Abstract
Sample preparation techniques allowing reliable and reproducible imaging of DNA with various structures, topologies and complexes with proteins are reviewed. The major emphasis is given to methods utilizing chemical functionalization of mica, enabling preparation of the surfaces with required characteristics. The methods are illustrated by examples of imaging of different DNA structures. Special attention is given to the possibility of AFM to image the dynamics of DNA at the nanoscale. The capabilities of time-lapse AFM in aqueous solutions are illustrated by imaging of dynamic processes as transitions of local alternative structures (transition of DNA between H and B forms). The application of AFM to studies of protein-DNA complexes is illustrated by a few examples of imaging site-specific complexes, as well as such systems as chromatin. The time-lapse AFM studies of protein-DNA complexes including very recent advances with the use of high-speed AFM are reviewed.
Keywords: AFM, surface functionalization, silanes, DNA, DNA supercoiling, alternative DNA structures, protein-DNA complexes, single molecule imaging
1. AFM: basic concepts
Sample preparation is a key step for any imaging method, and particularly for scanning probe microscopy (SPM). The prototype scanning tunneling microscope (STM) instrument was conceived by Binnig and Rohrer (Binnig et al., 1982; Binning and Rohrer, 1982), an invention for which the authors were awarded the 1986 Nobel Prize in Physics. The atomic force microscope (AFM) was invented in 1986 (Binnig et al., 1986) and its development by the Hansma group (Hansma et al., 1988) resulted in the commercial production of the AFM available to the biological community.
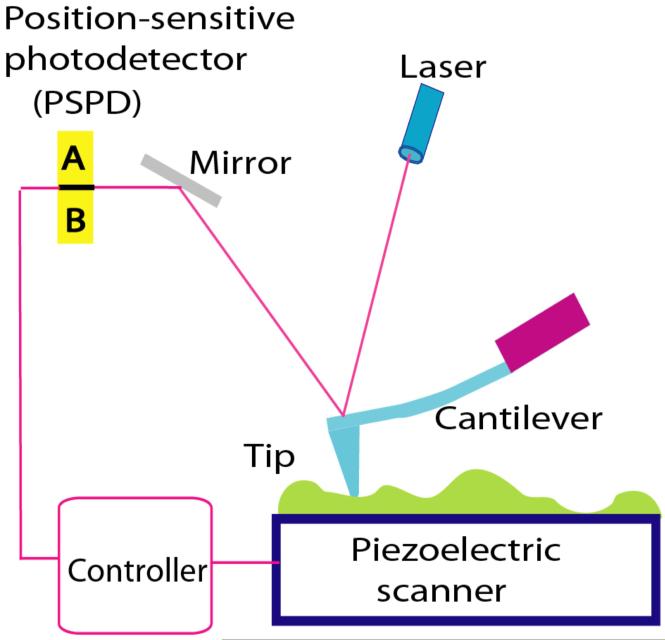
The schematic illustrating principles of the AFM operation is shown in Fig. 1. A sharp stylus (AFM tip shown as a blue triangle) reads the profile of the sample (shown as a green bumpy profile) by scanning over the sample. The tip is attached to a cantilever that works as a spring pressing the tip against the sample to reproduce the surface profile. The vertical position of the tip is measured by a laser light reflected from the cantilever to the position sensitive photodetector (PSPD). In this particular scheme, the tip is stationary, but the sample mounted on the piezoelectric scanner moves. There are a number of important features of the AFM instrument listed below. First, the position of the sample relative to the tip is controlled by the scanner and it can be done with accuracy better than 1 nm. Second, the tip can be atomically sharp. Third, the displacement of the tip relative to the surface is determined with sub-nanometer accuracy. These three major factors lay the foundation for the capability of AFM to provide the topographic image with atomic accuracy. Indeed, the atomic resolution for AFM was achieved in the early paper Ohnesorge and Binnig (1993) in which the atomic-scale periodicities of calcite as well as the expected relative positions of the atoms within each unit cell were obtained. Importantly, attractive forces on the order of 10 pico Newton (pN) acting between single atomic sites on the sample and the front atoms of the tip were directly measured. Later both calcium and oxygen atoms at the cleavage plane of CaSO4 surface were identified (Sokolov et al., 1999). A breakthrough in the high-resolution AFM imaging was made with implementation of the so-called non-contact mode (NC-AFM; see Lauritsen and Reichling (2010) and references therein), offering a unique tool for real space atomic-scale studies of surfaces, nanoparticles, as well as thin films, irrespective of the substrate being electrically conducting or non-conducting. Note that atomic resolution of STM was demonstrated in the early works of Binnig and Rohrer (Binnig et al., 1982; Binning and Rohrer, 1982). The ability of STM to provide atomic resolution more easily than AFM is explained by the steeper dependence of the output signal on the tip-sample distance for STM compared to AFM. In STM, the tip-sample distance is measured by the tunneling current that decays exponentially with the interatomic distance, whereas the contrast in the AFM is provided by the interatomic interactions that decay much weaker with the tip-sample distance. Therefore, additional improvements of AFM instrumentation and sample purification were needed to achieve atomic resolution. Note, however, that STM requires electric conductivity that is not an issue for AFM. Superatomic resolution has been achieved recently for isolated organic molecules imaged with AFM and STM (Gross et al., 2010). Imaging was performed with a combined AFM/STM system (5 K and high vacuum) and AFM resolution was superior to the one achieved with STM.
Figure 1.
Schematic explaining the principles of AFM operation. The position of the tip relative to the sample is controlled by a piezoelectric scanner. The vertical displacement of the tip during scanning is detected using the optical lever principle, in which the position of the light spot on the PSPD is measured.
2. AFM operation modes
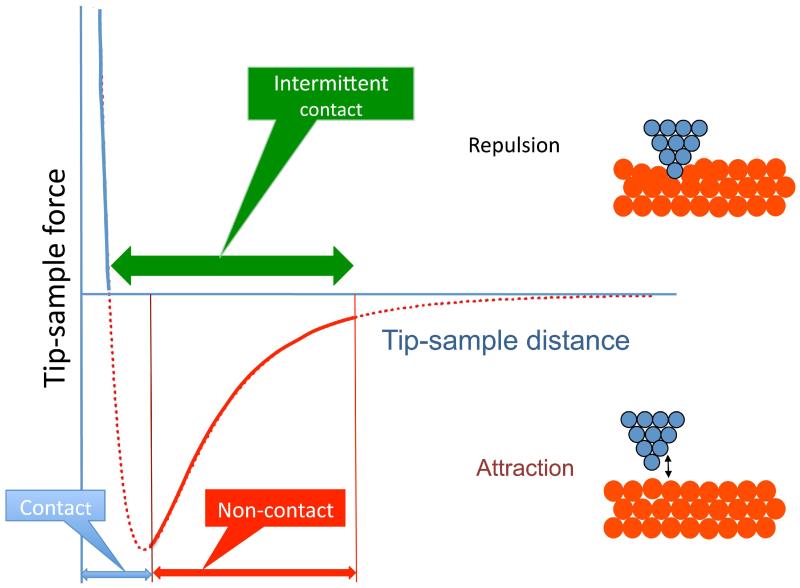
Fig. 2 illustrates the major modes of the AFM operation. The regime of the microscope operation corresponding to the attraction part of the tip-sample interaction potential is termed non-contact mode (NC-AFM). After passing the minimum on the potential curve, the repulsion interaction rises very fast, achieving large positive values in the very short range of the tip-sample distances. This scanning regime is termed contact mode. The interaction of the tip and the surface is strong, so the deformation of a soft sample typically occurs. Schematically, this deformation is illustrated in the top right cartoon by displacement of the red spheres. For hard samples at small tip-surface separation, the pressure created by the tip at the apex is so strong that atoms can jump from tip to surface and vice versa. Therefore, contact mode of the AFM operation does not allow for obtaining reliable and stable images at atomic resolution. In addition, damage or deformation of soft biological samples can be made to the tip during approach or scanning.
Figure 2.
Scheme explaining the modes of the AFM operation. A curve in the scheme shows the change of the tip-sample interaction depending on the distance between the apex of the tip and the sample.
Efficient in numerous biological applications was an intermediate or intermittent contact mode (IC mode). Initially, this mode was termed Tapping mode (Zhong et al., 1993), but the name has been trademarked by the AFM manufacturer Veeco. Therefore, other manufacturers utilize this methodology under different names such as alternating contact (AC). Schematically, the range of operation of AC/IC/TM mode is indicated in Fig. 2 with a thick horizontal double arrow. In the contact mode, the tip-sample distance is maintained via measuring the deflection of the tip cantilever determined by the van der Waals repulsion forces (Fig. 1), but the detection principle for the intermediate mode is different. In AC/IC/ /TM-AFM, a cantilever is deliberately vibrated at the frequency close to the cantilever resonant frequency by a piezoelectric modulator with very small amplitude. As the tip approaches a surface, the van der Waals attractive force between the tip and the sample changes both the amplitude and the phase of the cantilever vibration. These changes are monitored by a Z-servo system feedback loop to control the tip-sample distance. The ability of the AC/IC/TM mode to image biological samples gently was the major attractive feature of this operating mode.
