Abstract
ID2 is a rhythmically expressed HLH transcriptional repressor. Deletion of Id2 in mice results in circadian phenotypes, highlighted by disrupted locomotor activity rhythms and an enhanced photoentrainment response. ID2 can suppress the transactivation potential of the positive elements of the clock, CLOCK-BMAL1, on mPer1 and clock-controlled gene (CCG) activity. Misregulation of CCGs is observed in Id2−/− liver, and mutant mice exhibit associated alterations in lipid homeostasis. These data suggest that ID2 contributes to both input and output components of the clock and that this may be via interaction with the bHLH clock proteins CLOCK and BMAL1. The aim of the present study was to explore this potential interaction. Coimmunoprecipitation analysis revealed the capability of ID2 to complex with both CLOCK and BMAL1, and mammalian two-hybrid analysis revealed direct interactions of ID2, ID1 and ID3 with CLOCK and BMAL1. Deletion of the ID2 HLH domain rendered ID2 ineffective at inhibiting CLOCK-BMAL1 transactivation, suggesting that interaction between the proteins is via the HLH region. Immunofluorescence analysis revealed overlapping localization of ID2 with CLOCK and BMAL1 in the cytoplasm. Overexpression of CLOCK and BMAL1 in the presence of ID2 resulted in a significant reduction in their nuclear localization, revealing that ID2 can sequester CLOCK and BMAL1 to the cytoplasm. Serum stimulation of Id2−/− mouse embryonic fibroblasts resulted in an enhanced induction of mPer1 expression. These data provide the basis for a molecular mechanism through which ID2 could regulate aspects of both clock input and output through a time-of-day specific interaction with CLOCK and BMAL1.
Keywords: Gene Expression, Gene Knock-out, Gene Regulation, Immunochemistry, Protein-Protein Interactions, Transcription Factors, Transcription Regulation, Transcription Repressor, Circadian Rhythm, Clock
Introduction
Circadian rhythms are 24-h temporal patterns of biochemistry, physiology, and behavior that are an integral component of eukaryotic life (1, 2). The molecular circadian clock is a cell autonomous system composed of three conceptual components: a self-sustainable pacemaker that generates 24-h rhythmicity; an input pathway that allows the clock to be reset and entrained by temporal cues in the environment; and mechanisms of output that regulate molecular and biochemical pathways and ultimately translate to rhythms in physiology and behavior. The master circadian pacemaker resides in the suprachiasmatic nucleus (SCN)2 of the hypothalamus (1). Additionally, peripheral tissues and cell lines contain self-sustaining clocks (3–6). The molecular clock is comprised of a series of transcriptional-translational feedback loops that take ∼24 h to complete. The positive loop contains the basic helix-loop-helix (bHLH)/PAS (Per-Arnt-Sim) proteins BMAL1 (brain and muscle ARNT-like 1) and CLOCK (Circadian locomotor output cycle kaput) and/or NPAS2 (neuronal PAS domain-containing protein 2). These transcription factors interact through their PAS and HLH regions and bind to the promoter region of their target genes of the negative loop at specific E-box regulatory elements (CACGTG), resulting in transcriptional activation. The negative loop is comprised of the period (mPer1, mPer2, mPer3) and cryptochrome genes (mCry1, mCry2). Rev-ERBα and RAR-related orphan receptor α RORα are additional components that close the interlocking loops, and additional loops have been identified (e.g. Dec1, Dec2), which are proposed to increase stability and precision of the clock (1, 2, 7–9).
