Abstract
The surprising observation that a 10 residue class G* peptide from apolipoprotein J, [113-122]apoJ, possesses anti-inflammatory and anti-atherogenic properties prompted us to delineate its structural characteristics in the presence of normal and oxidized lipid. Towards this, we have determined high resolution structure of [113-122]apoJ in solution using nuclear magnetic resonance (NMR) spectroscopy and studied its interaction with lipids, including oxidized lipids, using a number of biophysical methods. Circular dichroism and NMR studies established that in the presence of dodecylphosphocholine (DPC) micelle this peptide adopts amphipathic α helical structure. The observed Nuclear Overhauser effects indicate that the amphipathic helical structure of the peptide is stabilized by the N-terminal acetyl and C-terminal amide blocking groups. We used isothermal titration calorimetry to measure binding enthalpy of the peptide with DPC micelle, an oxidized lipid, 1-(palmitoyl)-2-(5-keto-6-octene-dioyl) phosphatidylcholine (KOdiA-PC), and the mixture of these two lipids (5mol% KOdiA-PC in DPC micelle). We find that the peptide binding with DPC micelle is associated with an enthalpy change (-16.75±0.16 Kcal/mol) much larger than that resulting from the binding with KodiA-PC (-3.67±0.13 Kcal/mol). Incorporation of a small amount of KOdiA-PC (5mol %) in DPC micelle also results in the lowering of peptide binding enthalpy (-13.43±0.18 Kcal/mol). These results are consistent with overall negative charge and altered conformational properties of oxidized sn-2 chain of KOdiA-PC. Our results have unambiguously established the amphipathic α helical structure of [113-122]apoJ peptide in the presence of DPC micelle as well as its ability to bind oxidized lipid. These in vitro results help explain the previously observed anti-inflammatory and anti-atherosclerotic properties of this peptide.
1. Introduction
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality throughout the world. A significant reduction in the LDL cholesterol (LDL-C) levels has been achieved through statin therapy. However, despite this reduction in LDL-C, the residual risk of CVD remains high [1]. This underscores the need for developing novel therapeutic modalities to further reduce CVD risk.
Epidemiological studies have identified an inverse association with the high density lipoprotein cholesterol (HDL-C) levels and the incidence of coronary heart disease in both men and women [2,3]. The mechanism through which HDL-C confers its beneficial effects has been attributed to its ability to mediate cholesterol removal from macrophages in a process termed as reverse cholesterol transport, as well as its anti-inflammatory, anti-apoptotic, anti-oxidative, and anti-thrombotic properties [4].
The effect of homologous plasma HDL on established atherosclerotic lesions was studied in cholesterol-fed rabbits in 1990 by Badimon et al. for the first time [5]. This study provided the impetus to study the anti-atherogenic potential of a number of HDL-C mimetic. Apolipoprotein A-I (apoA-I) is the major apolipoprotein of HDL comprising of ~70% of all the apolipoproteins present in HDL. Apolipoprotein A-IMilano (apoA-IMilano) is a genetic variant of apoA-I with an arginine-to-cysteine substitution at position 173 present in individuals with a very low HDL-C level and no obvious signs of atherosclerosis [6,7]. A recombinant ApoA-IMilano/phospholipid complex (ETC-216; recombinant apoA-IMilano/1-palmitoyl-2-oleoyl phosphatidylcholine complexes) administered intravenously for 5 doses at weekly intervals produced significant regression of coronary atherosclerosis as measured by intravascular ultrasound [8]. Short-term infusions of reconstituted HDL (CSL-111; CSL-111 is reconstituted HDL consisting of apolipoprotein A-I from human plasma combined with soybean phosphatidylcholine) in the ‘Effect of rHDL on Atherosclerosis-Safety and Efficacy’ (ERASE) trial resulted in no significant reductions in percentage change in atheroma volume or nominal change in plaque volume compared with placebo but did result in statistically significant improvement in the plaque characterization index and coronary score on quantitative coronary angiography [9].
ApoA-I is a 243 amino acid residue protein and, therefore, difficult and expensive to produce in large quantities required for therapeutic applications. For this reason, there has been a great deal of interest in developing apoA-I mimetic peptides that are much shorter in length and, therefore, easy to synthesize in much larger quantities. Our lab has pioneered the development of such peptides. Garber et al. [10] showed for the first time that an 18-residue apoA-I mimetic class A amphipathic helical peptide, named 5F (because of the presence of five Phe residues on the non polar face), has anti-atherosclerotic properties in vivo. When an apoA-I mimetic peptide synthesized from D-amino acids (D-4F; so named because of the presence of four Phe residues on the non polar face) was given orally to LDL receptor-null mice on a Western diet, atherosclerotic lesions decreased by 79% [11]. It was shown later that the mechanism of action of oral D-4F in apoE-null mice involves rapid formation of small cholesterol-containing particles of 7 to 8 nm, with pre-β mobility enriched in apoA-I and paraoxonase activity. As a result, lipoprotein lipid hydroperoxide are reduced, HDL becomes anti-inflammatory, and HDL-mediated cholesterol efflux and reverse cholesterol transport from macrophages are stimulated [12]. Preliminary clinical data suggest that, in patients with established coronary artery disease, a single oral dose of D-4F is both safe and tolerable and reduces the HDL inflammatory index [13].
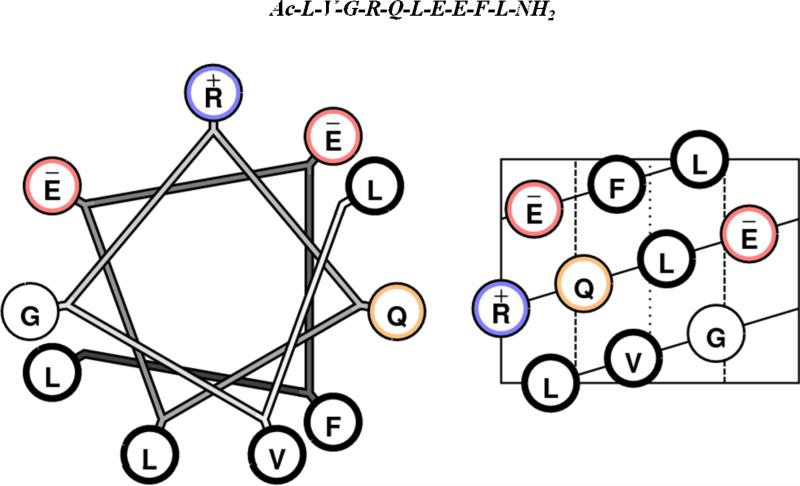
Both 5F and 4F peptides are de novo designed peptides with no sequence homology to apoA-I or any other apolipoprotein. Both these peptides are class A amphipathic helical peptides [14]. The ability of these peptides to inhibit atherosclerosis in mouse models has been closely correlated with their ability to inhibit LDL-induced monocyte chemotactic activity (MCA) in a human artery wall co-culture [15]. An approach to the design of smaller anti-atherogenic peptides comes from the analysis of naturally occurring anti-atherogenic apolipoprotein sequences. Apolipoprotein J (apoJ) is a glycoprotein and exists in the plasma associated with HDL [16]. ApoJ has been shown to bind inflammatory lipids and inactivate them; LDL added to human artery wall cells in the presence of apoJ could not induce MCA [17]. To test the hypothesis that short amphipathic helical sequences in apoJ could mimic its activity, the apoJ sequence was analyzed using an in house developed computer program called LOCATE [18]. This program identified 17 potential class G* amphipathic helices in the mature apoJ protein, including [113-122]apoJ [19]. It is important to note that all of the exchangeable apolipoproteins except apolipoproteins A-II and C-I, possess class G* helices. Class G* helices are characterized by a lack of clustering of positively charged amino acids at the polar – non polar interface and negatively charged amino acids at the center of polar face as present in class A amphipathic helices, a high mean hydrophobic moment, and moderately high nonpolar face hydrophobicity [14]. Figure 1 shows the helical wheel and helical net diagrams of [113-122]apoJ. Unlike class A amphipathic helices, the lipid associating properties of class G* helices have not been studied in detail. This peptide, synthesized using D-amino acids, renders HDL anti-inflammatory in both mice and monkeys and dramatically reduces atherosclerosis in apoE-null mice [19]. It was further shown that co-administration of L-[113-122]apoJ and niclosamide improved the ability of HDL to inhibit LDL-induced monocyte chemotactic activity, known as HDL-inflammatory index, in endothelial cell cultures to a degree similar to L-4F given with niclosamide and achieved an HDL-inflammatory index comparable to that obtained with normal human anti-inflammatory HDL [20]. Niclosamide (a chlorinated salicylanilide used as molluscicide, antihelminthic, and lampricide) has been shown to associate with L-4F and prevent its trypsin digestion [20].
Figure 1. [113-122]apoJ is a class G* helix.
