Abstract
Objective
The aim of the present study has been to establish serum free culture conditions for the ex vivo expansion and differentiation of human CD34+ cells into erythroid lineage and to study the chromatin structure, gene expression and transcription factor recruitment at the α–globin locus in the developing erythron.
Methods
A basal IMDM cell culture medium with 1% bovine serum albumin as a serum replacement and a combination of cytokines and growth factors was used for the expansion and differentiation of the CD34+ cells. Expression patterns of the alpha and beta like genes at various stages of erythropoiesis was studied by Reverse transcriptase (RT)-qPCR analysis, profile of key erythroid transcription factors was investigated by western blotting, and the chromatin structure and transcription factor recruitment at the alpha globin locus was investigated by ChIP-qPCR analysis.
Results
Human CD34+ cells in the serum free medium undergo near synchronous erythroid differentiation to yield large amount of cells at different differentiation stages. We observe distinct patterns of the histone modifications and transcription factor binding at the α-globin locus during erythroid differentiation of CD34+ cells. NF-E2 was present at upstream activator sites even before addition of erythropoietin (Epo), while bound GATA-1 was only detectable after Epo treatment. After seven days of erythropoietin treatment, H3K4Me2 modification uniformly increases throughout the α–globin locus. Acetylation at H3K9 and binding of Pol II, NF-E2 and GATA-1 were restricted to certain HS sites of the enhancer and theta gene, and were conspicuously low at the α-like globin promoters. Rearrangement of the insulator binding factor CTCF took place at and around the α-globin locus as CD34+ cells differentiated into erythroid pathway.
Conclusion
Our results indicate that remodeling of the upstream elements may be the primary event in activation of α–globin gene expression. Activation of α-globin genes upon Epo treatment involves initial binding of Pol II, down-regulation of pre-existing factors like NF-E2, removal of CTCF from the locus, then rebinding of CTCF in an altered pattern, and concurrent or subsequent binding of transcription factors like GATA-1.
INTRODUCTION
Erythropoiesis is one of the most established cell differentiation systems and is amenable to the study of chromatin structure and gene transcription in health and disease. During normal erythropoiesis, there is a burst of erythroid specific gene expression followed by gradual silencing of the transcriptome. EKLF and GATA1 are among the critical lineage restricted transcription factors responsible for the erythropoiesis as well as globin gene expression[1, 2].
Erythropoiesis in healthy individuals involves balanced high levels of transcription of alpha and beta like globin genes. In disease states, mutations in the alpha and beta like globin genes and their regulatory sequences may lead to the imbalance in the production of these gene products causing disorders such as thalassemia. The human alpha and beta like globin genes are situated not only on different chromosomes, but also in different chromatin environments. On chromosome 11, the β-globin locus is surrounded by olfactory receptor genes situated in a transcriptionally repressive environment in most cell types including erythroid cells[3]. On the other hand, on chromosome 16, the α-globin locus is surrounded by several house keeping genes in a transcriptionally active region in most cell types including erythroid cells[4].
The arrangement of the genes on the alpha globin locus is 5′-zeta(ζ) pseudozeta(ψζ) Mu, pseudoalpha (ψα)-alpha-2(α-2)-alpha-1(α-1) theta (θ)-3′. The ζ, α-1 and α-2 are the major alpha like genes (For recent review see Higgs et al [4]). The ζ gene is expressed in embryos, while the α-1 and α-2 genes are expressed throughout fetal and adult life. These genes are controlled by erythroid specific DNase hypersensitive sites (HS) HS-10, HS-33, HS-40 and HS-48 situated upstream of the 5′ end of the ζ gene located in the introns of the C16orf35 gene. The C16ORF35 gene itself is broadly expressed in non-erythroid as well as in erythroid cells. Among these hypersensitive sites, HS-40 functions as a strong erythroid specific enhancer of the α-like genes[5, 6]. The enhancer function of HS-40 in the α–like globin expression was established by measuring the α–globin promoter activity in the presence and absence of HS-40[7]. A naturally occurring deletion in the HS-40 and HS-33 region caused α-thalassemia trait [8]. In another case, a patient with α–thalassemia had a deletion of HS-40 and HS-48 sequences with the rest of the locus intact [9] and in transgenic mice deletion of HS-48 did not have a significant effect on the expression of the α-globin genes [10]. These data indicate that HS-40 is the major enhancer of the α-globin genes. In erythroid cells, both DNase hypersensitive sites (HS sites) and the α-globin gene promoters are bound by specific combinations of hematopoietic and other transcription factors such as GATA-1, NF-E2, EKLF, SCL and NFY [4, 11]. The acetylation and methylation patterns of histones, intergenic transcription, and nucleosome remodeling activity of the β-globin locus have been studied in some detail in leukemic cell lines and in transgenic mice [12, 13]. In vivo association of transcription factors, methylation of histone H3 at Lysine 4 and acetylation of histones H3 and H4 at the α–globin locus have been reported in murine primary erythroid cells and in erythroid cells derived from peripheral blood monouclear cells in culture [11, 14]. However, there are few studies on the chromatin structure and regulation of transcription including α-globin locus during differentiation of human CD34+ cells into the erythroid lineage.
The molecular mechanisms of primitive and definitive erythropoiesis and globin gene switching have been studied in erythroleukemic cell lines, in vitro culture of erythroid precursor cells and transgenic mice[3, 15–20]. In recent years breakthroughs were made in converting embryonic stem cells and adult hematopoietic CD34+ cells into erythroid cells[21–27]. However, these in vitro stem cell systems use serum-based media and/or co-culturing with stromal cells or macrophages that may have unspecific and varying effects on the progression of erythropoiesis. CD34+ cells derived from cord blood were expanded in serum free and Epo free medium[28]. However, upon Epo induction and growth in serum free medium, few cells could get past the basophilic erythroblast stage. Moreover, these cultures yield low amounts of cells at early stages of erythroid differentiation that hampers the biochemical and genome wide functional genomic and proteomic investigations. So far, to our knowledge, expansion of bone marrow CD34+ cells in serum-free medium followed by their terminal differentiation into erythroid lineage by Epo treatment has not been described. Keeping these issues in mind, in this study, we have developed an in vitro serum free human adult CD34+ cell expansion and erythroid differentiation system that yields large amount of cells from very early to late stages of differentiation. Using this cell system, we demonstrate the structural and functional properties of the α-globin locus, and pattern of gene expression during early to late erythropoiesis.