The clock is reset by Zeitgeber stimuli through input pathways that transiently induce or suppress levels of period gene transcripts (1, 2, 10, 11). In the case of the SCN, light activation of retinal photoreceptors and the retinal hypothalamic tract results in stimulation of SCN neurons. Activation of several intracellular signaling pathways culminates in the phosphorylation of Ca2+/cAMP response element binding protein (CREB) (12–15). Phospho-CREB (pCREB) can then bind to its response element, the CRE, within the promoter region of target genes, such as the immediate early gene c-fos and immediate early gene/clock genes mPer1 and mPer2. Phase shifts of the SCN clock by elements of the photic signaling pathways results in elevation of mPer1 (and sometimes mPer2). Nonphotic (arousal) generated phase shifts result in suppression of mPer1 (1, 11). Furthermore, phase shifts of clocks in peripheral tissues and cell lines by serum, hormone, and nutrient signaling elicit similar signal transduction cascades that modify mPer1 and/or mPer2 expression (2, 5, 6, 16–18). mPer1 is required for photic-induced phase shifts and appears to be a terminal step in the signaling pathway to the clock-resetting mechanism (1, 2). The CLOCK-BMAL1 heterodimer binds to the E-box element(s) within the period gene promoter region, and in addition to its function in driving rhythmic patterns of period gene expression (12), may additionally contribute, in a permissive manner, to the gating of pCREB induction of transcription (19, 20).
The circadian pacemaker regulates clock output through the regulation of CCGs, genes that are defined as rhythmically expressed and driven by the pacemaker but are not essential for sustained pacemaker function (1, 2); CCGs represent 2–13% of the transcriptome within a given tissue (21).
ID (Inhibitor of DNA binding) (ID1–4) proteins are a family of HLH transcription factors previously implicated in the regulation of cell cycle, apoptosis, and development and whose misregulation is associated with tumorigenesis (22–25). Their mechanistic role within these processes is as a transcriptional repressor, by binding to partner bHLH proteins, but as they do not contain the basic domain necessary to bind to DNA, transcription is not elicited (22–25). ID proteins are small (16–18 kDa) and contain a highly conserved HLH domain and variable C and N termini (23, 26). Whereas Id genes continue to be expressed throughout the body in the adult animal, few roles in normal postmitotic adult tissues have been described (23, 25, 27).
DNA microarray analysis and quantitative RT-PCR (qRT-PCR) reveal Id2 as rhythmically expressed in multiple tissues including the SCN, heart, liver, and fibroblasts (6, 27, 28). Moreover, ID2 protein is rhythmically expressed in the liver (29). The absence of Id2 results in alterations in each of the three components of the circadian clock. Data recently published by our laboratory (27) revealed that Id2−/− mice exhibit a more rapid re-entrainment to a large change in their photoschedule along with greater phase shifts compared with their wild type counterparts. This data suggests that Id2 is involved in entrainment within the mammalian clock. Furthermore, a significant proportion of mutant mice display an arrhythmic phenotype. At the molecular level, ID1, ID2, and ID3 demonstrated a marked dose-dependent suppression of CLOCK-BMAL1-induced activation of the clock gene mPer1 and CCG arginine vasopression (AVP) (27). This suggests a role for the ID proteins as transcriptional repressors within the mammalian clock (27). The absence of the Id2 gene also results in loss of normal rhythmic profiles of CCGs in the liver (29), including genes associated with fat metabolism. Additionally, mutant mice show corresponding physiological disturbances in lipid regulation. These phenotypes reveal a role of ID2 in clock resetting, photic entrainment, regulation of clock output, and connecting circadian and metabolic processes. Furthermore, disruption of circadian locomotor activity also indicates either a disturbance to output and/or pacemaker function (27, 29).
Based on these observations, we hypothesize that ID2 contributes to the regulation of the circadian clock at the levels of input, pacemaker, and output. Our transcriptional assay data, which reveal a potent inhibitory effect of ID2 upon CLOCK-BMAL1 transactivation of mPer1 and AVP genes, suggest that these phenotypes may be achieved in part through a direct action of ID2 upon CLOCK and/or BMAL1 activity. We propose that ID2 interferes with the interaction between CLOCK and BMAL1 at the level of the core oscillator and therefore disrupts their transactivation potential of genes involved in clock resetting and entrainment. To test this hypothesis, we examined the ability of ID2 to physically interact with CLOCK and BMAL1 and whether this might have functional consequences. In this study, we provide evidence for a direct interaction of ID2 with both CLOCK and BMAL1 and that suppression of CLOCK-BMAL1 transactivation requires the ID2 HLH domain. We provide evidence for the capability of ID2 to sequester CLOCK and BMAL1 to the cytoplasm. These data reveal a potential role for ID2 in modulating components of the core oscillator. Analysis of mPer1 gene induction following serum stimulation of Id2−/− MEFs reveal an enhanced response. As mPer1 is a state variable of the clock and a key contributor to clock resetting (1, 2, 10), these data correlate with our in vivo entrainment studies. A mechanistic model for these findings is provided.
