Abstract
Mitochondrial evolution has given rise to a complex array of organelles, ranging from classical aerobic mitochondria to mitochondrial remnants known as hydrogenosomes and mitosomes. The latter are found in anaerobic eukaryotes, and these highly derived organelles often retain only scant evidence of their mitochondrial origins. Intermediate evolutionary stages have also been reported as facultatively or even strictly anaerobic mitochondria, and hydrogenosomes that still retain some mitochondrial features. However, the diversity among these organelles with transitional features remains rather unclear and barely studied. Here, we report the sequence, structure, and gene content of the mitochondrial DNA of the anaerobic stramenopile Proteromonas lacertae. It has a linear genome with a unique central region flanked by two identical large inverted repeats containing numerous genes and “telomeres” with short inverted repeats. Comparison with the organelle genome of the strictly anaerobic human parasite Blastocystis reveals that, despite the close similarity of the sequences, features such as the genome structure display striking differences. It remains unclear whether the virtually identical gene repertoires are the result of convergence or descent.
Keywords: stramenopile, hydrogenosome, mitosome, mitochondrion, linear genome, Proteromonas
Introduction
One of the key features defining the eukaryotic cell is the presence of a series of organelles in the cytosolic compartment. Some of these are restricted to certain organisms, such as the chloroplasts, whereas others seem to be universally distributed, although with a large degree of variation and secondary modifications. The latter is the case for the mitochondria, organelles originating in a single event of acquisition of an alpha-proteobacterium by the ancestor of all the known eukaryotes. Although the majority of eukaryotes carry classical aerobic mitochondria capable of carrying out electron transport and adenosine triphosphate (ATP) generation not all mitochondria conform to this textbook view (Tielens and van Hellemond 2007), and many organisms, mainly unicellular, have undergone a secondary reduction of their mitochondria as a result of adaptation to anaerobiosis. The Archezoa hypothesis postulated that some of the “amitochondriates” were primitive eukaryotes, diverging prior to the establishment of the mitochondrial symbiosis (Cavalier-Smith 1983, 1998); however, we now know that none of those organisms have completely lost their mitochondria but instead carry highly derived organelles of mitochondrial origin that lack an organelle genome (Embley et al. 2003; Hrdý et al. 2004; Gray 2005; van der Giezen 2009). Such organisms include the parasitic trichomonad and diplomonad flagellates (Lindmark and Müller 1973; Tovar et al. 2003), anaerobic ciliates (Yarlett et al. 1981) and fungi (Yarlett et al. 1986), enteric and anaerobic amoebae (Tovar et al. 1999; Gill et al. 2007), and certain apicomplexa (Riordan et al. 2003) and microsporidia (Williams et al. 2002). Traditionally, such organelles have been classified into two categories according to their capacity to generate energy as a result of their metabolism. Those producing H2 and ATP are known as hydrogenosomes, the rest are called mitosomes. The more recent discovery in some anaerobic eukaryotes, such as Blastocystis sp. and Nyctotherus ovalis, of organelles retaining a genome has allowed us to confirm a direct link with aerobic mitochondria (Matsumoto et al. 1987; Nasirudeen and Tan 2004; Boxma et al. 2005; van der Giezen et al. 2005). Of particular interest are the organelles found in the human parasitic stramenopile (syn. heterokont) Blastocystis because they show a mixture of features shared with true mitochondria (presence of cristae, transmembrane potential, and DNA) and with hydrogenosomes and mitosomes (lack of cytochrome-mediated electron transport [Zierdt 1986] and presence of a hydrogenase [Stechmann et al. 2008]). Sequencing of the genome of this organelle (Pérez-Brocal and Clark 2008; Wawrzyniak et al. 2008) revealed the absence of all genes encoding components of complexes III and IV of the electron transport chain as well as the F0–F1-ATPase. Despite these losses, the set of genes and metabolic pathways retained by the Blastocystis mitochondrion-like organelle (MLO) (Stechmann et al. 2008) point to a less degenerate organelle when compared with typical hydrogenosomes and mitosomes. Therefore, this organelle is being classified as an obligately anaerobic mitochondrion.
Blastocystis is an atypical stramenopile with a highly derived morphology, so it is possible that the unusual features of its organellar genome are unique to this organism. Because no organelles in organisms specifically and closely related to Blastocystis have been studied so far, the answer to the question of its uniqueness remained unclear.
