Abstract
Cardiac hypertrophy occurs in association with heart diseases and ultimately results in cardiac dysfunction and heart failure. Histone deacetylases (HDACs) are post-translational modifying enzymes that can deacetylate histones and non-histone proteins. Research with HDAC inhibitors has provided evidence that the class I HDACs are pro-hypertrophic. Among the class I HDACs, HDAC2 is activated by hypertrophic stresses in association with the induction of heat shock protein 70. Activated HDAC2 triggers hypertrophy by inhibiting the signal cascades of either Krüppel like factor 4 (KLF4) or inositol polyphosphate-5-phosphatase f (Inpp5f). Thus, modulators of HDAC2 enzymes, such as selective HDAC inhibitors, are considered to be an important target for heart diseases, especially for preventing cardiac hypertrophy. In contrast, class IIa HDACs have been shown to repress cardiac hypertrophy by inhibiting cardiac-specific transcription factors such as myocyte enhancer factor 2 (MEF2), GATA4, and NFAT in the heart. Studies of class IIa HDACs have shown that the underlying mechanism is regulated by nucleo-cytoplasm shuttling in response to a variety of stress signals. In this review, we focus on the class I and IIa HDACs that play critical roles in mediating cardiac hypertrophy and discuss the non-histone targets of HDACs in heart disease.
1. Introduction
Cardiac hypertrophy is an adaptive response to an initial exogenous hypertrophic stimulus that leads to a maladaptive state when the stress is prolonged [1]. Cardiac hypertrophy is characterized by increased cell size, enhanced protein synthesis, and heightened organization of the sarcomere. In this state, fetal genes, such as natriuretic peptide precursor type A (Nppa), myosin heavy polypeptide 7 (Myh7), and skeletal alpha-actin, are reactivated whereas cardiac contractile proteins like myosin heavy polypeptide 6 (Myh6) and calcium-handling proteins are repressed [2]. In addition, immediate-early genes encoding c-fos, c-jun, and heat shock proteins are upregulated [3, 4]. In humans, stresses such as chronic hypertension and myocardial infarction can trigger the heart to undergo remodeling processes characterized by myocyte hypertrophy, myocyte death, and fibrosis, often resulting in pathological heart diseases including reduced cardiac function, cardiomyopathy, and heart failure [5–8]. Although it is believed that hypertrophy can be an adaptive response to exogenous stresses, advanced cardiac hypertrophy itself is associated with an increased risk of morbidity and mortality [9, 10].
Histones have long N-terminal tails and are subject to diverse post-translational modifications such as acetylation, methylation, phosphorylation, ubiquitination, and sumoylation. A well-characterized post-translational modification is acetylation, which occurs at the ε amino groups of lysine residues in the core histone. Acetylation of chromatin plays a central role in the epigenetic regulation of gene expression in eukaryotic cells. Acetylation is regulated by two opposing families of proteins, histone acetyltransferase (HAT), and histone deacetylases (HDACs). Recent evidence has indicated that different HDACs participate in a variety of heart diseases, such as arrhythmia, heart failure, and acute coronary syndromes, as well as in cardiac hypertrophy [11–19].
In mammals, there are four major classes of HDACs. Class I HDACs (HDAC1, 2, 3, and 8) are widely expressed and consist mainly of a catalytic domain. Class II HDACs are divided into two subclasses, IIa (HDAC4, 5, 7, and 9) and IIb (HDAC6 and 10). Class III HDACs are NAD(+)-dependent and are referred to as sirtuins (SIRT1-7). Most class IIa HDACs show cell-type-restricted expression patterns. Although many HDACs have a highly conserved domain, recent studies show that class I and IIa HDACs have opposing roles in regulating cardiac hypertrophy, and evidence for the mechanisms by which the distinct classes of HDACs act to control cardiac hypertrophy is growing. In this paper, we focus on the pathophysiological roles of class I and IIa HDACs in cardiac hypertrophy.
2. Heart Diseases Regulated by Class I HDACs: Cardiac Growth, Proliferation, Differentiation, Fibrosis, Ischemic Heart Disease, and Arrhythmia
HDACs are implicated as a regulator in various pathological heart diseases such as fibrosis, arrhythmia, ischemic heart diseases, and heart failure. Cardiac arrhythmia is related to a variety of cardiac stressors such as ischemia and an increase in wall stress. It is also associated with the renin-angiotensin-aldosterone system. A recent study indicated that the HDAC inhibitor, TSA, inhibits atrial fibrosis and arrhythmic inducibility and partially normalizes connexin 40 expression without changes in the angiotensin level in the Hopx transgenic mouse cardiac hypertrophy model [12].
Our group and others have demonstrated that myocardial fibrosis is reduced by HDAC inhibitors such as TSA and sodium valproate either in mice with left ventricular hypertrophy induced by aortic banding or in rats with right ventricular hypertrophy induced by pulmonary artery banding [15, 20, 21]. In addition, chemical HDAC inhibition was shown to reduce infarct size and improve ventricular function recovery in a model of myocardial ischemia and reperfusion injury, which suggests a novel therapeutic target for acute coronary syndromes [16, 17]. Sustained cardiac hypertrophic stimuli may lead to cardiomyopathy and heart failure. Likewise, heart failure with high mortality was prevented by apicidin derivatives with class I HDAC specificity in mice with heart failure induced by thoracic aortic constriction [13].