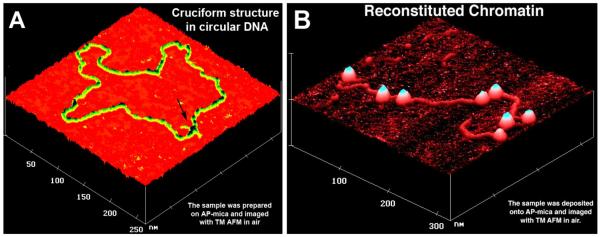
Fig. 3A shows images of two biological samples to illustrate the power of Tapping Mode for biological applications. An image of circular DNA with extruded cruciform structure (indicated with an arrow) is shown in this figure (Shlyakhtenko et al., 2000a). The DNA appears as a filament with a uniform thickness. Note that transmission electron microscopy was not that efficient in the structural study of alternative DNA structures. Therefore, the ability of AFM to image alternative DNA structures makes this microscopy very attractive in the field of molecular and structural genetics.
Figure 3.
AFM images of (A) circular pUC8F14C plasmid DNA with the cruciform protrusion (indicated with an arrow) and (B) reconstituted chromatin. The images were acquired in air with a NanoScope III microscope operated in the Tapping Mode.
Fig. 3B shows the AFM image of reconstituted chromatin assembled on purified histone octamers (Yodh et al., 2002). Nucleosomal particles formed by wrapping of DNA around the histone octamers are unambiguously seen as bright globular features (blobs). In a majority of cases, the sizes of the blobs are almost identical and no damage to the nucleosomes is seen, confirming the appropriateness of Tapping Mode for imaging of such large biomolecular complexes. Electron microscopy was initially applied to prove the beads on a string morphology of isolated or reconstituted chromatin, but reliable imaging required tedious sample preparation including staining of the sample. The images shown here in frame B were obtained without any staining. The sample was crosslinked with glutaraldehyde, but the recent advances in the sample preparation for AFM made it possible to obtain images of chromatin, omitting the crosslinking step (Lyubchenko and Shlyakhtenko, 2009; Shlyakhtenko et al., 2009).
Note that oscillating tip is utilized in NC-AFM as well. According to Fig. 2, the major requirement for the reliable operation of AFM in NC mode is keeping the tip-sample distance in the range of a nanometer with sub-nanometer accuracy, which is accomplished with the use of frequency modulation mode originally proposed in Martin et al. (1987). Atomic resolution was achieved with the use of this AFM mode for solid-state materials-like crystals and the scanning is typically performed in vacuum. Imaging in liquid can also be performed in the non-contact regime, and also requires a special design of the instrument or substantial upgrade of a commercial AFM. For example, in Rode et al. (2009), a home-built controller was used and a special liquid cell was designed, so they were able to keep the cantilever oscillation amplitude constant at a level of 0.2-2.0 nm and acquire images with amplitudes in the range of 0.2-0.5 nm.
3. AFM resolution
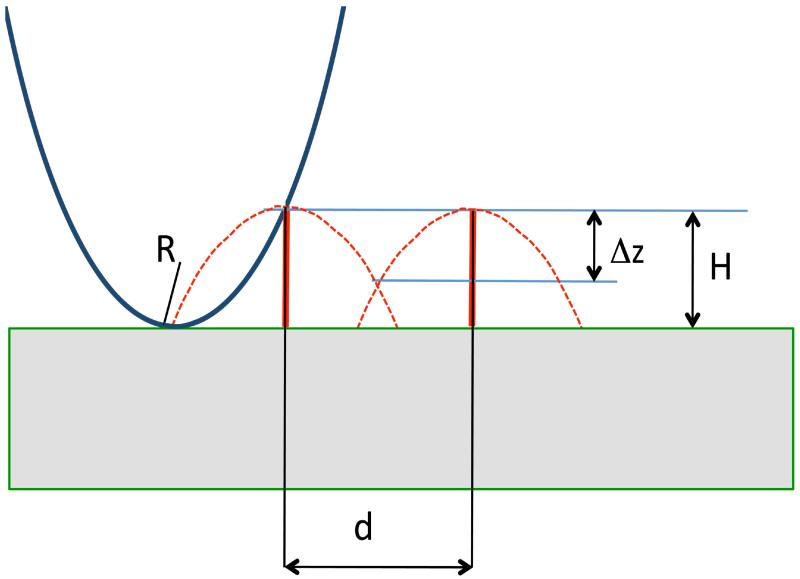
We will follow Bustamante et al. (1997) to illustrate the resolution principle for SPM using the scheme shown in Fig. 4. In this figure, the tip of smooth shape has at the apex radius of curvature, R. This tip performs scanning of the surface with very narrow spikes, with height H and distance d between them. Dotted lines show the tip trajectories that are in fact inverted profiles of the probe. Two major conclusions emerge from this scheme. The first one is the resolution criteria. If Δz is a vertical distance between the object top and the intersection of the two dotted profiles (a dimple in the profile), the following expression linking the value with the distance between the profile maxima (d) and the height of objects H was obtained in (Bustamante et al., 1997):
| (eq. 1) |
Figure 4.
Scheme explaining the resolution principles for SPM. See the text for explanation.
For the resolution limit, we can use the Rayleigh criterion according to which minimal distance dmin corresponds to Δz=0.2 H, the following formula can be obtained:
| (eq. 2) |
If R and H are equal to 0.1 nm, we can obtain dmin value ≈0. 12 nm and this example corresponds to imaging of atoms (size 0.1 nm), in which the image is generated by the very terminal atom of the tip (see Fig. 2, schemes on the right). In the case of double stranded DNA with the 2nm diameter and for the tip radius of curvature 10 nm, which is a typical nominal value for regular AFM tips, the value dmin ≈5.7 nm can be obtained. Thus, it is problematic to resolve the 3.4 nm helical periodicity of the DNA double helix using such blunt tips. Much sharper tips are required for the high resolution AFM imaging. Note, however, that regular AFM tips with 10 nm nominal radius of curvature can be much sharper, enabling imaging of the DNA helical periodicity (Lyubchenko and Shlyakhtenko, 1997; Mou et al., 1995) and the protein two-dimensional arrays with the subnanometer resolution (Engel et al., 1999). Recent advances in fabrication of ultrasharp AFM tips (radius of curvature ~ 1 nm) made it possible to perform high-resolution studies of nucleic acids of different types and conformations (Klinov et al., 2007). Note as well that the pitch for synthetic poly(dG)-poly(dC) was resolved using high resolution STM (Shapir et al., 2006). Nicks in the synthetic duplexes were identified on the images. However, a special sample preparation procedure was developed for reliable imaging of DNA. In addition, the scanning was performed at low temperatures (78K) to reliably resolve the pitch, because usually the sample is easily displaced by the STM tip during scanning. Note the AFM study Mou et al. (1995) in which the DNA pitch was resolved for the DNA array imaged at room temperature and in aqueous solution.
Another feature of the AFM is the difference in the sizes of real objects and their AFM images that are enlarged compared to the sizes of real object. The effect, called tip convolution, is graphically illustrated in Fig. 4, showing that initially thin vertical lines are transformed into wide bell-shape images after scanning. For the model in which a large tip with radius of curvature R scans over a small sample with radius r, the following simple equation for the calculating the size of the AFM image D at conditions R»r can be obtained (Bustamante et al., 1992):
| (eq. 3) |
This equation for the diameter of the DNA image (r = 1 nm) generated by the tip with R=10 nm yields D = 12.6 nm, a value typically obtained in routine imaging of DNA (see Fig. 1). The value drops 3 times for sharp tips and this effect is also in line with the high-resolution AFM images in which DNA appears as thin as 3-4 nm (Lyubchenko and Shlyakhtenko, 1997) (Klinov et al., 2007).
4. Factors influencing the AFM resolution
The major criteria for high-resolution imaging are the sharpness of the tip and the ability to operate the microscope at sub-nanometer distances corresponding to non-contact mode. Specifics can be found in Schwarz et al. (2000). According to eq. 2 for the tip with radius curvature 1 nm and DNA molecule with H = 2 nm, the expected resolution is ≈1.8 nm. The value is two times less for molecules that are 4 times lower. Indeed, the high-resolution data for self-assembled protein arrays characterizing by the height in the range of 0.5 nm (Engel et al., 1999; Muller et al., 2006; Scheuring and Dufrene, 2010; Scheuring and Sturgis, 2009) are generally in line with these estimates. Note, however, that the images were obtained with regular tips selected by their high-resolution capabilities, suggesting that these tips were terminated with sharp spikes (asperities) providing the necessary high-resolution topographs. This assumption was tested in Sheng et al. (1999) in which direct cryo-AFM analysis of regular AFM tips was performed.