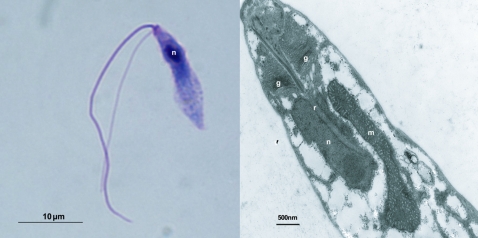
Proteromonas lacertae is an obligately anaerobic stramenopile that lives as a commensal in the posterior intestinal tract of lizards. The sequence of its small subunit ribosomal RNA (SSU rRNA) gene identifies it as a member of the Opalinata and closely related to Blastocystis (Kostka et al. 2004; Hoevers and Snowden 2005) but P. lacertae morphology is much less derived because it retains typical features defining the stramenopiles that have been lost by Blastocystis such as flagella (fig. 1A) and tripartite hair-like structures known as mastigonemes. Adjacent to the nucleus it has a single large mitochondrion with several lobes, tubular cristae, and a sometimes very dense matrix (fig. 1B). DNA has not been reported.
FIG. 1.—
Structure of Proteromonas lacertae. Left panel: Light micrograph of P. lacertae. The Giemsa-stained cell has a size of ∼13 × 3 μm. The anterior of the cell bears two flagella, one thicker and longer than the other. The single nucleus (n) is visible at the anterior pole of the cell. Right panel: Transmission electron micrograph of P. lacertae. This section of the anterior of the cell shows the rhizoplast (r) passing through the Golgi apparatus (g) and into the nucleus (n). The single large mitochondrion (m), in which cristae are visible, is adjacent to the nucleus. For a more detailed description see Brugerolle and Mignot (1990).
The relative ease with which P. lacertae can be grown axenically makes it a good candidate for investigation and thus for gaining insight into the origin of the Blastocystis organelle genome. We here report the sequence and describe the structure of the P. lacertae mitochondrial genome.
Materials and Methods
P. lacertae Culture and DNA Extraction
Proteromonas lacertae strain LA (Kulda 1973) was kindly provided by Prof. Jaroslav Kulda (Faculty of Science, Charles University, Prague, Czech Republic). Cells were cultivated in trypticase-yeast extract-maltose medium (Clark and Diamond 2002) pH7.2, supplemented with 10% heat inactivated horse serum (Sigma-Aldrich Ltd), 22.9 μg/ml ferric ammonium citrate (brown form; Sigma-Aldrich Ltd.), and 0.05% low-melting point agarose (Invitrogen Ltd.). Free-swimming cells were harvested from above the agarose layer and pelleted in a centrifuge (275 × g, 5 min). Total DNA extraction from the resulting pellet was carried out using the Gentra Puregene Cell kit (Qiagen Ltd).
Degenerate PCR
The initial sequences from the P. lacertae mitochondrial DNA (mtDNA) were obtained by direct polymerase chain reaction (PCR) using degenerate primers designed using aligned NADH dehydrogenase (nad) subunit gene sequences from other stramenopiles. Sets of degenerate primers were combined using total DNA from P. lacertae as a template. Standard touchdown PCR reactions were performed with Biomix (Bioline Ltd), using the following conditions: 94 °C for 2 min, followed by 10 cycles of 94 °C for 15 s, 55 °C (decreasing by 0.5 °C/cycle) for 30 s, and 68 °C for 2 min, followed by 20 cycles of 94 °C for 15 s, 50 °C for 30 s, and 68 °C for 2 min.
Inverse PCR
The products of the degenerate PCR reactions were sequenced and the resulting data used to design species-specific primers for amplification of templates consisting of self-ligated restriction fragments generated by digestion of total DNA with the restriction enzymes AluI, HindIII, MfeI, Sau3AI, and TaqI. Inverse touchdown PCR conditions consisted of 94 °C for 2 min, followed by 10 cycles of 94 °C for 15 s, 60 °C (decreasing by 0.5 °C/cycle) for 30 s, and 68 °C for 1–3 min for normal PCR, or 4–6 min for long PCR, followed by 20 cycles of 94 °C for 15 s, 55 °C for 30 s, and 68 °C for 1–3 min for normal PCR or 4–6 min for long PCR (increasing by 5 s/cycle). Following sequencing of the resulting products, the process was repeated.
Sequencing, Assembly, and Genome Analysis
PCR products were purified (QIAquick Gel Extraction kit, Qiagen Ltd or GeneJet PCR purification kit, Fermentas Life Sciences) and directly sequenced with ABI Prism BigDye Terminator v3.1 reagents (Applied Biosystems), followed by analysis on an ABI3730 capillary sequencer. Sequence assembly and analysis, Blast searches to identify genes, pseudogenes and structural RNAs, tRNA gene prediction, G + C content, guanine/cytosine (GC) skew, screening for group I and II introns, and tandem and inverted repeat searches were conducted as in Pérez-Brocal and Clark (2008). The complete P. lacertae mitochondrial genome analyzed in the present work has been deposited in GenBank with the accession number GU563431.
