Abstract
Human α-tocopherol (α-T) transfer protein (ATTP) plays a central role in vitamin E homeostasis, preventing degradation of α-T by routing this lipophilic molecule for secretion by hepatocytes. Mutations in the gene encoding ATTP have been shown to cause a severe deficiency in α-T, which results in a progressive neurode-generative spinocerebellar ataxia, known as ataxia with vitamin E deficiency (AVED). We have determined the high-resolution crystal structure of human ATTP with (2R,4′R,8′R)-α-T in the binding pocket. Surprisingly, the ligand is sequestered deep in the hydrophobic core of the protein, implicating a large structural rearrangement for the entry and release of α-T. A comparison to the structure of a related protein, Sec14p, crystallized without a bona fide ligand, shows a possibly relevant open conformation for this family of proteins. Furthermore, of the known mutations that cause AVED, one mutation, L183P, is located directly in the binding pocket. Finally, three mutations associated with AVED involve arginine residues that are grouped together on the surface of ATTP. We propose that this positively charged surface may serve to orient an interacting protein, which might function to regulate the release of α-T through an induced change in conformation of ATTP.
Alpha-tocopherol (α-T) transfer protein (ATTP) is a member of the family of lipid-binding proteins containing two CRAL-TRIO domains, pfam03765 (residues 11–83) and pfam00650 (residues 89–275), so-called because the domains common to this family were first recognized in cellular retinal-binding protein (CRALBP) and the Trio protein (1, 2). Other notable family members include Sec14p, a yeast phosphatidyl inositol (PI)/phosphatidyl choline transfer protein essential for the secretory pathway (2), and supernatant protein factor, which stimulates the epoxidation of squalene through an incompletely understood mechanism (3). The sequence identity over the conserved region of ATTP ranges from 28% (Sec14p) to 32% (CRALBP). All of the family members are cytosolic proteins, and both ATTP and CRALBP participate in the intercellular transfer of lipid-like ligands.
There are eight forms of vitamin E (α-δ tocopherol and α-δ tocotrienol), which differ in the degree of methylation (decreasing from α-T to δ-T) of the chroman ring and saturation of the phytyl tail; all forms are potent lipophilic antioxidants, but α-T is the most biologically potent because of the action of ATTP (4). Of the eight possible stereoisomers of α-T, only (2R,4′R,8′R)-α-T occurs naturally. Synthetic vitamin E supplements are racemic mixtures, but ATTP is sensitive to stereochemistry and particularly so at the C2 stereocenter (5). The importance of ATTP in α-T homeostasis became apparent when mutations in ATTP were found to cause ataxia with vitamin E deficiency (AVED), an autosomal recessive disease in which degeneration of neurons results in a progressive spinocerebellar ataxia (6). Mutations in ATTP were also found to cause retinitis pigmentosa (7). ATTP is a cytosolic protein expressed mainly in the liver (8), and heterologous expression of ATTP in cell culture confers the ability to secrete α-T through a secretory pathway that is brefeldin A insensitive (9). This activity prevents the metabolism of α-T and excretion as 2,5,7,8-tetramethyl-2(2′carboxyethyl)-6-hydroxychroman (4). Patients with AVED absorb α-T from their diets normally, but have nearly immeasurable steady-state plasma levels caused by defective incorporation of α-T for transport with very low-density lipoprotein in hepatocytes (10). A transgenic mouse in which the ATTP gene was deleted replicated the phenotype of the human disease (11).
We have now determined the crystal structure of ATTP in a complex with (2R,4′R,8′R)-α-T. The protein was expressed as recombinant selenomethionyl protein in Escherichia coli, and (2R,4′R,8′R)-α-T was added exogenously. A crystal of selenomethionyl protein was produced, and phases were determined to 2.1 Å by multiwavelength anomalous diffraction (MAD). The structure was refined to 1.5 Å by using an additional single wavelength data set. The structure reveals α-T bound in the hydrophobic core of ATTP. Extensive interactions between ATTP and α-T explain the exquisite specificity of the protein for its ligand. Comparisons to the reported structure of Sec14p and an analysis of disease-causing mutations led us to propose a mechanism by which the release of α-T may be mediated through a dramatic conformational change in ATTP.