EXPERIMENTAL PROCEDURES
Plasmids
Mouse CLOCK-HA, BMAL1-HA, CLOCK-FLAG, and BMAL1-FLAG in pcDNA3 gift plasmids from Dr. M. Antoch (30) were used for coimmunoprecipitation analysis. Human ID2-FLAG in pCβF (27, 29) was used for co-immunoprecipitation, mPer1 promoter reporter, and co-immunofluorescence experiments. For mammalian two-hybrid experiments, the following plasmids were used: mouse pBIND-ID1 and pACT-MyoD as a positive control (Promega); human ID1-GAL4, ID3-GAL4 (31), mCRY1, mouse BMAL1, and human CLOCK in pACT and pBIND, and pGL5-Luc were gifts from Dr. J. Hogenesch. Human ID1, ID2, and ID3 in both pBIND and pACT were provided by Dr. H. Axelson (32). For mPer1 reporter experiments: mPer1-Luc reporter, β-gal-CMV, and mouse CLOCK, BMAL1, and Id2 in pcDNA3, as described previously (27, 33, 34). Human Id2-ΔHLH in pHCMV and Id2-ΔN in pcDNA3 (N terminus region missing (amino acids 1–28) and missing last 14 amino acids of C terminus) were provided by Dr. M. Israel (35, 36), and mouse Id2-ΔC in pEMSV was provided by Dr. R. Lim (missing last 42 amino acids of C terminus (amino acids 93–134)) (37).
Coimmunoprecipitation and Western Blotting Procedure
NIH3T3 and HEK293 cells were maintained in DMEM supplemented with 5% fetal bovine serum and 100 units/ml of penicillin and streptomycin (Hyclone, Logan, UT). Cells were maintained at 37 °C in a humidified atmosphere containing 10% CO2. Cells were transiently transfected in 100-mm dishes using Lipofectamine PLUS reagents (Invitrogen). 3 μg of plasmid DNA was added to each dish, and pcDNA3 empty vector was included to ensure all combinations contained equal amounts of DNA. Transfected combinations were ID2-FLAG+BMAL1-HA, ID2-FLAG+CLOCK-HA, BMAL1-HA+pcDNA3, and CLOCK-HA+pcDNA3. 48 h following transfection, whole cell lysates were harvested in Nonidet P-40 lysis buffer (20 mm Tris-HCl, 150 mm NaCl, 10 nm MgCl2, 2 mm EDTA, 10% glycerol, 1% Nonidet P-40 plus 0.1% protease/phosphatase inhibiters). Lysates were normalized by measuring total protein concentration using the Pierce Micro BCA Protein Assay. For coimmunoprecipitation with ID2-FLAG, 1000 μg of total protein were precleared with mouse IgG-agarose for 30 min at room temperature. Lysates were centrifuged at 1500 rpm, and the precleared lysate was transferred to a separate tube where it was incubated with anti-FLAG antibody (1:200, Cell Signaling, Danvers, MA) for 1 h at room temperature. 50 μl of protein A-agarose beads (Bio-Rad) were added to the lysate and allowed to rotate overnight at 4 °C. Beads were washed twice in lysis buffer and eluted using SDS loading buffer. Samples were boiled at 95 °C for 5 min to elute and denature the protein. Proteins were separated on a 6% SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked for 1 h at room temperature in 5% milk/0.05% Tween 20 in TBS. BMAL1-HA and CLOCK-HA tagged proteins were detected using an anti-HA antibody (1:750, Cell Signaling, catalog no. 2367) in 5% milk/0.05% Tween 20 in TBS overnight at 4 °C, and with a secondary HRP-conjugated anti-mouse antibody (1:2000, Sigma Aldrich). 10% of unprocessed cell lysate used for pulldown (preimmunoprecipitation input) was separated on a SDS-PAGE and ID2-FLAG protein detected using an anti-ID2 monoclonal antibody (1:1000; M04, clone 2C11; Abnova, Walnut, CA) and secondary HRP-conjugated anti-mouse antibody. Proteins were visualized with an ECL detection system (Super Signal West Dura Extended Duration Substrate; Pierce). Protein sizes were determined using an Invitrogen benchmark prestained protein ladder, which was run alongside samples in each gel.