Helical wheel (left) and helical net (right) diagrams of [113-122]apoJ. The primary amino acid sequence of [113-122]apoJ peptide is shown above helical wheel and helical net diagrams. Ac (acetyl) and NH2 (amide) are blocking groups. Color codes are as follows: acidic, red; basic, blue; hydrophobic, thick black; neutral, orange.
Many of the known anti-inflammatory and anti-atherogenic peptides are amphipathic helical peptides [21-23]. The present studies were undertaken to correlate the biological activities of [113-122]apoJ with its structure and ability to bind lipids, including oxidized lipids. Specifically, since [113-122]apoJ is only ten residue long, we asked the question if this peptide could also adopt an amphipathic α helical structure and associate with lipids, particularly oxidized lipids.
2. Experimental Methods
2.1. Materials
The sequence for [113-122]apoJ is Ac-Leu-Val-Gly-Arg-Gln-Leu-Glu-Glu-Phe-Leu-NH2. The peptide is blocked at the N- and C- termini with acetyl (Ac-) and amide (-NH2) groups, respectively. [113-122]apoJ was synthesized from all L-amino acids by automated solid-phase synthesis using the Fmoc strategy, and purified by reverse-phase HPLC as previously described [19]. The purity of the peptide was confirmed by analytical reverse-phase HPLC and MALDI-TOF mass spectrometry.
Protonated dodecylphosphocholine (DPC) was obtained from Avanti Polar Lipids, Inc. (Alabaster, Alabama). Perdeuterated DPC (D38, 98%) was obtained from Cambridge Isotope Laboratories (Andover, MA). 1-(Palmitoyl)-2-(5-keto-6-octene-dioyl) phosphatidylcholine (KOdiA-PC), was obtained from Cayman Chemical Company (Ann Arbor, MI) and used without further purification.
2.2. Circular Dichroism (CD) spectroscopy
The CD spectra were recorded using JASCO J-815 CD spectrometer equipped with a Peltier type temperature control system (JASCO model PTC-423S/15) and interfaced to a personal computer. The instrument was calibrated with (1S)-(+)-10-camphorsulfonic acid. The CD spectra were measured from 260 to 190 nm every 0.5 nm with 4 s averaging per point and a 2 nm bandwidth. A 0.1 cm path length cell was used for obtaining the spectra. The CD spectra were signal averaged by adding four scans, baseline corrected, and smoothed. All the CD spectra were recorded at 37°C. Peptide concentration of 0.1 mg/ml was used for obtaining CD spectra. The mean residue ellipticity, [Θ]MRE(deg cm2 dmol-1), was calculated using the following equation:
where, MRW is the mean residue weight (molecular weight of the peptide divided by the number of amino acids in the peptide), Θ is the observed ellipticity in degrees, c is the concentration of the peptide in grams per milliliter, and l is the path length of the cell in centimeters. The secondary structure of the peptide was analyzed using the programs CDPro (http://lamar.colostate.edu/~sreeram/CDPro) and K2D (http://www.emblheidelberg.de/~andrade/k2d.html) [24].
2.3. Isothermal Titration Calorimetry (ITC)
ITC was used to measure the binding enthalpy (ΔHbind) of peptide with DPC micelle, KOdiA-PC, and a mixture of KOdiA-PC (5mol %) and DPC micelle using a VP-ITC microcalorimeter (Microcal, Northampton, MA). Measurements were made at 37°C and pH 7. An excess site method was employed wherein a high concentration of DPC, KOdiA-PC, or the mixture of the two were placed in the sample cell and titrated with a low concentration of peptide. This method eliminates the saturation of binding sites of the lipid and results in the full heat of binding, (ΔHbind), evolved upon each injection. Specifically, titrations were performed by injecting 10 μL aliquots of the peptide solution at a concentration of typically 200–250 μM into the calorimeter cell containing the lipid at a concentration of 5–15 mM with 200 seconds resting time between each injection. The heats of dilution for the peptide was determined by injecting the peptide solution into pure buffer and subtracted from the data prior to analysis. The data was analyzed by integrating the area of each injection peak and plotted using the Origin 7.0 software integrated into the VP-ITC instrument software. The first data point was removed and ΔHbind was calculated by taking the averaged integrated value for the remaining injections.
2.4. NMR sample preparation
Protonated and perdeuterated DPC micelle were prepared in 5 mM KH2PO4 (pH 5.5). KodiA-PC was prepared by evaporating ethanloic solution of the lipid under a stream of nitrogen gas. Residual solvent was removed by placing the sample under high vacuum at room temperature overnight. Dried lipid sample was suspended in 5 mM KH2PO4 (pH 5.5). Lyophilized dry peptide was dissolved separately in 5 mM KH2PO4 (pH 5.5). An appropriate volume of peptide solution was added to either DPC micelle or KOdiA-PC and the mixture was incubated overnight at 37°C. The pH of the final solution was 5.5. The final peptide concentration in solution was between 1-2 mM. The lipid to peptide molar ratio (125:1 for DPC and 25:1 for KOdiA-PC) was identical with that used in the CD studies. This lipid to peptide ratio was selected based on the CD studies that indicated no further change in the secondary structure of the peptide above this ratio.
2.5. NMR measurements and structure calculations
1D- and 2D- 1H NMR experiments were performed either on a Bruker Avance-600 NMR system with a TCI CryoProbe or an Avance-500 NMR spectrometer at 37°C. Mixing times of 200 msec or 300 msec were used in NOESY, and 80 msec spin-lock time was used in total correlation spectroscopy (TOCSY) measurements. 2D-data sets were collected with 1024 complex t2 points with 512 t1 increments. States-time proportional phase incrementation (TPPI) was employed for frequency discrimination in the indirect dimension. Chemical shifts were referenced with respect to sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) (0.0 ppm) used as an internal standard. Water suppression was accomplished either by presaturation or by using the WATERGATE (WATER suppression by GrAdient-Tailored Excitation) pulse sequence [25].
31P NMR experiments were conducted using Bruker Avance 500 MHz system equipped with a TBI broadband probe. 2048 scans were used to acquire each 31P spectrum, at a temperature of 310 K. The samples were allowed to equilibrate in the probe for 30 minutes before acquisition was begun. The acquired spectra were referenced using 85% phosphoric acid as an external standard (0.0 ppm).
The NMR data were processed offline using the program FELIX (version 2007) (Felix NMR Inc., San Diego, CA). NMR structures were calculated using X-PLOR (online) version 3.851. Structure calculation protocol involved (a) generation of a “template” coordinate set, (b) bound smoothing, full structure embedding, and regularization to produce a family of 200 distance geometry (DG) structures, (c) simulated annealing regularization and refinement for embedded distance geometry structures, and (d) simulated annealing refinement, as described previously [26]. The best 100 structures were selected for further analysis on the basis of their least restraint violation, low energy and Ramachandran plot quality. Average coordinates of the accepted structures were energy minimized using 200 cycles of conjugate gradient energy minimization. All the NMR constraints were enforced during energy minimization.
On the basis of cross-peak intensities in the NOESY spectra, recorded with mixing times of 200 msec, the NOE distance constraints were classified as following. For NOEs involving intra-residue and sequential NH, αH, and βH protons, a distance constraint of 2.0–2.5 Å for strong, 2.0-3.0 Å for medium, and 2.0-4.0 Å for weak NOEs was used; for medium- or long-range NOEs involving NH, and αH protons, a distance constraint of 2.0-4.0Å was used; for NOEs involving medium- or long-range sidechain protons, a distance constraint of 2.0-5.0Å was used. Pseudoatom corrections, where appropriate, were added to the upper distance bounds as described previously [27]. Based on the difference (Δδ) in the observed and “coil” NH proton chemical shifts, H-bond (H-O) distances were calculated as described earlier [28,29]. These H-bond distances were included as additional constraints during structure calculation [30]. Dihedral angles Φ and Ψ were based on the difference in the observed and “coil” CαH proton chemical shifts [28,29]; a minimum deviation of 40° was allowed in the dihedral constraints.
2.6. Molecular modeling
Molecular modeling was performed using the program SYBYL (version 7.2) (Tripos, Inc., St. Louis, MO).
