP and facilitates its proteasomal degradation [100].
Several regulating pathways, biological targets, and/or interactors of HATs/HDACs have been identified to date. This knowledge might be taken advantage of to develop therapeutic strategies based on the use of HDAC inhibitors in conjunction with other agents to obtain synergistic results.
6. Conclusion
The identification of a large number of acetylated targets has uncovered new players involved in the acetylation balance. Nevertheless, it must be considered that acetyltransferases and deacetylases act primarily in protein complexes containing multiple cofactors and other enzymes responsible for a variety of posttranslational modifications and that cell processes are driven by the coordinated action of such complexes. The presence of one or another enzyme in a multiprotein complex is determined by signaling pathways activated by different external stimuli. Thus, a better understanding of the players involved in the response to these stimuli might allow specific pharmacological interventions aimed at preserving the physiological equilibrium of acetylation.
Acknowledgments
The authors thank Dr. Francesco Paolo Jori for his helpful discussion during the preparation of the paper and editorial assistance. This work was partially supported by a “My First Grant” (to C. Simone) from the Italian Association for Cancer Research.
References
- 1.Vidali G, Gershey EL, Allfrey VG. Chemical studies of histone acetylation. The distribution of epsilon-N-acetyllysine in calf thymus histones. Journal of Biological Chemistry. 1968;243(24):6361–6366. [PubMed] [Google Scholar]
- 2.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast historic H4 acetyltransferase. Journal of Biological Chemistry. 1995;270(42):24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 3.Spange S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. International Journal of Biochemistry and Cell Biology. 2009;41(1):185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 5.Kim SC, Sprung R, Chen Y, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 6.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minucci S, Nervi C, Lo Coco F, Pelicci PG. Histone deacetylases: a common molecular target for differentiation treatment of acute myeloid leukemias? Oncogene. 2001;20(24):3110–3115. doi: 10.1038/sj.onc.1204336. [DOI] [PubMed] [Google Scholar]
- 8.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death and Differentiation. 2006;13(4):539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224(5216):234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 10.Matsui T, Segall J, Weil PA, Roeder RG. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. Journal of Biological Chemistry. 1980;255(24):11992–11996. [PubMed] [Google Scholar]
- 11.Segall J, Matsui T, Roeder RG. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. Journal of Biological Chemistry. 1980;255(24):11986–11991. [PubMed] [Google Scholar]
- 12.Gregoretti IV, Lee Y-M, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. Journal of Molecular Biology. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 13.De Ruijter AJM, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochemical Journal. 2003;370(3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aiging, and calorie restriction. Genes and Development. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 15.Xue-Franzén Y, Johnsson A, Brodin D, Henriksson J, Bürglin TR, Wright APH. Genome-wide characterisation of the Gcn5 histone acetyltransferase in budding yeast during stress adaptation reveals evolutionarily conserved and diverged roles. BMC Genomics. 2010;11(1, article no. 200) doi: 10.1186/1471-2164-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(12):6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinosa MC, Rehman MA, Chisamore-Robert P, Jeffery D, Yankulov K. GCN5 is a positive regulator of origins of DNA replication in Saccharomyces cerevisiae. PLoS ONE. 2010;5(1, article no. e8964):1–11. doi: 10.1371/journal.pone.0008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartorelli V, Puri PL. The link between chromatin structure, protein acetylation and cellular differentiation. Frontiers in Bioscience. 2001;6:D1024–D1047. doi: 10.2741/sartorel. [DOI] [PubMed] [Google Scholar]
- 19.Simone C, Stiegler P, Forcales SV, et al. Deacetylase recruitment by the C/H3 domain of the acetyltransferase p300. Oncogene. 2004;23(12):2177–2187. doi: 10.1038/sj.onc.1207327. [DOI] [PubMed] [Google Scholar]
- 20.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. Journal of Cell Science. 2001;114(13):2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 21.Gupta MP, Samant SA, Smith SH, Shroff SG. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. Journal of Biological Chemistry. 2008;283(15):10135–10146. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao YS, Hubbert CC, Yao TP. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. Journal of Biological Chemistry. 2010;285(15):11219–11226. doi: 10.1074/jbc.M109.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(14):7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316(5827):1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi T, Cubizolles F, Zhang Y, et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes and Development. 2010;24(5):455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhl M, Csernok A, Aydin S, Kreienberg R, Wiesmüller L, Gatz SA. Role of SIRT1 in homologous recombination. DNA Repair. 2010;9(4):383–393. doi: 10.1016/j.dnarep.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Hageman J, Rujano MA, van Waarde MAWH, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Molecular Cell. 2010;37(3):355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nature Neuroscience. 1999;2(10):867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 29.Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes and Development. 2000;14(1):55–66. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger SL. Histone modifications in transcriptional regulation. Current Opinion in Genetics and Development. 2002;12(2):142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 32.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 33.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Current Opinion in Cell Biology. 2003;15(2):172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 34.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 35.Simone C. SWI/SNF: the crossroads where extracellular signaling pathways meet chromatin. Journal of Cellular Physiology. 2006;207(2):309–314. doi: 10.1002/jcp.20514. [DOI] [PubMed] [Google Scholar]