We [14] and other research groups [15, 20, 22] reported that class I and II broad HDAC inhibitors could prevent cardiac hypertrophy in animal models. We demonstrated that class I HDACs are required for the hypertrophic response in aortic banding or angiotensin II infusion-induced hypertrophy animal models with class I HDAC-selective HDAC inhibitor. Chemical HDAC inhibitors such as TSA or valproate induced the partial regression of pre-established cardiac hypertrophy. We were the first to show that class I HDACs may play a pro-hypertrophic role in the heart. Recently, another group reported similar results that broad-spectrum HDAC inhibitors such as TSA or scriptaid blunt the cardiac hypertrophy induced by aortic banding [15]. In rat neonatal cardiomyocytes, HDAC inhibition by TSA was also reported to blunt a stress-induced hypertrophic marker [22]. In addition, our group reported that sodium valproate, an alternate HDAC inhibitor, prevents right ventricular hypertrophy induced by pulmonary artery banding in rats [21]. Considering that class II HDACs work as anti-hypertrophic mediators, prevention of cardiac hypertrophy with nonspecific HDAC inhibitors strongly suggests that class I HDAC may function as a pro-hypertrophic regulator in the heart. These suggestions would be further supported by the reports that class I HDAC-selective inhibitors still show anti-hypertrophic effects [13, 14].
Recent studies have indicated that the class I HDACs (HDAC1, 2, 3, and 8) are involved in the control of cardiac growth, proliferation, and differentiation. Table 1 summarizes the phenotype and function of HDAC1, HDAC2, and HDAC3 in the heart. HDAC1 was reported to be a regulator of cardiomyogenesis, cell proliferation, and embryonic development. During cardiomyogenesis, downregulation of HDAC1 promotes the expression of Nkx2.5, a critical regulator of cardiac gene expression and heart development [23]. Disruption of the HDAC1 gene results in embryonic death before E10.5 as the result of severe proliferation defects [24]. These data suggest that HDAC1 is an important mediator in cardiac differentiation and early embryonic development.
Table 1.
The physiological role of class I and class IIa HDACs in cardiac development and heart diseases.
| HDAC subtype | Model | Phenotype | Disease functions in the heart | References |
|---|---|---|---|---|
| HDAC1 | P19CL16 cells | Differentiation | HDAC1 protein was decreased during cardiomyogenesis | Liu et al. [23] |
| HDAC1-deficient mice | Proliferation | Embryo lethality because of proliferation defects | Lagger et al. [24], Montgomery et al. [25] |
|
| HDAC2 | HDAC2 knockout mice | Proliferation | Proliferation rates of cardiac myocytes in HDAC2 knockout mice were elevated | Trivedi et al. [26] |
| HDAC2 knockout mice | Proliferation | Increase in proliferation at P1Lethality after P1 | Montgomery et al. [25] | |
| HDAC2 knockout mice | Hypertrophy | HDAC2 knockout mice are resistant to cardiac hypertrophy | Trivedi et al. [26] | |
| HDAC2 transgenic mice | Hypertrophy | HDAC2 transgenic mice show cardiac hypertrophy | Trivedi et al. [26] | |
| Aortic banding mice | Hypertrophy | HDAC2 and HSP70 cause cardiac hypertrophy | Kee et al. [14] | |
| Hsp70 knockout mice | ||||
| HDAC3 | HDAC3 transgenic mice | Proliferation | HDAC3 transgenic mice show postnatal cardiac myocyte proliferation | Trivedi et al. [27] |
| HDAC3 knockout mice | Hypertrophy | HDAC3 knockout mice show massive cardiac hypertrophy | Montgomery et al. [28] | |
| HDAC3 knockout mice | Metabolism | HDAC3 regulates cardiac energy metabolism | Montgomery et al. [28] | |
| HDAC4 | HDAC4 knockout mice | HDAC4 knockout mice die perinatally because of abnormal chondrocyte hypertrophy | Vega et al. [29] | |
| HDAC5 | HDAC5 knockout mice | Hypertrophy | Mice lacking HDAC5 develop enlarged hearts in response to pressure overload | Chang et al. [30] |
| HDAC9 | HDAC9 knockout mice | Hypertrophy | HDAC9 mutant mice develop cardiac hypertrophy | Zhang et al. [31] |
Recent evidence implicates HDAC2 as a positive regulator of the hypertrophic response during embryonic development and in the adult heart. For example, HDAC2-deficient mice generated by a gene-trap technique show partial postnatal lethality, increased numbers of mitochondria, and abnormal sarcomere structure [26]. In addition, loss of HDAC2 makes the heart resistant to hypertrophic stimuli through the increased expression of inositol polyphosphate-5-phosphatase f (Inpp5f). The increased expression of Inpp5f is associated with activation of glycogen synthase kinase 3β (GSK3β). In contrast, transgenic mice overexpressing HDAC2 in the heart are sensitive to hypertrophic stimuli [26].
However, another group recently reported a different finding that HDAC2-null mice generated by use of the Cre/loxP system die within 24 hours after birth [25]. The most interesting difference between the studies of Trivedi et al. [26] and Montgomery et al. is that HDAC2 deletion by lacZ insertional mutation removes the region with exons 9–14, in the C-terminus of HDAC2 whereas the conditional deletion regions of HDAC2 include exons 2–4. Although HDAC2 consists mainly of the HDAC catalytic domain, the catalytic domain extends from partial exon 2 to exon 9. Therefore, the catalytic domain of HDAC2 seems to be implicated in postnatal viability. The different approaches to HDAC2 mutation resulted in distinct findings in response to hypertrophic stimuli such as chronic isoproterenol or aortic banding. For example, heart-specific deletion of HDAC2 did not result in an increased response to hypertrophic stimuli whereas HDAC2 disruption by lacZ insertional mutation prevented cardiac hypertrophy in response to hypertrophic stimuli [25].
HDAC3 is suggested to function as a regulator of cardiac myocyte proliferation during cardiac development [27]. For example, transgenic hearts with myocyte-specific overexpression of HDAC3 show thickening in the ventricular wall and interventricular septum and an increase in phospho-histone H3 staining at neonatal day 1 (P1). The mRNA expression of several cyclin-dependent kinase inhibitors (p21, p27, p57, p16, and p15) is repressed in HDAC3 transgenic mice at P1. However, at 2 months of age, HDAC3 transgenic mice do not show an increase in the proliferation index. The results of this study imply that HDAC3 is related to cardiomyocyte proliferation rather than to cardiac hypertrophy. Recently, another group demonstrated a different role and phenotype of HDAC3 in the heart. In that study, although global deletion of HDAC3 resulted in embryonic death, heart-specific deletion of HDAC3 resulted in severe cardiac hypertrophy and increased expression of mitochondrial uncoupling genes at 3 months of age, and the mice survived until 4 months of age [28]. HDAC3 knockout mice elicited a significant increase in p21 expression [28].