Another resolution limiting factor is a thermal motion of molecules. Sub-molecular resolution for the protein array was primarily due to limiting the motion of the molecules by their packing. The implementation of cryo AFM by pioneering works of Z. Shao made it possible to dramatically improve the resolution of isolated molecules to the nanometer level (Zhang et al., 1996). Cooling the sample down to 85K resulted in resolving the domain structure of isolated immunoglobulin IgA protein in various states. Note that low temperature scanning was a factor in reaching nanometer resolution in high-resolution STM imaging of DNA (Shapir et al., 2006).
Capillary effect is another factor dramatically influencing AFM resolution. When a surface is exposed to air, it absorbs water from vapors and forms a thin water layer over the surface. Mica is one of the traditional AFM substrates that are very hydrophilic, retaining strong water molecules that can be removed in very high vacuum. AFM tips, made from either silicon or silicon nitride, are quite hydrophilic as well. Therefore, upon approaching the AFM tip to the surface, a water bridge is formed, and such a system, due to a high surface tension, is characterized by a strong interaction force termed a capillary force. The forces are in the dozens of nano Newton (nN) range (Freund et al., 1999); therefore, the sample can be dragged over the surface, deteriorating the imaging process. Drying of the sample and imaging at low relative humidity helps in reducing the effect of capillary forces. The use of oscillating tip can break the capillary and this was the major factor facilitating the imaging of biological samples that are typically are hydrophilic (Zhong et al., 1993). The capillary effect can be reduced if scanning is performed in vacuum, which is a typical operating regime for the high-resolution AFM or STM studies in the field of material science, but vacuum is not attractive for imaging of biological samples. At the same time, there are no capillary forces if the imaging is performed in aqueous solution. Given the fact that imaging in aqueous solutions is the best way to preserve biological samples, imaging in liquids is very attractive for biological studies. Indeed, the images of DNA with highest resolution were obtained for images acquired in aqueous solutions (Lyubchenko and Shlyakhtenko, 1997; Mou et al., 1995). Note that in Lyubchenko and Shlyakhtenko (1997), high-resolution images were obtained for individual DNA molecules rather than tightly packed DNA molecules (Mou et al., 1995). High-resolution STM images of DNA are primarily obtained for dried samples, although the electrochemical approach was useful for imaging of short DNA molecules (Jing et al., 1993). However, covalent bonding to the surface using thiolated DNA was needed to enhance the resolution of STM images of DNA (Rekesh et al., 1996).
5. Sample preparation techniques for AFM imaging of nucleic acids
The scanning tip can move or even sweep the samples that are weakly bound to the surface. Ignoring the sweeping effect led to a number of scanning artifacts in early attempts to image DNA with STM (Arscott et al., 1989; Beebe et al., 1989; Clemmer and Beebe, 1991; Lee et al., 1989). Therefore, the sample preparation procedure received special attention in the studies of biological samples with AFM, and there was a slow start with the biological applications of AFM.
The first reliable AFM imaging of long DNA molecules was made in the laboratory of C. Bustamante (Bustamante et al., 1992; Vesenka et al., 1992) in which the method of ionic treatment of mica suggested earlier for EM sample preparation (Brack, 1981) was implemented. In this approach, the mica surface is treated with Mg2+ to increase the affinity of negatively charged mica surface to DNA. As a result, the DNA molecules were held in place strongly enough to permit reliable imaging by AFM. Later studies showed that metal cations such as Co2+, La3+, and Zr4+ can be used for mica pretreatment to obtain images of DNA (Thundat et al., 1992). Later experiments showed that pretreatment of mica with cations is not necessary (Bezanilla et al., 1995; Bustamante and Rivetti, 1996; Bustamante et al., 1997; Hansma et al., 1993a). The adhesion of DNA takes place if Mg2+ cations are present in the buffer. In addition to imaging DNA, this cation-assisted method was successfully applied to studies of DNA complexes with proteins of various sizes and functions (e.g., (Bustamante and Rivetti, 1996; Bustamante et al., 1997; Dame et al., 2003; Erie et al., 1994; Guthold et al., 1994; Modesti et al., 2007; Rippe et al., 1997a; van der Linden et al., 2009; Wyman et al., 1997; Zlatanova et al., 1994)). Note the unique capability of AFM in elucidating protein stoichiometry in the protein-DNA complexes via measuring protein volume (Mohd-Sarip et al., 2006; Ratcliff and Erie, 2001; van der Linden et al., 2009).
In the laboratory of Z. Shao, a different approach utilizing a modification of the well-known electron microscopic procedure for imaging DNA has been developed (Shao et al., 1995; Yang and Shao, 1993; Yang et al., 1992). This method involves spreading DNA onto a carbon-coated mica substrate due to the cytochrome c denaturation at an air-water interface. The same group showed that DNA can be adsorbed on a supported cationic bilayer surface and imaged with AFM in aqueous buffers (Mou et al., 1995). The authors, using densely and uniformly packed DNA filaments, enabled them to resolve a periodic lateral modulation of 3.4 nm ± 0.4 nm that was in an excellent agreement with the known pitch of the double helix.
Gold substrates can be activated by self-assembled monolayers of thiols for reliable imaging of DNA (Hegner et al., 1993). Non-treated cover glass appeared to be a good substrate for binding chromatin, although the rinsing step (to remove unbound material and salt components) should be done gently and the dried sample should be imaged immediately (Allen et al., 1993). Among these methods, the cation-assisted technique has become the most widely used due to the simplicity of sample preparation. However, the requirement for the multivalent cations is mandatory, and therefore limits the range of experimental conditions to buffers with a defined concentration of the cations.
In parallel with the cation-assisted method, the approaches based on chemical functionalization of the mica surface with appropriate alkoxysilanes widely used for functionalization of silicon surfaces (Plueddemann, 1991) were developed. 3-Aminopropyltriethoxy silane (APTES) is one of the appropriate reagents that couples to glass or silica through the silane moieties and brings amino groups to the surface. As a result, functionalized surfaces remain positively charged at pH below pKa values (pKa =10.4) and are capable of binding to negatively charged DNA in the pH range of the DNA duplex stability. A procedure for the mica functionalization in vapors of APTES (Lyubchenko, 2001) yields a smooth aminopropyl-mica (AP-mica) surface capable of binding DNA and RNA of various lengths not only in air, but also in air and in liquids including aqueous solutions (Lindsay et al., 1992; Lyubchenko et al., 1993a; Lyubchenko et al., 1995; Lyubchenko et al., 1993b). Images shown in Fig. 3 were obtained by AP-mica methodology. DNA binding to AP-mica is insensitive to the type of buffer and presence of Mg or other di- and multivalent cations required for the cation-assisted technique and the preparation of sample can be done in a wide variety of pHs and over a wide range of temperatures (Lyubchenko, 2001). These are attractive features of the AP-mica procedure, enabling sample preparation in a broad range of conditions. None of the techniques reviewed above have these features.
Technical comments
Any type of commercially available mica product can be used. Asheville-Schoonmaker Mica Co. (Newport News, VA) provides mica sheets of various sizes that can be cut into smaller pieces.
3-Aminopropyltriethoxy silane (Fluka, Chemika-BioChemika, Switzerland, Aldrich, USA, United Chemical Technology, USA) can be used, but it is recommended that the reagent is vacuum distilled before use. See Lyubchenko (2001) for specifics.
A useful alternative to AP-mica is a surface functionalization procedure with the use of 1-(3-aminopropyl)silatrane (APS) (Lyubchenko et al., 2009; Shlyakhtenko et al., 2003a; Shlyakhtenko et al., 2000b; Tiner et al., 2001). APS is a water-soluble reagent, enabling the modification of the hydrophilic mica surface in aqueous solutions (APS-mica). The APS-mica is also smooth and has all the characteristics of AP-mica described above. AFM images of short DNA fragments (130 bp) deposited on APS-mica are shown in Fig. 5. The DNA molecules are spread over the surface, illustrating a uniform modification of the surface. Importantly, APS chemistry can be performed on the AFM probes as well allowing functionalization of the probe for various force spectroscopy applications (Lyubchenko and Shlyakhtenko, 2009; Lyubchenko
Figure 5.
