Abstract
Studies of the size and morphology of anatomical structures rely on accurate and reproducible delineation of the structures, obtained either by human raters or automatic segmentation algorithms. Measures of reproducibility and variability are vital aspects of such studies and are usually acquired using repeated scans and repeated delineations (in the case of human raters). Methods exist for simultaneously estimating the true structure and rater performance parameters from multiple segmentations and have been demonstrated on volumetric images. In this work, we extend the application of previous methods onto two-dimensional surfaces parameterized as triangle meshes. Label homogeneity is enforced using a Markov random field formulated with an energy that addresses the challenges introduced by the surface parameterization. The method was explored using both simulated raters and surface labels obtained from an atlas registration. Simulated raters are computed using a global error as well as a novel and more realistic boundary error model. We study the impact of raters and their accuracy based on both models, and show how effectively this method estimates the true segmentation on simulated and real surfaces.
Keywords: Rater evaluation, labeling, label fusion, surface mesh, statistics, STAPLE
1. INTRODUCTION
Assessment of structural and morphological anatomical characteristics plays an essential role in the application of medical imaging in clinical research. Such assessments depend upon the ability to accurately and precisely label structures in multidimensional images. The labeling process may be carried out by human raters or by automated segmentation algorithms. Statistically motivated approaches1,2 have been developed (e.g., Simultaneous Truth and Performance Level Estimation, STAPLE ) to fuse or combine labels from multiple individuals or algorithms into an estimate of a truth label set that can be made more reliable than any of the individual, underlying labelings. These approaches have been applied to volumetric studies, and additionally provide measures of rater or algorithm performance. Although typically acquired in two or three dimensions, medical images may be processed and/or analyzed to form representations on manifolds (e.g., cortical surfaces). In this paper, we extend the STAPLE algorithm1 to two-dimensional surfaces represented by triangle meshes in the STAPLE Surface (STAPLES) method. To characterize this approach, we develop a new, realistic model of “simulated rater” behavior and explore application of STAPLES in an atlas-based approach for determining gyral labels on brain cortical surfaces.
2. METHODS
STAPLE1 simultaneously estimates a true segmentation and a reliability characterization for each rater. It was originally presented on a voxel-wise basis for multiple-label, volumetric images, and included a Markov Random Field (MRF) to model spatially correlated structures. Here we modify the original approach to operate on surface labels. First, the update equations of the STAPLE algorithm are applied vertex-wise to the triangle mesh (Section 2.1). Second, a new mesh-based MRF accounts for spatial consistency with multiple labels (Section 2.2).
2.1 STAPLE on Triangle Meshes
In this formulation, labels exist on a surface parameterized as a triangle mesh with vertices , i ∊{1 … N} and labels edges ej ∊ (i, i’), j ∊ {1 … M}. A label is to be assigned each vertex, Ti ∊ {0, 1, …L,} given the labels from R raters Di = Dir, r ∊ {1 … R}. As well, we seek to estimate rater performances, modeled as a set of confusion matrices Θ = θrs’s, s, s’ ∊ {1 … L}.For any vertex vi, θrs’s denotes the probability that rater r assigns label s when s’ is the true label.
The algorithm is initialized by setting the performance parameters equal to a matrix close to the identity for all raters as suggested in Warfield et al.1 At iteration k, the segmentation given the performance parameters is then estimated using the update equation
| (1) |
Next, the rater performance parameters for the (k + 1)th iteration are estimated with
| (2) |
These equations are identical to the conventional STAPLE algorithm since at this level the specifics of the locations of the points on which labels are defined are irrelevant. The segmentation and performance estimation are iterated until a convergence criterion is met.
2.2 Markov Random Field
As with the original approach, we utilize a Markov Random Field (MRF) to model the spatial homogeneity of labels. However, a reformulation of the MRF model was required due to two major challenges of the mesh parameterization not present in the conventional pixel or voxel image representation. First, vertices need not be equally spaced nor regularly connected as they are in the volumetric framework. A Gaussian kernel was applied to account for this, as the relative contribution to the local conditional probability from vertices within the clique are weighted by the distance of the clique vertex to the vertex itself. Second, cliques need not be of the same order (i.e. vertices are connected by different numbers of edges with different angular separation). Here we have addressed via simple normalization, but more sophisticated approaches will be explored in the future. Multiple labels are addressed by penalizing any “wrong” labels equally. The necessary changes were incorporated by defining the following MRF potential function
| (3) |
where T is the full label configuration, dij is the distance between vertices i and j, d0 is fixed and controls the size of the kernel. The interaction weight, βii’ = β/Mi if i and i’ are neighbors where β is fixed and Mi denotes the number of neighbors of vertex i. If vertices i and i’ are not neighbors, βii’ = 0. Optimization was achieved using an iterative conditional modes (ICM) scheme,3 which successfully converged for the presented problems.