Although HDACs have enzymatic activity to remove acetyl groups from protein, their existence alone is also important, because they can interact with numerous binding partners including transcription factors to form large complexes. For example, the enzyme activity and binding to MEF2 of class IIa HDACs play a critical role in a variety of diseases. In particular, MITR, a splice variant of HDAC9 that lacks a deacetylase domain, is known to have anti-hypertrophic action [31]. In this regard, studies should be taken with caution to determine which mechanism of either existence or activity is critical for the biological actions of HDACs. Our groups have demonstrated that the enzymatic activation of HDAC2 preceded the substantial hypertrophic phenotype in response to hypertrophic stimuli whereas the mutant form of HDAC2 did not result in a substantial hypertrophic phenotype. These results suggest that HDAC2 activation rather than its presence is important to induce cardiac hypertrophy [32].
Like the class IIa HDACs, the activities of the class I HDACs are regulated by post-translational modifications. The activity of HDAC1, HDAC2, and HDAC3 is mainly regulated by the phosphorylation of the casein kinase CK2 [33–35]. HDAC8 is phosphorylated by PKA [36]. For a better understanding of pathological heart diseases, further experiments are necessary to determine whether the phosphorylation status of the HDACs is modulated by phosphatases.
In summary, although both are class I HDACs, HDAC2 and HDAC3 have different functions in the regulation of cardiac hypertrophy and proliferation. HDAC2 works to cause cardiac hypertrophy accompanied by a repression of proliferation whereas HDAC3 acts to inhibit cardiac hypertrophy with increasing proliferation. We discuss the mechanism by which HDAC2 regulates cardiac hypertrophy further in the next section.
3. HDAC2, a Class I HDAC, in Cardiac Hypertrophy
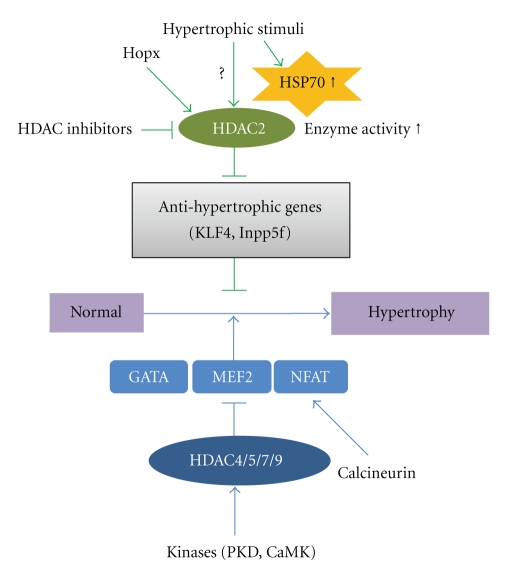
Recently, by use of HDAC2 knockout mice or transgenic mice and cardiomyocytes, our group and others have identified that HDAC2, a class I HDAC, positively regulates cardiac hypertrophy. Figure 1 shows the Hopx/HDAC2/HSP70/KLF4 signal cascades that regulate prohypertrophy pathways in the heart. Our hypothesis is supported by our previous study with transgenic Hopx mice. Hopx, a cardiac-specific regulator of gene transcription, is an unusual homeodomain protein that is expressed in embryonic and postnatal cardiac myocytes. Inactivation of Hopx causes severe developmental cardiac defects with cardiac proliferation. Hopx transgenic mice exhibit cardiac hypertrophy and cardiac fibrosis resulting from recruitment of HDAC2 whereas Hopx mutants do not develop hypertrophy or recruit HDAC2. The cardiac hypertrophy induced in Hopx transgenic mice can be prevented by HDAC inhibitors such as TSA and valproate [20, 37]. Although previous studies showed an increase in HDAC activity in Hopx transgenic mice compared with wild-type mice, little is known about changes in the activity of each of the HDACs in cardiac hypertrophy. Thus, our group focused on determining HDAC activity in response to hypertrophic stimuli among the class I HDACs. We observed that only HDAC2 activity was induced in response to hypertrophic stimuli.
Figure 1.
The signaling cascade of class I and IIa HDACs in the development of cardiac hypertrophy. Class I HDACs can be regulated by the Hopx/HDAC2/HSP70/KLF4 pathway. Cardiac hypertrophy is induced in Hopx transgenic mice through the recruitment of HDAC2. Hypertrophic stresses including pressure overload or agonist-induced HSP70 expression are followed by its association with HDAC2. The resultant increase in HDAC2 activity decreases the expression of anti-hypertrophic mediators including KLF4 and Inpp5f. This diagram is adapted from the works of Kook et al. [20] and Kee et al. [32, 38] (JCI, 2003; Circulation Research, 2008; JMCC, 2009). Class II HDACs can be activated by PKD or CaMK. They interact with MEF2. MEF2 also associates with GATA4 and NFAT transcription factor. NFAT can be triggered by calcineurin. Class IIa HDACs can repress cardiac hypertrophy through MEF2 either directly or indirectly.
Heat shock proteins (HSPs) are induced by hypertrophic stimuli [3]. Thus, we determined whether the induction of HSPs in response to hypertrophic stresses induces cardiac hypertrophy. We found that the novel HDAC2-interacting partner HSP70 activates HDAC2 and induces cardiac hypertrophy. HDAC2 increased the cardiomyocyte cell size. In addition, cardiac hypertrophy and HDAC2 activation were blunted in HSP70 knockout mice [32]. These data strongly suggested that the HSP70/HDAC2 axis is an important mediator in the regulation of cardiac hypertrophy and highlighted HSP70 as a regulator of HDAC2 activity in response to hypertrophic stresses.