Materials and Methods
1. Cell culture and antibodies
The human CD34+ cells, isolated from the peripheral blood of G-CSF mobilized healthy volunteers, were obtained from the Yale Center of Excellence in Molecular Hematology. These CD34+ cells were expanded in StemSpan medium containing IMDM medium and 1% BSA, 10μg/ml human recombinant insulin, 200 μg/ml human transferrin, 0.1 mM β–mercaptoethanol and 2mM L-glutamine (Stem cell Technologies, Cat # 09650) supplemented with 100 ng/ml Flt-3, 100ng/ml Stem Cell Factor (SCF), 20ng/ml IL3 and 20ng/ml IL6 (StemSpan Cytokine Cocktail, Stem Cell Technologies Cat # 02690). For erythroid differentiation, the expanded CD34+ cells were grown in StemSpan medium supplemented with 20 ng/ml SCF, 5ng/ml IL3, 2μM dexamethasone, 0.2 μM estradiol and 1 unit/ml Epo (Amgen Inc. Catalogue # 606-10-432-4) in a CO2 incubator at 37°C. Throughout the CD34+ cell expansion and during their erythroid differentiation, the cell concentration was maintained at 0.5–0.65×106 cells/ml culture medium. As needed, appropriate numbers of cells were withdrawn for further studies at different time points.
The following antibodies were used in this study: H3K9Me3 (Abcam Inc., Cat # ab8898), H3K4Me2 (Upstate, Cat # 07-030), H3K9Ac (Upstate, Cat # 07-352), Pol II (Covance, Cat # 8WG 16), Pol II S2 (AbCam Cat # an-5095, Pol II S5 (AbCam Cat # ab-5131), CTCF (Santa Cruz Inc., Cat # sc15914). The fluorescent dye conjugated antibodies were from BD Biosciences: PE-conjugated-CD34+(Cat # 555822), FITC-conjugated CD14 (Cat # 555937), FITC-conjugated human CD71 (Cat # 555536) and PE-conjugated glycophorin-A (Cat # 555570). The rabbit polyclonal GATA-1 and NF-E2 antibodies were generous gifts from Dr. Emery Bresnick (Department of Pharmacology, University of Wisconsin Medical School, Madison, WI, USA.).
2. Chromatin immunoprecipitation (ChIP)
The ChIP protocol was adapted from a previously described procedure[29]. Briefly, the hCD34+ cells and their erythroid derivatives were crosslinked with 1% formaldehyde for 10 minutes at room temperature and nuclei were isolated after Dounce homogenization using a B type pestle. The crosslinked nuclei were sonicated to obtain chromatin containing ~500 bp average size DNA fragments. For each ChIP 5 μg of antibody and appropriate control IgG species were used. The antigen-antibody complex was captured on protein-G beads, washed four times with RIPA buffer and finally with PBS. The DNA-protein complex from the protein-G beads was eluted by incubating with 1% SDS for 20 minutes at 65°C. The formaldehyde cross linking of the eluted DNA-protein adducts was reversed by overnight incubation at 65°C. After proteinase K and RNase digestion of the sample, the DNA was extracted with phenol:chloroform, precipitated with ethanol, suspended in 50 μl of 10 mM TE pH8.0 and used for the quantitative polymerase chain reaction (qPCR). The primer sets used in the ChIP-qPCR are listed in sTable-1.
3. RNA, Reverse transcription and PCR
RNA from CD34+ cells and their erythroid derivatives were isolated with an RNeasy kit (Qiagen Inc) that included in-column DNase digestion. The cDNA was prepared from 2 μg of total RNA in a 20 μl reaction mixture using the superscript II (Invitrogen Inc) reverse transcriptase protocol provided by the manufacturer. The cDNA was then digested with DNase free RNase (Roche Inc) and phenol:chloroform extracted, ethanol precipitated and subjected to qPCR analysis. Equal amounts of cDNA from each sample were used for qPCR, and sample values were normalized with actin as an internal control. The qPCR was carried out on a BioRad iCycler machine with the CYBR-green PCR mix (Cat # 170-8882). The protocol was followed as per the manufacturer’s instructions. The primer sets used in the RT-qPCR are listed in sTable-2.
4. Protein extraction and western blots
Total protein from expanded and Epo treated CD34+ cells was extracted with RIPA buffer (10mM Tris-HCl pH 8.0, 140 mM NaCl, 1% Triton X100, 0.1% SDS, 1% deoxycholic acid, 1 mM DTT, Complete Protease Inhibitor cocktail from Roche diagnostics and 0.1mM Diisopropyl fluorophosphate from Sigma-Aldrich). For western blots, 50 μg protein extract per lane was resolved on 4–15% SDS-polyacrylamide gradient gels (Bio-Rad Cat # 161-1158) and transferred onto Immobilon transfer membranes (Millipore, Catalog Number IPVH15150). After protein transfer, membranes were blocked with 5% non-fat milk and probed with primary and corresponding secondary antibodies at appropriate concentrations. The blots were developed with the Super Signal West Pico kit supplied by Pierce (Catalog Number 34080).
Results
1. Expansion and differentiation of human CD34+ cells
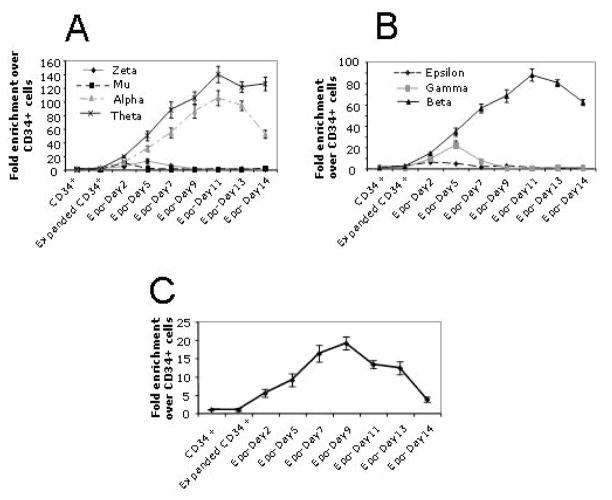
In the present study, we report establishment of a two-step process for large scale production of erythroid cells in culture. In the first step the CD34+ cells were expanded before treatment with erythropoietin (Epo). In the presence of IL3, IL6, SCF, Flt-3 and Insulin in IMDM medium containing 1% BSA, the number of CD34+ cells increased ~200 fold in two weeks (Fig 1A). We monitored for the retention of the CD34+ marker and appearance of glycophorin-A (GYPA) marker in these expanding cells by FACS analysis. We found that during expansion, CD34+ expression declines and from day-13 onwards less than 5 % of the cells were CD34+. With concomitant reduction of CD34+ mark, we observed appearance of the GYPA+ mark on a maximum of 60% of cells at day-8 in expansion culture, after which it declined rapidly and disappeared from day-13 onwards (Fig 1B). Even after disappearance of CD34 and GYPA surface markers these cells divided rapidly up to 15 days, after which their growth started declining. We estimated the mean fluorescence intensity (MFI) of PE labeled anti GYPA antibody coated cells expanded for days 6–11 in culture (Fig 1C), and found that it was markedly less than in the same cells treated with Epo for 5 days and more. Thus, the CD34+ cells expanded to day 6–11 in culture are GYPA(low) and Epo treated cells at day-5 in culture and beyond are GYPA(high). Although NF-E2 was expressed in the GYPA(low) cells (Fig 4), we could not detect α and β like globin transcripts, nor the erythroid specific ALAS2 enzyme which is a rate limiting enzyme in heme biosynthesis (Fig 3). Similarly, GATA-1 was present at a very low level as determined by western blotting (Fig 4). In the second step, when these cells at day-6 to day-11 in expansion medium were shifted to erythroid differentiation medium consisting of Epo, SCF, IL3, dexamethasone, insulin, transferrin and estradiol in IMDM medium containing 1% BSA, they become GYPA(high) at day 5 (Fig 1C) and beyond in culture. More than 85% of the total cells were GYPA(high) at day 5 of Epo treatment and this persisted until the late stages of differentiation (Fig 2B). The differentiating cells proliferated rapidly until day-10 of Epo treatment, beyond which we observed a gradual decline in the growth curve (Fig 2A). The morphology of the cells and FACS analysis by treating the cells with fluorescent dye labeled CD71 and GYPA antibodies at various stages of differentiation suggested that from day 5 onwards the differentiation followed fairly synchronous stages (Fig 2C and Supplemental sFig 1). As early as at day-2 of Epo treatment, the ε-globin from the β-locus and Mu globin from the αlocus were expressed, and were replaced by γ and ζ globin peaks at day-5 of Epo treatment (Fig 3). Subsequently, expression of these fetal genes decreased rapidly to basal level with concomitant increase in the expression of adult type α and β globin genes. Interestingly, in this cell system we find high amounts of θ-globin transcripts that peak along with the α–globin transcript and persist at late stages of differentiation. We further tested if the production of high levels of θ-globin transcripts is unique to this cell system. We hybridized the Epo treated peripheral blood mononuclear cell cDNA on a tiling ENCODE array (supplemental sFig 3). These peripheral blood mononuclear cells were grown for 8 days in the presence of Epo in a serum based medium described previously[26]. We found high levels of θ-globin message in these cells suggesting that the transcription of θ-globin gene may generally occur in cultured erythroid cells. We also confirmed the presence of full length θ-globin transcript in these cells by RT-PCR analysis (data not shown).