DNA Tailing
3′-termini of total DNA were tailed with dGTP and terminal deoxynucleotidyl transferase (Fermentas Life Sciences) according to the manufacturer’s specifications. The resulting tailed molecules were used as templates for a primary PCR with an anchor primer 5′ CTCTTGCTTGGATCCGGACCCCCCCCCCCCCDN 3′ plus a specific internal primer from close to the end of the known sequence. Secondary PCRs were carried out using a second specific internal primer plus the primer 5′ CTTGGATCCGGACCCC 3′, complementary to the anchor. Standard touchdown PCR conditions as described above were used.
Direct PCR and Southern Blots
A series of, on average, 2-kbp overlapping PCR products were generated by direct PCR using a set of primers designed to cover the entire mtDNA sequence. The resulting bands were separated in a 0.8% agarose gel to test for the presence of single or multiple bands (indicating the absence/presence of insertions–deletions in the molecule).
To further verify the structure of the P. lacertae mitochondrial genome, sequential hybridization of Southern blots of total DNA digested with MfeI was carried out using several probes from across the unique and repeated regions. The DIG-Hi Prime Labeling and Detection kit (Roche Diagnostics) was used for the probe labeling, hybridization, and detection following DNA transfer to BioDyne A membranes (Gibco-BRL).
Phylogenetic Analyses
Maximum likelihood and Bayesian inference analyses were carried out as in Pérez-Brocal and Clark (2008), with the nad gene alignment extended and updated by the inclusion of new sequences from P. lacertae, Synedra acus (GenBank accession number: GU002153), Saccharina sp. (GenBank accession number: AP0114998, Yotsukura et al. 2009), and a third Blastocystis subtype (subtype 7, GenBank accession number: CU914152, Wawrzyniak et al. 2008).
Results
Sequencing
Sequences of the inverse PCR products obtained using primers designed internally on partial sequence of the Proteromonas nad4 and nad5 genes revealed in each case that they were adjacent to the 5′ end of a nad11 gene. To verify that nad11 exists in two copies in the genome, direct long PCRs were carried out, in each case giving positive results. Subsequent analyses confirmed the existence of a long repeated region starting with the nad11 gene and just downstream of the nad4 and nad5 genes. Long direct PCR products of up to 7.5 kbp were produced by combining specific forward primers in the nad4 and nad5 genes with reverse primers from within the repeat region. Sequencing of these long products showed not only that identical genes were present but also that the repeats were identical in sequence.
In order to establish that the remainder of the two repeat regions were identical, at least in length and restriction sites, overlapping direct PCR products across the repeated region were obtained and all showed single bands when separated in an agarose gel (data not shown). This indicates the absence of detectable insertions or deletions between the two copies. Southern blots using different probes (data not shown) further confirmed this by revealing the bands expected based on the predicted restriction map. No extra bands that could indicate differences in size between the ends of the repeated regions were observed. Finally, sequencing of all PCR products from within the repeat region gave unambiguous results. We conclude that the two inverted repeat regions are identical in sequence.
Ends of the Chromosome
An inverted repeat of at least 11 bp (TATAATAAATT) located just after the tRNA-Gln gene coincided with an abrupt end to the inverse PCR strategy. All the attempts to read further using inverse PCR products generated with different restriction enzymes failed. Sequencing across the ligation junction indicated variability in the exact location at which the known restriction site was joined to the downstream sequence. This suggested ligation to the end of a linear molecule with slightly “ragged” ends. To verify this, a strategy of end tailing the genome with terminal deoxynucleotidyl transferase was carried out several times, as described in the Materials and Methods. The results were consistent with the hypothesis that this region is at the end of a linear molecule, because the direct sequencing of the PCR products showed a reproducible location for the run of Gs with only slight positional variation, likely due to factors such as the effect of nucleases during DNA purification. This indicated that the G tail was added not randomly at internal breaks but to the end of a molecule. The exact structure of the end of the repeat domain remains unknown but its location is likely to be very close to that indicated in our final sequence.
