Abstract
Thiols affect a variety of cell functions, an effect known as redox regulation. We show here that treatment (1–2 h) of cells with 0.1–5 mM N-acetyl-l-cysteine (NAC) increases surface protein thiol expression in human peripheral blood mononuclear cells. This effect is not associated with changes in cellular glutathione (GSH) and is also observed with a non-GSH precursor thiol N-acetyl-d-cysteine or with GSH itself, which is not cell-permeable, suggesting a direct reducing action. NAC did not augment protein SH in the cytosol, indicating that they are already maximally reduced under normal, nonstressed, conditions. By using labeling with a non permeable, biotinylated SH reagent followed by two-dimensional gel electrophoresis and analysis by MS, we identified some of the proteins associated with the membrane that are reduced by NAC. These proteins include the following: integrin α-4, myosin heavy chain (nonmuscle type A), myosin light-chain alkali (nonmuscle isoform), and β-actin. NAC pretreatment augmented integrin α-4-dependent fibronectin adhesion and aggregation of Jurkat cells without changing its expression by fluorescence-activated cell sorter, suggesting that reduction of surface disulfides can affect proteins function. We postulate that some of the activities of NAC or other thiol antioxidants may not only be due to free radical scavenging or increase of intracellular GSH and subsequent effects on transcription factors, but could modify the redox state of functional membrane proteins with exofacial SH critical for their activity.
There is a growing interest in the role of thiol-disulfide oxidoreduction as a mechanism of redox regulation of cellular functions and gene expression (1, 2). This concept is based on the cellular effects of oxidative stress, glutathione (GSH) depletion, or by the use of thiol antioxidants or precursors of GSH synthesis, including N-acetyl-l-cysteine (NAC).
However, some effects of NAC seem not mediated by an increase of GSH, as suggested by experiments with inhibitors of GSH synthesis or the d-stereoisomer of NAC (d-NAC) that cannot be converted to GSH. These effects include inhibition of apoptosis (3), antiproliferative effect (4), inhibition of epidermal growth factor receptor activation (5), or enhancement of IL-1-induced iNOS expression (6).
Recent interest has focused on oxidoreduction of proteins' cysteines as a means of redox regulation of their function. In particular, protein SHs can be reversibly oxidized to form intra- or interchain disulfides, glutathionylated proteins (7, 8), or S-nitrosothiols (9). It is possible that thiol antioxidants like NAC interfere with these mechanisms by acting as reductants (4).
Whereas most of the attention has focused on intracellular targets or redox regulation, including transcription factors (10), or signaling molecules (11), surface thiols have also been suggested to be regulated in lymphocytes and are increased in HIV-infected patients (12). Among the membrane proteins that can be modified by oxidoreduction there are ion channels and the N-methyl-d-aspartate (NMDA) receptor (13, 14).
In the present study, we investigated protein thiols at the cell surface as possible candidate targets of redox regulation. To this end, we studied the effect of NAC on the expression of surface SH in human peripheral blood mononuclear cells (PBMCs). We used two different methods of detection of surface SH. A first method, which is more quantitative, relies on the use of Ellman's reagent, 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). Incubation of cells with DTNB, which is not cell-permeable, will cause release of the chromogenic product reflecting the amount of surface SH (6, 15).
A second method utilizes a cell-impermeable, biotinylated SH reagent N-(biotinoyl)-N′-(iodoacetyl)ethylendiamine (BIAM) to label the cell-surface proteins (16, 17). We applied this method to proteomic analysis to identify protein SH specifically reduced in NAC-treated cells. As one of the proteins identified as targets of NAC-reducing action was an integrin, we also studied the effect of NAC on integrin-mediated cell adhesion.
Materials and Methods
Cells and Treatments. Human PBMCs, which were isolated by using standard Ficoll/Hypaque gradients from buffy coat of healthy donors, were plated at 2.5 × 106 in 3 ml in 25 cm2 tissue culture flasks for DTNB experiments. For experiments using BIAM labeling and membrane purification, cells were plated in 75 cm2 of tissue culture flasks (≈5 × 107 cells per flask). Cells were treated as indicated in Results, with any of the following additions: NAC, buthionine sulfoximine (BSO), H2O2, GSH (all from Sigma), and d-NAC (Research Organics), which were washed twice with PBS, and used for biochemical analyses as described below. Jurkat cells were cultured in RPMI medium 1640 supplemented with 10% FBS and treated as described below.