Materials and Methods
Expression and Purification of ATTP. Analysis of an irreproducible crystal with full-length ATTP indicated that an N-terminal deletion favored crystallization. Therefore, we cloned residues 21–278 of human ATTP by means of PCR amplification from a cDNA plasmid (kind gift of T. Siddique, Northwestern University, Evanston, IL) into a modified version of the pET21 vector that contained the cloning sites of pET28 (Novagen). The resulting gene encoded an N-terminal hexahistidine tag and thrombin-cleavage site followed by ATTP residues 21–278. This protein was expressed in E. coli BL21 (DE3) under control of the T7 promoter with overnight induction by 0.4 mM isopropyl β-d-thiogalactoside at 20°C for natural and selenomethionyl proteins. Selenomethionyl protein was produced by a nonauxotrophic protocol (12). All purification steps were performed at 4°C unless otherwise noted. Soluble recombinant ATTP was obtained by sonication of cells in lysis buffer (50 mM Tris·HCl, pH 8.0/300 mM NaCl) supplemented by Protease Inhibitor Tablets (Roche Molecular Biochemicals) and 4 μM all-rac-α-T (Fluka). After clarification, the extract was applied to a nickelchelation column (Qiagen, Chatsworth, CA) and eluted with a stepwise gradient of 250 mM imidazole in lysis buffer. An aliquot of (2R,4′R,8′R)-α-T (Fluka) in ethanol was added in equimolar amounts to recombinant ATTP as determined by the method of Bradford (13), and the protein was applied to a Superdex 75 HK26/60 column (Amersham Pharmacia) in column buffer (20 mM Tris·HCl, pH 8.0/150 mM NaCl/1 mM DTT). Peak fractions were pooled and supplemented with an equimolar amount of (2R,4′R,8′R)-α-T. The purified protein was digested with 2 units of thrombin (Amersham Pharmacia) per mg of recombinant protein for 90 min at 25°C. The product contained the residues GSHM followed by residues 21–278 of ATTP. After addition of 0.1 mM PMSF, cleaved recombinant ATTP was again applied to a Superdex 75 column, and peak fractions were pooled and concentrated to ≈5 mg/ml by ultrafiltration.
Crystallography. Crystals were grown by hanging-drop vapor diffusion against 5% (wt/vol) PEG-4000 (Fluka) in 100 mM Tris·HCl, pH 8.0–8.4 at 20°C, starting from 1 μl of purified ATTP and 1 μl of reservoir buffer. Microseeding with crystals of native protein was required to produce crystals of selenomethionyl protein. Typical crystals had dimensions of 0.30 × 0.15 × 0.03 mm. They are in spacegroup P212121 with a = 40.1 Å, b = 77.2 Å, and c = 85.4 Å and contain one molecule per asymmetric unit with a solvent content of 37.2%. Crystals were frozen in a cryoprotectant composed by the addition of 25% (vol/vol) PEG400 (Aldrich) in reservoir buffer. MAD data were measured at three wavelengths near the Se K edge by using a Q4R charge-coupled device detector system at beamline X4A of the National Synchrotron Light Source, Brookhaven, NY. hkl (14) was used for data processing, and solve (15) was used to find four of five potential Se sites and calculate experimental phases. The MAD data set was combined with a single-wavelength data set and submitted to arp/warp (16) for automated model building. The initial model contained main-chain and side-chain atoms for residues 25–273, except for a gap between residues 37 and 46. Residues were manually added to the model by using o (17) to fill the gap, extend the C terminus, and add α-T during refinement with cns (18) against all data from 50 to 1.5 Å. The α-T model was constructed by combining the chroman ring from Cambridge Structural Database (19) entry MOPHLB01 and the phytyl chain from entry YORDEO. The final model contained residues 25–275, α-T, and 253 water molecules.
Structure Comparisons. The coordinates of ATTP were compared to existing structural data with dali (20). The only strong match was 1AUA, which contains the coordinates of the Sec14p crystal structure. The N-terminal domain of ATTP was separately aligned in lsqkab (21) by matching Cα atoms of residues 28–37 and 48–81 to residues 37–46 and 54–87 from Sec14p, respectively. Similarly, the C-terminal domain alignment was optimized by using residues primarily from the β-sheet, 100–104, 112–118, 154–160, 190–198, and 221–227 of ATTP to 108–112, 119–125, 173–179, 208–216, and 239–245 of Sec14p, respectively.