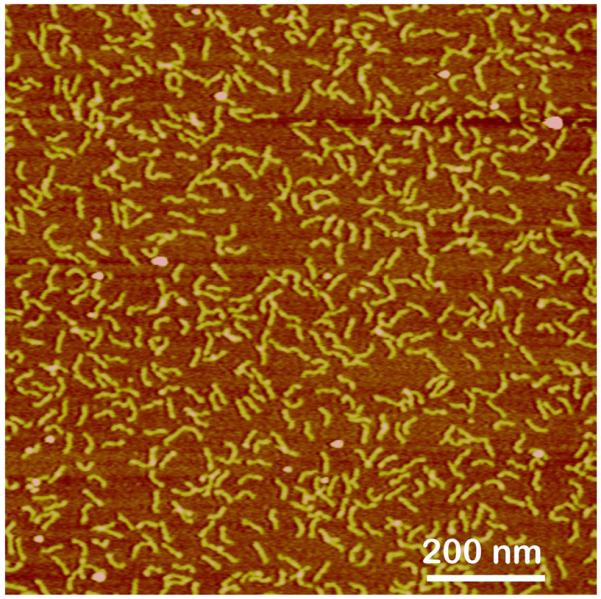
AFM image taken in air of DNA fragment (130 bp) sample deposited on APS-mica and imaged in air. The images were acquired in air with a NanoScope III microscope operated in Tapping Mode.
Technical comments
1-(3-aminopropyl)silatrane (APS) is not available commercially and needs to be synthesized. The procedure for the APS synthesis, purification, storage and use is described in details in Lyubchenko et al. (2009) and Shlyakhtenko et al. (2003a).
For the mica functionalization, freshly cleaved pieces of mica are immersed into the APS solution in water, incubated for 30 min at room temperature, rinsed with double distilled water, and dried in the gas flow. The substrates remain active after storage in Ar filled tubes for a week (Lyubchenko et al., 2009; Shlyakhtenko et al., 2003a).
6. DNA structure preservation - AFM imaging of supercoiled DNA
Binding of DNA molecules to surfaces leads to distortion of its structure, but the extent of such distortion depends on the physical and chemical characteristics of the surface and the forces between the DNA and surfaces. An appropriate test system for such effects is supercoiled DNA morphology, which is very sensitive to environmental conditions. These structures are very dynamic, and their formation in supercoiled DNA as a function of environmental conditions can be monitored by well-developed techniques such as gel electrophoresis and chemical and enzymatic probing (Sinden, 1994).
Dramatic changes of the conformation of supercoiled DNA morphology were observed when a cationic lipid bilayer was used as a substrate for imaging circular plasmid DNA (Mou et al., 1995). Initially, supercoiled DNA molecules with a typical plectonemic superhelical morphology were, upon addition of EDTA, transformed into a more extended morphology with many overlaps, and eventually a densely packed array was formed. DNA packing was further improved when the bilayer with the adsorbed DNA (in the presence of EDTA) was incubated at about 50°C for an hour and images of plectonemes were obtained. Using this procedure, the pitch of the DNA double helix was resolved. However, the change of DNA tertiary structure should be taken into a careful consideration if this methodology is used.
The Mg-assisted immobilization approach was applied to supercoiled DNA starting from the initial presentation of this technique (Bustamante et al., 1992; Vesenka et al., 1992) and was further applied by others (Hansma et al., 1995; Thundat et al., 1992). Surprisingly, the expected plectonemic shape of supercoiled DNA has not been revealed. Rather, molecules with a low number of intersections or even topologically relaxed molecules were the predominant conformations of plasmid DNA on the surface (Bezanilla et al., 1995). In cases when plectonemic conformations of supercoiled DNA were obtained, the chirality of superhelices was resolved (Samori et al., 1993). Later studies with the use of a modified Mg-assisted technique allowed the acquisition of images of supercoiled DNA with a plectonemic shape, but the formation of large loops between very tightly twisted segments of the plectonemic superhelix was a typical feature of such images (Jett et al., 2000; Nagami et al., 2002; Pfannschmidt and Langowski, 1998; Pfannschmidt et al., 1996; Rippe et al., 1997b). It is very unlikely that such large loops are present in DNA in solution; therefore, the appearances of large loops in supercoiled DNA are probably induced by the immobilization procedure. Systematic studies of the mechanism of DNA deposition onto a mica surface in the presence of Mg2+ cations showed that DNA molecules equilibrate on the surface, behaving as flexible polymers by persistence lengths larger than the persistence length of DNA in solution (Rivetti et al., 1996). Stiffening of a DNA molecule during the equilibration step may be the reason for the enlargement of supercoiling loops and the accompanying tight winding of the rest of the molecule. Modification of the Mg-assisted technique, in which deposition of the DNA solution is followed by a rinse with a diluted solution of uranyl acetate instead of water, reduced the looping-out morphology in supercoiled DNA (Cherny et al., 1998; Nagami et al., 2002).
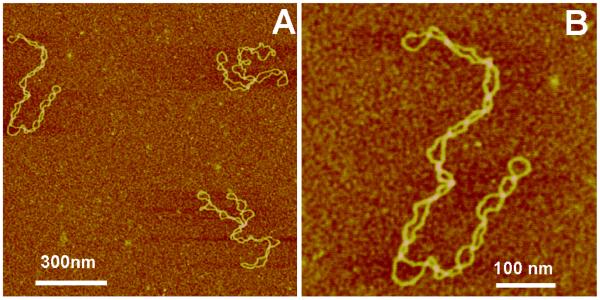
AFM images of plasmid DNA (5.6 kB) prepared by the use of AP-mica procedure are shown in Fig. 6. The plectonemic morphology of supercoiled DNA molecules is clearly seen in the images. Some of them are branched that is typical for molecules of this size. It was shown that the morphology of supercoiled DNA depends on ionic conditions (Lyubchenko and Shlyakhtenko, 1997; Lyubchenko et al., 1997; Shlyakhtenko et al., 2003b) and these observations are fully consistent with the data obtained in solution (Rybenkov et al., 1997a, b) as well as the results of theoretical analysis of conformations of supercoiled DNA (Vologodskii and Cozzarelli, 1996). Mg2+ cations further increase interwinding of plectonemic molecules ((Lyubchenko and Shlyakhtenko, 1997; Lyubchenko et al., 1997; Shlyakhtenko et al., 2003b), which is in agreement with experimental studies in solution (Shaw and Wang, 1993). Thus AP-mica procedure preserves the structure of supercoiled DNA. Therefore, it is an appropriate to use this technique for imaging of global DNA conformations and the structure and dynamics of negative DNA supercoiling, which is a natural state of DNA at physiological conditions and within cells.
Figure 6.
AFM images of supercoiled DNA (supercoiling density σ= − 0.075). (A) A large-scale image and (B) high resolution image obtained by rescanning around the molecule on the left. The samples after the deposition onto AP-mica were rinsed with water and argon-dried. The images were acquired in air with a NanoScope III microscope operated in Tapping Mode.
Technical comments
Samples prepared on AP-mica were imaged on variety of commercially available instruments, for example, the microscopes manufactured by Asylum Research, Agilent, Veeco.
With the Veeco MultiMode system, any types of probes designed for non-contact imaging can be used. NanoProbe TESP tips (Veeco) and conical sharp silicon probes of K-TEK International (Portland, OR) work well. Typical tapping frequency 240-380 KHz; scanning rate 2-3 Hz allows one to obtain stable images.
7. AFM imaging of alternative DNA conformations
Local conformational dynamics is a fundamental property of DNA, enabling it to accomplish various genetic functions. Local changes refer to the transition of a short DNA region from a canonical B conformation into one of the so-called alternative structures, such as left-handed Z-DNA, cruciforms, intramolecular triple helices and unwound regions, the biological role of which is widely discussed (e.g., (Hatfield and Benham, 2002; Sinden, 1994; Wells, 2008, 2009)). The formation of such structures was detected and analyzed using various chemical and enzymatic probe techniques and gel electrophoresis (Hatfield and Benham, 2002; Sinden, 1994), but these methods do not provide the information on the structure of local conformations within a long DNA molecule. Powerful structural techniques such as electron microscopy failed to reliably detect these alternative DNA structures, which is very likely due to a severe sample preparation procedure. The application of AFM to this problem was very successful and a brief review of the data obtained for cruciforms and the intramolecular triple helix (H-DNA) is given below.
7.1. Imaging of cruciforms
AP-mica procedure was used for imaging of the cruciform in plasmid DNA containing a 106-bp inverted repeat with an expected length of 53 bp for arms (Shlyakhtenko et al., 1998). The AFM images of two DNA molecules with cruciforms for the sample prepared by deposition from a low salt solution (Tris-EDTA buffer) are shown in Fig. 7. The cruciforms appear as clear-cut extrusions; they are indicated on these images with arrows. The sizes of the arms for extended extrusions are 15-20 nm, in full agreement with the expected length of the hairpins containing 53 bp (18 nm for B-helix DNA geometry). The appearance of two classes of cruciforms discovered by the AFM imaging led to the hypothesis on a novel regulatory role of cruciforms in global DNA dynamics (Shlyakhtenko et al., 2000a).
Figure 7.