We next investigated whether HDAC2 could regulate downstream targets in cardiomyocytes. In the cardiomyocytes, Krüppel-like factor 4 (KLF4) was shown to be a target of HDAC2 and to repress Nppa, leading to the inhibition of cardiac hypertrophy [38]. KLF4 overexpression blocked the increase in Nppa transcript levels and [3H]-leucine incorporation induced by 20% FBS. In contrast, KLF4 knockdown stimulated hypertrophic phenotypes. The results of this study suggested that KLF4 is a novel anti-hypertrophic regulator. Recently, Liao et al. reported that KLF4 functions as a negative regulator of cardiac hypertrophy by use of the cardiomyocyte-specific KLF4 knockout mice [39].
Like KLF4, Inpp5f is known to be a downstream target of HDAC2 in mediating cardiac hypertrophy. HDAC2-regulation of Inpp5f is mediated through the phosphatidylinositol 3-kinase- (PI3K-) Akt-Gsk3β pathway, which is important for growth control in heart. Overexpression of HDAC2 suppresses Inpp5f expression, and accumulation of phosphatidyl inositol-3,4,5-trisphosphate (PIP3) leads to the recruitment of protein kinase Akt. Activated Akt inactivates Gsk3β by its phosphorylation. Suppression of Gsk3β activity relieves the inhibition of some hypertrophic signaling pathways, such as c-Myc, GATA4, and β-catenin, leading to cardiac hypertrophy [26]. In the case of HDAC2 deficiency, the decrease in Akt activity and increase in Gsk3β activity result in resistance to cardiac hypertrophy. Furthermore, hypertrophic responsiveness was shown to be reduced in mice with Inpp5f overexpression whereas knockout mice showed augmented hypertrophy [40].
4. Negative Regulation of Cardiac Hypertrophy and Heart Failure by Class IIa HDACs
The class IIa HDACs (HDAC4, HDAC5, HDAC7, and HDAC9) are expressed in muscles and brain and have an N-terminal regulatory domain that mediates interactions with transcription factors, coactivators, and corepressors [41–43]. The underlying mechanism and functional role of the class IIa HDACs in hypertrophy has been intensively investigated by the Olson group by use of knockout mice. Diverse hypertrophic stimuli lead to the activation of cardiac HDAC kinases such as protein kinase C (PKC), protein kinase D (PKD), and calcium/calmodulin-dependent protein kinase (CaMK). In this section, we describe how each of the class II HDACs regulates and affects cardiac hypertrophy through signaling pathways and associated proteins.
HDAC4 is phosphorylated by activated CaMKII in response to phenylephrine. Phosphorylated HDAC4 is translocated into the cytoplasm by 14-3-3 protein, resulting in derepression of the MEF2 gene, which leads to hypertrophic growth [44]. HDAC4 appears to interact with and inhibit the activity of runt-related transcription factor 2 (Runx2). HDAC4-null mice show skeletal defects and chondrocyte hypertrophy, whereas overexpression of HDAC4 inhibits chondrocyte hypertrophy and shows Runx2 loss of function of phenotypes [29].
HDAC5, an alternate class IIa HDAC, has similar actions in regulating cardiac hypertrophy. Phosphorylation of HDAC5 by PKC or PKD leads this protein to bind to 14-3-3 protein, which stimulates the nuclear export of HDAC5. In cardiomyocytes, an HDAC5 phosphorylation mutant was shown to be resistant to the PKC signaling pathway and to block cardiomyocyte hypertrophy [45]. Chang et al. demonstrated that HDAC5 knockout mice develop cardiac hypertrophy with age in response to pressure overload and calcineurin signaling whereas overexpression of HDAC5 blocks hypertrophy [30]. Although little is known about the role of HDAC7 in cardiac hypertrophy, this HDAC is also regulated by shuttling [46]. By contrast, the role of HDAC9 has been well scrutinized in cardiac hypertrophy by use of knockout mice. Class IIa HDACs and MEF2-interacting transcriptional repressor (MITR), a splice variant of HDAC9, have MEF2 binding sites in the N-terminus and an HDAC catalytic domain in the C-terminal region. HDAC9 mutant mice show cardiac hypertrophy with advanced age in vivo and are hypersensitive to hypertrophic stimuli such as thoracic aortic banding [31]. HDAC5/9 double knockout mice show an even greater degree of cardiac hypertrophy than mice lacking either HDAC5 or HDAC9 [30]. In addition, other pro-hypertrophic transcription factors including GATA4 and NFAT have been shown to be repressed indirectly by class IIa HDACs [47]. These transcription factors have been shown to be associated with MEF2 [48]. Therefore, class IIa HDACs also can repress the activity of these cardiac-specific transcription factors though MEF2.Taken together, these findings suggest that the class IIa HDACs function as negative regulators of cardiac hypertrophy by repressing MEF2/GATA4/NFAT-mediated gene expression [31, 49].
Although most of the class IIa HDACs can be shuttled from the nucleus to the cytoplasm by phosphorylation in the control of cardiac hypertrophy [50, 51], recent evidence has shown that HDAC4 is subject to oxidative modification in the mediation of cardiac hypertrophy [52]. When HDAC4 oxidation is induced by hypertrophic stimuli, thioredoxin1, a 12-kDa antioxidant, regulates the nucleocytoplasmic shuttling of HDAC4. In addition, cysteine residues in HDAC2 are subject to nitrosylation modification in neurons [53, 54]. Therefore, like phosphorylation, other forms of post-translational modification may be critical mechanisms in the regulation of cardiac hypertrophy.