3. Results
3.1. Circular dichroism studies
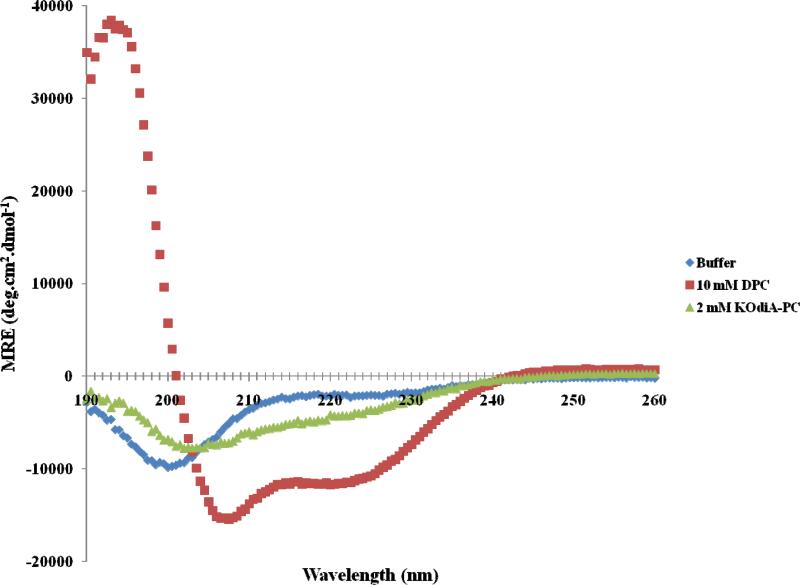
Figure 2 shows the far-UV CD spectra of [113-122]apoJ in PBS and in the presence of DPC micelle (lipid to peptide molar ration 125:1) and KodiA-PC (lipid to peptide molar ratio 25:1). There was no further change in the CD spectra upon increasing the above lipid to peptide molar ratios (results not shown). The secondary structure of [113-122]apoJ was analyzed using CDPro and K2D. The results are shown in Table 1. Based on the analysis of CD spectra, it is evident that [113-122]apoJ adopts a largely helical structure in the presence of DPC micelle (Table 1). However, the helical content of [113-122]apoJ is much reduced in the presence of KOdiA-PC (Figure 2, Table 1).
Figure 2. [113-122]apoJ forms helical structure in the presence of DPC micelle; helicity is reduced in the presence of KOdiA-PC.
Far-UV CD Spectra of [113-122]apoJ in phosphate buffered saline (PBS, pH 7.4) and in the presence of DPC (lipid to peptide molar ratio, 125:1) and KOdiA-PC (lipid to peptide molar ratio, 25:1) at 37°C. There was no change in the CD spectra above the lipid to peptide molar ratios used. Peptide concentration was 0.1 mg/ml.
Table 1.
Secondary structure analysis of [113-122]apoJ in buffer and in the presence of DPC (lipid to peptide molar ratio, 125:1) and KOdiA-PC (lipid to peptide molar ratio, 25:1).
| CDPro | |||||||
|---|---|---|---|---|---|---|---|
| SELCON3 | |||||||
| H(r) | H(d) | S(r) | S(d) | Trn | Unrd | RMSD | |
| [113-122]apoJ:Buffer | 0.058 | 0.129 | 0.068 | 0.07 | 0.246 | 0.401 | 1.956 |
| [113-122]apoJ:DPC (10 mM) | 0.752 | 0.101 | 0.017 | 0.04 | 0.035 | 0.04 | 2.37 |
| [113-122]apoJ:KOdiA-PC (2 mM) | 0.124 | 0.119 | 0.156 | 0.098 | 0.228 | 0.306 | 0.337 |
| Continll | |||||||
| [113-122]apoJ:Buffer | 0.048 | 0.141 | 0.058 | 0.08 | 0.268 | 0.399 | 0.179 |
| [113-122]apoJ:DPC (10 mM) | 0.69 | 0.271 | 0 | 0 | 0.004 | 0.031 | 0.408 |
| [113-122]apoJ:KOdiA-PC (2 mM) | 0.162 | 0.173 | 0.007 | 0.044 | 0.271 | 0.343 | 0.237 |
| CDSSTR | |||||||
| [113-122]apoJ:Buffer | 0.074 | 0.135 | 0.133 | 0.08 | 0.253 | 0.299 | 0.467 |
| [113-122]apoJ:DPC (10 mM) | 0.585 | 0.19 | 0.034 | 0.03 | 0.043 | 0.108 | 0.676 |
| [113-122]apoJ:KOdiA-PC (2 mM) | 0.313 | 0.139 | 0.124 | 0.0085 | 0.128 | 0.214 | 0.595 |
| K2D | ||
|---|---|---|
| Alpha Helix | Beta Strand | |
| [113-122]apoJ:Buffer | 0.081 | 0.241 |
| [113-122]apoJ:DPC (10 mM) | 0.849 | 0.006 |
| [113-122]apoJ:KOdiA-PC (2 mM) | 0.105 | 0.291 |
H(r), regular helix; H(d), distorted helix; S(r), regular sheet; S(d), distorted sheet; Trn, turn; Unrd, unordered; RMSD, root mean square deviation between the calculated and experimental CD spectra.
3.2. Isothermal titration calorimetry
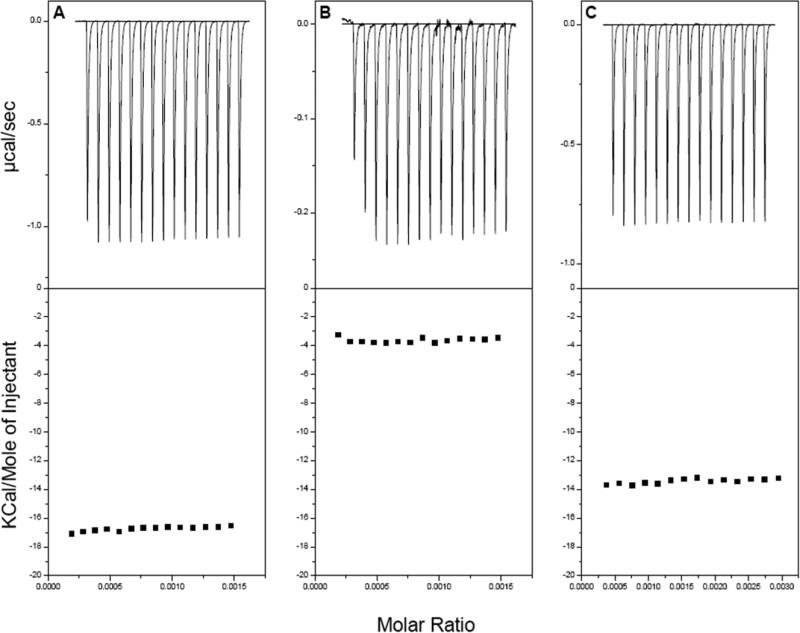
Binding of [113-122]apoJ to DPC, KOdiA-PC, and a mixture of the two lipids (5mol% KOdiA-PC in DPC micelle) was further probed using ITC (Figure 3). The peptide was titrated into a concentrated solution of lipid. In this method, saturation is not reached and each injection results in the total enthalpy of reaction (ΔH) for each injection. The ΔH for the reaction was found to be -16.7±0.16 , -3.7±0.13, and -13.4±0.18 Kcal/mole for DPC, KOdiA-PC, and 5mol% KOdiA-PC in DPC, respectively, by taking the average of 15 injections (Figure 3).
Figure 3. [113-122] apoJ binding with DPC and KodiA-PC is associated with a favorable enthalpy change.
Enthalpy of [113-122] apoJ binding with DPC micelle (A), KOdiA-PC (B), and 5mol% KOdiA-PC in DPC micelle (C), determined using isothermal titration calorimetry (ITC).
3.3. Nuclear magnetic resonance studies
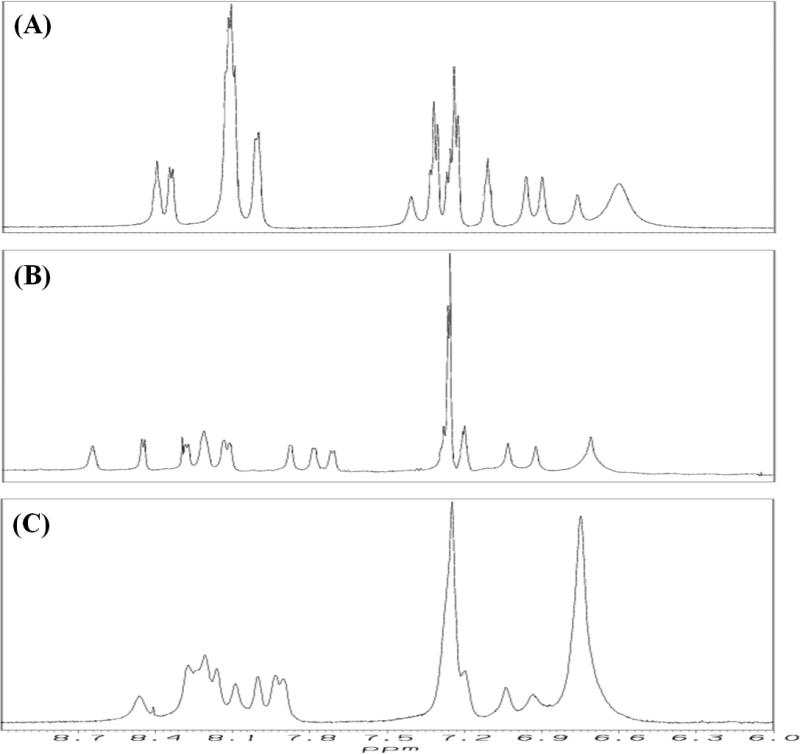
Figure 4 shows the aromatic/amide region of the 1H NMR spectra of [113-122]apoJ in buffer (A), DPC micelle (B), and in the presence of KOdiA-PC (C). All the 1H NMR spectra were obtained under identical conditions (500 MHz, 37°C) and processed using identical window functions. There is a greater dispersion of amide protons (7.6 – 8.7 ppm) in the presence of DPC micelle (B) than in buffer (A). It is interesting to note that the amide protons are less dispersed in the presence of KOdiA-PC (C) than in the presence of DPC micelle (B), but more dispersed than in buffer (A), indicating that the peptide adopts different conformations in different environments.