Determination of Cell-Surface Protein Thiols with DTNB. DTNB was added to the cells to a final concentration of 200 μM in PBS. After 20 min at room temperature, the supernatant was harvested and its absorbance read at 412 nm. Results are expressed as nanomoles SH per 106 cells calculated from a standard curve with NAC.
Determination of Cell-Surface Protein Thiols with BIAM. Cells were resuspended at 1 × 106 per ml in medium and treated for 15 min at 37°C with 100 μM BIAM, then plasma membranes were isolated essentially as described (18). Briefly, the cells were homogenized in 250 mM sucrose containing 10 mM Hepes NaOH (pH 7.5) and 1 mM EDTA by using a Potter Teflon homogenizer. The homogenate was centrifuged at 700 × g for 10 min, and the resulting supernatant fraction was centrifuged again at 100,000 × g for 35 min. The pellet was then resuspended in PBS containing 2% Zwittergent 3–10 (Calbiochem; ref. 19), and the lysate was clarified by centrifugation.
Electrophoretic Separation of Proteins and Detection of Protein SH by Immunoblotting. A minimum of 40 μg of membrane proteins were reduced with 5% 2-mercaptoethanol, and two-dimensional gel electrophoresis (2-DE) was performed essentially as in ref. 8, except that 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate was added to the sample buffer. Proteins were then transferred to a nitrocellulose membrane and probed with streptavidin-peroxidase. Parallel gels were stained with Coomassie blue and used for protein identification.
Protein Identification. Protein spots were excised from 2-DE gels, destained for few hours in 25 mM ammonium bicarbonate/40% ethanol, and washed with a sequential increasing percentage of acetonitrile. Proteins were in gel-digested overnight at 30°C with trypsin (Promega) at a concentration of 12 ng/ml in a 25 mM ammonium bicarbonate/10% acetonitrile solution. Peptide mass fingerprinting (PMF) was performed on a Bruker ReflexIII matrix-assisted laser desorption/ionization (MALDI) mass spectrometer by using α-cyano-4-hydroxycinnamic acid (Bruker, Billerica, MA) as a matrix. The mass spectra were externally calibrated with a mixture of seven standard peptides in the range between 1,000 and 3,000 Da. Data generated were subjected to database (NCBInr) searching by using as programs mascot (http://www.matrixscience.com) and profound (http://prowl.rockefeller.edu), allowing up to one missed trypsin cleavage and a mass tolerance of ± 0.2 Da. Postsource decay (PSD) MALDI MS of selected peptide ions and analysis with the MS-Tag searching algorithm (http://prospector.ucsf.edu) were necessary to confirm identifications in some cases. All of the identified proteins were in the expected size range and pI based on position in the gel.
GSH Assay. Cells (1 × 106 per sample) were deproteinized with 100 μl of 10% sulfosalicylic acid, left for 30 min on ice, centrifuged at 17,000 × g for 5 min at 4°C, and GSH measured according to the methods of Tietze (20).
Cell Adhesion Assay. Seven μg/ml of human fibronectin (Sigma) was used to coat 48-well plates for2hat37°C; then, plates were saturated with 2% BSA for 30 min at 37°C and washed twice with PBS. Jurkat cells were then seeded onto the wells for 2h at 37°C; then, nonadherent cells were aspirated, and the wells were rinsed with PBS. Adhering cells were fixed overnight with 2% formaldehyde and stained with eosin Y for 30 min. Eosin Y was then extracted by addition of a mixture of 1% of glacial acetic acid and 50% ethanol, and absorbance was measured at 540 nm.
Integrin α-4 (VLA-4) Expression. VLA-4 antigen expression was detected on Jurkat cells by indirect immunofluorescence with a monoclonal anti-VLA-4 (clone HP1/7) and flow cytometry. Staining of cells was performed according to standard protocols and flow cytometry analysis was performed by using a FACS-Calibur cytometer (Becton Dickinson).