5. Class III HDACs: Aging and Stress
Class III HDACs consist of seven Sirt isoforms (Sirt 1 to Sirt 7). The Sirts, known as sirtuins, are NAD(+)-dependent acetyl-lysine deacetylases. The Sirts have been shown to be involved in aging and protection against oxidative stress in the heart [55, 56]. Heart-specific moderate overexpression of Sirt 1 in mice results in the retardation of aging and a protective response to oxidative stress [57]. As for Sirt 1, similar results have been shown for Sirt 3 and Sirt7 in regulating oxidative stress-mediated cell death in cardiomyocytes [58, 59]. Considering the beneficial effects of Sirts, intensive studies of Sirt activators will enhance our understanding of the underlying mechanism in heart diseases. Sirt activators are being developed, and clinical trials have been conduced for some metabolic diseases such as diabetes [60]. We suggest that Sirt may be a critical target in mediating cardiovascular diseases.
6. Deacetylation of Nonhistone Proteins by HDACs: Transcription Factors and Cytoskeleton Proteins
Non-histone proteins have been identified as substrates for HDACs [61]. Many studies have demonstrated that different HDACs target various transcription factors in cancer cells, during protein repair, in immune reactions, and in redox regulation. For example, p53, the most commonly mutated gene in cancer cells, is deacetylated by HDAC1 and SIRT1 (Sir2 alpha) [62, 63]. SRY, a sex-determining region on the Y chromosome, is deacetylated by HDAC3 [64]. STAT3 (signal transducers and activators of transcription) is completely deacetylated by HDAC3 and, to a lesser extent, by HDAC1 and HDAC2 [65]. SHP, orphan nuclear receptor, recruits HDAC1, HDAC3, HDAC6, and SIRT1 [66, 67]. Currently, however, the HDACs responsible for the deacetylation of SHP are unknown. E2F1 transcription factor, which organizes early cell cycle progression, can be deacetylated by HDAC1 via an indirect interaction mediated by Rb [68]. RelA, a class II NF-kB family member (nuclear factor kappa-light chain enhancer of activated B cells) that acts to regulate the immune response, can be deacetylated by HDAC3 and SIRT1 [69, 70]. Peroxiredoxins (I and II), Hsp90, and α-tubulin are a specific target of HDAC6 [71]. Ku70, a repair protein, is deacetylated by SIRT1 [72]. GCM (glial cell missing) transcription factor can be deacetylated by HDAC1, HDAC3, HDAC4, and HDAC5 [73].
Deacetylation of non-histone proteins can alter protein stability, localization, protein-protein interaction, DNA-binding affinity, and transcriptional activity. In addition, deacetylation of non-histone proteins promotes ubiquitination and decreases the half-life of the protein [74]. Cardiac-specific transcription factors such as GATA4, NFAT, NFκB, MEF2, SRF, Smads, Nkx2-5, YY1, Hand, Egr-1, and CREB are reported to be involved in heart diseases. Among these, some transcription factors may be a direct target of HDACs in the heart. For example, the MEF2D family of transcription factors, which mediate cardiac hypertrophy, is deacetylated by HDAC3 [75]. In addition, SIRT1, an NAD+-dependent deacetylase, induces the acetylation of MEF2 [51]. In contrast, although HDAC4 and HDAC5 are known partners of MEF2, they do not function as a MEF2 deacetylase. MyoD, a skeletal muscle-specific transcription factor, is deacetylated by HDAC1 and SIRT1 [76].
Among the six GATA transcription factors, GATA4 has been extensively studied as a regulator of cardiac development and differentiation as well as in hypertrophic growth in adult heart [77–79]. Many studies have indicated that GATAs are mainly acetylated by p300, PCAF, and CBP with HAT activity [80]. It has been reported that GATAs can interact with different HDACs although little is known about whether these GATAs are substrates of the HDACs. It is suggested that GATA4 may be deacetylated by class I and IIa HDACs because the acetylation of GATA4 is augmented by treatment with TSA [81].
Recent data show that the TGFβ-Smad signaling pathway can regulate the development of cardiomyocyte hypertrophy and cardiac fibrosis. For example, Smad2 overexpression inhibits agonist-induced hypertrophy in culture, and Smad4-deficient mice develop cardiac hypertrophy. Lysine residues of Smad7, an inhibitory Smad, are deacetylated by SIRT1 (class III histone deacetylase) [82]. Smad7 overexpression suppresses collagen type I and III whereas decreased Sma7 expression in cardiac fibroblasts is found in the infarcted rat heart [83].
The homeobox transcription factor Nkx2.5 is also implicated in the hypertrophic response and in heart development. Little is known about the direct deacetylation of Nkx2.5 by HDACs. However, associated HDAC5 was found in a complex with acetylated Nkx2.5 in cardiomyocytes [84]. Thus, Nkx2.5 is expected to be a direct target of HDACs.
YY1 is a multifunctional transcription factor that can repress or activate the transcription of many genes. YY1 residues acetylated by p300 and PCAF are deacetylated by HDAC1 and HDAC2 in Hela cells [85]. Recent evidence demonstrated that YY1 interacts with HDAC5 in cardiac myocytes and plays an anti-hypertrophic role in pathological hypertrophy [86]. Similarly, YY1 interacts with HDAC5 in differentiated H9c2 cells and functions as a regulator in muscle differentiation [87]. In addition, YY1 was shown to associate with HDAC2 and to activate the expression of BNP, which is a marker related to cardiac myocyte hypertrophy [88].
In addition to transcription factors, other non-histone proteins can be regulated by deacetylation. For example, α-tubulin, another marker of cardiac hypertrophy, is increased in a congestive heart failure animal model. α-Tubulin was found to be deacetylated by HDAC6 and SIRT2 [89–91].
In summary, the epigenetic modification of diverse proteins including heart-specific transcription factors can play vital roles in a variety of heart diseases. The finding of direct HDAC targets will provide us a new understanding of pathological heart diseases and insight into promising drug development for the treatment of cardiac hypertrophy.