Figure 4. [113-122]apoJ adopts different conformation under different solvent environments.
Aromatic/amide region of the 1H NMR spectrum of [113-122]apoJ in 5mM potassium phosphate (pH 5.5) (A), DPC (lipid to peptide molar ratio, 125:1) (B), and KOdiA-PC (lipid to peptide molar ratio, 25:1) (C). All the spectra were obtained at 500 MHz and 37°C and processed in an identical manner.
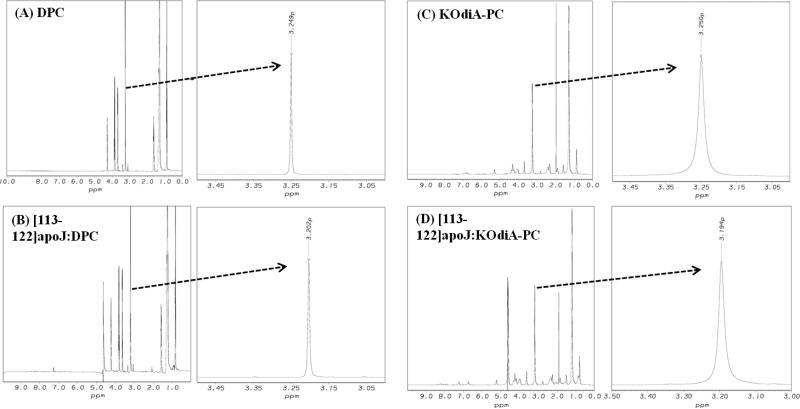
Figure 5 compares the 1H NMR spectra of DPC micelle alone (A), [113-122]apoJ in the presence of DPC micelle (B), KodiA-PC alone (C) and [113-122]apoJ in the presence of KOdiA-PC (D). The choline methyl 1H NMR signal of DPC molecules shows upfield shift in the presence of [113-122]apoJ (from 3.249 ppm to 3.202 ppm) (Figure 5, A and B). The upfield shift of the choline methyl proton signal, indicative of a more hydrophobic environment, is consistent with the binding of the [113-122]apoJ with the DPC micelle. The choline methyl 1H NMR signal of KOdiA-PC, similar to that of DPC micelle (Figure 5, A and B), shows upfield shift in the presence of [113-122]apoJ (from 3.250 ppm to 3.194 ppm) (Figure 5, C and D). The upfield shift of the choline methyl proton signal is consistent with the binding of [113-122]apoJ with KOdiA-PC.
Figure 5. [113-122]apoJ binding results in an upfield shift of the methy proton signal of lipid choline headgroup.
Methyl proton resonance of DPC (A and B) and KodiA-PC (C and D) choline headgroups shift upfield as a result of [113-122]apoJ binding. 1H NMR spectra were obtained and processed as described in Figure 4.
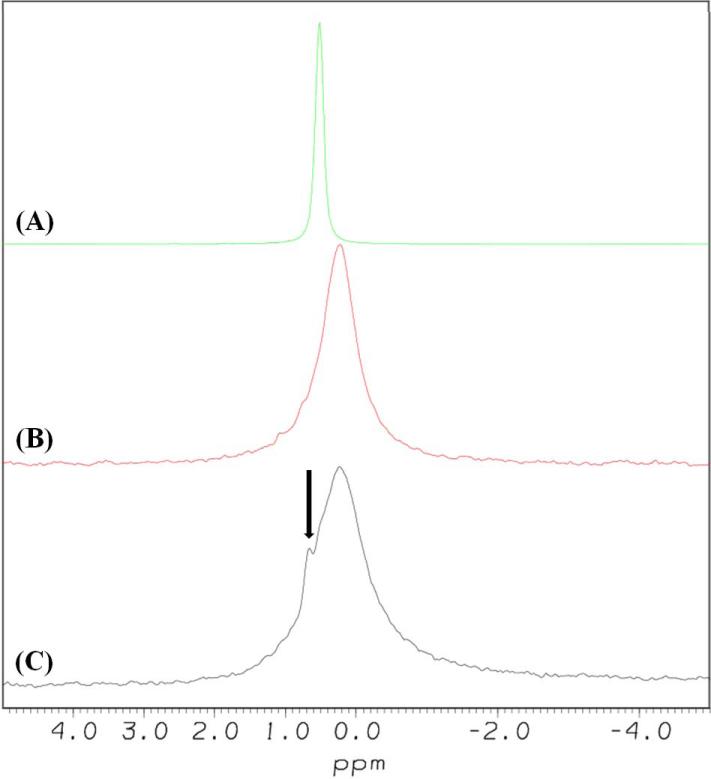
Interaction of [113-122]apoJ with KOdiA-PC was further probed using 31P NMR spectroscopy. 31P NMR spectra of DPC micelle, KOdiA-PC, and a mixture (lipid to peptide molar ratio, 25:1) of KOdiA-PC and [113-122]apoJ are compared in Figure 6. Compared to DPC micelles, KOdiA-PC exhibits broader 31P NMR spectrum (Figure 6). This is presumably because of the close spatial proximity of the truncated sn-2 chain with the phosphate group [31] resulting in the restricted mobility of the phosphate group in KodiA-PC compared to that in DPC micelle. Addition of [113-122]apoJ to KOdiA-PC results in the appearance of an isotropic narrow 31P signal, superimposed on the broad KOdiA-PC signal (Figure 6). The narrow 31P signal presumably arises from the KOdiA-PC complexed with [113-122]apoJ.
Figure 6. Addition of [113-122]apoJ to KOdiA-PC results in the appearance of an isotropic narrow 31P signal.
31P NMR spectra of DPC micelle (A), KOdiA-PC (B), and mixture of [113-122]apoJ and KOdiA-PC (lipid to peptide molar ratio, 25:1) (C). 2048 scans were used to acquire each 31P spectrum, at a temperature of 310 K. The samples were allowed to equilibrate in the probe for 30 minutes before acquisition was begun. The spectra were processed with exponential multiplier window functions, using a line broadening value of 10 Hz. The spectra were referenced using 85% phosphoric acid (0.0 ppm). The narrow 31P signal, indicated by an arrow (C), presumably arises from the KOdiA-PC complexed with [113-122]apoJ.
A combination of two dimensional 1H total correlation spectroscopy (TOCSY; spectrum not shown) and nuclear Overhauser spectroscopy (NOESY) was used to make sequence-specific resonance assignments of the individual amino acids in [113-122]apoJ [32].
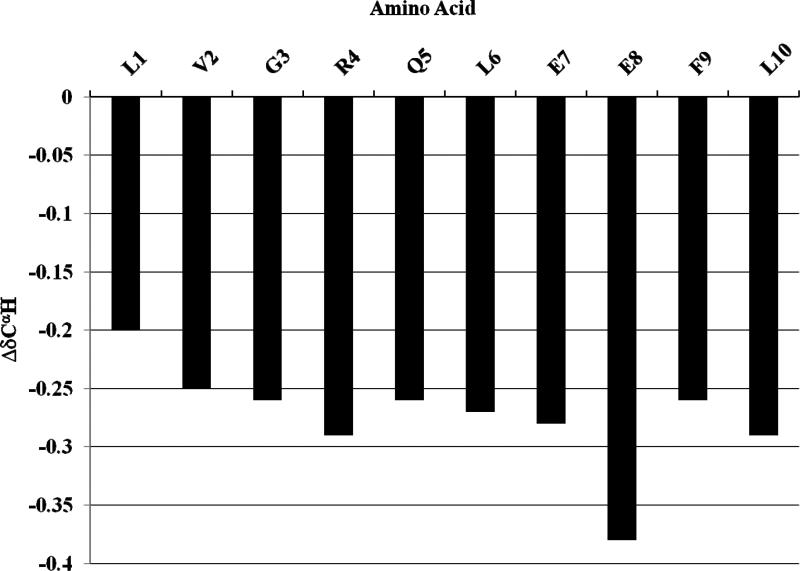
Proton chemical shifts are related to the secondary structures of peptides and proteins [28,29]. The plots of the difference between the observed chemical shift and the coil chemical shift (Δδ = δobserved - δcoil) for the CαH in [113-122]apoJ in the presence of DPC micelle are shown in Figure 7. It is evident that all of the CαH protons show an upfield shift compared to the random coil values, indicating a predominantly α helical secondary structure of [113-122]apoJ in the presence of DPC micelle.