Gene Content
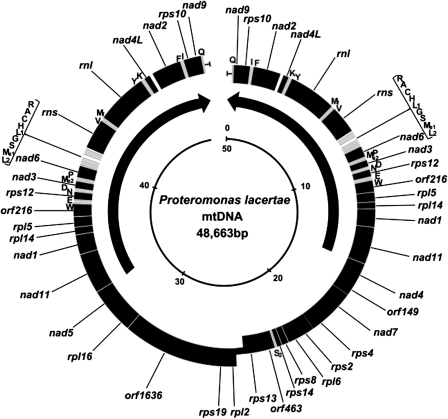
Following confirmation of the linear structure of the mitochondrial genome of P. lacertae LA, we undertook its annotation to elucidate the gene content (fig. 2). The most distinctive feature displayed by this genome is the presence of two identical large inverted repeat regions of 15,586 bp each, flanking a central unique region of 17,491 bp. This implies two identical copies of virtually all the structural RNA genes (both rRNAs and all tRNAs except one, tRNA-Ser2) as well as 12 out of the 27 protein-coding genes. Other features, such as the total length, G + C content and gene number are displayed in table 1. Unsurprisingly, the large flanking repeats have a G + C content significantly higher (26.3%) than the central region (16.1%) because the structural RNAs are characterized by a higher G + C content and most are encoded within the repeated region. The 52 different genes that make up the repertoire of this genome include 25 structural RNA genes, 23 of them tRNA genes. There are 27 protein-encoding genes, 10 of which encode NADH dehydrogenase subunits and 13 encode ribosomal proteins, with four open reading frames (ORFs) currently unidentified: orf149, orf216, orf463, and orf1636. No atp or cob/cox genes were found. There are eight examples of overlapping genes, six that overlap by just one base, whereas the others overlap by 8 bases (orf463-rps13) and 14 bases (rpl2-rps19).
FIG. 2.—
Circular representation of the gene and physical map of the linear Proteromonas lacertae organellar genome. Black blocks represent genes and ORFs and are transcribed from different strands in the right and left halves of the molecule. The interior arrows identify the two repeated regions and show their transcriptional direction. Gray blocks represent tRNA genes, and these are identified by their amino acid using the single-letter code. Mf and Me1/Me2 are initiator and elongator methionyl tRNAs, respectively. Short segments at each end of the molecule represent the putative “telomeric” region. The inner circle shows the size scale. The map was created using GenomeViz 1.1 (Ghai et al. 2004). The inner circle shows the size scale (kbp).
Table 1.
Characteristics of the Proteromonas lacertae mtDNA
| P. lacertaea | Blastocystis sp. | Other Stramenopilesb | |
| Total length (bp) | 48,663 | 27,719–29,270 | 31,617–58,507 |
| G + C content (%) | |||
| Total | 20.9 (22.7) | 19.9–21.9 | 22.3–38.0 |
| Intergenic regions | 13.2 (12.0) | 10.7–10.9 | 6.8–37.4 |
| Protein-coding regions | 18.0 (19.0) | 17.9–20.7 | 20.9–37.1 |
| Structural RNA genes | 35.4 (35.4) | 28.7–30.0 | 33.9–45.2 |
| Gene number | 52 (88) | 45 | 58–79 |
| Protein coding | 27 (39) | 27 | 34–52 |
| Structural RNAs | 25 (49) | 18 | 24–34 |
| Average length ORFs (nt) | 955 (886) | 793–835 | 645–1,049 |
| Average length IGRs (nt) | 34 (31) | 28–37 | 20–160 |
NOTE.—Various genome features of the complete organelle genome of P. lacertae, Blastocystis, and other stramenopiles are compared. IGR, intergenic region.
In parentheses are the values for G + C content, gene number etc., in P. lacertae when both copies of the repeated region are included.
Data for all other stramenopile complete mitochondrial genome sequences have been summarized in this column, namely those shown in figure 3 plus six additional Saccharina species.
Genome Comparison and Blocks of Genes
Comparisons of the genome features and gene content in P. lacertae with those of other stramenopile mitochondrial genomes are displayed in tables 1 and 2, respectively. It is noteworthy that all the genes in each half of the genome are transcribed from the same strand of DNA. At present we do not know if polycistronic transcription occurs (as seen in Blastocystis [Stechmann et al. 2008]) but this seems likely. Although the total length of the P. lacertae mtDNA makes this the second largest of the stramenopile mitochondrial genomes to date, this is misleading because if we include only a single copy of the repeated region the length is reduced to ∼33 kbp, still longer than that of Blastocystis sp. but at the lower end of the size range among stramenopiles. Similarly, the G + C content and gene number are not as drastically reduced as in the Blastocystis MLO genome but fall between Blastocystis and other stramenopiles.
Table 2.