7. Summary and Further Suggestions
The functional roles of the HDACs in heart development and in heart diseases including cardiac hypertrophy and heart failure have been reviewed. Class I and IIa HDACs have been shown to have opposing roles in the regulation of cardiac myocyte growth and pathological heart disease. HDAC2, a class I HDAC, induces cardiac hypertrophy in response to early hypertrophic stimuli whereas class IIa HDACs including HDAC4, HDAC5, and HDAC9 suppress cardiac hypertrophy. Several lines of evidence with HDAC inhibitors show that class I HDACs play a more important role than class IIa HDACs in cardiac hypertrophy. Recent work has demonstrated that broad-spectrum HDAC inhibitors as well as class I or II HDAC-specific inhibitors prevent cardiac hypertrophy, fibrosis, and ischemic heart diseases. Unlike class I and IIa HDACs, class III HDACs play an important role in regulating oxidative stress in heart diseases with metabolic syndromes. Future studies to develop the agonists of class III HDACs could be a good strategy for treating and protecting against the cardiovascular diseases associated with aging.
HDAC inhibitors are being highlighted as cancer therapy. SAHA, known as Vorinostat, has been approved by the Food and Drug Administration [92], and several other HDAC inhibitors are currently being investigated in clinical trials [93, 94]. Thus, one can easily assume that inhibition of HDAC may be applied for the treatment of cardiac diseases as well as various cancers. For example, if heart-specific HDAC inhibitors are developed and if the regulation of HDAC activity or HDAC-interacting protein interaction can be modulated by specific chemicals, the application of such drugs will be of great benefit. Actually, several pharmaceutical companies are developing chemicals to regulate HDAC itself or its target-specific protein to treat pathological diseases such as in heart and muscle.
Because of their involvement in many pathological heart diseases, further investigation of the HDACs is vital to understand the underlying mechanism of their action, their biological roles, and their direct substrates. The enzymatic activity of HDACs can be regulated by phosphorylation/dephosphorylation by protein kinases and phosphatases or by protein -protein interaction. Therefore, to improve our understanding of the mechanism by which HDAC2 acts in cardiac hypertrophy, we will need to search for HDAC2 kinase and other post-translational modifications such as sumoylation, ubiquitination, S-nitrosylation, carbonylation (alkylation), and glycosylation. In this regard, further studies are necessary to elucidate the mechanism and effect of novel epigenetic regulator-mediated cardiac diseases. Further experimental evidence should be provided to develop selective pharmacological drugs such as inhibitors of pathology-associated HDACs or HDAC modifying enzyme to initiate clinical trials for the treatment of cardiac diseases. We must critically consider the non-histone targets of HDACs. These targets contain many important transcription factors. Knowledge from the targets of HDAC regulation is expected to improve our comprehension of HDAC biological activity and functions.
Further studies of both the downstream and the upstream targets of HDAC2 will extend our knowledge of the biological role of the HDACs and may suggest novel combined therapeutic strategies for heart diseases.
Acknowledgments
This study was supported by the Korea Science and Engineering Foundation through the Medical Research Center for Gene Regulation (R13-2002-013-05001-0) and in part by a National Research Foundation of Korea (NRF) grant funded by the Korean Government (MEST, no. 2010-0015012).
References
- 1.Hunter JJ, Chien KR. Signaling pathways for cardiac hypertrophy and failure. New England Journal of Medicine. 1999;341(17):1276–1283. doi: 10.1056/NEJM199910213411706. [DOI] [PubMed] [Google Scholar]
- 2.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Failure Reviews. 2007;12(3-4):331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 3.Osaki J, Haneda T, Kashiwagi Y, et al. Pressure-induced expression of heat shock protein 70 mRNA in adult rat heart is coupled both to protein kinase A-dependent and protein kinase C-dependent systems. Journal of Hypertension. 1998;16(8):1193–1200. doi: 10.1097/00004872-199816080-00013. [DOI] [PubMed] [Google Scholar]
- 4.Sadoshima J-I, Takahashi T, Jahn L, Izumo S. Roles of mechano-sensitive ion channels, cytoskeleton, and contractile activity in stretch-induced immediate-early gene expression and hypertrophy of cardiac myocytes. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(20):9905–9909. doi: 10.1073/pnas.89.20.9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gradman AH, Alfayoumi F. From left ventricular hypertrophy to congestive heart failure: management of hypertensive heart disease. Progress in Cardiovascular Diseases. 2006;48(5):326–341. doi: 10.1016/j.pcad.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Dobaczewski M, Bujak M, Li N, et al. Smad3 signaling critically regulates fibroblast phenotype and function in healing myocardial infarction. Circulation research. 2010;107(3):418–428. doi: 10.1161/CIRCRESAHA.109.216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramaniam V, Lip GYH. Hypertension to heart failure: a pathophysiological spectrum relating blood pressure, drug treatments and stroke. Expert Review of Cardiovascular Therapy. 2009;7(6):703–713. doi: 10.1586/erc.09.43. [DOI] [PubMed] [Google Scholar]
- 8.Kahan T. The importance of left ventricular hypertrophy in human hypertension. Journal of Hypertension, Supplement. 1998;16(7):S23–S29. [PubMed] [Google Scholar]
- 9.Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. American Heart Journal. 2001;141(3):334–341. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 10.Levy D, Anderson KM, Savage DD, Kannel WB, Christiansen JC, Castelli WP. Echocardiographically detected left ventricular hypertrophy: prevalence and risk factors. The Framingham heart study. Annals of Internal Medicine. 1988;108(1):7–13. doi: 10.7326/0003-4819-108-1-7. [DOI] [PubMed] [Google Scholar]
- 11.Hamamori Y, Schneider MD. HATs off to Hop: recruitment of a class I histone deacetylase incriminates a novel transcriptional pathway that opposes cardiac hypertrophy. Journal of Clinical Investigation. 2003;112(6):824–826. doi: 10.1172/JCI19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Levin MD, Petrenko NB, et al. Histone-deacetylase inhibition reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy independent of angiotensin. Journal of Molecular and Cellular Cardiology. 2008;45(6):715–723. doi: 10.1016/j.yjmcc.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo P, Latronico MVG, Gallo P, et al. Inhibition of class I histone deacetylase with an apicidin derivative prevents cardiac hypertrophy and failure. Cardiovascular Research. 2008;80(3):416–424. doi: 10.1093/cvr/cvn215. [DOI] [PubMed] [Google Scholar]