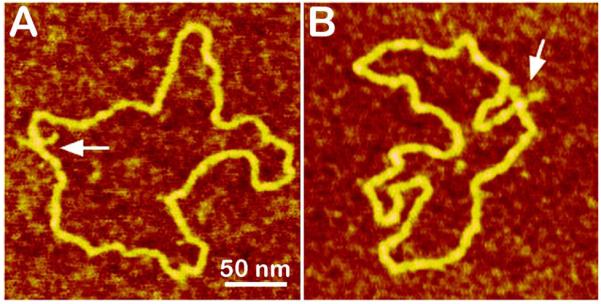
AFM images of pUC8F14C plasmid DNA containing 106 bp inverted repeat forming cruciforms under appropriate supercoiling density. Two molecules with different conformations of the cruciform are shown in plates (A) and (B). The images of the sample deposited onto AP-mica and dried were taken in air with a NanoScope III microscope operated in Tapping Mode. Cruciforms are indicated with arrows. See Shlyakhtenko et al. (1998) for more images.
7.2. AFM imaging of an intramolecular DNA triplex, H-DNA
An intramolecular DNA triplex or H-DNA formed by homopurine-homopyrimidine (Pu·Py) tracts is a biologically important alternative DNA structure (Soyfer and Potaman, 1996). Structural studies of intramolecular triplexes are instrumental in understanding the mechanism of how H-DNA is involved in genetic processes. The challenge in imaging of H-DNA is the requirement of low pH and this requirement was met in a paper (Tiner et al., 2001) in which APS-mica was implemented. A high-resolution AFM image of pUC19 plasmid with a 46 bp long purine-pyrimidine repeat prepared at acidic pH is shown in Fig. 8. A distinct feature of the molecules prepared at acidic pH is the formation of a clear kink with a short protrusion indicated by an arrow. The formation of a sharp kink is fully consistent with the model of the intramolecular DNA triplex (Potaman et al., 1996; Soyfer and Potaman, 1996) inserted in Fig. 8. The same technique was instrumental in detecting of H-DNA conformation in long imperfect Py-Pu inserts (Kato et al., 2002).
Figure 8.
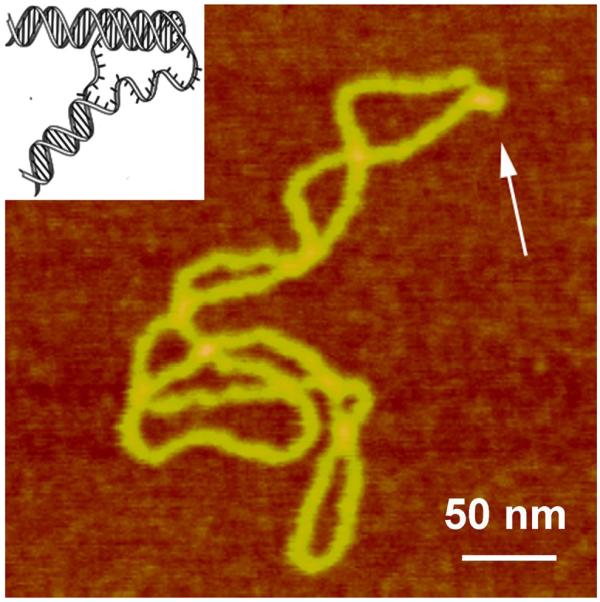
AFM images of plasmid pCW2966 containing a 46 bp mirror repeat forming H-DNA at acidic pH. H-DNA regions are indicated with arrows. The DNA sample was incubated in 50 mM Na acetate buffer at pH 5.0 and then deposited onto APS-mica in the same buffer. The images for plasmid containing H-DNA were acquired in air with a NanoScope III microscope operated in Tapping Mode. A sharp kink at the base of a thick protrusion indicated with an arrow. Insert shows the model for H-DNA.
8. Imaging in aqueous solutions. Time-lapse AFM studies
The potential of AFM for direct imaging of actual molecular processes demonstrated in the group of P. Hansma in the very early stages of the AFM development, in which visualization of fibrin polymerization was observed (Drake et al., 1989; Hansma et al., 1991). Introduction of Tapping Mode imaging (Hansma et al., 1993b) considerably eased the problem of the sample movement by the tip. Note again that imaging in aqueous solutions eliminates resolution limiting capillary effect, so the DNA molecules with parameters very close to crystallographic ones can be obtained (Lyubchenko and Shlyakhtenko, 1997), (Mou et al., 1995).
8.1 Time lapse imaging of protein-DNA complexes
The time-lapse AFM technique was successfully applied by a number of groups actively working with DNA. Early joint efforts of the Hansma and Bustamante groups demonstrated the ability to image the assembly process of RNA polymerase-DNA complexes in a series of time-lapse images (Guthold et al., 1994). Later, these groups visualized the sliding of the polymerase along DNA (Kasas et al., 1997), concluding that diffusion of RNA polymerase along DNA constitutes a mechanism for accelerated promoter location. Later studies in the Bustamante group (Rivetti et al., 1999) showed that during transcription initiation, the promoter DNA wraps nearly 300 degrees around the polymerase. These advances were partially possible due to substantial improvements in the DNA immobilization procedure. From one point of view, DNA molecules should be stably bound to the surface for the reliable identification of the molecules. At the same time, the imaging of dynamics at the single molecule level requires loose DNA binding, permitting the molecule to move relatively freely at the surface. The divalent cations of different kinds helped to reconcile these conflicting requirements. A similar immobilization approach enabled AFM observation of interaction of DNA with photolyase (van Noort et al., 1998). Nonspecific photolyase DNA complexes were visualized, showing association, dissociation, and movement of photolyase over the DNA. The latter result suggests a sliding mechanism by which photolyase can scan DNA for damaged sites. Time-lapse AFM was applied to imaging of the human Rad50/Mre11/Nbs1 complex (Moreno-Herrero et al., 2005). High-resolution AFM imaging revealed the unusual morphology of the protein complex, the structure and dynamics of which changes upon binding to DNA. Mg-assisted immobilization technique was used in these studies.
The dynamics of the nucleosome is a key property of chromatin, providing access to DNA wrapped around the histone core. The recent single molecule studies using fluorescence and time resolved techniques detected local dissociation of DNA from nucleosomes in the absence of remodeling proteins (e.g., (Bucceri et al., 2006; Koopmans et al., 2007; Li et al., 2005; Tims and Widom, 2007)), but they did not provide the answer to the spatial range of such dynamics. The time-lapse AFM was instrumental in the direct observation of the dynamics of nucleosomes assembled on a 353 bp DNA substrate containing a 147 bp nucleosome positioning sequence (Lyubchenko and Shlyakhtenko, 2009; Shlyakhtenko et al., 2009). Fig. 9 shows that a nucleosome starting as a complex with 2 turns of the DNA around the histone octamers (frame 1), unwraps on frame 2 losing one DNA turn and finally undergoes full dissociation in frame 3. APS-mica methodology was used in these studies.
Figure 9.
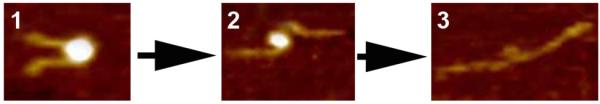
Time-lapse AFM images of reconstituted mononucleosome particles taken in aqueous solution. Three frames are shown. APS-mica was used as a substrate and scanning was performed in buffer containing 10 mM Tris-HCl (pH 7.5) and 4 mM MgCl2. More information and animated images can be found in Shlyakhtenko et al. (2009).
8.2. Dynamics of protein-DNA complexes with high-speed AFM
The major drawback of the AFM time-lapse studies described above is the slow (minute-scale) data acquisition rate of a conventional AFM instrument, so most system dynamics remain undetectable. Ando’s group has developed a high speed AFM (HS-AFM) instrument operating almost 1000 times faster and capable of acquiring the data with a subsecond rate (Ando et al., 2001; Ando et al., 2008; Ando et al., 2007). With this instrument, Ando’s group was able to visualize the dynamics of individual myosin molecules with a 100 millisecond temporal resolution (Ando et al. 2008; Ando et al. 2007). Takeyasu’s group applied this instrumentation to study the dynamics of site-specific DNA binding proteins in the subsecond time scale (Crampton et al., 2007) and dynamics of reconstituted chromatin (Suzuki et al., 2010). The latter paper revealed a number of dynamic properties of chromatin not amenable to the traditional AFM instrumentation. In these studies, the sliding mechanism was characterized with fluctuation within approximately 50 nm along the DNA strand. In addition, the histone dissociation was visualized and two distinct ways were identified. This emerging novel instrumentation has also recently applied to study the process of searching specific sites by site-specific EcoRII protein (Gilmore et al., 2009). Fig. 10 displays 4 frames of one of the search events, spanning 20 s. The initial state (5 s) is a complex with a small loop that gradually increases over time (10 s). After 15 s, the protein stops moving and binds to another specific DNA site, as indicated by DNA length measurements. The change in the loop length occurs over a period of about 10 s, covering a distance of about 300 bp (102 nm) until the EcoRII complex stops at another recognition site, which corresponds to a rate of about 30 bp/s (10.2 nm/s). These data suggest that EcoRII binds to one recognition site and searches for another site by threading the DNA filament through the complex until the enzyme finds the second recognition site, upon which it forms a stable synaptic complex.