Comparative Gene Content in 25 Completely Sequenced Stramenopile mtDNAsa
| Genesb | Bacillariophyta [2c] | Cafeteria roenbergensis | Chrysodydimus synuroideus | Ochromonas danica | Peronosporomycetes [4] | Phaeophyceae [12] | Blastocystis sp. [3] | Proteromonas lacertae |
| rns, rnl | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| trnA-W | 23–25 | 22 | 23 | 24 (29) | 25 (30) | 23–25 | 15 (16) | 23 (45) |
| atp6,8 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | □ |
| cob | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | □ |
| cox1-3 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | □ |
| nad1-7,9 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| nad11 | ▪ | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ |
| atp1 | □ | ▪ | □ | □ | ▪ | □ | □ | □ |
| atp9 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | □ |
| rps2 | ▪□ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | ▪ |
| rps3 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| rps4 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪b | ▪ |
| rps7 | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪b | □ |
| rps8 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rps10 | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rps11 | ▪ | □ | ▪ | ▪ | ▪ | ▪ | ▪ | □ |
| rps12,14,19 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rpl2 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rpl5 | ▪ | □ | □ | □ | ▪ | ▪ | ▪b | ▪ |
| rpl6,14 | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| rpl16 | ▪□ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ |
| ymf16 | □ | □ | ▪ | □ | ▪□ | □ | □ | □ |
| ORFs | 1–5 | 4 | 5 | 10 (12) | 4 | 3–16 | 1–4 (*) | 4 |
NOTE.—Filled boxes, genes present; open boxes, genes absent. Figures in parentheses represent the total gene number when genes present in two copies are included.
Based on the gene content reported by Pérez-Brocal and Clark (2008). In square brackets is the number of taxa within the group.
Genes identified as present in Blastocystis by Wawrzyniak et al. (2008) who report only one ORF (*) but considered as unidentified ORFs by Pérez-Brocal and Clark (2008) who therefore have 4 (*).
Square brackets indicate the number of taxa within the group.
Interestingly, the overall gene repertoire in P. lacertae resembles that of Blastocystis sp. in having lost all the genes encoding cytochrome oxidase subunits (cox1-3), cytochrome b (cob), and all the F0F1-ATPase subunits (atp6, 8, 9, and in some species atp1), encoded by the mtDNA in other stramenopiles. Also, Blastocystis and Proteromonas both encode two different elongator tRNA-Met genes instead of the single copy found in all other stramenopiles, as well both lacking a tRNA-Arg gene. In total, 41 genes are shared by the two organisms. However, some significant differences are present. Notably, P. lacertae retains a more complete set of tRNA genes, and in this it resembles more distantly related stramenopiles. Also, although the number of protein-coding genes is 27 in both cases, there is no evidence of homology, or even detectable similarity, between orf160, rps3, rps7, and rps11 of Blastocystis and orf149, orf216, orf463, and orf1636 of P. lacertae.
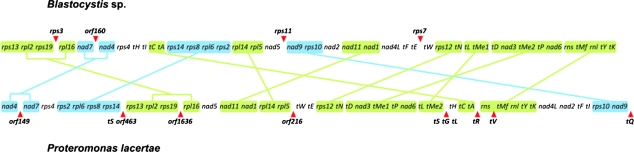
Small syntenic blocks of genes are observed when the mtDNA of P. lacertae and MLO genomes of Blastocystis sp. are compared (fig. 3), with up to 11 examples being observed comprising between 2 and 5 genes each. Interestingly, synteny suggests that orf1636 and orf 149 of P. lacertae may be homologous to rps3 and orf160 of Blastocystis, respectively, despite the lack of detectable similarity in the protein sequences.
FIG. 3.—
Gene order comparison between the mtDNA of Blastocystis sp. (linearized) and Proteromonas lacertae. For simplicity, only one of the repeated regions in P. lacertae has been depicted, and for this comparison, the beginning of the Blastocystis sp. MLO genome has been placed at rps13. The Blastocystis sp. gene nomenclature of Wawrzyniak et al. (2008) was used. “tX” indicates a tRNA gene, identified by its amino acid using the single-letter code. Solid lines connect the conserved gene blocks. Green: conserved block of genes with identical order and forward orientation. Blue: conserved block of genes in inverted orientation. Red arrowheads: genes apparently absent in one of the species or with no clear homologue.
Phylogenetic Analyses
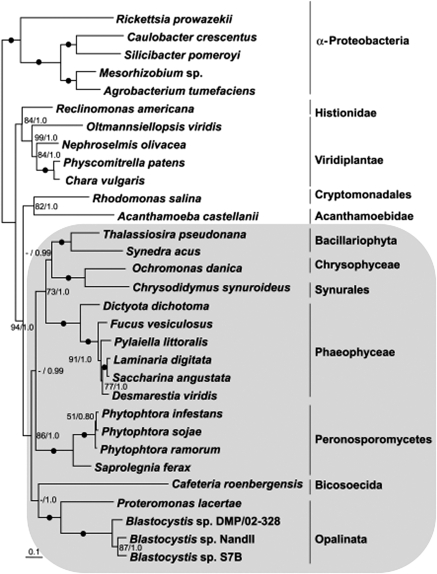
Using an alignment of nine nad genes, there is strong support for Proteromonas clustering with Blastocystis within the stramenopiles, by both maximum likelihood and Bayesian inference (fig. 4). This confirms the relationship inferred from analyses using nuclear SSU rRNA gene sequences (Kostka et al. 2004; Hoevers and Snowden 2005). The addition to the analysis of the Proteromonas and three additional stramenopile organelle genome sequences (from Synedra, Saccharina, and the third Blastocystis) has, however, led to some significant differences in the relationships observed between stramenopile groups when compared with our previous analysis (Pérez-Brocal and Clark 2008). In particular, the sister group of the Opalinata is no longer the Peronosporomycetes but the bicosoecid Cafeteria, in agreement with the results of Kostka et al. (2007) based on SSU rRNA. Other differences with the previous nad-based results are also seen, suggesting that the relationships deduced from nad gene analysis are not yet stable.