Mammalian Two-hybrid and Luciferase Reporter Assay
Cells were transiently transfected in 24-well plates and allowed to proliferate to 70% confluency, with each condition plated in three wells for statistical analysis. Cells were transfected with 150 ng of each expression plasmid using Lipofectamine PLUS reagents (Invitrogen). A total of 25 ng of β-gal plasmid was added to normalize transfection efficiency. All transfection combinations contained 150 ng pGL5-Luc reporter plasmid. Following a 48-h incubation, cells were harvested using 1X Passive Lysis Buffer (Promega) and firefly luciferase activity assayed with Luciferase Assay reagent (Promega) and by luminometry (Veritas Luminometer, Turner BioSystems, Sunnyvale, CA). β-gal activity was measured using Z-buffer reagent and by measurement of optical density by spectrophotometry. Luciferase activity was corrected for transfection efficiency by normalizing to β-gal activity. All experiments were performed at least twice, and samples were measured in triplicate. Statistical analysis was performed by GraphPad Prism (version 4, GraphPad Software, San Diego, CA). One-factor ANOVA was conducted on each experimental versus control measurements, with Bonferroni post hoc t tests (p < 0.05).
mPer1 Gene Promoter Luciferase Reporter Studies
NIH3T3 cells were transiently transfected with Lipofectamine PLUS reagent in 24-well plates with 125 ng of each expression vector. The plasmids encoding full-length (ID2 and ID2-FLAG) or truncated forms of ID2 (ID2-ΔHLH, ID2-ΔN, and ID2-ΔC) were individually co-expressed with CLOCK and BMAL1 and the mPer1 luciferase promoter reporter gene (10 ng/reaction). β-gal (25 ng) was used to normalize data and assess transfection efficiency. Western blot analysis was conducted to test for the efficiency of protein expression using methods described above and outlined previously (27, 29) using anti-Id2 antibody (M04, Abnova) and to control for protein loading, an anti-β-actin monoclonal antibody (1:5000, A1978, Sigma Aldrich), visualized with a secondary antibody at 1:5000.
Immunofluorescence Experiments
HEK293 and NIH3T3 cells were transiently transfected in six-well plates. Sterile coverslips were added to each well for imaging purposes. Cells were transfected with 330 ng of each plasmid using CLOCK-HA, BMAL1, and ID2-FLAG and with pcDNA3-empty vector allowing for equal amounts of total DNA. After 48 h, cells were fixed with 5% paraformaldehyde/phosphate buffer for 20 min at 37 °C. Cells were washed three times with ice-cold PBS and blocked for 1 h at room temperature in 3% BSA/PBS. Primary antibodies were delivered in 3% BSA/PBS and were as follows: anti-HA (1:500, Cell Signaling, catalog no. 2367) to visualize CLOCK-HA, anti-BMAL1 (1:50, catalog no. sc-8550, Santa Cruz Biotechnology, Santa Cruz, CA), and anti-ID2 (1:50, catalog no. sc-489, C20, Santa Cruz Biotechnology). CLOCK was also visualized with anti-CLOCK (CLOCK H-276, catalog no. sc-25361, Santa Cruz Biotechnology; CLO11, Alpha Diagnostic International, San Antonio, TX), but due to concerns with confidence in the localization