Results
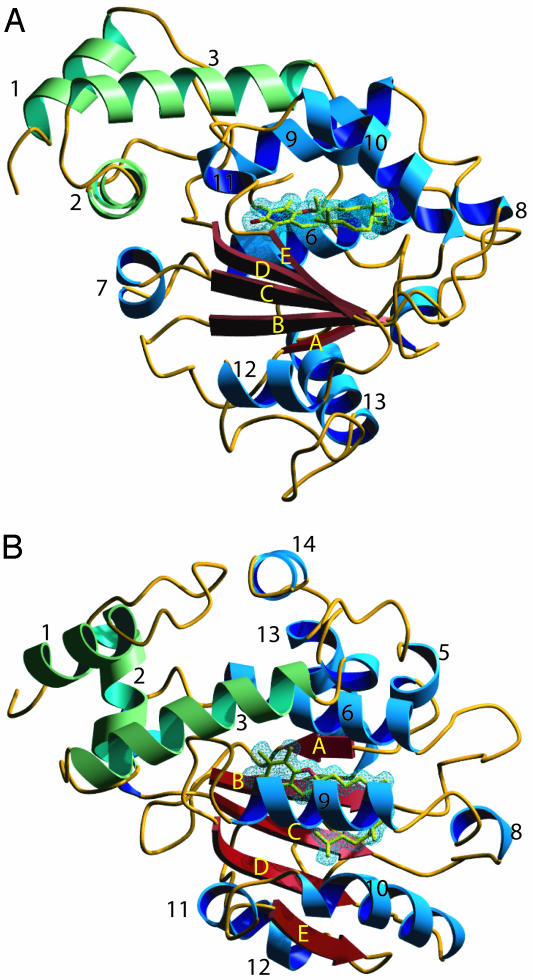
Overall Fold. The crystal structure of the complex of ATTP and (2R,4′R,8′R)-α-T was solved by phasing with MAD data from a single selenomethionyl crystal (Tables 1, 2, 3). An atomic model containing residues 25–275 and α-T was refined to 1.5-Å resolution. The structure of ATTP is composed of two domains, an N-terminal all-helical domain and a C-terminal domain, which at its core is composed of a βαβαβαβ fold (Fig. 1). Most of the interactions between the domains involve hydrophobic contacts from the packing of helix 3 of the N-terminal domain against helices 6 and 9 of the C-terminal domain. In addition, there is a hydrogen bond between the side chains of Y73 of helix 3 and E141 of helix 6, another hydrogen bond between the side chain of N72 of helix 3 and both the side chain and carbonyl oxygen of D185 from helix 9, and a covalent bond between C80 of helix 3 and K178 of helix 9 involving the terminal sulfur atom and epsilon amino group. There was no evidence for the presence of the covalent bond in a sample of purified protein subjected to tryptic digestion and mass spectroscopic analysis (data not shown). The formation of this bond may be specific to the milieu of the protein crystals and is unlikely to play a role in the function of ATTP.
Table 1. MAD diffraction data statistics.
| λ1 (0.9686) | λ2 (0.9789) | λ3 (0.9792) | |
|---|---|---|---|
| Resolution, Å | 20 > d > 2.1 | 20 > d > 2.1 | 20 > d > 2.1 |
| Reflections | |||
| Total | 138,081 | 137,224 | 137,835 |
| Unique | 28,105 | 28,371 | 28,374 |
| Completeness, % | 95.9 (83.7) | 96.8 (83.3) | 96.8 (82.9) |
| 〈I/σ(I)〉 | 23.6 (5.3) | 25.6 (7.8) | 30.7 (9.2) |
| Rsym, % | 9.1 (33.8) | 7.7 (18.8) | 6.4 (17.6) |
Numbers in parentheses refer to the values for the outer shell.
Table 2. MAD diffraction difference ratios (20 > d > 2.1).
| λ1 | λ2 | λ3 | |
|---|---|---|---|
| λ1 | 0.051 | 0.033 | 0.043 |
| λ2 | 0.064 | 0.027 | |
| λ3 | 0.039 |
Table 3. Refinement statistics, 50 > d > 1.5 Å (λ = 0.9918).
| Reflections, total | 325,760 | |
| Work/test | 71,973 | 7,829 |
| Completeness, Rsym, % | 97.4 (89.2) | 7.4 (21.3) |
| 〈I/σ(I)〉 | 24.5 (8.4) | |
| Atoms, total | 2,332 | |
| Protein | 2,048 | |
| Ligand | 31 | |
| Water | 253 | |
| R/Rfree, % | 18.7 | 21.3 |
| rms bond ideality | 0.0154 | |
| rms B bonds/angles Å2 | Main chain | 2.14/2.97 |
| Side chain | 2.87/4.05 |
Numbers in parentheses refer to the values for the outer shell.
Fig. 1.
Secondary structure of ATTP. (A) Ribbon diagram of ATTP viewed down the β-sheet. (B) View after rotation of A by 90° on horizontal axis. α-T is colored in yellow. The N-terminal domain helices are indicated in green, and the C-terminal domain helices are indicated in blue and strands are indicated in red. The 2 Fo – Fc electron density that covers α-T is drawn as a blue mesh at 1 σ. Images were generated with povscript (36) and rendered with povray.