Figure 1.
Expansion of CD34+ cells and changes in lineage marks. The conditions of CD34+ cell expansion are described in Materials and Methods. (A) Growth curve of CD34+ cells in expansion medium. The plot depicts total number of cells at different days of culture. The starting cell number is 1 million. (B) Status of CD34, GYPA and CD14 cell surface markers during expansion of CD34+ cells. Percentage of cells stained with fluorescent labeled antibodies against CD34 (PE labeled), GYPA (PE labeled) and CD14 (FITC labeled) were determined by FACS analysis. Note that throughout expansion there was no appreciable elevation in CD14 cell surface mark that is mainly associated with monocytes. (C) Mean Fluorescence Intensity (MFI) of PE-conjugated anti Glycophorin A (GYPA) antibody bound to expanding CD34+ cells and Epo treated cells after 6 days in expansion culture. The MFI was calculated by FACS analysis at different days of expansion and Epo induced erythroid differentiation. The data in all the graphs are the averages of two independent biological replicates.
Figure 4.
Dimethyl Lysine 4 modification of Histone H3 (H3K4Me2) at α–globin locus. Chromatin immunoprecipitations (ChIP) using anti H3K4Me2 antibody were performed with sonicated formaldehyde cross linked chromatin prepared from Day-6 expanded CD34+ cells and after their erythroid differentiation for 7 days. Each ChIP-qPCR set contained equal amounts (10 nanograms) of ChIP DNA and IgG control DNA. The α-promoter qPCR primer set is common for alpha-1 and alpha-2 genes to yield identical PCR products. The data presented in the bar diagrams is the average of two independent biological replicates done in duplicate.
Figure 3.
Reverse transcriptase-quantitative PCR (RT-qPCR) analysis for transcription status of genes from alpha (A) and beta (B) globin loci and erythroid specific delta aminolaevulinic acid synthase-2 (ALAS2) enzyme (C) involved in heme biosynthesis. The unexpanded CD34+ cells that lacked erythroid specific GYPA cell surface marker was used as a reference to measure the relative increase in the messenger RNA during expansion and erythroid differentiation by Epo treatment. Ten nanograms each of reverse transcribed cDNA (see Materials and Methods) from unexpanded CD34+ cells, day-6 expanded CD34+ cells and at various days of Epo treatment was used for qPCR. The data are the averages of two independent biological replicates. The γ globin qPCR primers are common for Aγ and Gγ genes to give rise to identical PCR products, and α–globin qPCR primer set is common for alpha-1 and alpha-2 genes that produce identical PCR products.
Figure 2.
Erythroid differentiation of expanded CD34+ cells. (A) Growth curve of expanded CD34+ cells during erythroid differentiation upon Epo treatment. The X-axis shows the days after shifting day-6 expanded CD34+ cells in Epo containing erythroid differentiation medium. (B) Status of cell surface marker proteins during erythroid differentiation. Unexpanded cells were 100% CD34+ that did not have any lineage markers such as GYPA and CD14. These CD34+ cells were expanded for 6 days before treating them with Epo (see Materials and Methods). The percentage of cells expressing CD34, GYPA and CD14 was calculated by FACS analysis of the fluorescence labeled cells with antibodies against CD14 (FITC conjugated), CD34 (PE conjugated) and GYPA (PE conjugated). (C) Giemsa staining of day-6 expanded CD34+ cells and during different stages of their differentiation into erythroid lineage. The images were captured using Olympus BX51 microscope with 60X/0.90 Objective lens, Olympus Q-Color5 camera (Olympus America, Center Valley, PA) and QCapturePro software (QImaging, Surrey, BC). The stages of depicted erythropoiesis are CFUe (Day-2), basophilic erythroblasts (Day-6), polychromatic erythroblasts (Day-11), orthochromatic erythroblasts (Day-16) and mixture of orthochromatic erythroblasts and orthochromatic erythrocytes (Day-19).
The transcript of ALAS2, an erythroid specific enzyme necessary for heme biosynthesis is synthesized rapidly upon Epo treatment and peaks at day-9 before the peak of globin transcription. We observed a sharp decline of ALAS2 message at later stages of erythropoiesis (Fig 3C). Cells expanded for 6 to 11 days in culture without Epo had the ability to differentiate into the erythroid lineage with equal efficiency. We have not tested the ability of CD34+ cells expanded beyond Day-11 to differentiate into erythroid cells. In all the experiments described in this study, we have used the CD34+ cells expanded up to day-6 in culture and their subsequent differentiation into the erythroid lineage.
2. Chromatin structure at the α–globin locus during erythropoiesis
2.1 Methylation of Histone H3 at lysine 4
H3K4 methylation is associated with transcriptionally active and/or poised genes in euchromatic regions of the genome[30]. As the α-globin lies in the neighborhood of constitutively expressing housekeeping genes, we investigated the status of H3K4Me2 at the α-globin locus before and during erythropoiesis (Fig 4). We observed significant amounts of H3K4Me2 modification at the HS-48 and HS-46 enhancer sites and at the Mu and θ globin genes in expanded CD34+ cells in which the α-globin locus is transcriptionally silent. This confirms earlier observations that the α-globin locus is not in a heterochromatic structure in non-erythroid cells[31]. Upon differentiation into the erythroid pathway, there was an overall increase in the dimethylation of H3K4 across the α-globin locus, except at the α and ζ promoters and at HS-46 and HS-40. There was complete absence of H3K4Me2 modification at the transcriptionally inactive ζ globin promoter before and after Epo treatment of the cells (Fig 4). We did not detect any change in the H3 methylation level at the HS-40 sequences upon erythroid differentiation, while there was a marginal decrease of H3K4Me2 levels at the HS-46 region. Thus, among the α-globin regulatory sequences that participate in active transcription, HS-40 was conspicuous by basal level of H3K4Me2 modification that remained static after Epo treatment, while the α-promoter had modest levels of increase of dimethylated H3K4 (Fig 4).