Figure 7. CαH proton chemical shifts indicate predominantly α helical secondary structure of [113-122]apoJ in the presence of DPC micelle.
A plot of the ΔδCαH (δobserved-δcoil) chemical shift for all the residues in [113-122]apoJ. Note that the CαH protons show an upfield shift compared to the corresponding coil values, indicating α-helical structure. Random coil chemical shifts used were those reported in: Wuthrich, K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York
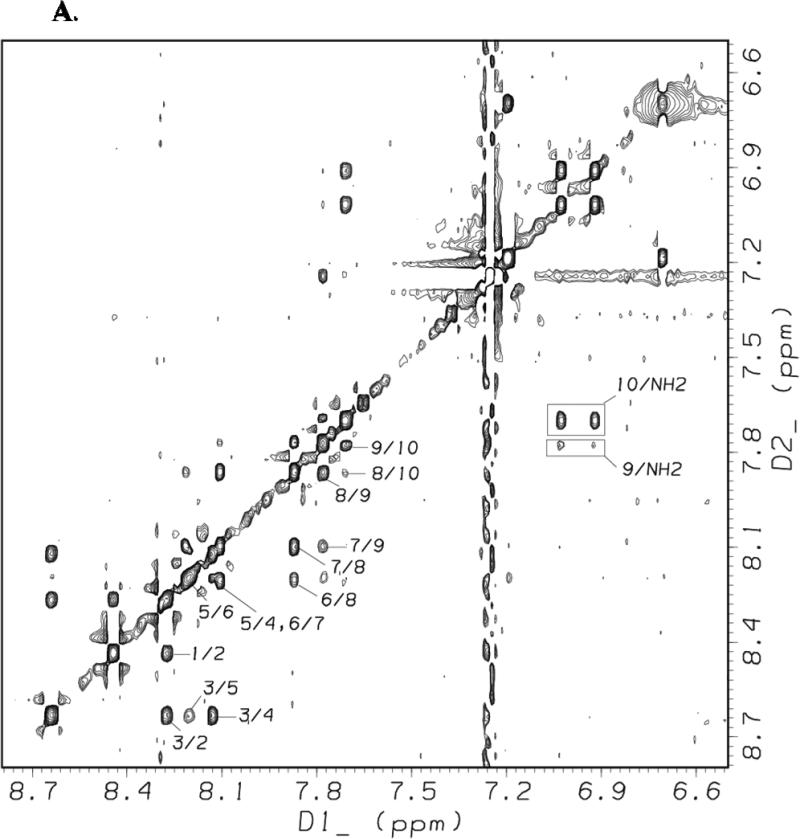
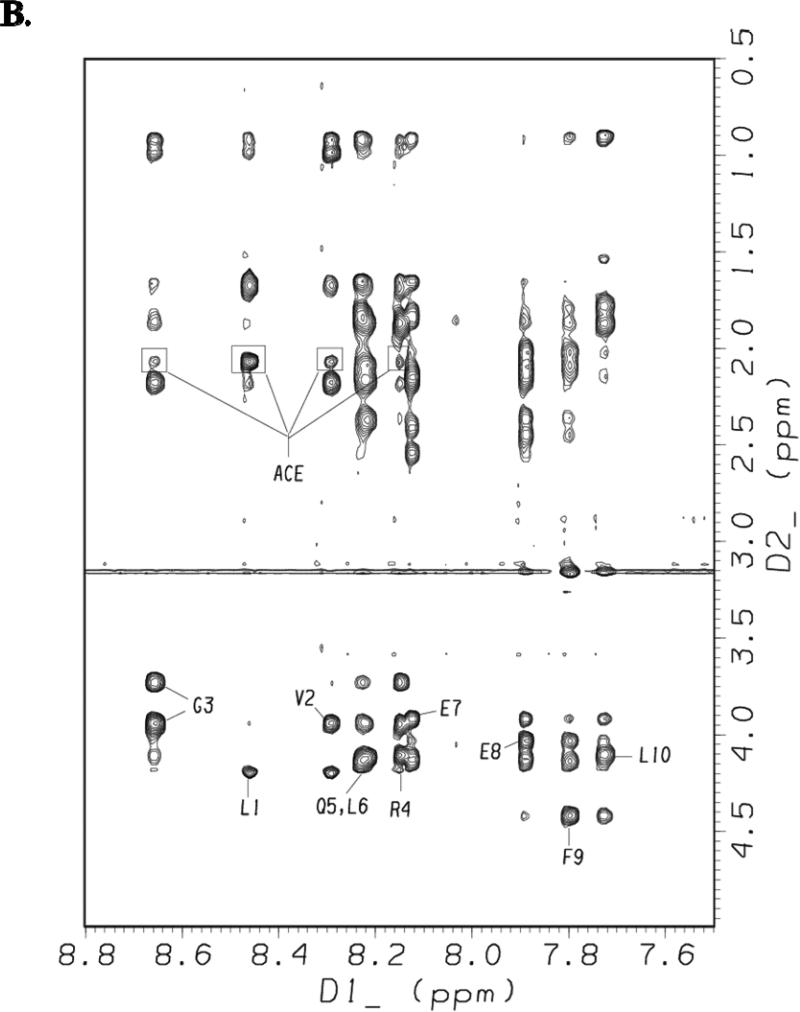
Figures 8A and 8B show the amide/aromatic proton region and the fingerprint region, respectively, of the NOESY spectrum of [113-122]apoJ obtained using a mixing time of 200 ms in the presence of DPC micelle. Sequential dNN(i, i + 1) NOESY cross-peaks for all the residues in [113-122]apoJ are identified in Figure 8A. In addition, dNN(i, i + 2) cross-peaks were observed for Gly3/Gln5, Leu6/Glu8, Glu7/Phe9, and Glu8/Leu10 pairs and are labeled in Figure 8A. In Figure 8B intraresidue HN/CαH NOEs for all the residues in [113-122]apoJ are labeled. The methyl protons of the N-terminal acetyl protecting group showed NOEs to Leu1, Val2, Gly3, and Arg4 (Figure 8B). The observed (i, i + 1), (i, i + 2), (i, i + 3), and (i, i + 4) NOEs are schematically shown in Supplement Figure 1.
Figure 8. NOESY spectrum of [113-122]apoJ in DPC micelle shows characteristic NOEs of an α helical structure.
Amide/aromatic (A) and fingerprint (B) regions of the NOESY spectrum (mixing time 200 msec) of [113-122]apoJ in DPC micelle in 5mM potassium phosphate, pH 5.5, at 37°C. The spectrum was obtained as described in Figure 4. In A, sequential dNN(i,i+1) and the observed dNN(i,i+2) NOEs are labeled. Also labeled are the observed NOEs between residues Phe9 and Leu10 and C-terminal amide (NH2) blocking group. In B, intraresidue HN/HA NOEs as well as observed NOEs between N-terminal acetyl group (ACE) and other peptide protons are labeled.
Because of the conformational equilibrium present in solution, NMR data cannot be interpreted in terms of a single structure as is the case with structure determined using x-ray crystallography. However, the calculation of a limited number of structures compatible with the observed NMR data is a useful way to visualize the features of the family of structures present in the conformational ensemble. The distance and dihedral constraints were used to calculate NMR structures of [113-122]apoJ in the presence of DPC micelle. Starting from a set of initial 300 extended structures of [113-122]apoJ, 100 structures were accepted based on the following criteria: (a) no NOE violations >0.5 Å, (b) no dihedral angle violations >5°, (c) r.m.s. difference for bond deviations from ideality <0.01 Å, and (d) r.m.s. difference for angle deviations from ideality <2°. To obtain the average NMR structure, average coordinates of the 100 accepted structures of [113-122]apoJ were energy-minimized. All the distance and the dihedral angle constraints were enforced during the energy minimization of the average coordinates. The structural statistics for the 100 accepted NMR structures and the energy-minimized average structure of [113-122]apoJ are summarized in Table 2. The 100 accepted structures were superimposed onto the energy-minimized average structure using MATCH (SYBYL, version 7.2). The program MATCH performs an automatic least squares fit of two molecules that differ only in the coordinates of their atoms. Figure 9 shows superposition of the Cα atoms of all the 100 accepted structures on the energy-minimized average NMR structure of [113-122]apoJ. The energy-minimized average structure is shown as a ribbon/tube (Figure 9). Side chains of the energy-minimized average structure of [113-122]apoJ are also shown in Figure 9.
Table 2.