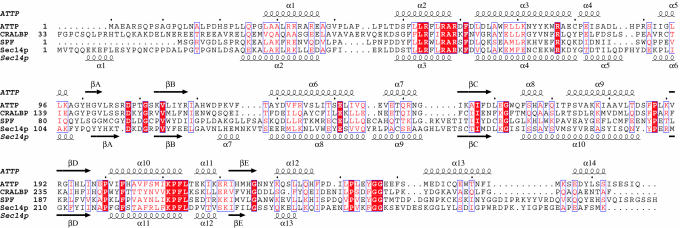
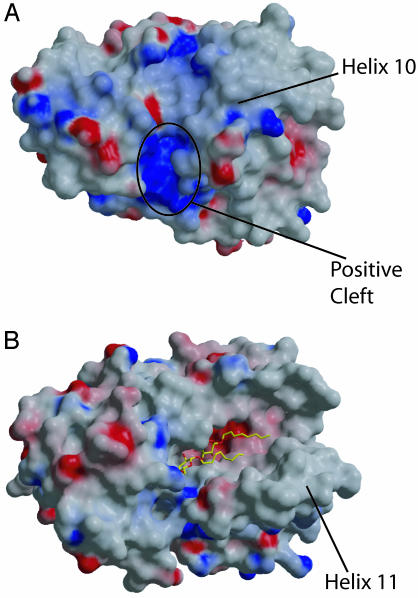
Crystal structures of two other CRAL-TRIO family members have been reported (3, 22, 23), but coordinates are available only for Sec14p. The Sec14p structure was determined with two molecules of β-octyl glucopyranoside (β-OG) in the presumed ligand-binding pocket. Although the overall fold of ATTP is similar to Sec14p (Figs. 1 and 2), a Cα superposition using residues from throughout the molecule with dali (20) resulted in a poor fit (rms deviation 4.7 Å). Separate alignments of the N-terminal domain (limited to helices 1, 2, and 3) and C-terminal domain (limited primarily to the β-sheet) resulted in a rms deviation of 0.6 Å over 44 residues and 0.8 Å over 28 residues, respectively. The N-terminal domain is rotated relative to the C-terminal domain in ATTP by 49.5° when compared to the Sec14p structure. The location of the two β-OG molecules was similar in the C-terminal domain to the location of α-T (Fig. 3 A and B). The most striking difference in the two structures relevant to the ligand-binding pocket is the arrangement of helices 9 and 10 of ATTP as compared to helices 10 and 11 of Sec14p. In the Sec14p structure, these helices are ≈25 Å apart, exposing the ligand-binding pocket (Figs. 3 and 4). In contrast, the equivalent helices are ≈10 Å apart in ATTP, and the ligand is buried from solvent.
Fig. 2.
Sequence alignment of four CRAL-TRIO proteins. The sequences of human ATTP and yeast Sec14p were aligned by using structural information, after which the sequences of human CRALBP and supernatant protein factor (SPF) over the relevant region were then also aligned. Secondary structure assignments are indicated above for ATTP and below for Sec14p. White lettering boxed with a red background indicates residues that are identical in all four proteins, and red lettering indicates similar residues. Figure was generated by espript (37).
Fig. 3.
Comparison of ATTP and Sec14p. (A) Stereo diagram of the Cα backbone of ATTP. α-T is shown in ball-and-stick representation in yellow. Helix 9 (residues 173–184) is in contact with helix 10 (residues 201–213), separated by ≈10 Å. (B) Stereo diagram of the Cα backbone of Sec14p oriented by alignment of the C-terminal domain to ATTP (see Materials and Methods). Two β-OG molecules are shown in ball-and-stick representation in yellow. Helix 10 (residues 184–204) does not have contact with helix 11 (residues 219–231), being separated by ≈25 Å, leading to exposure of the ligand-binding pocket. (C) Stereodiagram of Cα backbone of ATTP in red (residues 100–239) superimposed with sec14p in black (residues 108–257). The amino-terminal domain is excluded for clarity. Images were generated by bobscript (38).
Fig. 4.
Comparison of molecular surfaces of ATTP and Sec14p. (A) Electrostatic potential surface map of ATTP. The position of helix 10 is indicated. A positively charged cleft is indicated where a number of disease-causing mutations are located. (B) Electrostatic potential surface map of Sec14p. Here, the exposure of the ligand-binding pocket is apparent from the difference in position of helix 11, which is equivalent to helix 10 of ATTP. Surface potentials were calculated in grasp and are indicated from red (–) to dark blue (+)(>+15 kT). Images were rendered with raster3d (39).