signals, we focused on detecting ectopically expressed CLOCK-HA using the anti-HA antibody. Cells were incubated overnight at 4 °C, and cells were washed three times with PBS for 5 min each. The following secondary antibodies were used at 1:300 dilutions: 705-165-147 Cy3-conjugated Donkey anti-goat, 711-095-152 FITC-conjugated donkey anti-rabbit, 715-165-150 Cy3-conjugated donkey anti-mouse, and 715-485-150 DyLight-conjugated donkey anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA). DAPI was used at 1:40,000 to visualize the nucleus. Secondary antibodies were delivered in 3% BSA/PBS, and cells were incubated in the dark for 2 h at room temperature. After washing with PBS, cells were mounted, and images were acquired using a camera (CoolSNAP HQ; Roper Scientific, Sarasota, FL) on a fluorescent microscope (Axiovert 200MOT; Carl Zeiss) and operated with Metamorph software (MDS Analytical Technologies, Sunnyvale, CA). Images were collected at room temperature using a 40× numerical aperture 1.0 objective, and raw data were maintained through image processing in Photoshop (Adobe Software, San Jose CA) and presented in a pseudo-color scale to convey relative image intensities. Control procedures were carried out on transfected and nontransfected cells processed with only secondary antisera and on nontransfected cells with both anti-HA and secondary antisera. Cell localization signals were assessed using the following criteria: staining intensity cytoplasm > nucleus; cytoplasm = nucleus; or nucleus > cytoplasm (38, 39).
Serum Stimulation of Mouse Embryonic Fibroblasts
Id2+/+ and Id2−/− MEFs were generated; mouse embryos from Id2+/− crosses on a C57BL/6 background were isolated at embryonic day 14, and cells isolated from individuals were immortalized by passing them for at least one month. Genotypes of mice were subsequently determined by PCR analysis using methods described previously (27). Wild type and Id2−/− MEFs were seeded in six-well plates in 5% FBS, 1× penicillin-streptomycin, and 1× DMEM. After the cells grew to confluency in ∼2 days, MEFs were serum reduced with 1% FBS, 1× penicillin-streptomycin, and 1× DMEM. After 2 days of serum starvation, cells were treated for 2 h with 50% horse serum (Invitrogen) followed by serum-free medium (5, 6). Cells were collected at 0 min, 30 min, 60 min, 90 min, 120 min, and 8 h after serum stimulation. Cells were first washed with 1× PBS, and collected with TRIzol reagent (Invitrogen). RNA was collected, and quantity was determined by spectrophotometry.
Real-time Quantitative RT-PCR
qRT-PCR was conducted as described previously and by using a standard curves method (6, 27). Primer sequences for mPer1, mPer2, and c-fos were as published (4, 40). For GAPDH analysis, the Taqman rodent GAPDH primers and probe were used (Applied Biosystems, proprietary sequences). GAPDH was used to normalize all gene profiles. A two-tailed unpaired Student's t test was performed at individual time points using the Statview program (Abacus Program Concepts) to determine statistical significance between genotypes (p < 0.05).