2.2 Modification of Histone H3 lysine 9
Acetylation and methylation of H3K9 are generally associated with transcriptionally active and inactive genes, respectively[30, 32, 33]. However, the transcriptionally active β–globin promoter was shown to exhibit H3K9Me3 modification[34]. Therefore, we examined the acetylation and methylation status of the H3K9 across α-globin locus. We found that the entire α-globin was devoid of any acetylation at the H3K9 site before Epo treatment (Fig 5B). However, upon erythroid differentiation by Epo treatment, H3K9 acetylation was exclusively associated with the α-globin enhancer sequences HS-48, HS-46 and HS-33. Among the α-like globin promoters we detected small amounts of acetylation only at the θ-globin promoter.
Figure 5.
ChIP-qPCR analysis for the modification of H3K9 at α-globin locus. ChIP were carried out using antibodies against H3K9Me3 (A) and H3K9Ac (B) and sonicated formaldehyde cross linked chromatin from Day-6 expanded CD34+ cells (CD34+) and upon their differentiation into erythroid lineage for 7 days (Epo-Day7). 10 nanograms each of ChIP DNA and IgG control DNA was used in qPCR. The qPCR primer set for α-promoter is common for alpha-1 and alpha-2 genes that give rise to identical PCR product. The data is the average of two independent biological replicates in duplicate.
In contrast to the acetylation of H3K9, H3K9Me3 in expanded CD34+ cells was present across the α-globin locus except at the α and ζ promoters, and at the HS-33 and HS-46 regions of the enhancer (Fig 5A). Upon erythroid differentiation, there was a marked decrease in the H3K9Me3 status at the HS-48 and HS-10 regions of the enhancer, while HS-46 and HS-48 were unaffected. On the other hand we detected a marked increase in H3K9Me3 at the Mu globin promoter and moderate increase over the α-globin prompter upon erythroid differentiation and marginal decrease over θ globin promoter. These data suggest that H3K9Me3 modification is dynamic and undergoes rearrangement upon erythroid differentiation of the expanded CD34+ cells. Further, a characteristic feature of the H3K9 modification upon Epo treatment is the increasing H3K9Me3 modification at the α-like globin promoter sequences and increasing H3K9Ac modification at the enhancer HS sites (Fig 5A & B).
3. Pattern of transcription factor appearance at various stages of erythropoiesis and their recruitment at the alpha globin locus
GATA-1 is an essential transcription factor for erythropoiesis as well as for the enhancement of globin gene transcription. It is reported to be present on promoter as well as enhancer sequences of the globin genes[2, 35, 36]. NF-E2 was shown to selectively bind to the HS2 site of the β-globin LCR and remotely load Pol II on its cognate β-globin promoter[37–39]. CTCF is a zinc finger containing sequence specific DNA binding factor that can act both as a transcription factor and as an essential component of boundary elements that block the activity of upstream enhancers on downstream promoters that include β–globin locus[40, 41]. During CD34+ erythroid differentiation in culture, we examined the appearance of these transcription factors at various stages of erythropoiesis and their distribution across the α-globin locus in CD34+ cells before and at day-7 after the addition of Epo.
3.1 Transcription factor profile during erythropoiesis
We found low but significant levels of NF-E2 and CTCF, and basal levels of GATA-1 in the extracts of expanded CD34+ cells (Fig 6). However, upon Epo treatment high levels of GATA-1 protein started appearing at day-4 and persisted until late stages of differentiation. NF-E2 protein levels declined initially until day-2 of Epo treatment, and from day-3 onwards reappeared in high amounts and, like GATA-1, persisted until late stages of differentiation. On the other hand, we did not see significant changes in CTCF levels until day-3 of Epo treatment. From day-4 onwards the CTCF levels increased to significantly higher levels and persisted. Thus, the pattern of appearance of these transcription factors in the initial stages of Epo treatment is distinct from their near uniform pattern of increase from day 4 of Epo treatment onwards (Fig 6).
Figure 6.
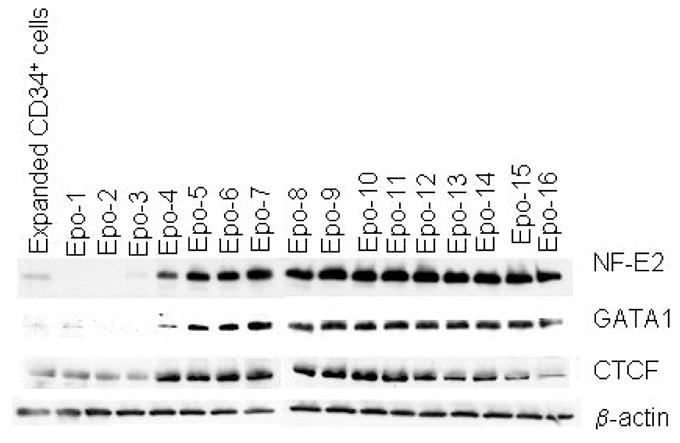
Western blot profile of transcription factor appearance during erythroid differentiation of expanded CD34+ cells. Aliquots of CD34+ cells expanded in culture for day 6 and shifted to Epo containing erythroid differentiation medium were taken for total protein extraction on each day. 50 μg of total protein from each sample was resolved on 4–15% SDS-PAGE gel to transfer on nylon membrane for western blotting. The Day-6 expanded CD34+ cells and their erythroid derivative at successive days are depicted at the top of the figure. β-actin served as a control as its expression was not significantly effected throughout the differentiation.
3.2 Recruitment of GATA-1 and NF-E2
In expanded CD34+ cells, there was no GATA-1 binding anywhere on the α-globin locus. However upon erythroid differentiation, we detected GATA-1 recruitment at the HS-10 and HS-46 sites of the α–globin enhancer (Fig 7A). There was little or no GATA-1 at the transcriptionally silent ζ and Mu genes. Interestingly, the transcriptionally active α and θ genes were also devoid of GATA-1. In these erythroid cells, at the time points examined, GATA-1 seems to be primarily associated with the enhancer HS-10 and HS-46 sequences of the α-globin locus.
Figure 7.