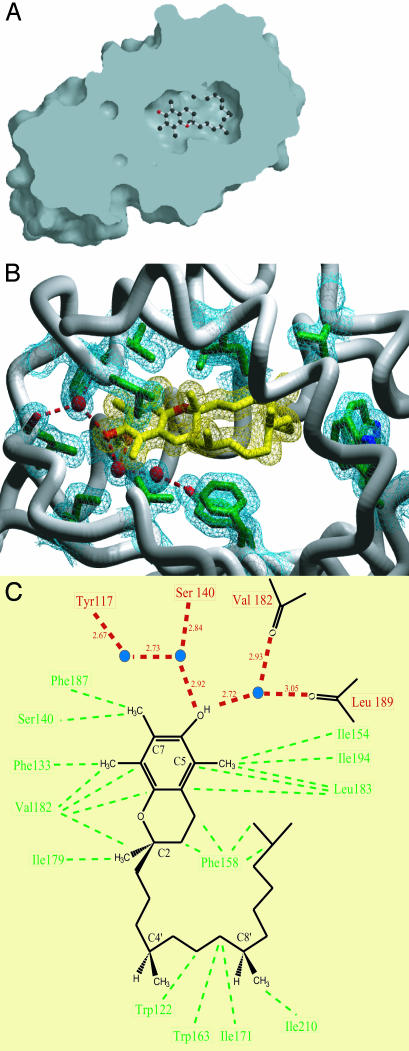
Interactions of ATTP and α-T. As seen from a cut molecular surface view in Fig. 5A, α-T is bound in a hydrophobic pocket that is entirely sequestered from bulk solvent. Large conformational changes would need to occur for entry or exit of α-T from the ligand-binding pocket. Several protein groups that line this pocket make van der Waals contacts with α-T (Fig. 5 B and C). These include side chains from β strands C (I154, F158) and D (I194); from helices 6 (F133, S136, S140), 9 (I179, V182, L183), and 10 (I210); and from several of the connecting loops, W122 between strand B and helix 6, W163 between strand C and helix 8, I171 between helices 8 and 9, and F187 between helix 9 and strand D. Four well-ordered water molecules are also enclosed in the binding pocket. Two of these are hydrogen-bonded to the hydroxyl group of the chroman ring; one is in turn hydrogen-bonded to the carbonyl oxygen atoms of V182 and L189, and the other is hydrogen-bonded to the side-chain hydroxyl group of S140 and a third water molecule. The fourth water molecule is near the phytyl tail but is not closely associated with α-T. The buried surface area of the complex of ATTP and α-T was calculated to be ≈1,200 Å2 by using grasp (24).
Fig. 5.
Ligand-binding pocket of ATTP. (A) Cut-surface view of the ligand-binding pocket of ATTP. α-T is shown in ball-and-stick representation. The ligand-binding pocket is buried deep from the exposed surface of the protein. Surface was generated in grasp and rendered in raster3d. (B) Close-up view of the ligand binding pocket. Side chains (in green) that form van der Waals contacts with α-T (in yellow) or participate in hydrogen-bonding networks in the ligand-binding pocket are shown. Three well-ordered water molecules located in the binding pocket are shown as red spheres. The protein backbone is shown as a worm model, excluding residues 216–220 and 249–275 for clarity. Electron densities that cover the residues of interest, α-T, and three water molecules are drawn as mesh at 1 σ in the 2 Fo – Fc map. Dashed red lines indicate hydrogen bonds. Image was generated in povscript (36) and rendered in povray. (C) Schematic representation of the ligand-binding pocket. α-T is shown with the C2, C4′, and C8′ stereocenters labeled. The C5 methyl is lacking in γ-T and δ-T, and the C7 methyl is lacking in β-T and δ-T. Tocotrienols contain double bonds at C3′,C7′, and C11′. The carbonyl oxygen atoms of Val-182 and Leu-189 are drawn to illustrate the hydrogen-bond interactions with one of the water molecules. Three well-ordered water molecules located in the binding pocket are depicted as blue circles. Dashed red lines indicate hydrogen bonds, and the lengths of the bonds are indicated. Dashed green lines indicate van der Waals interactions.
Discussion
Specificity of the Ligand-Binding Pocket. Recently the binding affinity of ATTP for tocopherols and related molecules was measured directly (25). The degree of methylation on the chroman ring affected the affinity of ATTP for ligand, with binding affinities in the order α-T ≫ β-T > γ-T > δ-T. The extensive van der Waals contacts between the methyl groups and protein likely explains the observed affinity differences. The C7 methyl group absent from β-T and δ-T interacts with the side chains of S140 and F187, whereas the C5 methyl group missing from γ-T and δ-T interacts with the side chains of I154, L183, and I194. In addition, it has been shown that a change in configuration at C2 to S lowers the binding affinity ≈20-fold. Residues F133, I179, V182, and L183 form a binding pocket for the methyl group at this position. Binding to the S configuration at C2 would require a major rearrangement of the ligand-binding pocket. In addition, modification of the hydroxyl group of the phenol ring dramatically affects the binding affinity of α-T analogs, likely because of the extensive hydrogen-bonding network involving the phenol group of the chroman ring. Finally, the lower affinity of ATTP for the tocotrienols can be explained by the remarkable way in which the phytyl chain of α-T is bent within the ligand-binding pocket, as this would not be as well supported by the unsaturated tail of the tocotrienols.