RESULTS
ID2 Can Interact with Both CLOCK and BMAL1 Complexes
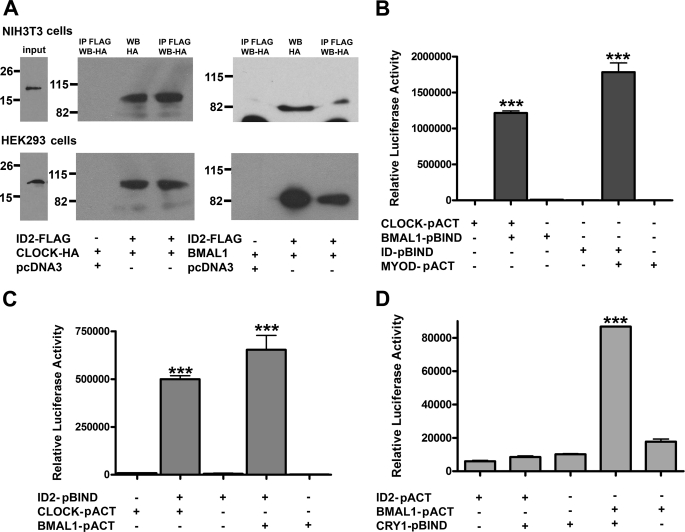
ID proteins are known to function by binding to bHLH transcription factors and thereby negatively regulate their effects on gene expression (23, 25, 41). We first sought to identify whether interactions occurred between ID2 and the positive elements of the circadian clock, CLOCK and BMAL1, and whether they would complex together. NIH3T3 and HEK293 cells were transiently transfected with different combinations of ID2-FLAG, CLOCK-HA, and BMAL1-HA. Co-immunoprecipitation was performed using an anti-FLAG antibody to pull down ID2 and probed for the presence of its binding partners by Western blot analysis. The presence of both CLOCK-HA and BMAL1-HA were detected using an anti-HA antibody in both cell lines (Fig. 1A). As a negative control, CLOCK-HA alone or BMAL1-HA alone was also subjected to co-immunoprecipitation with an anti-FLAG antibody. As expected, there was little or no signal detected for the presence of CLOCK-HA or BMAL1-HA upon probing. A positive control included CLOCK-HA- and BMAL1-HA-transfected cell lysates subjected to a straight immunoblot. This revealed a distinct band at the expected size of 105–120 kDa and 80–90 kDa for CLOCK and BMAL1, respectively (Fig. 1A).
FIGURE 1.
ID2 can interact directly with both CLOCK and BMAL1. A, NIH3T3 and HEK293 cells were cotransfected with ID2-FLAG and either CLOCK-HA or BMAL1-HA. Total cell extracts (middle lanes) or immunoprecipitates (IP, outer lanes) using an anti-FLAG antibody were subjected to Western blot (WB) analysis using anti-HA antiserum. Preimmunoprecipitation inputs for the respective immunoprecipitates are indicated (left lanes), representing 10% of input proteins and showing the presence of ID2-FLAG. ID2-FLAG signal was not detected in the negative control (not shown). Representative results are shown from one experiment. All co-immunoprecipitations were repeated at least twice in both cell lines. B–D, NIH3T3 cells were co-transfected with pGL5-Luc, β-gal, and the indicated expression plasmids. Shown are positive controls of known robustly interacting binding partners, CLOCK and BMAL1 (34, 46) and ID1 and MyoD (60) (B) and testing ID2 interaction with CLOCK and BMAL1 (C). D, as a negative control, the ability for ID2 to interact with CRY1 was tested, and a positive interaction signal was observed for BMAL1 with CRY1 (46). Luciferase reporter measurements were normalized to β-gal signals. Data are mean ± S.E. for four samples per treatment group. Differences determined by one-factor ANOVA with Bonferroni post hoc t tests. ***, p < 0.001. GAL4, DNA binding domain (pBIND plasmid); VP16, activation domain (pACT plasmid). Data are representative of three independent experiments.
ID1, ID2, and ID3 Can Interact Directly with Both CLOCK and BMAL1
As co-immunopreciptation analysis revealed that ID2 could complex with both CLOCK and BMAL1, we next sought to determine whether these interactions were through direct protein-protein binding. NIH3T3 cells were co-transfected in various combinations with plasmids containing BMAL1, CLOCK, and ID2 fused to either the pBIND (GAL4 DNA binding domain) or pACT (activation domain of the viral protein VP16). Within this mammalian two-hybrid system, an interaction between the two proteins of interest allows the activation domain and the DNA binding domain to come into close proximity, producing a functional transcriptional activator. This results in the activation of the luciferase reporter (pGL5-Luc) under the control of a GAL4-based promoter. For ID2-GAL4+CLOCK-VP16, a 58-fold increase in luciferase activity was observed (F3,8 = 730, p < 0.0001). For ID2-GAL4+BMAL1-VP16, a 119-fold induction was observed (F3,7 = 139, p < 0.0001) (Fig. 1C). To ensure that the activity we observed was due to a true interaction, we performed control assays. Positive controls were the established CLOCK-BMAL1 interaction, showing a 139-fold induction (F3,7 = 3263, p < 0.001) and a positive interaction between ID1-GAL4 and MyoD-VP16 with a fold induction of 646 (F3,7 = 357, p < 0.0001; Fig. 1B). Minimal activation of the luciferase reporter was observed when CLOCK-VP16, BMAL1-VP16, or ID2-GAL4 proteins were expressed alone (Fig. 1B). A negative control assaying for the interaction between ID2 and CRY1, revealed no interaction signal. Similar levels of activity were observed when both ID2-VP16 and CRY1-GAL4 were expressed together, as compared with when they were expressed alone. In contrast, the known interaction between BMAL1-VP16 and CRY1-GAL4 demonstrated an expected increase in luciferase activity (F3,7 =, p < 0.001; Fig. 1D).