- 14.Kee HJ, Sohn IS, Nam KI, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113(1):51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 15.Kong Y, Tannous P, Lu G, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113(22):2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao TC, Cheng G, Zhang LX, Tseng YT, Padbury JF. Inhibition of histone deacetylases triggers pharmacologic preconditioning effects against myocardial ischemic injury. Cardiovascular Research. 2007;76(3):473–481. doi: 10.1016/j.cardiores.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Granger A, Abdullah I, Huebner F, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB Journal. 2008;22(10):3549–3560. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee T-M, Lin M-S, Chang N-C. Inhibition of histone deacetylase on ventricular remodeling in infarcted rats. American Journal of Physiology. 2007;293(2):H968–H977. doi: 10.1152/ajpheart.00891.2006. [DOI] [PubMed] [Google Scholar]
- 19.Berry JM, Cao DJ, Rothermel BA, Hill JA. Histone deacetylase inhibition in the treatment of heart disease. Expert Opinion on Drug Safety. 2008;7(1):53–67. doi: 10.1517/14740338.7.1.53. [DOI] [PubMed] [Google Scholar]
- 20.Kook H, Lepore JJ, Gitler AD, et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. Journal of Clinical Investigation. 2003;112(6):863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho YK, Eom GH, Kee HJ, et al. Sodium valproate, a histone deacetylase inhibitor, but not captopril, prevents right ventricular hypertrophy in rats. Circulation Journal. 2010;74(4):760–770. doi: 10.1253/circj.cj-09-0580. [DOI] [PubMed] [Google Scholar]
- 22.Antos CL, McKinsey TA, Dreitz M, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. Journal of Biological Chemistry. 2003;278(31):28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Li T, Liu Y, et al. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochimica et Biophysica Acta. 2009;1793(2):300–311. doi: 10.1016/j.bbamcr.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Lagger G, O’Carroll D, Rembold M, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO Journal. 2002;21(11):2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes and Development. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trivedi CM, Luo Y, Yin Z, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3β activity. Nature Medicine. 2007;13(3):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 27.Trivedi CM, Min ML, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. Journal of Biological Chemistry. 2008;283(39):26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery RL, Potthoff MJ, Haberland M, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. Journal of Clinical Investigation. 2008;118(11):3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Molecular and Cellular Biology. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110(4):479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kee HJ, Eom GH, Joung H, et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circulation Research. 2008;103(11):1259–1269. doi: 10.1161/01.RES.0000338570.27156.84. [DOI] [PubMed] [Google Scholar]
- 33.Pflum MKH, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. Journal of Biological Chemistry. 2001;276(50):47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 34.Sun J-M, Chen HY, Moniwa M, Litchfield DW, Seto E, Davie JR. The transcriptional repressor Sp3 is associated with CK2-phosphorylated histone deacetylase 2. Journal of Biological Chemistry. 2002;277(39):35783–35786. doi: 10.1074/jbc.C200378200. [DOI] [PubMed] [Google Scholar]
- 35.Tsai S-C, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. Journal of Biological Chemistry. 2002;277(35):31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 36.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Molecular and Cellular Biology. 2004;24(2):765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen F, Kook H, Milewski R, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110(6):713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 38.Kee HJ, Kook H. Krüppel-like factor 4 mediates histone deacetylase inhibitor-induced prevention of cardiac hypertrophy. Journal of Molecular and Cellular Cardiology. 2009;47(6):770–780. doi: 10.1016/j.yjmcc.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Liao X, Haldar SM, Lu Y, et al. Krüppel-like factor 4 regulates pressure-induced cardiac hypertrophy. Journal of Molecular and Cellular Cardiology. 2010;49(2):334–338. doi: 10.1016/j.yjmcc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu W, Trivedi CM, Zhou D, Yuan L, Lu MM, Epstein JA. Inpp5f Is a polyphosphoinositide phosphatase that regulates cardiac hypertrophic responsiveness. Circulation Research. 2009;105(12):1240–1247. doi: 10.1161/CIRCRESAHA.109.208785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO Journal. 1999;18(18):5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. Journal of Clinical Investigation. 2006;116(7):1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vega RB, Harrison BC, Meadows E, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Molecular and Cellular Biology. 2004;24(19):8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margariti A, Zampetaki A, Xiao Q, et al. Histone deacetylase 7 controls endothelial cell growth through modulation of β-Catenin. Circulation Research. 2010;106(7):1202–1211. doi: 10.1161/CIRCRESAHA.109.213165. [DOI] [PubMed] [Google Scholar]
- 47.McKinsey TA, Olson EN. Cardiac histone acetylation—therapeutic opportunities abound. Trends in Genetics. 2004;20(4):206–213. doi: 10.1016/j.tig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Morin S, Charron F, Robitaille L, Nemer M. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO Journal. 2000;19(9):2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(14):7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirsh O, Seeler J-S, Pichler A, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO Journal. 2002;21(11):2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao T-P. Regulation of MEF2 by histone beacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Molecular and Cellular Biology. 2005;25(19):8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ago T, Liu T, Zhai P, et al. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133(6):978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 53.Watson PMD, Riccio A. Nitric oxide and histone deacetylases a new relationship between old molecules. Communitative and Integrative Biology. 2009;2(1):11–13. doi: 10.4161/cib.2.1.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455(7211):411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 55.Hsu C-P, Odewale I, Alcendor RR, Sadoshima J. Sirt1 protects the heart from aging and stress. Biological Chemistry. 2008;389(3):221–231. doi: 10.1515/BC.2008.032. [DOI] [PubMed] [Google Scholar]