Recruitment of GATA1 (A) and NF-E2 (B) at the α–globin locus. ChIPs were carried out using sonicated crosslinked chromatin prepared from Day-6 expanded CD34+ cells (CD34+) and after their differentiation into erythroid lineage for 7 days (Epo-Day7), and antibodies against GATA-1 and NF-E2. 10 nanograms of each of ChIP DNA and control IgG DNA was used for qPCR. The α-promoter qPCR primer set is common for alpha-1 and alpha -2 gene promoters that produce identical PCR products. Each of the ChIP-qPCR values in the figure is the average of two independent biological replicates done in duplicate.
On the other hand we detected robust presence of NF-E2 at several HS sites of the α-globin enhancer sequences in the expanded CD34+ cells in which the α-globin genes are transcriptionally silent (Fig 7B). Upon erythroid differentiation of these cells, we did not detect any redistribution of NF-E2 on these enhancer HS sites, although there were differences in the ChIP signals at various HS sites. We did not see NF-E2 occupancy either at the actively transcribing α and θ genes promoters, or at the silent ζ promoter either before or after erythroid differentiation (Fig 7B). However, in the case of the Mu gene we see a substantial increase in NF-E2 occupancy of the promoter even though this gene is transcriptionally inactive at day-7 of differentiation (Fig 3).
3.3 Recruitment of Pol II
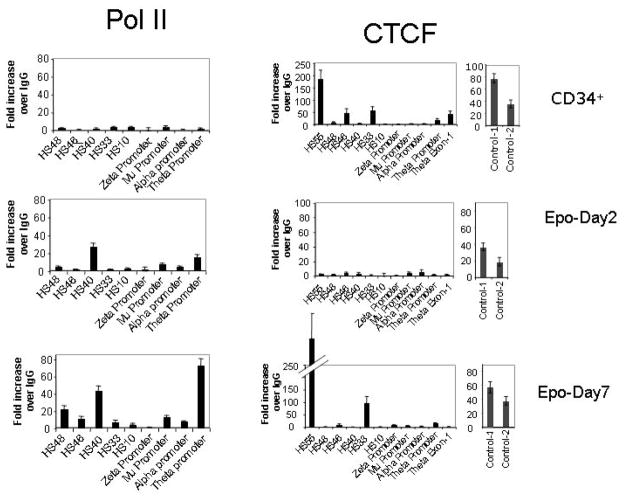
We further analyzed for the binding of the dephosphorylated form of pol II at carboxy terminal domain of the large subunit (CTD) that is generally associated with transcriptionally active or poised promoters[42]. In the expanded CD34+ cells we did not observe any Pol II binding at the α-globin locus suggesting that the locus in these cells is transcriptionally silent (Fig 8). Upon induction of erythropoiesis, we observed appearance of Pol II at the HS-40 enhancer site as early as day-2 of Epo treatment. At day-7, Pol II occupied other HS sites, namely HS-46 and HS-48. Interestingly we did not observe any Pol II at the α–promoter either in expanded CD34+ cells or in their erythroid derivatives. However we see a strong signal of Pol II at the θ globin promoter, suggesting that transcription of α and θ globin genes are differently regulated (Fig 8). Among the elongating forms of Pol II phosphorylated at serine-5 (Pol II S5) and serine-2 (Pol II S2) of the C-terminal domain repeat region, we detected Pol II S5 specifically at the α-globin promoter. Pol II S2 was present at HS-46 and at the body of the α and θ globin genes (Supplemental sFig 2).
Figure 8.
Pol II and CTCF recruitment profile on the α–globin locus during erythroid differentiation. Sonicated crosslinked chromatin isolated from 6 days-expanded CD34+ cells and after 2 days (Epo-Day2) and 7 days (Epo-Day7) into erythroid differentiation medium were used in ChIP-qPCR experiments along antibodies against Pol II and CTCF. 10 nanograms of each ChIP DNA and control IgG DNA was used for qPCR. Due to the extensive homology between alpha-1 and alpha-2 genes a common promoter qPCR primer set is designed to produce one identical PCR product. Control-1 and Control-2 are the two qPCR primer sets from the imprinted region upstream of IGF2 that served as positive control for CTCF ChIP-qPCR.
3.4 Rearrangement of CTCF
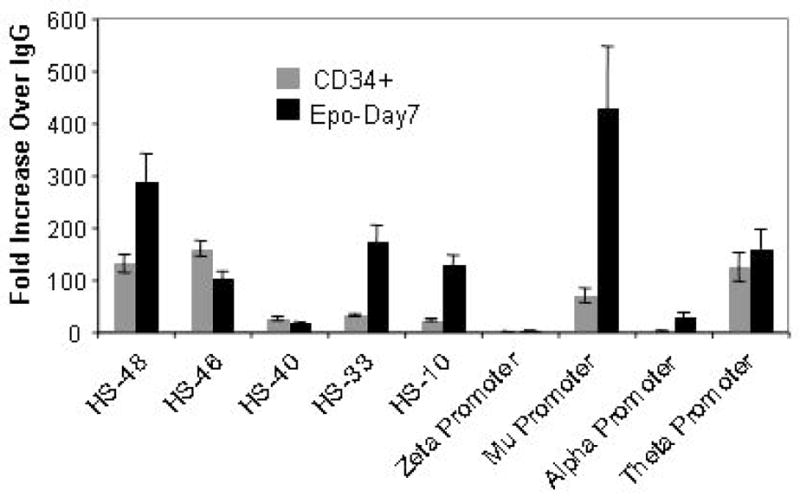
The erythroid specific α–globin locus is positioned between transcriptionally active housekeeping genes in the majority of cell types including erythroid cells. Hence, insulation of α–globin locus from the surrounding transcriptional activity may be required in erythroid as well as non-erythroid environments. Therefore, we investigated the positioning of the CTCF across the α-globin locus during erythropoiesis of CD34+ cells (Fig 8). In expanded CD34+ cells we found robust association of CTCF at HS-33 and HS-46 surrounding the core HS-40 enhancer site and at HS-55, which is a constitutive DNase hypersensitive site in various cell types including erythroid cells. In these cells, CTCF was also present at θ globin gene situated at the 3′ end of the locus. Thus, prior to erythropoiesis, the α–globin locus is flanked by CTCF binding at the 5′ end enhancer HS sites and at 3′ end of the locus at θ globin gene. Remarkably, at day-2 of differentiation there was almost complete disappearance of CTCF from the binding sites in the α–globin cluster, even though there was little or no change in binding to another chromosomal imprinting region between MRPL23 and IGF2 genes (Fig. 8). At day 2 of Epo treatment, we find mixture of CD71+ & GPA+; CD71+; GPA+; and CD71− & GPA− populations (supplemental sFig 1). However, the disappearance of CTCF from the HS-40 region is complete, and therefore must occur in all these cell populations. It will also be interesting to find out if the removal of CTCF binding precedes the appearance of erythroid surface marks. By Day-7, CTCF binding had been restored although there were some differences from the pattern seen at expanded CD34+ cells (Fig 8). Most notably, there was a severe reduction in binding at the HS-46 site at the 5′ end and at the θ globin gene at the 3′ end of the cluster.