Comparison to the Sec14p Structure: A Possible Second Conformation. The original interpretation of the Sec14p structure was that the β-OG molecules were bound in a manner analogous to one of its cognate ligands, PI. Given the structure of ATTP, it seems instead that the presence of these detergent molecules may have induced a conformational change to an “open” state (Fig. 4). Such a conformational change was previously proposed by others when comparing the structure of human supernatant protein factor to that of Sec14p (3) and would be required for entry and exit of α-T from the binding pocket of ATTP. As discussed below, we feel that this conformational change is likely regulated to target the release of the ligand to a specific destination within the cell.
Mutations Associated with AVED. Several mutations in the gene encoding ATTP, which result in a late-onset spinocerebellar ataxia, have been described (6, 7, 26–31). As the phenotype is recessive, the simplest explanation is that the disease results in a loss of function of ATTP. In addition to mutations that result in truncations or prevent the translation of the protein, there are several missense mutations that may provide a deeper understanding of the function of ATTP (Fig. 6). Biochemical characterization of mutant ATTP has been limited to the H101Q mutation. This variant is deficient in its ability to transfer α-T from liposomes to mitochondrial membranes, but it is not known whether its affinity for α-T is low or whether its ability to release ligand is defective (6).
Fig. 6.
Missense mutations associated with AVED. The side chain of residues that are mutated in AVED are shown as stick representations on a worm model of the protein backbone. α-T is shown in yellow. Image was generated in povscript and rendered with povray.
Of the eight missense mutations reported, only one involves a change in the ligand-binding pocket itself, L183P. This side chain, in combination with those of V191 and I194, forms a hydrophobic pocket for the methyl group at C5 on the chroman ring of α-T (Fig. 5). Residue L183 is located in the middle of helix 9, which forms a number of van der Waals contacts with the ligand, and a mutation to proline would be expected to kink the helix and disrupt several interactions between ATTP and α-T.
Two other mutations are located toward the interior of ATTP, E141K and H101Q. The side chain of E141 forms a hydrogen bond with Y73, which stabilizes the arrangement of helix 6 against helix 3. The introduction of a lysine residue at this position would likely alter this interaction and might cause a rearrangement of two nearby arginine residues, R54 and R77, strictly conserved among the four CRAL-TRIO family members (Fig. 1). The clinical phenotype of this mutation is severe, with affected individuals having disease onset before the age of 10 (29).
In contrast, the relatively conservative mutation of H101Q in strand A results in a mild phenotype (6, 7). Both nitrogen atoms of the imidazole ring participate in hydrogen bonds. One is a donor to the carbonyl oxygen of I116 of strand B and the other is an acceptor of the hydroxyl group of T139 of helix 6, which is located near the ligand-binding pocket. A glutamine residue at this position would not be capable of forming both hydrogen bonds and may lead to some destabilization of the protein. Despite the relatively conservative nature of the mutation, the H101Q mutant protein was ≈11% as active as WT in an α-T transfer assay when expressed in mammalian cell culture (6). We have attempted initial evaluations of these three mutants, H101Q, E141K, and L183P, but all three resulted in aggregated protein when expressed in E. coli (data not shown).
The rest of the disease-associated mutations map to the surface of ATTP. Of these, three involve arginine residues that contribute to a positively charged cleft in ATTP (Fig. 4A). Two of the mutations, R59W and R221W, are associated with severe phenotypes, whereas the third mutation, R192H, is relatively conservative and results in a milder phenotype (29). R59 is conserved among four of the CRAL-TRIO proteins as shown in Fig. 1 and forms a salt bridge with D185. The positive charge at position 221 is conserved within this family, and at position 192, three of four members have a positively charged residue at this position (Fig. 1).
Comparisons to CRALBP: Potential Clues to Regulation of Ligand Release. Three missense mutations of CRALBP, R150Q, M225K, and R233W, have been implicated in hereditary retinopathies in humans (32, 33). Although the crystal structure of this protein is yet to be determined, the biochemical properties of the mutant proteins have been investigated in detail and may thus provide some insight into the function of ATTP (32, 33). The R150Q mutation results in a highly insoluble protein that is unable to bind 11-cis-retinal when expressed in E. coli. The equivalent residue in ATTP is R107, which forms hydrogen bonds to the carbonyl oxygen of residue G111 in the turn between strands A and B and with the hydroxyl group of T149 of helix 7. These interactions might cause the observed destabilization. The M225K mutation completely abolishes the ability to bind to ligand. In ATTP, the equivalent residue, V182, is involved in a hydrogen bond network to α-T through its carbonyl oxygen, and the side chain forms a number of van der Waals contacts with the ligand (Fig. 5).