FIG. 4.—
Maximum likelihood-based phylogenetic tree of nad proteins. This tree is based on the concatenated amino acid sequences of 9 NADH dehydrogenase genes present in all the stramenopiles. Numbers beside the internal nodes are the maximum likelihood bootstrap values from 500 resamplings obtained with Phyml and the Bayesian Markov chain Monte Carlo posterior probability values. Black circles indicate 100% bootstrap support and 1.0 posterior probability values. Support values over 50% are shown adjacent to the corresponding nodes. Values below 50% are omitted. The clade containing all the stramenopiles is shaded. The scale bar represents the number of amino acids substitutions per residue.
Discussion
Mitochondrial genomes are known to vary extensively in size, structure, and gene content across eukaryotes. Data accumulated over recent years reveal a growing number of unrelated organisms that possess a linear mtDNA. For example: In Metazoa, the cnidarian Aurelia aurita (Shao et al. 2006); in green algae, Chlamydomonas reinhardtii (Ryan et al. 1978) and Polytomella capuana (Smith and Lee 2008); in ciliates, Tetrahymena pyriformis (Suyama and Miura 1968; Burger et al. 2000), Paramecium aurelia (Goddard and Cummings 1975; Pritchard et al. 1990), Euplotes minuta and E. crassus (de Graaf et al. 2009); in Apicomplexa, for example, Babesia spp. and Theileria spp. (Hikosaka et al. 2009); and in many yeast species (Fukuhara et al. 1993). The mitochondrial genome of P. lacertae represents the second linear mtDNA identified among the stramenopiles, after the genome the chrysophycean alga Ochromonas danica (Coleman et al. 2007). However, the gene content, gene order, organization, and structure of the two genomes all differ greatly, and because the two organisms are only distantly related it is likely that linearity arose independently. The mtDNA of P. lacertae shows a unique structure not only among the stramenopiles but also among all known mitochondrial genomes—the large terminal inverted repeats containing many genes. The biological significance of this topology remains unknown. The apparently radical divergence in architecture of the organelle genomes of Blastocystis and Proteromonas may not be surprising because the mitochondrial genomes of very closely related species of yeast can differ with respect to linearity/circularity (Fukuhara et al. 1993; Nosek et al. 1998; Laflamme and Lee 2003). There is even intraspecies occurrence of both linear- and circular-mapping genomes in the yeast Candida parapsilosis (Ryčovská et al. 2004), indicating that the two types of mitochondrial genome do not differ greatly from the organism’s viewpoint. In Candida, the organization and sequence of the coding regions appear to be highly conserved among linear- and circular-mapping mitochondrial genomes, indicating that both forms originated from a common ancestor via a relatively simple mechanism. At present, we cannot draw any conclusions as to whether the linear-mapping mtDNA of P. lacertae represents an ancestral or a derived form in this lineage. The sequencing of a more extensive range of mtDNAs from other Opalinata would help clarify this.
We have not been able to determine definitively the structure of the ends of the linear genome or its exact size, but based on our current evidence the presence of inverted terminal repeats is the most plausible organization. Various types of “telomere” are found in other linear mtDNAs (Nosek et al. 1998) some of which also involve inverted terminal repeats, as found in yeasts of the genera Williopsis (Drissi et al. 1994) and Pichia (Fukuhara et al. 1993), for example.
As for the origin of replication, we tentatively suggest the noncoding region between the rps13 and rpl2 genes as the most likely candidate, for two reasons: first, it is an unusually large noncoding region for this compact genome (420 bp) and second, the gene orientation and therefore the direction of transcription switches in this region. Analysis of the GC skew also shows a change in pattern around this region (data not shown).
Our phylogenetic analyses based on nine nad genes agree with those based on rRNA genes (Kostka et al. 2004; Hoevers and Snowden 2005) in placing P. lacertae and Blastocystis as sister taxa. Although the closest relatives of Proteromonas are probably other Slopalinida such as Protoopalina, Opalina, and especially the amphibian commensal Karotomorpha sp. (Kostka et al. 2007), until the organelle genomes of these organisms are sequenced, Blastocystis stands as the closest relative for which the mtDNA sequence is available.