Discussion
The system for in vitro expansion and differentiation of human CD34+ cells described in this study has several advantages including the generation of sufficient numbers of untransformed cells for various biochemical studies, and the relatively synchronous nature of the differentiation that makes it possible to determine the sequence of events during very early to late stages of erythroid differentiation (Fig 1 & 2 and sFig 1). The serum free culture conditions described in our protocol provides good control over the growth and differentiation of CD34+ cells and eliminates unknown effects associated with serum and co-culture based cultures. Induction of erythropoiesis of the expanded CD34+ cells converts more than 85% of the cells into GYPA+ cells by day-5 of Epo treatment, and these cells progressively differentiate in a relatively synchronous manner to produce large number of cells at different stages of erythropoiesis (Fig 2 and sFig 1). One caveat is that the conditions for differentiation are not fully physiologic, both with respect to the types of cytokines and cell to cell signaling, and environmental differences such as ambient rather than physiologic oxygen concentrations. Nevertheless, our data on the chromatin structure and transcription factor recruitment at the α–globin locus and the pattern of appearance of key erythroid transcription factors such as GATA1 and NF-E2 during erythropoiesis suggest that the system reflects many aspects of normal in vivo differentiation.
Although the expanded CD34+ cells express low levels of glycophorin-A until Day-11 in culture, they lack erythroid properties as evidenced by the silent α-globin locus and very low levels of GATA-1 and absence of other key erythroid specific genes such as β-globin and ALAS2 (Fig 3). These are still uncommitted multipotent cells that can be differentiated into either erythroid (Fig 1 & 2) or myeloid lineages (data not shown). The order of peak appearance of the α and β like globin gene mRNAs during erythroid development of expanded CD34+ cells in our experiments follows the same temporal pattern as does their appearance during ontogeny (Fig 3). Previously, expression of γ globin genes in erythroid precursor cells has been reported that is consistent with our observation in this study[18–20]. However, we see early appearance of Mu globin, which peaks at day 2 of Epo treatments. This timing of Mu expression is different from the previous report wherein it was observed to peak at day-10 of Epo induction[43]. In addition, we also detected unusually high amounts of θ globin message that persists at late stages of erythropoiesis (Fig 3 and sFig 3). Earlier studies have reported that θ globin is expressed in fetal/adult stage and its expression is about 50 fold less than alpha message in the fetal erythroid stage[44]. However, in bone marrow cells of patients with thalassemia and sickle cell disease, significantly higher amounts of θ-globin transcripts were detected[45]. This increase was attributed to the accelerated erythropoiesis in the bone marrow of these patients. In this in vitro cell culture system, cells are stimulated with non-physiological doses of cytokines at ambient oxygen levels, which may mimic accelerated erythropoiesis under stress, leading to the high levels of θ-globin transcript (Fig 3). Further insights into possible functions of the θ-gene must await identification and characterization of its presumptive protein product.
Our observation of the absence of NF-E2 in very early stages of induced erythropoiesis and its subsequent steep induction until late stages (Fig 6) is consistent with the earlier studies[18, 46, 47]. However, we detected significant levels of NF-E2 protein in uncommitted, multipotent, expanded CD34+ cells in culture. Interestingly, upon Epo treatment NF-E2 disappears initially to again reappear from day-3 onwards. Further studies are needed to understand the role of down regulation of NF-E2 in early erythropoiesis. ChIP experiments show recruitment of NF-E2 prior to erythropoiesis and at day-7 of epo treatment that suggest role of NF-E2 in α-globin gene regulation in erythroid as well as in non-erythroid cells (Fig 7). Previously, presence of NF-E2 was demonstrated at the HS-26 sequence of the α–globin locus and at the HS2 sequence of the β-globin locus in murine FDCP-mix cells and NF-E2 expressing HeLa cells, respectively[31, 48]. Forced expression of NF-E2 in HeLa cells was observed to generate DNase hypersensitive regions at the β-globin HS2 sequences that suggest the role of NF-E2 in chromatin modification[49]. Indeed, in erythroid cells NF-E2 was shown to mediate trimethylation of H3K4 by its association with MLL2 methyltransferase[50]. The prior presence of NF-E2 at the α–globin locus in expanded CD34+ cells and its continued presence at the same sites, albeit at different levels, suggests that NF-E2 may keep the α-globin enhancer in a poised state prior to erythroid differentiation. Further studies are needed to understand the role of NF-E2 in chromatin modification and α-globin transcription before and after the onset of erythroid differentiation of CD34+ cells. Identification of all the NF-E2 interacting proteins in expanded CD34+ cells and their erythroid derivatives will be helpful in understanding the differential role of NF-E2 at the α-globin locus. Curiously, we have seen high levels of NF-E2 recruitment at the transcriptionally inactive Mu globin promoter and absence of detectable NF-E2 at ζ, α and θ promoter regions.
Basal levels of GATA-1 protein in expanded CD34+ cells and their exponential increase from day-4 of Epo treatment co-insides with the lack of GATA-1 binding at the α-globin locus prior to onset of erythropoiesis and its subsequent binding at day-7 of Epo treatment (Fig 6 & 7). Association of GATA-1 at HS-10 and HS-46 in erythroid cells is consistent with earlier observations in murine CFUe cells[31]. However, in humanized mouse erythroid cells GATA-1 was shown to be present primarily at HS-40 [14]. Interestingly, like NF-E2, significant level of GATA-1 was present at the transcriptionally inactive Mu globin promoter (Fig 7). It remains to be seen if the presence of GATA-1 and NF-E2 at Mu globin promoter has any influence on the activity of surrounding α–like globin genes.
The initiating form of Pol II (recognized by monoclonal antibody 8WG16) is associated with transcriptionally active as well as poised promoters[42]. The transcriptionally silent α-globin locus prior to erythropoietin seems to be devoid of Pol II binding (Fig 8). At the onset of erythropoiesis, as early as day-2, when GATA-1 and NF-E2 are not at detectable levels and CTCF is stripped off the locus, Pol II appears on the HS-40, then spreads to other regions at the later stages of erythropoiesis (Fig 8). Thus HS-40 seems to be the earliest recruitment site for Pol II. At Day-2 of Epo treatment we find a mixture of CD71+ and GYPA+, CD71+ and CD71− cells (sFig 1). It will be interesting to find out which of these cell populations show the appearance of Pol II at HS-40. The presence of the initiating form of Pol II at the enhancer region (Fig 8) and of phospho-S5 Pol II at the α-globin promoter (sFig 2) suggests that the α-globin enhancer may be a nucleus for formation of the initiation complex. In addition, presence of Pol II-phospho S2 at the HS-46 region (sFig 2) of the enhancer suggests the participation of the α-globin enhancer in the elongation of transcription also. Similarly, a transcription elongation function was suggested for the β-globin LCR[51]. However, De Gobbi et al and Anguita et al found Pol II prominently at the α-globin promoters in the peripheral blood mononuclear cell derived erythroid cells and murine TER119+ cells using antibodies against the N-terminal region of the large subunit of the Pol II[14, 31]. The antibodies raised against the N-terminal region of the Pol II would not distinguish between the initiating and elongating forms. Thus, the discrepancy in the Pol II binding could be related to the cell type, or choice of antibody used. Interestingly, we detected high levels of Pol II at the θ globin promoter in erythroid cells, which may reflect a difference in the regulation of α and θ promoters.