Results
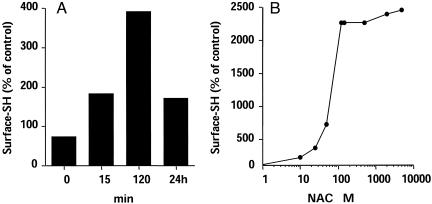
NAC Treatment Preferentially Reduces Surface Protein Disulfides. To study the effect of NAC on the levels of exofacial protein thiols, as measured by using the DTNB method, cells were treated with NAC and then extensively washed with PBS until no SH could be detected in the washings by DTNB assay. The cells were then incubated with DTNB for measuring cell-surface SH. As shown in Fig. 1A, NAC rapidly increased surface SH levels (by >5-fold in the experiment shown), reaching a plateau within 2 h. Fig. 1B shows the results of dose-response experiments, indicating that maximal effect of NAC was observed at 0.5 mM concentration, with an approximate EC50 of 0.1 mM.
Fig. 1.
Time course (A) and dose response (B) of NAC-induced increase of surface SH expression. PBMCs (106 cells per ml) were cultured with 5 mM NAC for the indicated time (A), after which surface thiols were quantified with DTNB. In the experiment shown in B, cells were incubated for 2 h with the indicated concentration of NAC. Data are the mean of duplicate experiments.
To confirm that the increase in DTNB reactivity was not an artifact due to a carryover of NAC, or to the fact that a small amount of DTNB might enter into the cells, we purified membranes, then we measured their SH. A comparable increase in free SH was found both on whole cells (control, 99 ± 18; NAC, 478 ± 27 nmol per 106 cells; mean ± SD, n = 3) and on purified membranes (control, 69 ± 3; NAC, 321 ± 12 nmol per 106 cells; mean ± SD, n = 3. P < 0.01 vs. control).
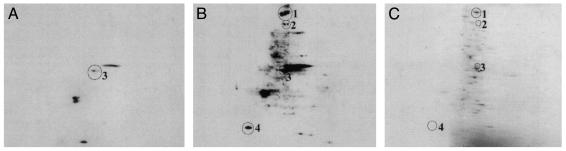
Identification of Surface Membrane Proteins Whose SH Expression Is Augmented by NAC. In these experiments, cells were incubated for 2 h with 5 mM NAC, then surface SHs were labeled with BIAM. Membranes were then purified, and proteins were analyzed by 2-DE, followed by immunoblotting with streptavidin. Fig. 2 shows that very little SH expression was present in control cells (Fig. 2 A), whereas several protein spots became labeled after NAC treatment (Fig. 2B). The protein spots indicated were identified as myosin heavy chain, nonmuscle type A (NMMHC-IIA) (spot no. 1), VLA-4 (spot no. 2), β-actin (spot no. 3), and myosin light-chain alkali, nonmuscle isoform (MLC) (spot no. 4; Table 1). The proteins were identified by PMF with significantly high probability-based Mowse and Z scores, except VLA-4, for which PSD analysis of the fragment ion at 969.7 m/z was necessary for unambiguous identification.
Fig. 2.
BIAM labeling pattern of membrane proteins from PBMCs under basal conditions (A) or treated with 5 mM NAC for 2 h (B). Proteins were separated by 2-DE based on pI (4–10 nonlinear gradient, left to right) and molecular weight (SDS/12% PAGE, top to bottom). (C) A Coomassie blue-stained gel from control cells.
Table 1. Surface membrane proteins sensitive to NAC treatment.
| Spot no. | Protein name | NCBI accession no. | PBM score | Z score | Percent sequence coverage | No. of matching peptides | MALDI MS identification methods |
|---|---|---|---|---|---|---|---|
| 1 | NMMHC-IIA | gi189036 | 171 | 2.43 | 24 | 27 | PMF |
| 2 | VLA-4 | gi4504749 | - | 0.12 | 7 | 6 | PSD |
| 3 | β-actin | gi4501885 | 82 | 2.1 | 37 | 13 | PMF |
| 4 | MLC | gi576 | 96 | 2.43 | 46 | 9 | PMF |
Spot nos. refer to Fig. 3. PBM and Z scores are the probability scores obtained with mascot and profound programs with PMF data. PSD analysis was used as confirmation when PMF data did not give significant scores (PBM score <63 and Z score <1.65). MALDI, matrix-assisted laser desorption/ionization.