3. DATA
3.1 Simulated Raters
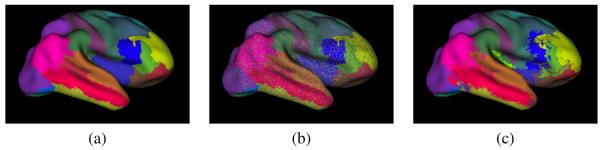
Simulated rater errors were modeled in two ways. First, we used confusion matrix in which the i, jth element indicates the probability that the rater will assign the jth label when the ith label is correct at any particular location. In this case, the identity matrix describes the “perfect” rater. In these simulations, confusion matrices were constructed such that each rater had equal expected performance for all labels, and errors were uniformly distributed among the remaining labels. Modeled errors are equally likely to occur throughout the image domain, i.e., every vertex is equally likely to be incorrectly labeled. An example of such a rater is shown in Figure 1(b).
Figure 1.
Examples of simulated cortical surface raters. (a) The true surface segmentation. Sample segmentations by a rater with true positive rates of 0.7 parameterized by (b) a confusion matrix and (c) boundary error rate.
Since in practice, raters are more likely to make errors near the object boundaries, we introduce a second rater error model that models this kind of behavior. Three vector parameters describe a rater’s performance: r, l, and b. The scalar r is the rater’s global true positive fraction. The vector l encodes the probability, given an error occurred, that it was at the ith boundary. Finally the vector b describes the error bias at every boundary. The unbiased rater has bi = 0.5 ∀i, while values of 0.0 or 1.0 indicate that one label or the other is always favored when an error occurs. A labeling produced by this type of simulated rater is shown in Figure 1(c). This is visually similar to the types of errors commonly observed in automatic gyral labeling approaches.
4. RESULTS
4.1 Simulations
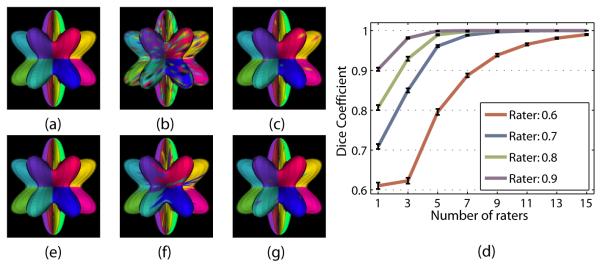
We first examined how the quality of the segmentation estimate produced with STAPLES varies with changes in the number of raters and their performance parameters. This was done by measuring the average label overlap (measured by the Dice Coefficient, defined as DC = 2ǀA ∩ Bǀ/(ǀAǀ + ǀBǀ)), between the true label configuration and estimated the true configuration. Figure 2 shows the results of 50 Monte Carlo iterations for the confusion matrix raters. We observe that the quality of the estimated true segmentation improves as the number of raters increases, and as the rater performance increases. A more detailed analysis will be provided in the full manuscript.
Figure 2.
Results for simulated surfaces: (a) The ground truth phantom parcellation, (b) simulated confusion matrix rater (true positive fraction = 0.7), (c) STAPLES result using 7 such raters without spatial correlation, (d) the results of 50 Monte Carlo iterations varying the expected rater performance and the number of raters, and (e) the STAPLES result using a MRF. Also shown are (f) a sample simulated boundary error rater (true positive fraction = 0.7), and (g) the STAPLES result using 7 such raters without spatial correlation.