- 36.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111(3):381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 37.Memedula S, Belmont AS. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Current Biology. 2003;13(3):241–246. doi: 10.1016/s0960-9822(03)00048-4. [DOI] [PubMed] [Google Scholar]
- 38.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Molecular Cell. 2002;10(2):227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 39.Robert F, Pokholok DK, Hannett NM, et al. Global position and recruitment of HATs and HDACs in the yeast genome. Molecular Cell. 2004;16(2):199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nature Genetics. 2002;31(3):248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 41.Park SH, Lee SR, Kim BC, et al. Transcriptional regulation of the transforming growth factor β type II receptor gene by histone acetyltransferase and deacetylase is mediated by NF-Y in human breast cancer cells. Journal of Biological Chemistry. 2002;277(7):5168–5174. doi: 10.1074/jbc.M106451200. [DOI] [PubMed] [Google Scholar]
- 42.Iezzi S, Di Padova M, Serra C, et al. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Developmental Cell. 2004;6(5):673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- 43.Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91(7):1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 44.Huang W, Zhao S, Ammanamanchi S, Brattain M, Venkatasubbarao K, Freeman JW. Trichostatin A induces transforming growth factor β type II receptor promoter activity and acetylation of Sp1 by recruitment of PCAF/p300 to a Sp1·NF-Y complex. Journal of Biological Chemistry. 2005;280(11):10047–10054. doi: 10.1074/jbc.M408680200. [DOI] [PubMed] [Google Scholar]
- 45.Johnsson A, Durand-Dubief M, Xue-Franzén Y, Rönnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Reports. 2009;10(9):1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408(6810):377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 47.Ge X, Jin Q, Zhang F, Yan T, Zhai Q. PCAF Acetylates β-catenin and improves its stability. Molecular Biology of the Cell. 2009;20(1):419–427. doi: 10.1091/mbc.E08-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonsson M, Heldin CH, Ericsson J, Grönroos E. The balance between acetylation and deacetylation controls Smad7 stability. Journal of Biological Chemistry. 2005;280(23):21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- 49.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90(2):306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 50.Caron C, Boyault C, Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in control of protein stability. BioEssays. 2005;27(4):408–415. doi: 10.1002/bies.20210. [DOI] [PubMed] [Google Scholar]
- 51.Li K, Wang R, Lozada E, Fan W, Orren DK, Luo J. Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. PLoS ONE. 2010;5(4, article no. e10341):1–11. doi: 10.1371/journal.pone.0010341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bannister AJ, Miska EA, Görlich D, Kouzarides T. Acetylation of importin-α nuclear import factors by CBP/p300. Current Biology. 2000;10(8):467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 53.Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26(37):5468–5476. doi: 10.1038/sj.onc.1210614. [DOI] [PubMed] [Google Scholar]
- 54.Boyault C, Zhang Y, Fritah S, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes and Development. 2007;21(17):2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthias P, Yoshida M, Khochbin S. HDAC6 a new cellular stress surveillance factor. Cell Cycle. 2008;7(1):7–10. doi: 10.4161/cc.7.1.5186. [DOI] [PubMed] [Google Scholar]
- 56.Han Y, Jin YH, Kim YJ, et al. Acetylation of Sirt2 by p300 attenuates its deacetylase activity. Biochemical and Biophysical Research Communications. 2008;375(4):576–580. doi: 10.1016/j.bbrc.2008.08.042. [DOI] [PubMed] [Google Scholar]
- 57.Han Y, Jeong HM, Jin YH, et al. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochemical and Biophysical Research Communications. 2009;383(1):88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]
- 58.Kovacs JJ, Murphy PJM, Gaillard S, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Molecular Cell. 2005;18(5):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 59.Yamagoe S, Kanno T, Kanno Y, et al. Interaction of histone acetylases and deacetylases in vivo. Molecular and Cellular Biology. 2003;23(3):1025–1033. doi: 10.1128/MCB.23.3.1025-1033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Covault J, Chalkley R. The identification of distinct populations of acetylated histone. Journal of Biological Chemistry. 1980;255(19):9110–9116. [PubMed] [Google Scholar]
- 61.Davie JR. Covalent modifications of histones: expression from chromatin templates. Current Opinion in Genetics and Development. 1998;8(2):173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 62.Eung JY, Chung JJ, Sung SC, Kang HOK, Jae BK. Down-regulation of histone deacetylases stimulates adipocyte differentiation. Journal of Biological Chemistry. 2006;281(10):6608–6615. doi: 10.1074/jbc.M508982200. [DOI] [PubMed] [Google Scholar]
- 63.Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, Di Giovanni S. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death and Differentiation. 2010;17:1392–1408. doi: 10.1038/cdd.2009.216. [DOI] [PubMed] [Google Scholar]