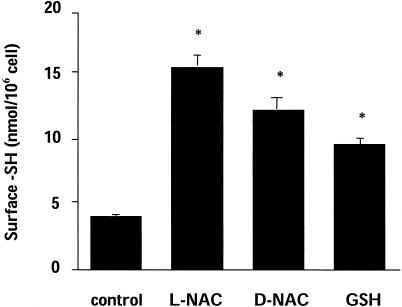
NAC Reduces Surface Proteins Directly Through Its Reducing Activity. To study whether the effect of NAC was mediated by GSH, we tested d-NAC, which is not a precursor for GSH synthesis, or GSH itself, which, unlike NAC, does not enter the cells. As shown in Fig. 3, all these thiol compounds, tested in the same experimental conditions used for NAC, increased surface SH, suggesting that they act through a direct reducing action.
Fig. 3.
Effect of NAC, d-NAC, or GSH on surface SH. Cells were treated for 2 h with a 5-mM concentration of the indicated compound. Surface SHs were quantitated by using the DTNB method. Data are mean ± SE (n = 3). *, P < 0.01 vs. control by Student's t test.
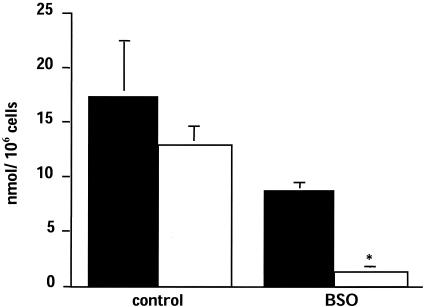
Role of Endogenous GSH in the Maintenance of Surface SH. To study whether intracellular GSH is implicated in maintaining surface SH under basal conditions (i.e., in the absence of any exogenous NAC or reducing agents), we treated cells overnight with the inhibitor of GSH synthesis, BSO. As shown in Fig. 4, BSO caused a 50% decrease in GSH, and this result was paralleled by an 85% decrease in surface SH.
Fig. 4.
Effect of prolonged treatment with BSO on intracellular GSH (black bars) and surface SH (white bars). Cells were treated with 1 mM BSO for 24 h. Surface SH was measured by using the DTNB method. Data are mean ± SE (n = 3). *, P < 0.05 vs. control.
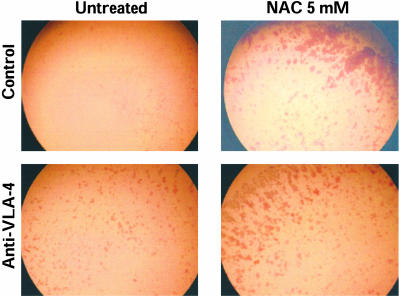
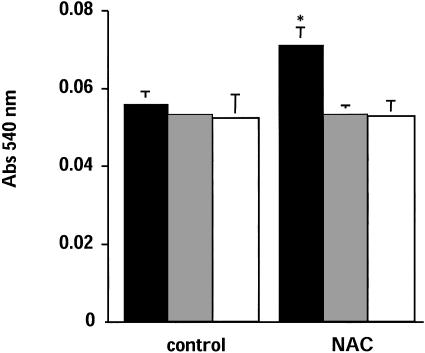
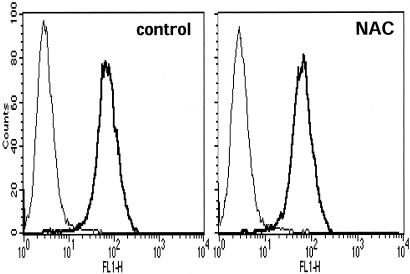
Effect of NAC and SH-Blocking Agents on VLA-4-Dependent Adhesion. To investigate the effect of NAC-induced augmentation of SH expression in VLA-4, we used human Jurkat cells that represent a well studied model for studying adhesion to fibronectin (21). Cells were treated with 5 mM NAC for 2 h, then we studied their adhesion to fibronectin. As shown in Fig. 5, NAC increases cell aggregations and adhesion. Incubation of the cells, after NAC treatment, with anti-VLA-4 antibody, inhibited both adhesion and aggregation, indicating that the effect of NAC is specific for VLA-4. Cell adhesion was then quantitated by staining the cells attached to fibronectin-coated plates with eosin Y and reading the absorbance in a microplate reader. As shown in Fig. 6, NAC pretreatment augmented cell adhesion by ≈35%, an effect that was abolished by anti-VLA-4. Under these experimental conditions, surface expression of VLA-4, as measured by flow cytometry, was unchanged (Fig. 7).