Structural statistics for the 100 accepted (<SA>) and the energy minimized average (<SA>r) NMR structures of [113-122]apoJ associated with DPC micelle
| Experimental restraints | ||
| Intraresidue NOEs | 41 | |
| Sequential NOEs | 23 | |
| Medium and long range NOEs | 19 | |
| (i,i+2; i,i+3; and i,i+4) | ||
| Other NOEs | 8 | |
| Hydrogen bonds (H-O distance) | 7 | |
| Φ angles | 9 | |
| Ψ angles | 8 | |
| Total | 115 | |
| Mean r.m.s.d.a from experimental restraints | <SA> ± SDb | |
| Distance restraints (Å) | 0.022±0.004 | |
| Dihedral angle (°) | 0.054±0.123 | |
| Mean r.m.s.d. from ideal geometry | <SA> ± SD | |
| Bonds (Å) | 0.001±0.000(4) | |
| Angles (°) | 0.473±0.029 | |
| Impropers (°) | 0.374±0.038 | |
| Ramachandran statistics from PROCHECK-NMRc | ||
| Residues in most favored α helical region | 100 % | |
| r.m.s.d. from the average structure (Å) | ||
| Backbone N, Cα, C | 0.296 | |
| All heavy atoms | 1.371 | |
| Pairwise r.m.s.d. analysis | ||
| Average backbone pairwise r.m.s.d. (Å) | 0.420 (±0.122) | |
| Average non-hydrogen atom pairwise r.m.s.d. (Å) | 1.875 (±0.264) | |
| X-PLOR potential energies (kcal/mol) | ||
| <SA> ± SD | (<SA>r)d | |
| Eoverall | 15.039±2.489 | 14.774 |
| Ebond | 0.378±0.147 | 0.408 |
| Eangle | 11.095±1.446 | 10.971 |
| Eimpropers | 1.969±0.424 | 1.809 |
| Evdw | 0.328±0.532 | 0.0 |
| ENOE | 1.248±0.522 | 1.585 |
| Ecdih | 0.018±0.072 | 0.0 |
Root mean square deviation.
Standard deviation.
Ref. [41]
200 cycles of conjugate gradient minimization were used. All of the NMR constraints were enforced during the energy minimization of the average coordinates.
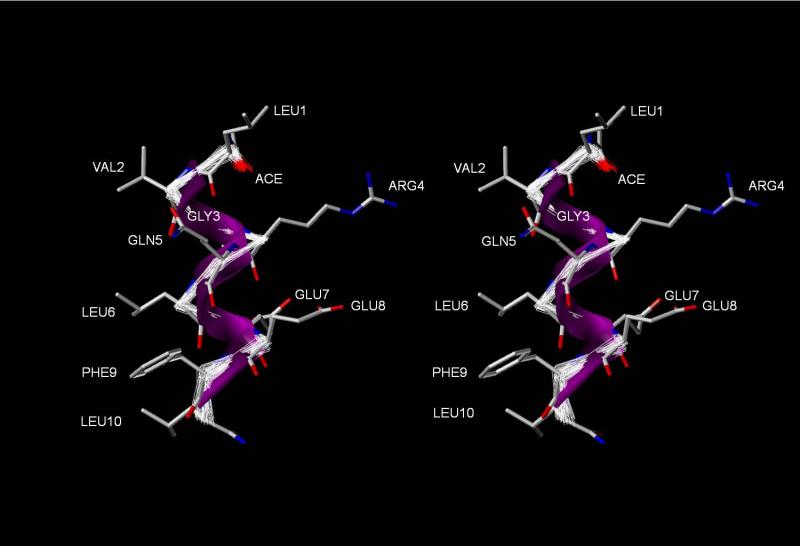
Figure 9. Amphipathic α helical structure of [113-122]apoJ in the presence of DPC micelle.
A relaxed eye stereo view of the 100 accepted NMR structures superimposed on the energy minimized average structure of [113-122]apoJ. N-terminus is towards the top and C-terminus is towards the bottom of the page. Note the distinct segregation of nonpolar and polar side chains along the long axis of the α helix.
4. Discussion
An understanding of detailed structure and lipid-associating properties of anti-inflammatory and anti-atherogenic peptides is expected to provide valuable information for the future design of highly active peptide analogs. LOCATE [18] analysis, as well as helical wheel and helical net diagrams (Figure 1), predict an amphipathic α helical structure of [113-122]apoJ. Results of CD analysis indicate that indeed in the presence of DPC micelle [113-122]apoJ adopts an α-helical structure (Figure 2, Table 1). An α-helical structure is stabilized by the formation of i + 4 → i hydrogen bonds. Short polypeptides generally do not adopt an α-helical structure in solution as the hydrogen bond formation does not occur because of the high entropic cost associated with the folding of the polypeptide chain. However, within a hydrophobic environment, e.g., within the hydrophobic pockets of proteins, the α-helical secondary structures of even small peptide fragments can be stabilized through local inter-residue interactions. The water-lipid interface of the DPC micelle in the present study provides such an environment for facilitating the formation and stabilization of α-helical structure of [113-122]apoJ peptide.
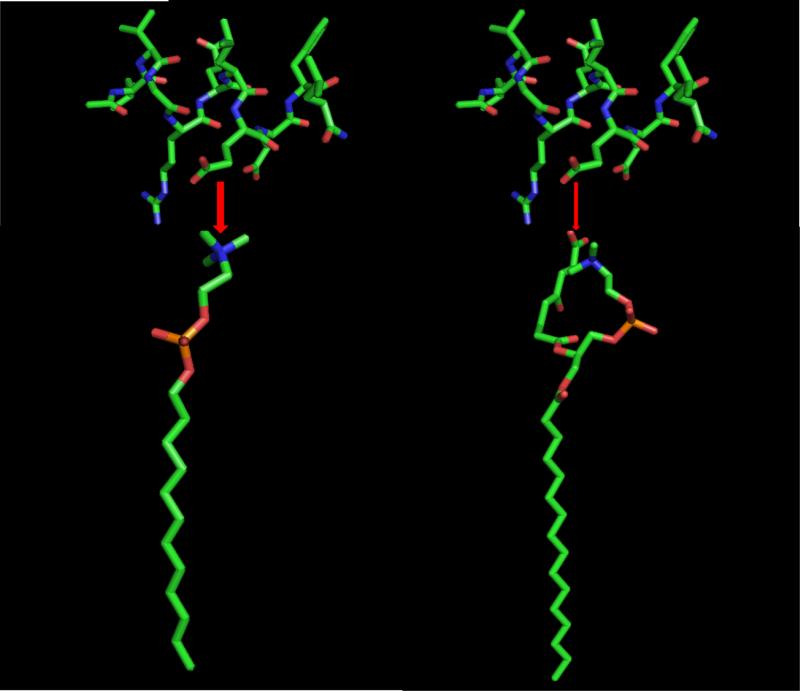
Results of ITC experiments indicate that binding of [113-122]apoJ with lipids is accompanied by a favorable enthalpy change (Figure 3). It is interesting to note, however, that the change in enthalpy is much smaller for the interaction of [113-122]apoJ with KOdiA-PC (Figure 3B). Even addition of 5mol% of KOdiA-PC to DPC micelle lowers the enthalpy of interaction (Figure 3C). It is important to note that the overall electrical charge of [113-122]apoJ at neutral pH is minus one (-1). The sn-2 side chain of KOdiA-PC which ends in a carboxyl group, according to the “Whisker” model [31], will produce a water-lipid interface that is overall negatively charged. This is in contrast to the zwitterionic nature of the water-lipid interface in case of DPC. An overall negatively charged peptide is likely to interact weakly with an overall negatively charged lipid surface compared to a zwitterionic surface. This is shown schematically in Figure 10.
Figure 10. A proposed model of initial interaction of [113-122]apoJ with DPC and KOdiA-PC.
A schematic representation of the binding of [113-122]apoJ with DPC (left) and KOdiA-PC (right). The thickness of the red arrow indicates strength of binding. Because of the overall negative charge of [113-122]apoJ, it is proposed to bind weakly with negatively charged KOdiA-PC than with zwitterionic DPC.
Although CD results indicated that [113-122]apoJ is predominantly α-helical in the presence of DPC micelle (Figure 2, Table 1), conformation of individual residues cannot by obtained using this method. To obtain a high resolution three dimensional structure of [113-122]apoJ in the presence of DPC micelle, two dimensional (2D) 1H NMR experiments were carried out. The 1H NMR chemical shifts in peptides and proteins are dependent on the secondary structure, e.g. CαH protons move upfield (relative to their random coil value) on helix formation [28,29]. An inspection of Figure 7 reveals that all the CαH protons in [113-122]apoJ show an upfield shift compared to random coil values consistent with an α-helical secondary structure.
The α-helical secondary structure of [113-122]apoJ was further evident by the observation of sequential and medium range (dNN(i, i+1) and dNN(i,i+2) NOEs (Figure 8A). In addition, several other long range (i, i+3) NOEs were also observed (Figure S1) consistent with the α-helical secondary structure of [113-122]apoJ in the presence of DPC micelle. The amphipathic α-helical structure of [113-122]apoJ peptide is evident in Figure 9, with the hydrophobic residues clustered on one face (non polar) of the helix and charged and polar residues clustered on the opposite face (polar) of the helix.