- 64.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(47):16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Current Opinion in Genetics and Development. 2004;14(5):461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 66.Liu A, Han YR, Li J, et al. The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. Journal of Neuroscience. 2007;27(27):7339–7343. doi: 10.1523/JNEUROSCI.1226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes and Development. 2004;18(23):2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen S, Sandoval J, Swiss VA, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nature Neuroscience. 2008;11(9):1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen M, Takano-Maruyama M, Pereira-Smith OM, Gaufo GO, Tominaga K. MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. Journal of Neuroscience Research. 2009;87(7):1522–1531. doi: 10.1002/jnr.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai Y, Jin J, Florens L, et al. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. Journal of Biological Chemistry. 2005;280(14):13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 71.Cai Y, Jin J, Tomomori-Sato C, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. Journal of Biological Chemistry. 2003;278(44):42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 72.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Molecular and Cellular Biology. 2004;24(5):1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hayakawa T, Ohtani Y, Hayakawa N, et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes to Cells. 2007;12(6):811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 74.Sardiu ME, Cai Y, Jin J, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and transducin-like enhancer of split. Molecular and Cellular Biology. 2002;22(22):7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassilev A, Yamauchi J, Kotani T, et al. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Molecular Cell. 1998;2(6):869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 77.Puri PL, Iezzi S, Stiegler P, et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Molecular Cell. 2001;8(4):885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 78.Luo J, Nikolaev AY, Imai SI, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 79.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO Journal. 2001;20(7):1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serra C, Palacios D, Mozzetta C, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Molecular Cell. 2007;28(2):200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Girdwood D, Bumpass D, Vaughan OA, et al. p300 transcriptional repression is mediated by SUMO modification. Molecular Cell. 2003;11(4):1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 82.Sartorelli V, Puri PL, Hamamori Y, et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Molecular Cell. 1999;4(5):725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 83.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annual Review of Biochemistry. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 84.Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Molecular and Cellular Biology. 2001;21(16):5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Puri PL, Sartorelli V, Yang XJ, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Molecular Cell. 1997;1(1):35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 86.Ait-Si-Ali S, Polesskaya A, Filleur S, et al. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19(20):2430–2437. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 87.Yao TP, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 88.Minetti GC, Colussi C, Adami R, et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nature Medicine. 2006;12(10):1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 89.Iezzi S, Cossu G, Nervi C, Sartorelli V, Puri PL. Stage-specific modulation of skeletal myogenesis by inhibitors of nuclear deacetylases. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(11):7757–7762. doi: 10.1073/pnas.112218599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guasconi V, Puri PL. Epigenetic drugs in the treatment of skeletal muscle atrophy. Current Opinion in Clinical Nutrition and Metabolic Care. 2008;11(3):233–241. doi: 10.1097/MCO.0b013e3282fa1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nebbioso A, Clarke N, Voltz E, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nature Medicine. 2005;11(1):77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 92.He LZ, Tolentino T, Grayson P, et al. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. Journal of Clinical Investigation. 2001;108(9):1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. Journal of the National Cancer Institute. 2000;92(15):1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 94.Vigushin DM, Coombes RC. Histone deacetylase inhibitors in cancer treatment. Anti-Cancer Drugs. 2002;13(1):1–13. doi: 10.1097/00001813-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 95.Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Histone acetylation and the cell-cycle in cancer. Frontiers in Bioscience. 2001;6:D610–D629. doi: 10.2741/1wang1. [DOI] [PubMed] [Google Scholar]
- 96.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nature Genetics. 2002;32(4):606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 97.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kontopoulos E, Parvin JD, Feany MB. α-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Human Molecular Genetics. 2006;15(20):3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 99.Haibing J, Nucifora FC, Ross CA, DeFranco DB. Cell death triggered by polyglutamine-expanded huntingtin in a neuronal cell line is associated with degradation of CREB-binding protein. Human Molecular Genetics. 2003;12(1):1–12. doi: 10.1093/hmg/ddg002. [DOI] [PubMed] [Google Scholar]
- 100.Marambaud P, Wen PH, Dutt A, et al. A CBP binding transcriptional repressor produced by the PS1/ ε-cleavage of N-Cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114(5):635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]