Fig. 5.
NAC increases the adhesions of Jurkat cells to fibronectin. The adhesion assay was performed by using control or NAC (5 mM for 2 h)-treated Jurkat cells, as described in Materials and Methods. The cells were photographed after staining with eosin Y.
Fig. 6.
Quantitation of the effect of NAC on cell adhesion. Experiments were performed exactly as in Fig. 5. Cells were solubilized and eosin Y and absorbance was quantitated at 540 nm. No antibody, white bars; anti-VLA-4 (10 ng/ml), gray bars; and anti-VLA-4 (10 μg/ml), black bars. Data are means ± SE (n = 3).*, P < 0.01 vs. control.
Fig. 7.
Surface expression of VLA-4 is unchanged by NAC. Jurkat cells were treated with NAC (5 mM for 2 h) or left untreated. The cells were then stained with HP1/7, fixed, and analyzed by FACS. VLA-4-positive cells were 77% in control and 78% in NAC-treated cells.
Discussion
Recent studies (12) indicate a role for surface thiols as targets of redox regulation in the immune system. We show that NAC augments the expression of surface SH on PBMCs. This effect was observed at concentrations of 0.1–5 mM, which was in the lower range of those used in vitro to study regulatory effects of NAC (often in the 10–30 mM range). This result indicates that redox-sensitive cysteines on the cell surface are mostly in an oxidized state, very likely because they are at the interface with the highly oxidizing extracellular environment, and can be easily reduced by mild reductants such as NAC or GSH. From these experiments, it is not known whether the protein cysteines reduced by NAC are oxidized as protein disulfides, or mixed disulfides with small molecular weight thiols. Garant et al. (22) have shown that NAC can reduce the insulin receptor α-subunit disulfides, indicating that some of the SH might actually derive from reduction of interchain disulfides. NAC could also reduce intrachain disulfides that will normally be in the oxidized state. Another possibility is that some cysteines on the cell surface are present in part as mixed disulfides with small molecular weight thiols (e.g., GSH, cysteine), as it was described for soluble proteins such as albumin (23).
The effect of NAC is probably due to a direct reducing effect. In fact, d-NAC, which is not a GSH precursor, and GSH itself, which is not cell-permeable, had a comparable effect on surface SH. On the other hand, in the absence of exogenous reductants, cellular GSH seems to be required for maintaining surface SH in a reduced state, as indicated by the fact that GSH depletion with BSO markedly decreased surface SH.
A second aspect of the present study was the identification of the surface proteins susceptible of redox regulation by using a proteomic approach. There is currently a paucity of data pertaining to the comprehensive analysis of surface membrane proteins, due to the difficulty in purifying and identifying them in addition to their relatively low abundance. Protein databases are rapidly increasing the number of the hits and the quality of their annotations; however, very little information is reported on posttranslational modifications such as glycosylation, which is a common modification on surface membrane proteins. Specific protein labeling has been used to selectively separate the cell-surface proteome (16, 24). These strategies have simplified the purification of membrane proteins, but have added more difficulties to the MS analysis of the labeled proteins. For example, in our PMF experiments, we did not detect any peptide labeled with BIAM, suggesting a poor ionization efficiency of the peptide containing such a modification. All in all, the identification of surface membrane proteins still remains a challenge.
Some of the proteins identified were cytoskeletal proteins, including NMMHC-IIA, MLC, and β-actin. All these proteins can be found associated with the cell membrane (25) and may become accessible to BIAM, given the fluid state of the membrane.