The most instructive CRALBP mutant is R233W, which is the equivalent of mutation R192H in ATTP. This mutation does not disrupt binding of ligand. Rather, this mutant was defective in presenting 11-cis-retinol to retinal deyhdrogenase for oxidation in a membrane-free assay system. CRALBP would be expected to have a similar positively charged cleft as was seen with ATTP, with an arginine at residues 102 and 262, which are equivalent to R59 and R221 in ATTP, respectively. Furthermore, other positively charged residues found in the positively charged cleft in ATTP are conserved for CRALBP.
These findings have led us to a model for release of ligand from these two proteins. The positively charged cleft may represent an area of protein interaction that orients a hydrophobic surface to disrupt the interactions of helix 9 with helix 10, similarly to the way β-OG appears to do so in the Sec14p structure. This could enable a conformational change in which helices 10 and 11 swing away from the core to expose the ligand-binding pocket. Such a regulated change in conformation could be important for the ability of these proteins to direct their ligands to the right pathway, in the case of ATTP to rescue α-T from degradation and direct it to a secretory pathway. There are no known proteins that directly interact with ATTP to cause the release of ligand, although one possible candidate would be ATP-binding cassette transporter A-1, which has been implicated in the cellular secretion of α-T (34). Alternatively, the positive surface could serve to dock ATTP onto cell membranes through interactions with phospholipid headgroups, with induction of the open conformation through interactions with lipid.
After we had completed the structure analysis of ATTP bound to α-T, another group reported the structure of ATTP (35). We have not made a detailed comparison because the atomic coordinates are not yet available, but the two structures are clearly similar despite differences between the constructs used for expression and different crystal lattices. In the other work, a detergent-bound form of ATTP was also crystallized, resulting in a conformation similar to what was reported for Sec14p, further strengthening the argument that the structure of Sec14p represents an alternative open conformation.
Acknowledgments
We thank E. Martinez, G. Gregorio, R. Abramowitz, and X. Yang for help at the synchrotron and present and former Hendrickson laboratory members for general support and advice. This work was supported in part by a Physician Postdoctoral Fellowship from the Howard Hughes Medical Institute (to K.C.M.) and a Career Development Award from the Leukemia and Lymphoma Society (to R.A.K.). Beamline X4A at the National Synchrotron Light Source, a Department of Energy facility, was supported by the Howard Hughes Medical Institute.
Abbreviations: α-T, α-tocopherol; ATTP, α-T transfer protein; AVED, ataxia with vitamin E deficiency; β-OG, β-octylglucopyranoside; CRALBP, cellular retinal-binding protein; MAD, multiwavelength anomalous diffraction; PI, phosphatidyl inositol.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1R5L).
References
- 1.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sha, B., Phillips, S. E., Bankaitis, V. A. & Luo, M. (1998) Nature 391, 506–510. [DOI] [PubMed] [Google Scholar]
- 3.Stocker, A., Tomizaki, T., Schulze-Briese, C. & Baumann, U. (2002) Structure (London) 10, 1533–1540. [DOI] [PubMed] [Google Scholar]
- 4.Brigelius-Flohe, R. & Traber, M. G. (1999) FASEB J. 13, 1145–1155. [PubMed] [Google Scholar]
- 5.Hosomi, A., Arita, M., Sato, Y., Kiyose, C., Ueda, T., Igarashi, O., Arai, H. & Inoue, K. (1997) FEBS Lett. 409, 105–108. [DOI] [PubMed] [Google Scholar]
- 6.Gotoda, T., Arita, M., Arai, H., Inoue, K., Yokota, T., Fukuo, Y., Yazaki, Y. & Yamada, N. (1995) N. Engl. J. Med. 333, 1313–1318. [DOI] [PubMed] [Google Scholar]