Figure 10.
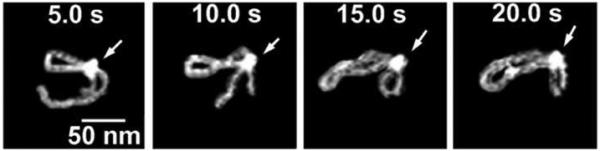
EcoRII translocation process analyzed by HS-AFM. Four frames out of 40 frames total taken over a time period of 20 s measured in 0.5 s intervals are shown. In the first frame (5s) looped structure stabilized by the protein (indicated with an arrow) bound to two specific sites on DNA is formed. The following frames illustrate sliding of the protein along the DNA strand leading to an increase in the loop size. The final image (20 s) in which a large loop is formed corresponds to binding of the protein to two distant binding sites on the DNA substrate. More information and animated images can be found in Gilmore et al. (2009).
Technical comments
The experiments described above were performed on the HS AFM instrument (the design of Dr. Ando) assembled in the laboratory of K. Takeyasu. Specifics for the preparation of the sample for HS AFM imaging are described in Crampton et al. (2007) and Gilmore et al. (2009).
Details on the instrument principle, design, operation and imaging process are provided in papers (Ando et al., 2008; Ando et al., 2007).
Conclusions
AFM as a technique offering unique advantages in the high resolution imaging of DNA, complexes with proteins and small ligands in the absence of stains, shadows and labels, is coming of age. Due to advances in the sample preparation procedures for DNA, AFM has already been successfully applied to structural studies of complexes of linear DNA with a number of nucleoprotein complexes. In addition to linear DNA, supercoiled DNA can be analyzed with AFM. Importantly, the conformational dynamics of this physiologically relevant state of double helical DNA can be assessed with AFM. Recent experimental studies and advanced theoretical analyses suggest that DNA dynamics rather than static DNA structure plays a critical role in the control of gene expression, DNA replication, recombination, and repair. Time-lapse AFM studies of the dynamics of cruciforms and H-DNA structure in supercoiled DNA allowed direct imaging of the interplay between local DNA conformations, global conformations, and dynamics of the entire molecule. Local and global DNA conformations are in a dynamic equilibrium, and competing transitions between different local structures accompanied by dynamic changes of global DNA conformations may have a profound role in various DNA functions. The unique possibility to image a sample while avoiding the drying step makes it possible to apply time-lapse AFM to follow DNA transactions. These advance open prospects for applying the time-lapse AFM technique to directly image critical genetic processes such as transcription, replication and recombination. Note in this regard recent advances in the high-speed AFM (HS AFM), so the technique is becoming available for a broad biomedical community. At the current stage of development, HS AFM is capable of imaging molecular dynamics with the rate approaching video optical microscopy, but providing a resolution at the nanometer level. The current HS AFM already operates in Tapping Mode with the oscillation amplitude in the range of a nanometer, thereby dramatically minimizing potential deformation effect of the tip. This technological advance is an important step for further improvement of the instrument, enabling AFM to operate in attractive tip-sample interaction regime. The latter will almost eliminate tip-induced modification of the sample. The tip geometry is the major resolution-limiting factor for AFM. Recent advances in tip manufacturing brought AFM probes with the tip radius as small as a few nanometers to the market compared to the typical probes with the radius of curvature as large as 10-20 nm. Fortunately, many such “blunt” tips have tiny asperities enabling the possibility to reach a nanometer-range resolution. Importantly, such high resolution was achieved during imaging in solution that is critical for reliable detection of the intramolecular dynamics. There is room for improvements in the instrumentation, such as fast scanning and the sample preparation techniques. Finally, the technological advances of AFM studies of DNA and protein-DNA complexes can be extended to other biomolecular systems and thus will provide structural biologists pursuing single molecule approaches with novel powerful nanoimaging tools.
ACKNOWLEDGMENTS
I am grateful to Luda Shlyakhtenko for valuable comments and critical reading of the manuscript, Alex Portillo for proofreading of the paper and useful suggestions, and current and former members of the group for their contribution to works incorporated into the manuscript. The work was supported by grants from NIH (GM 62235), NSF (NSF-EPSCOR EPS-0701892, PHY-061590), DOE (DE-FG02-08ER64579), NATO (CBN.NR.NRSFP 983204) and Nebraska Research Initiative (NRI).
Abbreviations
- AFM
atomic force microscope
- AP-mica
aminopropyl mica
- APTES
aminopropyltriethoxy silane
- APS
aminopropyltriethoxy silatrane
- APS-mica
aminopropylsilatrane mica
- DNA
deoxyribonucleic acid
- SPM
scanning probe microscope
- STM
scanning tunneling microscope
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen MJ, Dong XF, O’Neill TE, Yau P, Kowalczykowski SC, Gatewood J, Balhorn R, Bradbury EM. Atomic force microscope measurements of nucleosome cores assembled along defined DNA sequences. Biochemistry. 1993;32:8390–8396. doi: 10.1021/bi00084a002. [DOI] [PubMed] [Google Scholar]
- Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proc Natl Acad Sci U S A. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Uchihashi T, Kodera N, Yamamoto D, Miyagi A, Taniguchi M, Yamashita H. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Arch. 2008;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- Ando T, Uchihashi T, Kodera N, Yamamoto D, Taniguchi M, Miyagi A, Yamashita H. High-speed atomic force microscopy for observing dynamic biomolecular processes. J Mol Recognit. 2007;20:448–458. doi: 10.1002/jmr.843. [DOI] [PubMed] [Google Scholar]
- Arscott PG, Lee G, Bloomfield VA, Evans DF. Scanning tunnelling microscopy of Z-DNA. Nature. 1989;339:484–486. doi: 10.1038/339484a0. [DOI] [PubMed] [Google Scholar]
- Beebe TP, Jr., Wilson TE, Ogletree DF, Katz JE, Balhorn R, Salmeron MB, Siekhaus WJ. Direct observation of native DNA structures with the scanning tunneling microscope. Science. 1989;243:370–372. doi: 10.1126/science.2911747. [DOI] [PubMed] [Google Scholar]
- Bezanilla M, Manne S, Laney DE, Lyubchenko YL, Hansma HG. Adsorption of DNA to mica, silylated mica and minerals: characterization by atomic force microscopy. Langmuir. 1995;11:655–659. [Google Scholar]
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Binnig G, Rohrer H, Gerber C, Weibel E. Surface studies by scanning tunneling microscopy. Phys. Rev. Lett. 1982;49:57–61. [Google Scholar]
- Binning G, Rohrer H. Scanning tunneling microscopy. Helvetica Phys. Acta. 1982;55:726–735. [Google Scholar]
- Brack C. DNA electron microscopy. CRC Crit Rev Biochem. 1981;10:113–169. doi: 10.3109/10409238109114551. [DOI] [PubMed] [Google Scholar]
- Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. Embo J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Rivetti C. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annu Rev Biophys Biomol Struct. 1996;25:395–429. doi: 10.1146/annurev.bb.25.060196.002143. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Rivetti C, Keller DJ. Scanning force microscopy under aqueous solutions. Curr Opin Struct Biol. 1997;7:709–716. doi: 10.1016/s0959-440x(97)80082-6. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Vesenka J, Tang CL, Rees W, Guthold M, Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry. 1992;31:22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- Cherny DI, Fourcade A, Svinarchuk F, Nielsen PE, Malvy C, Delain E. Analysis of various sequence-specific triplexes by electron and atomic force microscopies. Biophys J. 1998;74:1015–1023. doi: 10.1016/S0006-3495(98)74026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmer CR, Beebe TP., Jr. Graphite: a mimic for DNA and other biomolecules in scanning tunneling microscope studies. Science. 1991;251:640–642. doi: 10.1126/science.1992517. [DOI] [PubMed] [Google Scholar]
- Crampton N, Yokokawa M, Dryden DT, Edwardson JM, Rao DN, Takeyasu K, Yoshimura SH, Henderson RM. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc Natl Acad Sci U S A. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N. Insights into the regulation of transcription by scanning force microscopy. J Microsc. 2003;212:244–253. doi: 10.1111/j.1365-2818.2003.01243.x. [DOI] [PubMed] [Google Scholar]