One protein identified was the VLA-4, which is clearly located on the cell surface. This protein has 25 cysteines, of which 18 are engaged in disulfide bonds and 7 as free SH, and previous studies (26) had shown that at least two of the free cysteines are important for VLA-4 ligand binding and cell adhesion. In our experimental conditions, NAC markedly augments the expression of free SH in VLA-4 and VLA-4-dependent adhesion. It is important to note that a 2-h treatment with NAC did not alter VLA-4 surface expression, as measured by fluorescence-activated cell sorter (FACS) analysis.
Reactive oxygen species are also known to regulate cell adhesion at various steps, for instance, by regulating protein phosphatases implicated in integrin signaling (27). Thiol antioxidants inhibit the induction of VCAM and ICAM expression and diminish inflammation in vivo (28, 29).
In conclusion, we demonstrated that thiols can affect the redox state of several membrane proteins. We hypothesize that some of the effects of exogenous NAC may not be mediated by an increase of intracellular GSH, its free radical scavenging properties, or a reduction of intracellular disulfides; but, rather, by a reduction of membrane protein disulfides. Oxidoreduction of membrane SH could result in the modification of receptors, transporters, or of proteins acting as redox sensor. In fact, whereas most of the redox-sensitive molecular targets are intracellular proteins, including transcription factors and signaling proteins (30), some membrane receptors, including ion channels, NMDA receptor, glucose transporter, GLUT1, and insulin receptor are known to be redox-sensitive (22, 30). This finding might be important, because it could provide a mechanism for the activity of extracellular, or exogenously administered, thiol compounds, in conditions where this cannot be explained by an increase in cellular GSH or a as a free radical scavenger. The identification of these redox sensitive targets is obviously important for characterizing these mechanisms at the molecular level.
Acknowledgments
This work was supported by the Zambon Group (Bresso, Milan), by Fondazione Cariplo (Milan) (P.G.), by Fondazione Mariani (Milan) Grant R-03-30 (to P.G.), and by Telethon-Italy (Milan) Grant TCP01010 (to V.B). V.B. is an Assistant Telethon Scientist.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VLA-4, integrin α-4; NAC, N-acetyl-l-cysteine; d-NAC, d-stereoisomer of NAC; GSH, glutathione; PBMC, peripheral blood mononuclear cell; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); BIAM, N-(biotinoyl-N′-(iodoacetyl)ethylendiamine; BSO, buthionine sulfoximine.
References
- 1.Pomposiello, P. J. & Demple, B. (2001) Trends Biotechnol. 19, 109–114. [DOI] [PubMed] [Google Scholar]
- 2.Hogg, P. J. (2003) Trends Biochem. Sci. 28, 210–214. [DOI] [PubMed] [Google Scholar]
- 3.Yan, C. Y., Ferrari, G. & Greene, L. A. (1995) J. Biol. Chem. 270, 26827–26832. [DOI] [PubMed] [Google Scholar]
- 4.Kim, K. Y., Rhim, T., Choi, I. & Kim, S. S. (2001) J. Biol. Chem. 276, 40591–40598. [DOI] [PubMed] [Google Scholar]
- 5.Kamata, H., Shibukawa, Y., Oka, S. I. & Hirata, H. (2000) Eur. J. Biochem. 267, 1933–1944. [DOI] [PubMed] [Google Scholar]
- 6.Jiang, X. M., Fitzgerald, M., Grant, C. M. & Hogg, P. J. (1999) J. Biol. Chem. 274, 2416–2423. [DOI] [PubMed] [Google Scholar]
- 7.Ghezzi, P., Romines, B., Fratelli, M., Eberini, I., Gianazza, E., Casagrande, S., Laragione, T., Mengozzi, M. & Herzenberg, L. A. (2002) Mol. Immunol. 38, 773–780. [DOI] [PubMed] [Google Scholar]