One of our aims in sequencing the mtDNA of Proteromonas was to gain an insight into the order of events leading to the reduced gene repertoire observed in the Blastocystis MLO genome compared with other stramenopiles. In this, we were only partially successful. The subunits of complexes III and IV of the electron transport chain and the F0–F1-ATPase genes may have been lost before the two lineages split, but it is equally possible that the losses occurred independently and the current situation has arisen by convergence. Differences in the gene repertoire between P. lacertae and Blastocystis include divergent unidentified ORFs and ribosomal protein-coding genes. Their divergence is such that gene homology cannot be confirmed or ruled out. The most convincing case for lineage-specific differences is provided by the tRNA genes, where the very limited tRNA gene repertoire in the Blastocystis MLO genome is most easily explained by additional gene transfers to the nucleus having occurred in this lineage compared with P. lacertae. This also suggests that loss of the genes encoding unnecessary proteins is likely to have been completed well before the transfer of tRNA genes to the nucleus, although the reason for this occurring in Blastocystis and not in other stramenopiles is unclear. To further investigate this point and in order to understand the events underlying the highly divergent structure of the organelle DNA in the two organisms, the mtDNAs of additional Opalinata need to be sequenced.
Acknowledgments
We would like to thank Dr Jaroslav Kulda, Charles University, Prague, Czech Republic, for providing us with the axenic culture of P. lacertae LA. We are also grateful to John Williams and Maria McCrossan for kindly providing us with the light and electron micrographs of P. lacertae, respectively. A portion of this work formed the basis of an MSc thesis submitted by R.S.G. This project was supported by grant 078566 from the Wellcome Trust awarded to C.G.C.
References
- Boxma B, et al. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- Brugerolle G, Mignot JP. Phylum Zoomastigina, class Proteromonadida. In: Margulis L, Corliss JO, Melkonian M, Chapman DJ, editors. Handbook of Protoctista. Boston (MA): Jones & Bartlett; 1990. pp. 246–251. [Google Scholar]
- Burger G, et al. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J Mol Biol. 2000;297:365–380. doi: 10.1006/jmbi.2000.3529. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. A 6-kingdom classification and a unified phylogeny. In: Schenk HEA, Schwemmler W, editors. Endocytobiology II: intracellular space as oligogenetic ecosystem. Berlin (Germany): Walter de Gruyter; 1983. pp. 1027–1034. [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biol Rev Camb Philos Soc. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- Clark CG, Diamond LS. Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev. 2002;15:329–341. doi: 10.1128/CMR.15.3.329-341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman A, Thompson WF, Coff LJ. Identification of the mitochondrial genome in the chrysophyte alga Ochromonas danica. J Eukaryot Microbiol. 2007;38:129–135. [Google Scholar]
- de Graaf RM, et al. The mitochondrial genomes of the ciliates Euplotes minuta and Euplotes crassus. BMC Genomics. 2009;10:514. doi: 10.1186/1471-2164-10-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi R, Sor F, Nosek J, Fukuhara H. Genes of the linear mitochondrial DNA of Williopsis mrakii: coding sequences for a maturase-like protein, a ribosomal protein VAR1 homologue, cytochrome oxidase subunit 2 and methionyl tRNA. Yeast. 1994;10:391–398. doi: 10.1002/yea.320100312. [DOI] [PubMed] [Google Scholar]
- Embley TM, et al. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- Fukuhara H, et al. Linear mitochondrial DNAs of yeasts: frequency of occurrence and general features. Mol Cell Biol. 1993;13:2309–2314. doi: 10.1128/mcb.13.4.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Hain T, Chakraborty T. GenomeViz: visualizing microbial genomes. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill EE, et al. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol Microbiol. 2007;66:1306–1320. doi: 10.1111/j.1365-2958.2007.05979.x. [DOI] [PubMed] [Google Scholar]
- Goddard JM, Cummings DJ. Structure and replication of mitochondrial DNA from Paramecium aurelia. J Mol Biol. 1975;97:593–609. doi: 10.1016/s0022-2836(75)80061-1. [DOI] [PubMed] [Google Scholar]
- Gray MW. The hydrogenosome's murky past. Nature. 2005;434:29–31. doi: 10.1038/434029a. [DOI] [PubMed] [Google Scholar]
- Hikosaka K, et al. Divergence of the mitochondrialgenome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol. 2010;27:1107–1116. doi: 10.1093/molbev/msp320. [DOI] [PubMed] [Google Scholar]