4.2 Multi-Atlas Parcellation
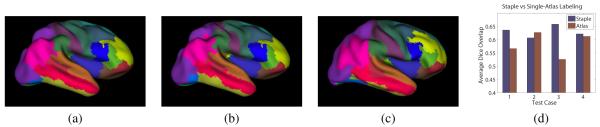
We also examined the applicability of STAPLES as part of a multi-atlas parcellation scheme in the spirit of Rohlfing et al.,2 using a leave-one-out experiment. Four subjects were drawn from the OASIS data7 (http://www.oasis-brains.org) and gyral labels were obtained using Freesurfer.5 These labels were transferred to cortical surfaces that were obtained using CRUISE.4 Three labeled surfaces were used as an atlas, and the remaining brain surface were considered as a target for the atlas-based parcellation. The parcellation was done by first partially inflating8 both the target and source surfaces. Next, the iterative closest point algorithm9 was used to register the target to the source using an affine tranformation. Labels were then transfered to vertices on the source surface from the nearest vertex on the target surface. This method was repeated to obtain a labelling of the source surface for each of the three atlases. These were combined using STAPLES to provide an estimate of the true labeling of the cortical gyri on the source surface, the results of which are shown in Figure 3. We see that a reasonable parcellation is obtained, despite the variability in the atlases and the simplicity of the registration and label transfer. Future work will include the incorporation of surface features such as curvature in this labelling scheme. Figure3(d) shows a quantitative, fair comparison of STAPLES against average individual atlas performance in our four test cases. Though not conclusive, these results suggest that STAPLES may improve the segmentation obtained relative to single-atlas methods. More thorough results with larger data pools will be provided in the full manuscript.
Figure 3.
An example of our method applied to a cortical gyral labeling task. Shown are (a) the true cortical parcellation, (b) the parcellation estimated from this multi-atlas / STAPLES approach, (c) one of the atlas subject surfaces, and (d) a comparison of STAPLES and single-atlas parcellation schemes.
5. CONCLUSION
This work introduced STAPLES, an extension of STAPLE that enables statistical label recombination on 2D surfaces parameterized as a triangle meshes. This enables the improvement of labels on surfaces obtained from biomedical images, including surface representations of the cortex, cerebellum, and subcortical structures, for example. It also suggests a further generalization to arbitrary higher dimensional manifolds such as time series, tensor spaces and the like. This type of generalization is easily accomplished for a point-wise STAPLE approach, as demonstrated in this work. Generalizing the MRF potential for any manifold, however, presents a significant challenge, but would be extremely beneficial to research involving these types of high-dimensional data.
6. ACKNOWLEDGEMENTS
This work was supported in part by NIH/NINDS grants R01NS56307 and R01NS37747. The OASIS project is supported in part by grants P50AG05681, P01AG03991, R01AG021910, P50MH071616, U24RR021382, and R01MH56584.
REFERENCES
- [1].Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE TMI. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rohlfing T, Russakoff DB, Maurer CR. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE TMI. 2004;23(8):983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- [3].Besag J. On the statistical analysis of dirty pictures. Journal of the Royal Statistical Society. 1986;48:259–302. [Google Scholar]
- [4].Han X, Pham DL, Tosun D, Rettman ME, Xu C, Prince JL. CRUISE: Cortical Reconstruction Using Implicit Surface Evolution. Neuroimage. 2004;23(3):997–1012. doi: 10.1016/j.neuroimage.2004.06.043. [DOI] [PubMed] [Google Scholar]
- [5].Desikan RS, Sgonne F, Fischl B, Albert MS, Killiany RJ, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- [6].Makris M, Schlerf JE, Hodge SM, Schmahmann D, et al. MRI-based surface-assisted parcellation of human cerebellar cortex: An anatomically specified method with estimate of reliability. Neuroimage. 2005;25(4):1146–1160. doi: 10.1016/j.neuroimage.2004.12.056. [DOI] [PubMed] [Google Scholar]
- [7].Marcus DS, Wang TH, Buckner RL, et al. Open Access Series of Imaging Studies (OASIS): Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. Journal of Cognitive Neuroscience. 2004;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- [8].Tosun D, Rettmann M, Prince JL. Mapping techniques for aligning sulci across multiple brains. Medical Image Analysis. 2004;8(3):295–309. doi: 10.1016/j.media.2004.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Besl PP, McKay NN. A Method for Registration of 3-D Shapes. IEEE Trans. PAMI. 1992;14:239–256. [Google Scholar]