- 8.Fratelli, M., Demol, H., Puype, M., Casagrande, S., Eberini, I., Salmona, M., Bonetto, V., Mengozzi, M., Duffieux, F., Miclet, E., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 3505–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallis, R. J. & Thomas, J. A. (2000) Arch. Biochem. Biophys. 383, 60–69. [DOI] [PubMed] [Google Scholar]
- 10.Schreck, R., Rieber, P. & Baeuerle, P. A. (1991) EMBO J. 10, 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le, F., Stomski, F., Woodcock, J. M., Lopez, A. F. & Gonda, T. J. (2000) J. Biol. Chem. 275, 5124–5130. [DOI] [PubMed] [Google Scholar]
- 12.Sahaf, B., Heydari, K. & Herzenberg, L. A. (2003) Proc. Natl. Acad. Sci. USA 100, 4001–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipton, S. A., Choi, Y. B., Takahashi, H., Zhang, D., Li, W., Godzik, A. & Bankston, L. A. (2002) Trends Neurosci. 25, 474–480. [DOI] [PubMed] [Google Scholar]
- 14.Zeng, X. H., Xia, X. M. & Lingle, C. J. (2003) Nat. Struct. Biol. 10, 448–454. [DOI] [PubMed] [Google Scholar]
- 15.Zai, A., Rudd, M. A., Scribner, A. W. & Loscalzo, J. (1999) J. Clin. Invest. 103, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominici, S., Valentini, M., Maellaro, E., Del Bello, B., Paolicchi, A., Lorenzini, E., Tongiani, R., Comporti, M. & Pompella, A. (1999) Free Radical Biol. Med. 27, 623–635. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. R., Yoon, H. W., Kwon, K. S., Lee, S. R. & Rhee, S. G. (2000) Anal. Biochem. 283, 214–221. [DOI] [PubMed] [Google Scholar]
- 18.Kuge, O., Yamakawa, Y. & Nishijima, M. (2001) J. Biol. Chem. 276, 23700–23706. [DOI] [PubMed] [Google Scholar]
- 19.Chevallet, M., Santoni, V., Poinas, A., Rouquie, D., Fuchs, A., Kieffer, S., Rossignol, M., Lunardi, J., Garin, J. & Rabilloud, T. (1998) Electrophoresis 19, 1901–1909. [DOI] [PubMed] [Google Scholar]
- 20.Tietze, F. (1969) Anal. Biochem. 27, 502–522. [DOI] [PubMed] [Google Scholar]
- 21.Campanero, M. R., Sanchez-Mateos, P., del Pozo, M. A. & Sanchez-Madrid, F. (1994) J. Cell Biol. 127, 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garant, M. J., Kole, S., Maksimova, E. M. & Bernier, M. (1999) Biochemistry 38, 5896–5904. [DOI] [PubMed] [Google Scholar]
- 23.Harrap, K. R., Jackson, R. C., Riches, P. G., Smith, C. A. & Hill, B. T. (1973) Biochim. Biophys. Acta 310, 104–110. [DOI] [PubMed] [Google Scholar]
- 24.Shin, B. K., Wang, H., Yim, A. M., Le Naour, F., Brichory, F., Jang, J. H., Zhao, R., Puravs, E., Tra, J., Michael, C. W., Misek, D. E. & Hanash, S. M. (2003) J. Biol. Chem. 278, 7607–7616. [DOI] [PubMed] [Google Scholar]
- 25.Rey, M., Vicente-Manzanares, M., Viedma, F., Yanez-Mo, M., Urzainqui, A., Barreiro, O., Vazquez, J. & Sanchez-Madrid, F. (2002) J. Immunol. 169, 5410–5414. [DOI] [PubMed] [Google Scholar]
- 26.Pujades, C., Teixido, J., Bazzoni, G. & Hemler, M. E. (1996) Biochem. J. 313, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiarugi, P., Pani, G., Giannoni, E., Taddei, L., Colavitti, R., Raugei, G., Symons, M., Borrello, S., Galeotti, T. & Ramponi, G. (2003) J. Cell Biol. 161, 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moynagh, P. N., Williams, D. C. & O'Neill, L. A. (1994) J. Immunol. 153, 2681–2690. [PubMed] [Google Scholar]
- 29.Khachigian, L. M., Collins, T. & Fries, J. W. (1997) Am. J. Pathol. 151, 1225–1229. [PMC free article] [PubMed] [Google Scholar]
- 30.Sen, C. K. (1998) Biochem. Pharmacol. 55, 1747–1758. [DOI] [PubMed] [Google Scholar]