- 56.Borradaile NM, Pickering G. NAD+, sirtuins, and cardiovascular disease. Current Pharmaceutical Design. 2009;15(1):110–117. doi: 10.2174/138161209787185742. [DOI] [PubMed] [Google Scholar]
- 57.Alcendor RR, Gao S, Zhai P, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circulation Research. 2007;100(10):1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 58.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Molecular and Cellular Biology. 2008;28(20):6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vakhrusheva O, Smolka C, Gajawada P, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circulation Research. 2008;102(6):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 60.Balcerczyk A, Pirola L. Therapeutic potential of activators and inhibitors of sirtuins. Biofactors. 2010;36(5):383–393. doi: 10.1002/biof.112. [DOI] [PubMed] [Google Scholar]
- 61.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363(1-2):15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 62.Luo J, Nikolaev AY, Imai S-I, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 63.Juan L-J, Shia W-J, Chen M-H, et al. Histone deacetylases specifically down-regulate p53-dependent gene activation. Journal of Biological Chemistry. 2000;275(27):20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 64.Thevenet L, Méjean C, Moniot B, et al. Regulation of human SRY subcellular distribution by its acetylation/deacetylation. EMBO Journal. 2004;23(16):3336–3345. doi: 10.1038/sj.emboj.7600352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan Z-L, Guan Y-J, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307(5707):269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 66.Chanda D, Xie Y-B, Choi H-S. Transcriptional corepressor shp recruits sirt1 histone deacetylase to inhibit LRH-1 transactivation. Nucleic Acids Research. 2010;38(14):4607–4619. doi: 10.1093/nar/gkq227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gobinet J, Carascossa S, Cavaillès V, Vignon F, Nicolas J-C, Jalaguier S. SHP represses transcriptional activity via recruitment of histone deacetylases. Biochemistry. 2005;44(16):6312–6320. doi: 10.1021/bi047308d. [DOI] [PubMed] [Google Scholar]
- 68.Martínez-Balbás MA, Bauer U-M, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO Journal. 2000;19(4):662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO Journal. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L-F, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293(5535):1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 71.Parmigiani RB, Xu WS, Venta-Perez G, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(28):9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jeong J, Juhn K, Lee H, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Experimental and Molecular Medicine. 2007;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 73.Chuang H-C, Chang C-W, Chang G-D, Yao T-P, Chen H. Histone deacetylase 3 binds to and regulates the GCMa transcription factor. Nucleic Acids Research. 2006;34(5):1459–1469. doi: 10.1093/nar/gkl048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grönroos E, Hellman U, Heldin C-H, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Molecular Cell. 2002;10(3):483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 75.Grégoire S, Xiao L, Nie J, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Molecular and Cellular Biology. 2007;27(4):1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO Journal. 2001;20(7):1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6: ubiquitously expressed regulators of tissue-specific gene expression. Journal of Biological Chemistry. 2000;275(50):38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 78.Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovascular Research. 2004;63(2):196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 79.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes and Development. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 80.Hayakawa F, Towatari M, Ozawa Y, Tomita A, Privalsky ML, Saito H. Functional regulation of GATA-2 by acetylation. Journal of Leukocyte Biology. 2004;75(3):529–540. doi: 10.1189/jlb.0603389. [DOI] [PubMed] [Google Scholar]
- 81.Kawamura T, Ono K, Morimoto T, et al. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. Journal of Biological Chemistry. 2005;280(20):19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 82.Kume S, Haneda M, Kanasaki K, et al. SIRT1 inhibits transforming growth factor β-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. Journal of Biological Chemistry. 2007;282(1):151–158. doi: 10.1074/jbc.M605904200. [DOI] [PubMed] [Google Scholar]
- 83.Wang B, Hao J, Jones SC, Yee M-S, Roth JC, Dixon IMC. Decreased Smad 7 expression contributes to cardiac fibrosis in the infarcted rat heart. American Journal of Physiology. 2002;282(5):H1685–H1696. doi: 10.1152/ajpheart.00266.2001. [DOI] [PubMed] [Google Scholar]
- 84.Chandrasekaran S, Peterson RE, Mani SK, et al. Histone deacetylases facilitate sodium/calcium exchanger up-regulation in adult cardiomyocytes. FASEB Journal. 2009;23(11):3851–3864. doi: 10.1096/fj.09-132415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao Y-L, Yang W-M, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Molecular and Cellular Biology. 2001;21(17):5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sucharov CC, Dockstader K, McKinsey TA. YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Molecular Biology of the Cell. 2008;19(10):4141–4153. doi: 10.1091/mbc.E07-12-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sucharov CC, Langer S, Bristow M, Leinwand L. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. American Journal of Physiology. 2006;291(5):C1029–C1037. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- 88.Glenn DJ, Wang F, Chen S, Nishimoto M, Gardner DG. Endothelin-stimulated human b-type natriuretic peptide gene expression is mediated by Yin Yang 1 in association with histone deacetylase 2. Hypertension. 2009;53(3):549–555. doi: 10.1161/HYPERTENSIONAHA.108.125088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Molecular Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO Journal. 2003;22(5):1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 92.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opinion on Investigational Drugs. 2007;16(7):1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 93.Glaser KB. HDAC inhibitors: clinical update and mechanism-based potential. Biochemical Pharmacology. 2007;74(5):659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Atmaca A, Al-Batran S-E, Maurer A, et al. Valproic acid (VPA) in patients with refractory advanced cancer: a dose escalating phase I clinical trial. British Journal of Cancer. 2007;97(2):177–182. doi: 10.1038/sj.bjc.6603851. [DOI] [PMC free article] [PubMed] [Google Scholar]