- Hoevers JD, Snowden KF. Analysis of the ITS region and partial SSU and LSU rRNA genes of Blastocystis and Proteromonas lacertae. Parasitology. 2005;131:187–196. doi: 10.1017/s0031182005007596. [DOI] [PubMed] [Google Scholar]
- Hrdý I, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Kostka M, Čepička I, Hampl V, Flegr J. Phylogenetic position of Karotomorpha and paraphyly of Proteromonadidae. Mol Phylogenet Evol. 2007;43:1167–1170. doi: 10.1016/j.ympev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Kostka M, Hampl V, Čepička I, Flegr J. Phylogenetic position of Protoopalina intestinalis based on SSU rRNA gene sequence. Mol Biol Evol. 2004;33:220–224. doi: 10.1016/j.ympev.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Kulda J. Axenic cultivation of Proteromonas lacertae-viridis (Grassi 1879) J Protozool. 1973;20:536. [Google Scholar]
- Laflamme M, Lee RW. Mitochondrial genome conformation among CW-group chlorophycean algae. J Phycol. 2003;39:213–220. [Google Scholar]
- Lindmark D, Müller M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem. 1973;248:7724–7728. [PubMed] [Google Scholar]
- Matsumoto Y, Yamada M, Yoshida Y. Light-microscopical appearance and ultrastructure of Blastocystis hominis, an intestinal parasite of man. Zentralbl Bakteriol Mikrobiol Hyg [A]. 1987;264:379–385. doi: 10.1016/s0176-6724(87)80059-7. [DOI] [PubMed] [Google Scholar]
- Nasirudeen AM, Tan KS. Isolation and characterization of the mitochondrion-like organelle from Blastocystis hominis. J Microbiol Methods. 2004;58:101–109. doi: 10.1016/j.mimet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Nosek J, Tomáška L, Fukuhara H, Suyama Y, Kováč L. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 1998;14:184–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- Pérez-Brocal V, Clark CG. Analysis of two genomes from the mitochondrion-like organelle of the intestinal parasite Blastocystis: complete sequences, gene content and genome organization. Mol Biol Evol. 2008;25:2475–2482. doi: 10.1093/molbev/msn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard AE, et al. Nucleotide sequence of the mitochondrial genome of Paramecium. Nucleic Acids Res. 1990;18:173–180. doi: 10.1093/nar/18.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan CE, Ault JG, Langreth SG, Keithly JS. Cryptosporidium parvum Cpn60 targets a relict organelle. Curr Genet. 2003;44:138–147. doi: 10.1007/s00294-003-0432-1. [DOI] [PubMed] [Google Scholar]
- Ryan R, Grant D, Chiang KS, Swift H. Isolation and characterization of mitochondrial DNA from Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1978;75:3268–3272. doi: 10.1073/pnas.75.7.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryčovská A, Valach M, Tomáška L, Bolotin-Fukuhara M, Nosek J. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology. 2004;150:1571–1580. doi: 10.1099/mic.0.26988-0. [DOI] [PubMed] [Google Scholar]
- Shao Z, Graf S, Chaga OY, Lavrov DV. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 2006;381:92–101. doi: 10.1016/j.gene.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW. Mitochondrial genome of the colorless green alga Polytomella capuana: a linear molecule with an unprecedented GC content. Mol Biol Evol. 2008;25:487–496. doi: 10.1093/molbev/msm245. [DOI] [PubMed] [Google Scholar]
- Stechmann A, et al. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama Y, Miura K. Size and structural variations of mitochondrial DNA. Proc Natl Acad Sci U S A. 1968;60:235–242. doi: 10.1073/pnas.60.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens AGM, van Hellemond JJ. Anaerobic mitochondria: properties and origins. In: Martin WF, Müller M, editors. Origin of mitochondria and hydrogenosomes. Berlin (Germany): Springer-Verlag; 2007. pp. 85–103. [Google Scholar]
- Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- Tovar J, et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J, Clark CG. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak I, et al. Complete circular DNA in the mitochondria-like organelles of Blastocystis hominis. Int J Parasitol. 2008;38:1377–1382. doi: 10.1016/j.ijpara.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Hann AC, Lloyd D, Williams A. Hydrogenosomes in the rumen protozoon Dasytricha ruminantium Schuberg. Biochem J. 1981;200:365–372. doi: 10.1042/bj2000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarlett N, Orpin CG, Munn EA, Yarlett NC, Greenwood CA. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem J. 1986;236:729–739. doi: 10.1042/bj2360729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsukura N, Shimizu T, Katayama T, Druehl L. Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. J Appl Phycol. 2009 doi: 10.1007/s10811-009-9452-7. [Google Scholar]
- Zierdt CH. Cytochrome-free mitochondria of an anaerobic protozoan: Blastocystis hominis. J Protozool. 1986;33:67–69. doi: 10.1111/j.1550-7408.1986.tb05559.x. [DOI] [PubMed] [Google Scholar]