We have shown previously that both N-terminal acetyl and C-terminal amide blocking groups stabilize α-helical structure in peptides by allowing the formation of two additional H-bonds (one at each end) between residues i (acceptor) and i + 4 (donor) compared with the unblocked peptide [26,33]. Consistent with our earlier observations, CD analysis of unblocked [113-122]apoJ peptide (without acetyl and amide blocking groups) revealed reduced helicity (based on K2D analysis, 44% in the presence of DPC micelle). In addition, we found that the blocking groups are involved in the inter-residue interactions as well (Figures 8A and 8B) presumably further stabilizing the α-helical structure of [113-122]apoJ. The N-terminal acetyl group shows NOEs to Leu1, Val2, Gly3, and Arg4 backbone NH protons (Figure 8B), whereas C-terminal amide group shows NOEs to Phe9 and Leu10 backbone NH protons (Figure 8A). It is worthy of note that, in addition to helix stabilization, N-terminal acetylation increases the overall hydrophobicity of the peptide also facilitating its partitioning from the aqueous phase into the lipid phase.
How will the information obtained in the present work help explain the observed anti-inflammatory and anti-atherosclerotic properties of [113-122]apoJ? Dyslipidemia is associated with oxidative stress and the generation of biologically active oxidized lipids. Biologically active oxidized phospholipids can initiate and modulate many of the cellular events attributed to the pathogenesis of atherosclerosis [34]. A novel family of atherogenic oxidized choline glycerophospholipids (oxPCCD36) that are formed during the oxidization of low density lipoprotein by multiple pathways and are present in vivo at sites of enhanced oxidative stress have been isolated and structurally characterized [35,36]. oxPCCD36 serve as high affinity ligands for the macrophage scavenger receptor CD36 and facilitate macrophage foam cell formation through recognition and uptake of oxidized low density lipoprotein by CD36 [37]. It has been suggested that the selectively blocking the interaction between oxPCCD36 and CD36 might be an useful therapeutic approach against atherothrombotic diseases [38]. The conformation of a prototypic high affinity CD36 ligand, KOdiA-PC, near the hydrophobic-hydrophilic interface within membrane bilayers was probed by determining multiple critical internuclear distances using NOESY and it was shown that the truncated oxidized sn-2 fatty acid chain of KOdiA-PC within membranes protrudes into the aqueous phase (the “whisker” model) [31]. Based on these observations, it was hypothesized that this allows a direct physical access of the KOdiA-PC to the cell surface macrophage CD36 receptor [31]. It was shown further that this unusual conformation for an oxidized phospholipid within a membrane may not be unique to KOdiA-PC but rather might represent a more global phenomenon of oxidized phospholipids, enabling direct physical contact between pattern recognition receptor and molecular pattern ligand [39,40].
Based on the results obtained using different techniques (CD, ITC, and 1H and 31P NMR), we have shown that [113-122]apoJ is able to bind KOdiA-PC, an oxidized lipid. It is likely that a potential mechanism of anti-atherogenic and anti-inflammatory property of [113-122]apoJ is due to its association with the oxidized lipids. As proposed earlier for 4F [33], this may allow the sequestration of the oxidized phospholipids by [113-122]apoJ, thereby preventing interaction of oxidized phospholipids with their receptor (e.g. CD36).
In summary, this study has significantly improved our knowledge about the structure and molecular interactions of an anti-atherogenic and anti-inflammatory class G* amphipathic helix, [133-122]apoJ, with normal as well as oxidized lipids. We have demonstrated unambiguously that even a relatively short 10 residue peptide can be induced into α-helix conformations in aqueous solution through binding to lipid. The amphipathic α-helical structure of the peptide and its ability to associate with oxidized lipids as demonstrated in the present work for the first time help explain the previously observed anti-inflammatory and anti-atherosclerotic properties. Refinement of this structure using new peptide analogs may yield novel highly effective and easily synthesizable peptides in large amounts for the inhibition and treatment of atherosclerosis and other inflammatory diseases.
Supplementary Material
Acknowledgements
We thank Professors David E. Graves and Donald D. Muccio and Dr. Michael J. Jablonsky of the Chemistry Department, UAB, for access to their instrument facility. We thank Axel T. Brünger for the access of online version 3.851 of X-PLOR. We thank Martin K. Jones for his help in the software use and maintenance.
The abbreviations used are
- ApoA-I
apolipoprotein A-I
- CSI
chemical shift index
- DPC
dodecyphosphocholine
- DSS
sodium 2,2-dimethyl-2-silapentane-5-sulfonate
- KOdiA-PC
1-palmitoyl-2-(5-keto-6-octene dioyl)phosphatidylcholine
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- TOCSY
total correlation spectroscopy
- r.m.s.d.
root mean square deviation
- TPPI
time proportional phase incrementation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This research is supported in part by grants PO1 HL34343 (NHLBI) and CA-13148 (NCI) of the National Institutes of Health. 600 MHz CryoProbe was funded by 1S10RR021064-01A1 (NIH/NCRR).
The atomic coordinates of NMR structures of [113-122]apoJ in DPC micelle, 1H chemical shifts, and distance and dihedral constraints have been deposited in Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu; accession number 20124 ).
Reference List
- 1.Hausenloy DJ, Yellon DM. Enhancing cardiovascular disease risk reduction: raising high-density lipoprotein levels. Current Opinion in Cardiology. 2009;24 doi: 10.1097/HCO.0b013e32832ebfe7. [DOI] [PubMed] [Google Scholar]
- 2.Castelli WP, Garrison RJ, Wilson PW, Abbott RD, Kalousdian S, Kannel WB. Incidence of coronary heart disease and lipoprotein cholesterol levels. The Framingham Study. JAMA. 1986;256:2835. [PubMed] [Google Scholar]
- 3.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J Med. 1977;62:707. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 4.Hausenloy DJ, Yellon DM. Targeting residual cardiovascular risk: raising high-density lipoprotein cholesterol levels. Heart. 2008;94:706. doi: 10.1136/hrt.2007.125401. [DOI] [PubMed] [Google Scholar]
- 5.Badimon JJ, Badimon L, Fuster V. Regression of atherosclerotic lesions by high density lipoprotein plasma fraction in the cholesterol-fed rabbit. J Clin Invest. 1990;85:1234. doi: 10.1172/JCI114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franceschini G, Sirtori CR, Capurso A, Weisgraber KH, Mahley RW. A-IMilano apoprotein. Decreased high density lipoprotein cholesterol levels with significant lipoprotein modifications and without clinical atherosclerosis in an Italian family. J Clin Invest. 1980;66:892. doi: 10.1172/JCI109956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisgraber KH, Bersot TP, Mahley RW, Franceschini G, Sirtori CR. A-Imilano apoprotein. Isolation and characterization of a cysteine-containing variant of the A-I apoprotein from human high density lipoproteins. J Clin Invest. 1980;66:901. doi: 10.1172/JCI109957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissen SE, Tsunoda T, Tuzcu EM, Schoenhagen P, Cooper CJ, Yasin M, Eaton GM, Lauer MA, Sheldon WS, Grines CL, Halpern S, Crowe T, Blankenship JC, Kerensky R. Effect of Recombinant ApoA-I Milano on Coronary Atherosclerosis in Patients With Acute Coronary Syndromes: A Randomized Controlled Trial. JAMA. 2003;290:2292. doi: 10.1001/jama.290.17.2292. [DOI] [PubMed] [Google Scholar]
- 9.Tardif JC, Gregoire J, L'Allier PL, Ibrahim R, Lesperance J, Heinonen TM, Kouz S, Berry C, Basser R, Lavoie MA, Guertin MC, Rodes-Cabau J, the Effect of rHDL on Atherosclerosis-Safety and Efficacy (ERASE) Investigators Effects of Reconstituted High-Density Lipoprotein Infusions on Coronary Atherosclerosis: A Randomized Controlled Trial. JAMA. 2007;297:1675. doi: 10.1001/jama.297.15.jpc70004. [DOI] [PubMed] [Google Scholar]