- 7.Yokota, T., Shiojiri, T., Gotoda, T., Arita, M., Arai, H., Ohga, T., Kanda, T., Suzuki, J., Imai, T., Matsumoto, H., et al. (1997) Ann. Neurol. 41, 826–832. [DOI] [PubMed] [Google Scholar]
- 8.Arita, M., Sato, Y., Miyata, A., Tanabe, T., Takahashi, E., Kayden, H. J., Arai, H. & Inoue, K. (1995) Biochem. J. 306, 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arita, M., Nomura, K., Arai, H. & Inoue, K. (1997) Proc. Natl. Acad. Sci. USA 94, 12437–12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traber, M. G., Sokol, R. J., Burton, G. W., Ingold, K. U., Papas, A. M., Huffaker, J. E. & Kayden, H. J. (1990) J. Clin. Invest. 85, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokota, T., Igarashi, K., Uchihara, T., Jishage, K., Tomita, H., Inaba, A., Li, Y., Arita, M., Suzuki, H., Mizusawa, H. & Arai, H. (2001) Proc. Natl. Acad. Sci. USA 98, 15185–15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doublie, S. (1997) Methods Enzymol. 276, 523–530. [PubMed] [Google Scholar]
- 13.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. A. & Kjeldgaard, M. O. (1997) Methods Enzymol. 277, 173–208. [DOI] [PubMed] [Google Scholar]
- 18.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 19.Allen, F. H. (2002) Acta Crystallogr. B 58, 380–388. [DOI] [PubMed] [Google Scholar]
- 20.Holm, L. & Sander, C. (1996) Science 273, 595–603. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch, W. (1976) Acta Crystallogr. A 32, 922–923. [Google Scholar]
- 22.Phillips, S. E., Sha, B., Topalof, L., Xie, Z., Alb, J. G., Klenchin, V. A., Swigart, P., Cockcroft, S., Martin, T. F., Luo, M., et al. (1999) Mol. Cell 4, 187–197. [DOI] [PubMed] [Google Scholar]
- 23.Stocker, A. & Baumann, U. (2003) J. Mol. Biol. 332, 759–765. [DOI] [PubMed] [Google Scholar]
- 24.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 25.Panagabko, C., Morley, S., Hernandez, M., Cassolato, P., Gordon, H., Parsons, R., Manor, D. & Atkinson, J. (2003) Biochemistry 42, 6467–6474. [DOI] [PubMed] [Google Scholar]
- 26.Ouahchi, K., Arita, M., Kayden, H., Hentati, F., Ben Hamida, M., Sokol, R., Arai, H., Inoue, K., Mandel, J. L. & Koenig, M. (1995) Nat. Genet. 9, 141–145. [DOI] [PubMed] [Google Scholar]
- 27.Hentati, A., Deng, H. X., Hung, W. Y., Nayer, M., Ahmed, M. S., He, X., Tim, R., Stumpf, D. A. & Siddique, T. (1996) Ann. Neurol. 39, 295–300. [DOI] [PubMed] [Google Scholar]
- 28.Shimohata, T., Date, H., Ishiguro, H., Suzuki, T., Takano, H., Tanaka, H., Tsuji, S. & Hirota, K. (1997) Ann. Neurol. 43, 273. [DOI] [PubMed] [Google Scholar]
- 29.Cavalier, L., Ouahchi, K., Kayden, H. J., Di Donato, S., Reutenauer, L., Mandel, J. L. & Koenig, M. (1998) Am. J. Hum. Genet. 62, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino, M., Masuda, N., Ito, Y., Murata, M., Goto, J., Sakurai, M. & Kanazawa, I. (1999) Ann. Neurol. 45, 809–812. [DOI] [PubMed] [Google Scholar]
- 31.Usuki, F. & Maruyama, K. (2000) J. Neurol. Neurosurg. Psychiatry 69, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maw, M. A., Kennedy, B., Knight, A., Bridges, R., Roth, K. E., Mani, E. J., Mukkadan, J. K., Nancarrow, D., Crabb, J. W. & Denton, M. J. (1997) Nat. Genet. 17, 198–200. [DOI] [PubMed] [Google Scholar]
- 33.Golovleva, I., Bhattacharya, S., Wu, Z., Shaw, N., Yang, Y., Andrabi, K., West, K. A., Burstedt, M. S., Forsman, K., Holmgren, G., et al. (2003) J. Biol. Chem. 278, 12397–12402. [DOI] [PubMed] [Google Scholar]
- 34.Oram, J. F., Vaughan, A. M. & Stocker, R. (2001) J. Biol. Chem. 276, 39898–39902. [DOI] [PubMed] [Google Scholar]
- 35.Meier, R., Tomizaki, T., Schulze-Briese, C., Baumann, U. & Stocker, A. (2003) J. Mol. Biol. 331, 725–734. [DOI] [PubMed] [Google Scholar]
- 36.Fenn, T., Ringe, D. & Petsko, G. A. (2003) J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- 37.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
- 38.Esnouf, R. M. (1999) Acta Crystallogr. D 55, 938–940. [DOI] [PubMed] [Google Scholar]
- 39.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]