As ID1 and ID3 can also suppress CLOCK-BMAL1 transactivation (27), we sought to test whether ID1 and ID3 could bind directly to CLOCK and/or BMAL1. Using GAL4 and VP16 fusion proteins generated from pACT and pBIND vectors, an increase in luciferase activity was also detected for both ID1+CLOCK and ID1+BMAL1: 6-fold (F3,7 = 13.6, p < 0.01) and 59-fold (F3,8 = 162.6, p < 0.001), respectively (Fig. 2A). Interactions between ID3+CLOCK and ID3+BMAL1 were measured; 3.5-fold (F3,7 = 39.5, p < 0.001) and 39-fold (F3,8 = 119.3, p < 0.001) changes were reported, respectively (Fig. 2C). We also performed interaction assays using alternative ID1-GAL4 and ID3-GAL4 constructs (pGAL4) and, consistent with the pBIND vectors, demonstrated significant interaction signals with both CLOCK and BMAL1 (ID1+CLOCK, F3,8 = 26.2, p < 0.01; ID1+BMAL1, F3,8 = 220, p < 0.0001; ID3+CLOCK, F3,7 = 103, p < 0.0001; ID3+BMAL1, F3,8 = 93, p < 0.0001; Fig. 2, B and D). All two-hybrid experiments were performed with proteins expressed in the opposite partner vectors where possible, i.e. ID proteins fused to VP16 and CLOCK and BMAL1 fused to GAL4. Similar interaction signals were observed (data not shown).
FIGURE 2.
ID1 and ID3 can interact with both CLOCK and BMAL1. A–D, NIH3T3 cells were co-transfected with pGL5-Luc, β-gal, and the indicated expression plasmids. Interaction between ID1 with CLOCK and with BMAL1 was tested using alternative ID1 plasmids with different vector backbones (A and B). Interaction between ID3 with CLOCK and with BMAL1 was also tested using two different ID3 plasmids (C and D). As a negative control, the ability for ID1 and ID3 to interact with CRY1 was tested (A and C). ID plasmids in A and C were pBIND, and ID plasmids in B and D were pGAL4. A positive interaction signal was observed for BMAL1 with CRY1 (see Fig. 1D). Luciferase reporter measurements were normalized to β-gal signals. Data are mean ± S.E. for four samples per treatment group. Differences determined by one factor ANOVA with Bonferroni post hoc t tests, *, p < 0.05; **, p < 0.01; and ***, p < 0.001. GAL4, DNA binding domain (pBIND or pGAL4 plasmid); VP16, activation domain (pACT or pVP16 plasmid). Data are representative of three independent experiments.