- 10.Garber DW, Datta G, Chaddha M, Palgunachari MN, Hama SY, Navab M, Fogelman AM, Segrest JP, Anantharamaiah GM. A new synthetic class A amphipathic peptide analogue protects mice from diet-induced atherosclerosis. J. Lipid Res. 2001;42:545. [PubMed] [Google Scholar]
- 11.Navab M, Anantharamaiah GM, Hama S, Garber DW, Chaddha M, Hough G, Lallone R, Fogelman AM. Oral Administration of an Apo A-I Mimetic Peptide Synthesized From D-Amino Acids Dramatically Reduces Atherosclerosis in Mice Independent of Plasma Cholesterol. Circulation. 2002;105:290. doi: 10.1161/hc0302.103711. [DOI] [PubMed] [Google Scholar]
- 12.Navab M, Anantharamaiah GM, Reddy ST, Hama S, Hough G, Grijalva VR, Wagner AC, Frank JS, Datta G, Garber D, Fogelman AM. Oral D-4F Causes Formation of Pre-{beta} High-Density Lipoprotein and Improves High-Density Lipoprotein-Mediated Cholesterol Efflux and Reverse Cholesterol Transport From Macrophages in Apolipoprotein E-Null Mice. Circulation. 2004;109:3215. doi: 10.1161/01.CIR.0000134275.90823.87. [DOI] [PubMed] [Google Scholar]
- 13.Bloedon LT, Dunbar R, Duffy D, Pinell-Salles P, Norris R, DeGroot BJ, Movva R, Navab M, Fogelman AM, Rader DJ. Safety, pharmacokinetics, and pharmacodynamics of oral apoA-I mimetic peptide D-4F in high-risk cardiovascular patients. J. Lipid Res. 2008;49:1344. doi: 10.1194/jlr.P800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141. [PubMed] [Google Scholar]
- 15.Datta G, Chaddha M, Hama S, Navab M, Fogelman AM, Garber DW, Mishra VK, Epand RM, Epand RF, Lund-Katz S, Phillips MC, Segrest JP, Anantharamaiah GM. Effects of increasing hydrophobicity on the physical-chemical and biological properties of a class A amphipathic helical peptide. J. Lipid Res. 2001;42:1096. [PubMed] [Google Scholar]
- 16.de Silva HV, Stuart WD, Duvic CR, Wetterau JR, Ray MJ, Ferguson DG, Albers HW, Smith WR, Harmony JA. A 70-kDa apolipoprotein designated ApoJ is a marker for subclasses of human plasma high density lipoproteins. J Biol Chem. 1990;265:13240. [PubMed] [Google Scholar]
- 17.Navab M, Hama-Levy S, Van Lenten BJ, Fonarow GC, Cardinez CJ, Castellani LW, Brennan ML, Lusis AJ, Fogelman AM, La Du BN. Mildly oxidized LDL induces an increased apolipoprotein J/paraoxonase ratio. J Clin Invest. 1997;99:2005. doi: 10.1172/JCI119369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segrest JP, Jones MK, Mishra VK, Anantharamaiah GM, Garber DW. apoB-100 has a pentapartite structure composed of three amphipathic alpha-helical domains alternating with two amphipathic beta-strand domains. Detection by the computer program LOCATE. Arterioscler Thromb. 1994;14:1674. doi: 10.1161/01.atv.14.10.1674. [DOI] [PubMed] [Google Scholar]
- 19.Navab M, Anantharamaiah GM, Reddy ST, Van Lenten BJ, Wagner AC, Hama S, Hough G, Bachini E, Garber DW, Mishra VK, Palgunachari MN, Fogelman AM. An Oral ApoJ Peptide Renders HDL Antiinflammatory in Mice and Monkeys and Dramatically Reduces Atherosclerosis in Apolipoprotein E-Null Mice. Arterioscler Thromb Vasc Biol. 2005;25:1932. doi: 10.1161/01.ATV.0000174589.70190.e2. [DOI] [PubMed] [Google Scholar]
- 20.Navab M, Ruchala P, Waring AJ, Lehrer RI, Hama S, Hough G, Palgunachari MN, Anantharamaiah GM, Fogelman AM. A novel method for oral delivery of apolipoprotein mimetic peptides synthesized from all L-amino acids 2. J. Lipid Res. 2009;50:1538. doi: 10.1194/jlr.M800539-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anantharamaiah GM, Mishra VK, Garber DW, Datta G, Handattu SP, Palgunachari MN, Chaddha M, Navab M, Reddy ST, Segrest JP, Fogelman AM. Structural requirements for antioxidative and anti-inflammatory properties of apolipoprotein A-I mimetic peptides 130. J. Lipid Res. 2007;48:1915. doi: 10.1194/jlr.R700010-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Amar MJ, D'Souza W, Turner S, Demosky S, Sviridov D, Stonik J, Luchoomun J, Voogt J, Sviridov D, Remaley AT. 5A Apolipoprotein Mimetic Peptide Promotes Cholesterol Efflux and Reduces Atherosclerosis in Mice 147. J Pharmacol Exp Ther. 2010 doi: 10.1124/jpet.110.167890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bielicki JK, Zhang H, Cortez Y, Zheng Y, Narayanaswami V, Patel A, Johansson J, Azhar S. A new HDL mimetic peptide that stimulates cellular cholesterol efflux with high efficiency greatly reduces atherosclerosis in mice 4. J. Lipid Res. 2010;51:1496. doi: 10.1194/jlr.M003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protocols. 2007;1:2876. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol. NMR. 1992;2:661. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 26.Mishra VK, Anantharamaiah GM, Segrest JP, Palgunachari MN, Chaddha M, Sham SWS, Krishna NR. Association of a Model Class A (Apolipoprotein) Amphipathic {alpha} Helical Peptide with Lipid: HIGH RESOLUTION NMR STUDIES OF PEPTIDE{middle dot}LIPID DISCOIDAL COMPLEXES 2. J. Biol. Chem. 2006;281:6511. doi: 10.1074/jbc.M511475200. [DOI] [PubMed] [Google Scholar]
- 27.Wuthrich K, Billeter M, Braun W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J Mol Biol. 1983;169:949. doi: 10.1016/s0022-2836(83)80144-2. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. Journal of Molecular Biology. 1991;222:311. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363. doi: 10.1016/s0076-6879(94)39014-2. 363-92. [DOI] [PubMed] [Google Scholar]
- 30.Gibbs AC, Kondejewski LH, Gronwald W, Nip AM, Hodges RS, Sykes BD, Wishart DS. Unusual beta-sheet periodicity in small cyclic peptides. Nat Struct Biol. 1998;5:284. doi: 10.1038/nsb0498-284. [DOI] [PubMed] [Google Scholar]
- 31.Li XM, Salomon RG, Qin J, Hazen SL. Conformation of an Endogenous Ligand in a Membrane Bilayer for the Macrophage Scavenger Receptor CD36ΓÇá. Biochemistry. 2007;46:5009. doi: 10.1021/bi700163y. [DOI] [PubMed] [Google Scholar]
- 32.Basus VJ. Proton nuclear magnetic resonance assignments. Methods Enzymol. 1989;177:132. doi: 10.1016/0076-6879(89)77009-9. 132-49. [DOI] [PubMed] [Google Scholar]
- 33.Mishra VK, Palgunachari MN, Krishna NR, Glushka J, Segrest JP, Anantharamaiah GM. Effect of Leucine to Phenylalanine Substitution on the Nonpolar Face of a Class A Amphipathic Helical Peptide on Its Interaction with Lipid: HIGH RESOLUTION SOLUTION NMR STUDIES OF 4FDIMYRISTOYLPHOSPHATIDYLCHOLINE DISCOIDAL COMPLEX 3. J. Biol. Chem. 2008;283:34393. doi: 10.1074/jbc.M806384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berliner JA, Watson AD. A Role for Oxidized Phospholipids in Atherosclerosis. N Engl J Med. 2005;353:9. doi: 10.1056/NEJMp058118. [DOI] [PubMed] [Google Scholar]
- 35.Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Febbraio M, Hajjar DP, Silverstein RL, Hoff HF, Salomon RG, Hazen SL. A novel family of atherogenic oxidized phospholipids promotes macrophage foam cell formation via the scavenger receptor CD36 and is enriched in atherosclerotic lesions. J Biol Chem. 2002;277:38517. doi: 10.1074/jbc.M205924200. [DOI] [PubMed] [Google Scholar]
- 36.Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Gugiu B, Fox PL, Hoff HF, Salomon RG, Hazen SL. Identification of a Novel Family of Oxidized Phospholipids That Serve as Ligands for the Macrophage Scavenger Receptor CD36. J. Biol. Chem. 2002;277:38503. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 37.Podrez EA, Febbraio M, Sheibani N, Schmitt D, Silverstein RL, Hajjar DP, Cohen PA, Frazier WA, Hoff HF, Hazen SL. Macrophage scavenger receptor CD36 is the major receptor for LDL modified by monocyte-generated reactive nitrogen species. J Clin Invest. 2000;105:1095. doi: 10.1172/JCI8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson SP, Calkin AC. The clot thickens--oxidized lipids and thrombosis. Nat Med. 2007;13:1015. doi: 10.1038/nm0907-1015. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg ME, Li XM, Gugiu BG, Gu X, Qin J, Salomon RG, Hazen SL. The Lipid Whisker Model of the Structure of Oxidized Cell Membranes. J. Biol. Chem. 2008;283:2385. doi: 10.1074/jbc.M707348200. [DOI] [PubMed] [Google Scholar]
- 40.Hazen SL. Oxidized Phospholipids as Endogenous Pattern Recognition Ligands in Innate Immunity. J. Biol. Chem. 2008;283:15527. doi: 10.1074/jbc.R700054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - A Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.