CTCF is a multifunctional protein that associates with insulators, causes looping of the chromatin, regulates noncoding RNA transcription and establishes local chromatin structure at the repetitive elements in the mammalian genome[52]. CTCF is also reported to activate transcription by directly interacting with large subunit of the Pol II complex[40]. Many binding sites for CTCF are constitutive, and the sites found in the α–globin cluster of undifferentiated CD34+ cells closely resemble those seen, for example, in non-erythroid HeLa cells (data not shown). The pattern of CTCF recruitment at the 5′ and 3′ ends of the α-globin locus and its subsequent rearrangement during erythropoiesis suggests dual roles of CTCF as an insulator prior to erythropoiesis and as a facilitator of transcription in erythroid cells (Fig 8). Prior to erythropoiesis, CTCF may insulate the α-globin locus from the activity of surrounding genes. Upon erythropoiesis, the insulator CTCF is stripped off of the α-globin locus and is rearranged on the erythroid specific HS-33 site, perhaps acting as a positive transcription factor or changing the looping structure of the local chromatin. Further detailed studies are needed to understand the roles of CTCF as an insulator and facilitator of α-globin gene transcription during erythroid development.
Dimethylation of H3K4 (H3K4Me2) is broadly associated with transcriptionally active genes as wells as their enhancers[53]. The H3K4Me2 modification pattern described in this study (Fig 4) is consistent with the earlier observations[14, 31]. However, our result for the H3K9Ac modification pattern at the α-globin locus differs from the earlier reports that show H3 histone H3 acetylation predominantly at the α-globin genes[14, 31]. This discrepancy could be due to the choice of antibodies. The antibody used in the pervious studies recognizes acetylation of histone H3 at K9 as well as K14 positions, where as in this study we have used the antibody that is specific to the H3K9Ac modification (Fig 5). Although doubly acetylated histone H3 at K9 and K14 [53, 54] as well as the acetylation at the single K9 site[55] are generally associated with the transcriptionally active promoters, not all transcriptionally active genes have H3K9Ac modification[56]. This is in conformity with lack of this modification at transcriptionally active α-globin promoters in the erythroid derivatives of CD34+ cells (Fig 5). Instead, we detected H3K9 specific acetylation at HS-33, HS-46 and HS-48 enhancer sites and elevated levels of H3K9Me3 at the α–like gene promoters (Fig 5). These site-specific H3K9 acetylations and methylations may be a way to modulate the correct recruitment of transcription factors (Fig 7 & 8) and promote enhancer-promoter interactions. We also observe high levels of H3K9Me3 modification at the transcriptionally inactive Mu globin promoter, consistent with the general association of this modification with inactive genes[55]. Overall, our data shows that when CD34+ cells are differentiated into erythroid lineage, the α-like globin promoters are trimethylated at H3K9 and the H3K9Ac modification is associated with upstream enhancer sequences. The extent of H3K9Me3 modification may vary between transcriptionally active and inactive genes as in case of α–globin and Mu globin gene promoters.
Taken together, our results indicate that there is a prominent change in the status of the upstream activator elements and less at the promoters of the α–globin genes themselves during the stages of erythroid differentiation that we have studied. Remodeling of the upstream elements may be the primary event in activation of α–globin gene expression at endogenous loci. Activation of α-globin genes upon Epo treatment involves initial binding of Pol II and removal of pre-existing factors like NF-E2 and CTCF, then rearrangements of CTCF and concurrent or subsequent binding of transcription factors like GATA-1 and NF-E2. The ζ-Globin gene was conspicuously devoid of chromatin modifications and transcription factor recruitment. A unique set of chromatin remodeling and transcription factor binding may be playing the role of inhibition of ζ-globin transcription in adult erythroid cells.
Perhaps the most unexpected result in the present studies was the finding that CTCF disappears from the α–globin locus after stimulation by erythropoietin, without a significant change in total cellular CTCF levels or in binding at a non-erythroid site (Fig 8). Subsequently, with the progression of erythropoiesis, CTCF is rebound to these regions with a different distribution than seen in non-erythroid cells. This would be consistent with a rearrangement of higher order chromatin structure in this region early in erythropoiesis.
Supplementary Material
Acknowledgments
We thank Drs. Bernard Forget and Patrick Gallaghar for helpful discussions, and Dr. Emory Bresnick for his generous gift of GATA-1 and NF-E2 antibodies. We would like to acknowledge the supply of hCD34+ cells, facilities and expert assistance provided by Sharon Lin, Yale Center of Excellence in Molecular Hematology. This work was supported by the grants from NIH R01 AG23111 and in part by the Yale Center of Excellence in Molecular Hematology, NIH DK072442.
Footnotes
Conflict of Interest Disclosure.
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basu P, Lung TK, Lemsaddek W, et al. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood. 2007;110:3417–3425. doi: 10.1182/blood-2006-11-057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgs DR, Vernimmen D, Wood B. Chapter 5 Long-Range Regulation of alpha-Globin Gene Expression. Adv Genet. 2008;61:143–173. doi: 10.1016/S0065-2660(07)00005-3. [DOI] [PubMed] [Google Scholar]
- 5.Jarman AP, Wood WG, Sharpe JA, Gourdon G, Ayyub H, Higgs DR. Characterization of the major regulatory element upstream of the human alpha-globin gene cluster. Mol Cell Biol. 1991;11:4679–4689. doi: 10.1128/mcb.11.9.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Lowrey CH, Stamatoyannopoulos G. Analysis of enhancer function of the HS-40 core sequence of the human alpha-globin cluster. Nucleic Acids Res. 1997;25:2917–2922. doi: 10.1093/nar/25.14.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernet A, Sabatier S, Picketts DJ, et al. Targeted inactivation of the major positive regulatory element (HS-40) of the human alpha-globin gene locus. Blood. 1995;86:1202–1211. [PubMed] [Google Scholar]
- 8.Viprakasit V, Kidd AM, Ayyub H, Horsley S, Hughes J, Higgs DR. De novo deletion within the telomeric region flanking the human alpha globin locus as a cause of alpha thalassaemia. Br J Haematol. 2003;120:867–875. doi: 10.1046/j.1365-2141.2003.04197.x. [DOI] [PubMed] [Google Scholar]
- 9.Viprakasit V, Harteveld CL, Ayyub H, et al. A novel deletion causing alpha thalassemia clarifies the importance of the major human alpha globin regulatory element. Blood. 2006;107:3811–3812. doi: 10.1182/blood-2005-12-4834. [DOI] [PubMed] [Google Scholar]