- Drake B, Prater CB, Weisenhorn AL, Gould SA, Albrecht TR, Quate CF, Cannell DS, Hansma HG, Hansma PK. Imaging crystals, polymers, and processes in water with the atomic force microscope. Science. 1989;243:1586–1589. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- Engel A, Lyubchenko Y, Muller D. Atomic force microscopy: a powerful tool to observe biomolecules at work. Trends Cell Biol. 1999;9:77–80. doi: 10.1016/s0962-8924(98)01415-9. [DOI] [PubMed] [Google Scholar]
- Erie DA, Yang G, Schultz HC, Bustamante C. DNA bending by Cro protein in specific and nonspecific complexes: implications for protein site recognition and specificity. Science. 1994;266:1562–1566. doi: 10.1126/science.7985026. see comments. [DOI] [PubMed] [Google Scholar]
- Freund J, Halbritter J, Horber JK. How dry are dried samples? Water adsorption measured by STM. Microsc Res Tech. 1999;44:327–338. doi: 10.1002/(SICI)1097-0029(19990301)44:5<327::AID-JEMT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Gilmore JL, Suzuki Y, Tamulaitis G, Siksnys V, Takeyasu K, Lyubchenko YL. Single-molecule dynamics of the DNA-EcoRII protein complexes revealed with high-speed atomic force microscopy. Biochemistry. 2009;48:10492–10498. doi: 10.1021/bi9010368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross L, Mohn F, Moll N, Meyer G, Ebel R, Abdel-Mageed WM, Jaspars M. Organic structure determination using atomic-resolution scanning probe microscopy. Nature Chemistry. 2010 doi: 10.1038/nchem.765. Published online 01 August 2010. [DOI] [PubMed] [Google Scholar]
- Guthold M, Bezanilla M, Erie DA, Jenkins B, Hansma HG, Bustamante C. Following the assembly of RNA polymerase-DNA complexes in aqueous solutions with the scanning force microscope. Proc Natl Acad Sci U S A. 1994;91:12927–12931. doi: 10.1073/pnas.91.26.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma HG, Bezanilla M, Zenhausern F, Adrian M, Sinsheimer RL. Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 1993a;21:505–512. doi: 10.1093/nar/21.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma HG, Laney DE, Bezanilla M, Sinsheimer RL, Hansma PK. Applications for atomic force microscopy of DNA. Biophys J. 1995;68:1672–1677. doi: 10.1016/S0006-3495(95)80343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma HG, Sinsheimer RL, Groppe J, Bruice TC, Elings V, Gurley G, Bezanilla M, Mastrangelo IA, Hough PV, Hansma PK. Recent advances in atomic force microscopy of DNA. Scanning. 1993b;15:296–299. doi: 10.1002/sca.4950150509. [DOI] [PubMed] [Google Scholar]
- Hansma HG, Weisenhorn AL, Edmundson AB, Gaub HE, Hansma PK. Atomic force microscopy: seeing molecules of lipid and immunoglobulin. Clin Chem. 1991;37:1497–1501. [PubMed] [Google Scholar]
- Hansma PK, Elings VB, Marti O, Bracker CE. Scanning tunneling microscopy and atomic force microscopy: application to biology and technology. Science. 1988;242:209–216. doi: 10.1126/science.3051380. [DOI] [PubMed] [Google Scholar]
- Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu Rev Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- Hegner M, Wagner P, Semenza G. Immobilizing DNA on gold via thiol modification for atomic force microscopy imaging in buffer solutions. FEBS Lett. 1993;336:452–456. doi: 10.1016/0014-5793(93)80854-n. [DOI] [PubMed] [Google Scholar]
- Jett SD, Cherny DI, Subramaniam V, Jovin TM. Scanning force microscopy of the complexes of p53 core domain with supercoiled DNA. J Mol Biol. 2000;299:585–592. doi: 10.1006/jmbi.2000.3759. [DOI] [PubMed] [Google Scholar]
- Jing TW, Jeffrey AM, DeRose JA, Lyubchenko YL, Shlyakhtenko LS, Harrington RE, Appella E, Larsen J, Vaught A, Rekesh D, et al. Structure of hydrated oligonucleotides studied by in situ scanning tunneling microscopy. Proc Natl Acad Sci U S A. 1993;90:8934–8938. doi: 10.1073/pnas.90.19.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasas S, Thomson NH, Smith BL, Hansma HG, Zhu X, Guthold M, Bustamante C, Kool ET, Kashlev M, Hansma PK. Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry. 1997;36:461–468. doi: 10.1021/bi9624402. [DOI] [PubMed] [Google Scholar]
- Kato M, McAllister CJ, Hokabe S, Shimizu N, Lyubchenko YL. Structural heterogeneity of pyrimidine/purine-biased DNA sequence analyzed by atomic force microscopy. Eur J Biochem. 2002;269:3632–3636. doi: 10.1046/j.1432-1033.2002.03063.x. [DOI] [PubMed] [Google Scholar]
- Klinov D, Dwir B, Kapon E, Borovok N, Molotsky T, Kotlyar A. High-resolution atomic force microscopy of duplex and triplex DNA molecules. Nanotechnology. 2007;18:225102. [Google Scholar]
- Koopmans WJ, Brehm A, Logie C, Schmidt T, van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. J Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen J, Reichling M. Atomic resolution non-contact atomic force microscopy of clean metal oxide surfaces. Journal of Physics: Condensed Matter. 2010;22 doi: 10.1088/0953-8984/22/26/263001. [DOI] [PubMed] [Google Scholar]
- Lee G, Arscott PG, Bloomfield VA, Evans DF. Scanning tunneling microscopy of nucleic acids. Science. 1989;244:475–477. doi: 10.1126/science.2470146. [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Lindsay SM, Lyubchenko YL, Gall AA, Shlyakhtenko LS, Harrington RE. Imaging DNA Molecules Chemically Bound to a Mica Surface. SPIE PROCEEDINGS. 1992;1639:84–90. [Google Scholar]
- Lyubchenko Y, Shlyakhtenko L, Harrington R, Oden P, Lindsay S. Atomic force microscopy of long DNA: imaging in air and under water. Proc Natl Acad Sci U S A. 1993a;90:2137–2140. doi: 10.1073/pnas.90.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Gall AA, Shlyakhtenko LS. Atomic Force Microscopy of DNA and Protein-DNA Complexes Using Functionalized Mica Substrates. In: Moss T, editor. Methods Mol. Biol. DNA-protein interactions; Principles and protocols. Humana Press; Totowa, NJ: 2001. pp. 569–578. [DOI] [PubMed] [Google Scholar]
- Lyubchenko YL, Jacobs BL, Lindsay SM, Stasiak A. Atomic force microscopy of nucleoprotein complexes. Scanning Microsc. 1995;9:705–724. discussion 724-707. [PubMed] [Google Scholar]
- Lyubchenko YL, Oden PI, Lampner D, Lindsay SM, Dunker KA. Atomic force microscopy of DNA and bacteriophage in air, water and propanol: the role of adhesion forces. Nucleic Acids Res. 1993b;21:1117–1123. doi: 10.1093/nar/21.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc Natl Acad Sci U S A. 1997;94:496–501. doi: 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 2009;47:206–213. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS, Aki T, Adhya S. Atomic force microscopic demonstration of DNA looping by GalR and HU. Nucleic Acids Res. 1997;25:873–876. doi: 10.1093/nar/25.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS, Gall AA. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol Biol. 2009;543:337–351. doi: 10.1007/978-1-60327-015-1_21. [DOI] [PubMed] [Google Scholar]
- Martin Y, Williams CC, Wickramasinghe HK. Atomic force microscope–force mapping and profiling on a sub 100□Å scale. Journal of Applied Physics. 1987;61:4723–4729. [Google Scholar]
- Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Mol Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Mou J, Czajkowsky DM, Zhang Y, Shao Z. High-resolution atomic-force microscopy of DNA: the pitch of the double helix. FEBS Lett. 1995;371:279–282. doi: 10.1016/0014-5793(95)00906-p. [DOI] [PubMed] [Google Scholar]
- Muller DJ, Sapra KT, Scheuring S, Kedrov A, Frederix PL, Fotiadis D, Engel A. Single-molecule studies of membrane proteins. Curr Opin Struct Biol. 2006;16:489–495. doi: 10.1016/j.sbi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Nagami F, Zuccheri G, Samori B, Kuroda R. Time-lapse imaging of conformational changes in supercoiled DNA by scanning force microscopy. Anal Biochem. 2002;300:170–176. doi: 10.1006/abio.2001.5435. [DOI] [PubMed] [Google Scholar]
- Ohnesorge F, Binnig G. True atomic resolution by atomic force microscopy through repulsive and attractive forces. Science. 1993;260:1451–1456. doi: 10.1126/science.260.5113.1451. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt C, Langowski J. Superhelix organization by DNA curvature as measured through site- specific labeling. J Mol Biol. 1998;275:601–611. doi: 10.1006/jmbi.1997.1476. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt C, Schaper A, Heim G, Jovin TM, Langowski J. Sequence-specific labeling of superhelical DNA by triple helix formation and psoralen crosslinking. Nucleic Acids Res. 1996;24:1702–1709. doi: 10.1093/nar/24.9.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plueddemann E. Silane coupling agents. Springer-Verlag; Berlin: 1991. [Google Scholar]