- 10.Tang XB, Feng DX, Di LJ, et al. HS-48 alone has no enhancement role on the expression of human alpha-globin gene cluster. Blood Cells Mol Dis. 2007;38:32–36. doi: 10.1016/j.bcmd.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. Embo J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan MC, Karmakar S, Weissman SM. Control of beta globin genes. J Cell Biochem. 2007;102:801–810. doi: 10.1002/jcb.21507. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero G, Delgado-Olguin P, Escamilla-Del-Arenal M, et al. Globin genes transcriptional switching, chromatin structure and linked lessons to epigenetics in cancer: a comparative overview. Comp Biochem Physiol A Mol Integr Physiol. 2007;147:750–760. doi: 10.1016/j.cbpa.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 14.De Gobbi M, Anguita E, Hughes J, et al. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood. 2007;110:4503–4510. doi: 10.1182/blood-2007-06-097964. [DOI] [PubMed] [Google Scholar]
- 15.Kingsley PD, Malik J, Emerson RL, et al. “Maturational” globin switching in primary primitive erythroid cells. Blood. 2006;107:1665–1672. doi: 10.1182/blood-2005-08-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Han H, Ye X, Stafford M, Barkess G, Stamatoyannopoulos G. Transcriptional potentials of the beta-like globin genes at different developmental stages in transgenic mice and hemoglobin switching. Blood Cells Mol Dis. 2004;33:318–325. doi: 10.1016/j.bcmd.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Gabbianelli M, Testa U, Massa A, et al. Hemoglobin switching in unicellular erythroid culture of sibling erythroid burst-forming units: kit ligand induces a dose-dependent fetal hemoglobin reactivation potentiated by sodium butyrate. Blood. 2000;95:3555–3561. [PubMed] [Google Scholar]
- 18.Pope SH, Fibach E, Sun J, Chin K, Rodgers GP. Two-phase liquid culture system models normal human adult erythropoiesis at the molecular level. Eur J Haematol. 2000;64:292–303. doi: 10.1034/j.1600-0609.2000.90032.x. [DOI] [PubMed] [Google Scholar]
- 19.Clarke BJ, Nathan DG, Alter BP, Forget BG, Hillman DG, Housman D. Hemoglobin synthesis in human BFU-E and CFU-E-derived erythroid colonies. Blood. 1979;54:805–817. [PubMed] [Google Scholar]
- 20.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu C, Olivier EN, Velho M, Bouhassira EE. Globin switches in yolk sac-like primitive and fetal-like definitive red blood cells produced from human embryonic stem cells. Blood. 2008;111:2400–2408. doi: 10.1182/blood-2007-07-102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douay L, Giarratana MC. Ex vivo generation of human red blood cells: a new advance in stem cell engineering. Methods Mol Biol. 2009;482:127–140. doi: 10.1007/978-1-59745-060-7_8. [DOI] [PubMed] [Google Scholar]
- 23.Fujimi A, Matsunaga T, Kobune M, et al. Ex vivo large-scale generation of human red blood cells from cord blood CD34+ cells by co-culturing with macrophages. Int J Hematol. 2008;87:339–350. doi: 10.1007/s12185-008-0062-y. [DOI] [PubMed] [Google Scholar]
- 24.Giarratana MC, Kobari L, Lapillonne H, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 25.Dorn I, Lazar-Karsten P, Boie S, et al. In vitro proliferation and differentiation of human CD34+ cells from peripheral blood into mature red blood cells with two different cell culture systems. Transfusion. 2008;48:1122–1132. doi: 10.1111/j.1537-2995.2008.01653.x. [DOI] [PubMed] [Google Scholar]
- 26.Migliaccio G, Di Pietro R, di Giacomo V, et al. In vitro mass production of human erythroid cells from the blood of normal donors and of thalassemic patients. Blood Cells Mol Dis. 2002;28:169–180. doi: 10.1006/bcmd.2002.0502. [DOI] [PubMed] [Google Scholar]
- 27.Wojda U, Noel P, Miller JL. Fetal and adult hemoglobin production during adult erythropoiesis: coordinate expression correlates with cell proliferation. Blood. 2002;99:3005–3013. [PubMed] [Google Scholar]
- 28.Neildez-Nguyen TM, Wajcman H, Marden MC, et al. Human erythroid cells produced ex vivo at large scale differentiate into red blood cells in vivo. Nat Biotechnol. 2002;20:467–472. doi: 10.1038/nbt0502-467. [DOI] [PubMed] [Google Scholar]
- 29.Euskirchen GM, Rozowsky JS, Wei CL, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 31.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. Embo J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin W, Barkess G, Fang X, et al. Histone acetylation at the human beta-globin locus changes with developmental age. Blood. 2007;110:4101–4107. doi: 10.1182/blood-2007-05-091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genomic Proteomic. 2006;5:209–221. doi: 10.1093/bfgp/ell028. [DOI] [PubMed] [Google Scholar]
- 34.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Horak CE, Mahajan MC, Luscombe NM, Gerstein M, Weissman SM, Snyder M. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc Natl Acad Sci U S A. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Escamilla-Del-Arenal M, Recillas-Targa F. GATA-1 modulates the chromatin structure and activity of the chicken alpha-globin 3′ enhancer. Mol Cell Biol. 2008;28:575–586. doi: 10.1128/MCB.00943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- 38.Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 39.Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci U S A. 2001;98:10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chernukhin I, Shamsuddin S, Kang SY, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goh SH, Lee YT, Bhanu NV, et al. A newly discovered human alpha-globin gene. Blood. 2005;106:1466–1472. doi: 10.1182/blood-2005-03-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albitar M, Care A, Peschle C, Liebhaber SA. Developmental switching of messenger RNA expression from the human alpha-globin cluster: fetal/adult pattern of theta-globin gene expression. Blood. 1992;80:1586–1591. [PubMed] [Google Scholar]
- 45.Ley TJ, Maloney KA, Gordon JI, Schwartz AL. Globin gene expression in erythroid human fetal liver cells. J Clin Invest. 1989;83:1032–1038. doi: 10.1172/JCI113944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng T, Shen H, Giokas D, Gere J, Tenen DG, Scadden DT. Temporal mapping of gene expression levels during the differentiation of individual primary hematopoietic cells. Proc Natl Acad Sci U S A. 1996;93:13158–13163. doi: 10.1073/pnas.93.23.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labbaye C, Valtieri M, Barberi T, et al. Differential expression and functional role of GATA-2, NF-E2, and GATA-1 in normal adult hematopoiesis. J Clin Invest. 1995;95:2346–2358. doi: 10.1172/JCI117927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onishi Y, Kiyama R. Interaction of NF-E2 in the human beta-globin locus control region before chromatin remodeling. J Biol Chem. 2003;278:8163–8171. doi: 10.1074/jbc.M209612200. [DOI] [PubMed] [Google Scholar]
- 49.Onishi Y, Kiyama R. Enhancer activity of HS2 of the human beta-LCR is modulated by distance from the key nucleosome. Nucleic Acids Res. 2001;29:3448–3457. doi: 10.1093/nar/29.16.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demers C, Chaturvedi CP, Ranish JA, et al. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawado T, Halow J, Bender MA, Groudine M. The beta -globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 53.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 54.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishida H, Suzuki T, Kondo S, Miura H, Fujimura Y, Hayashizaki Y. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res. 2006;14:203–211. doi: 10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.