- Potaman VN, Ussery DW, Sinden RR. Formation of a combined H-DNA/open TATA box structure in the promoter sequence of the human Na,K-ATPase alpha2 gene. J Biol Chem. 1996;271:13441–13447. doi: 10.1074/jbc.271.23.13441. [DOI] [PubMed] [Google Scholar]
- Ratcliff GC, Erie DA. A novel single-molecule study to determine protein--protein association constants. J Am Chem Soc. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- Rekesh D, Lyubchenko Y, Shlyakhtenko LS, Lindsay SM. Scanning tunneling microscopy of mercapto-hexyl-oligonucleotides attached to gold. Biophys J. 1996;71:1079–1086. doi: 10.1016/S0006-3495(96)79308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K, Guthold M, von Hippel PH, Bustamante C. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase x sigma 54 holoenzyme by scanning force microscopy. J Mol Biol. 1997a;270:125–138. doi: 10.1006/jmbi.1997.1079. [DOI] [PubMed] [Google Scholar]
- Rippe K, Mucke N, Langowski J. Superhelix dimensions of a 1868 base pair plasmid determined by scanning force microscopy in air and in aqueous solution. Nucleic Acids Res. 1997b;25:1736–1744. doi: 10.1093/nar/25.9.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti C, Guthold M, Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J Mol Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E.coli RNA polymerase open promoter complex. Embo J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode S, Oyabu N, Kobayashi K, Yamada H, Kuhnle A. True atomic-resolution imaging of (1014) calcite in aqueous solution by frequency modulation atomic force microscopy. Langmuir. 2009;25:2850–2853. doi: 10.1021/la803448v. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on DNA helical repeat, effective diameter and free energy of supercoiling. Nucleic Acids Res. 1997a;25:1412–1418. doi: 10.1093/nar/25.7.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on the conformations of supercoiled DNA. I. Sedimentation analysis. J Mol Biol. 1997b;267:299–311. doi: 10.1006/jmbi.1996.0876. [DOI] [PubMed] [Google Scholar]
- Samori B, Siligardi G, Quagliariello C, Weisenhorn AL, Vesenka J, Bustamante CJ. Chirality of DNA supercoiling assigned by scanning force microscopy. Proc Natl Acad Sci U S A. 1993;90:3598–3601. doi: 10.1073/pnas.90.8.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring S, Dufrene YF. Atomic force microscopy: probing the spatial organization, interactions and elasticity of microbial cell envelopes at molecular resolution. Mol Microbiol. 2010;75:1327–1336. doi: 10.1111/j.1365-2958.2010.07064.x. [DOI] [PubMed] [Google Scholar]
- Scheuring S, Sturgis JN. Atomic force microscopy of the bacterial photosynthetic apparatus: plain pictures of an elaborate machinery. Photosynth Res. 2009 doi: 10.1007/s11120-009-9413-7. [DOI] [PubMed] [Google Scholar]
- Schwarz U, Hölscher H, Wiesendanger R. Phys. Rev. B. 2000;62:13089–13097. [Google Scholar]
- Shao Z, Yang J, Somlyo AP. Biological atomic force microscopy: from microns to nanometers and beyond. Annu Rev Cell Dev Biol. 1995;11:241–265. doi: 10.1146/annurev.cb.11.110195.001325. [DOI] [PubMed] [Google Scholar]
- Shapir E, Cohen H, Borovok N, Kotlyar AB, Porath D. High-resolution STM imaging of novel poly(G)-poly(C) DNA molecules. J Phys Chem B. 2006;110:4430–4433. doi: 10.1021/jp0569638. [DOI] [PubMed] [Google Scholar]
- Shaw SY, Wang JC. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- Sheng S, Czajkowsky DM, Shao Z. AFM tips: how sharp are they? J Microsc. 1999;196:1–5. [PubMed] [Google Scholar]
- Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003a;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Hsieh P, Grigoriev M, Potaman VN, Sinden RR, Lyubchenko YL. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J Mol Biol. 2000a;296:1169–1173. doi: 10.1006/jmbi.2000.3542. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Miloseska L, Potaman VN, Sinden RR, Lyubchenko YL. Intersegmental interactions in supercoiled DNA: atomic force microscope study. Ultramicroscopy. 2003b;97:263–270. doi: 10.1016/S0304-3991(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Potaman VN, Sinden RR, Gall AA, Lyubchenko YL. Structure and dynamics of three-way DNA junctions: atomic force microscopy studies. Nucleic Acids Res. 2000b;28:3472–3477. doi: 10.1093/nar/28.18.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Potaman VN, Sinden RR, Lyubchenko YL. Structure and dynamics of supercoil-stabilized DNA cruciforms. J Mol Biol. 1998;280:61–72. doi: 10.1006/jmbi.1998.1855. [DOI] [PubMed] [Google Scholar]
- Sinden RR. DNA structure and Function. Academic Press; San Diego: 1994. [Google Scholar]
- Sokolov IY, Henderson GS, Wicks FJ. Theoretical and experimental evidence for “true” atomic resolution under non-vacuum conditions. Journal of Applied Physics. 1999;86:5537–5540. [Google Scholar]
- Soyfer VN, Potaman VN. Triple-helical nucleic acids. Springer; New York: 1996. [Google Scholar]
- Suzuki Y, Higuchi Y, Hizume K, Yokokawa M, Yoshimura SH, Yoshikawa K, Takeyasu K. Molecular dynamics of DNA and nucleosomes in solution studied by fast-scanning atomic force microscopy. Ultramicroscopy. 2010 doi: 10.1016/j.ultramic.2010.02.032. [DOI] [PubMed] [Google Scholar]
- Thundat T, Allison DP, Warmack RJ, Brown GM, Jacobson KB, Schrick JJ, Ferrell TL. Atomic force microscopy of DNA on mica and chemically modified mica. Scanning Microsc. 1992;6:911–918. [PubMed] [Google Scholar]
- Tims HS, Widom J. Stopped-flow fluorescence resonance energy transfer for analysis of nucleosome dynamics. Methods. 2007;41:296–303. doi: 10.1016/j.ymeth.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiner WJ, Sr., Potaman VN, Sinden RR, Lyubchenko YL. The structure of intramolecular triplex DNA: atomic force microscopy study. J Mol Biol. 2001;314:353–357. doi: 10.1006/jmbi.2001.5174. [DOI] [PubMed] [Google Scholar]
- van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res. 2009;37:1580–1588. doi: 10.1093/nar/gkn1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort SJ, van der Werf KO, Eker AP, Wyman C, de Grooth BG, van Hulst NF, Greve J. Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophys J. 1998;74:2840–2849. doi: 10.1016/S0006-3495(98)77991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesenka J, Guthold M, Tang CL, Keller D, Delaine E, Bustamante C. Substrate preparation for reliable imaging of DNA molecules with the scanning force microscope. Ultramicroscopy. 1992;42-44:1243–1249. doi: 10.1016/0304-3991(92)90430-r. [DOI] [PubMed] [Google Scholar]
- Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD. DNA triplexes and Friedreich ataxia. FASEB J. 2008;22:1625–1634. doi: 10.1096/fj.07-097857. [DOI] [PubMed] [Google Scholar]
- Wells RD. Discovery of the role of non-B DNA structures in mutagenesis and human genomic disorders. J Biol Chem. 2009;284:8997–9009. doi: 10.1074/jbc.X800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Rombel I, North AK, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. see comments. [DOI] [PubMed] [Google Scholar]
- Yang J, Shao Z. Effect of probe force on the resolution of atomic force microscopy of DNA. Ultramicroscopy. 1993;50:157–170. doi: 10.1016/0304-3991(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Yang J, Takeyasu K, Shao Z. Atomic force microscopy of DNA molecules. FEBS Lett. 1992;301:173–176. doi: 10.1016/0014-5793(92)81241-d. [DOI] [PubMed] [Google Scholar]
- Yodh JG, Woodbury N, Shlyakhtenko LS, Lyubchenko YL, Lohr D. Mapping nucleosome locations on the 208-12 by AFM provides clear evidence for cooperativity in array occupation. Biochemistry. 2002;41:3565–3574. doi: 10.1021/bi011612e. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sheng S, Shao Z. Imaging biological structures with the cryo atomic force microscope. Biophys J. 1996;71:2168–2176. doi: 10.1016/S0006-3495(96)79418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Inniss D, Kjoller K, Elings VB. Fractured polymer/silica fiber surface studied by tapping mode atomic force microscopy. Surf. Sci. Lett. 1993;290:L688–L692. [Google Scholar]
- Zlatanova J, Leuba SH, Yang G, Bustamante C, van Holde K. Linker DNA accessibility in chromatin fibers of different conformations: a reevaluation. Proc Natl Acad Sci U S A. 1994;91:5277–5280. doi: 10.1073/pnas.91.12.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]