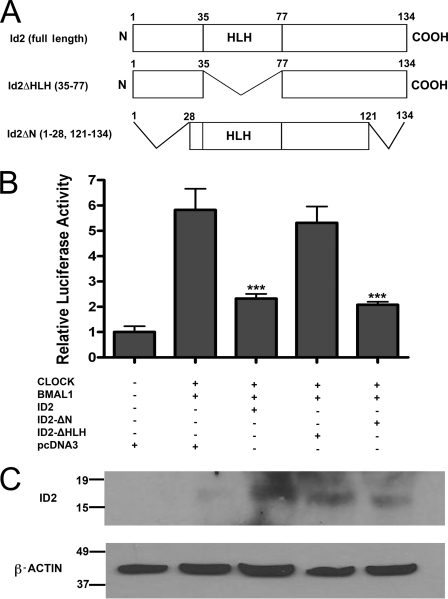
HLH Domain of ID2 Is Required for ID2 Suppression of CLOCK-BMAL1 Transactivation of mPer1 Promoter
Using truncated forms of the ID2 protein, we tested whether the ID2 HLH domain is required for the suppression of CLOCK-BMAL1 transactivation activity (27). Plasmids expressing CLOCK and BMAL1, an mPer1-Luc reporter plasmid, and one of each of ID2 full-length, ID2-ΔHLH, or ID2-ΔN plasmids were transfected into NIH3T3 cells (Fig. 3A). As a control, one set of cells was transfected with pcDNA3 empty vector alone, and as expected, little luciferase activity was observed (Fig. 3B). When CLOCK and BMAL1 are co-transfected, their heterodimer activity on the mPer1 promoter resulted in a 6-fold increase in luciferase activity (Fig. 3B). In concurrence with our previous data (27), when CLOCK and BMAL1 are co-transfected with full-length ID2, repression of the mPer1 promoter is observed (F5,23 = 16.9, p < 0.001; post hoc t test, p < 0.001 versus CLOCK-BMAL1-induced luciferase activity; Fig. 3B). Comparable repression was obtained using an ID2-FLAG tagged protein (data not shown). An equivalent level of inhibition is noted when ID2-ΔN was used, indicating that the N terminus is not necessary for the ID2 interaction (p < 0.001; Fig. 3B). However, when ID2 is missing its HLH domain, no repression of the mPer1 promoter was observed (Fig. 3B). Western blot analysis was employed to determine that the truncated plasmids generated translated ID2 protein, which was detected for ID2, ID2-ΔHLH, and ID2-ΔN (Fig. 3C).
FIGURE 3.
The helix-loop-helix domain of ID2 is the protein-binding site necessary for inhibition of CLOCK-BMAL1 transactivation. The effect of co-transfection of CLOCK and BMAL1 with ID2 expression plasmids on transactivation of the mPer1 promoter. NIH3T3 cells were co-transfected with mPer1:luciferase promoter reporter, β-gal, and the indicated expression plasmids. A, amino acid maps of full-length and truncated ID2 proteins encoded by DNA plasmids, with deleted residues noted in parentheses. B, mean ± S.E. luciferase reporter measurements normalized to β-gal, with five samples per treatment group. All groups, except pcDNA3.1 empty vector, were transfected with equal amount of CLOCK and BMAL1 expression vector (125 ng each). Values obtained for cells transfected with empty vector were set to 1.0. Differences determined by one-factor ANOVA with Bonferroni post hoc t tests. ***, p < 0.001 versus CLOCK-BMAL1-pcDNA3 group. Data are representative of four independent experiments. Results for inhibition of transativation by full-length ID2 is consistent with previous results (27). C, Western blot analysis of transfected cell lysates revealing protein bands for full-length ID2, ID2-ΔHLH, and ID2-ΔN truncated plasmids. Plasmid combinations for each lane are as described above in B.
Co-immunofluorescence Demonstrates That ID2 Co-exists in Subcellular Compartments with Both CLOCK and BMAL1
Subcellular locations of CLOCK, BMAL1, and ID2 were examined by immunofluorescence analysis in nontransfected and transfected HEK293 cells. Endogenous expression of CLOCK, BMAL1, and ID2, and overexpression of CLOCK-HA, BMAL1, and ID2 was examined using protein-specific antibodies and an anti-HA antibody to detect tagged CLOCK. Due to concerns about antibody specificity, we disregarded the endogenous CLOCK protein data. The subcellular distributions were classified into one of three categories: cytoplasm-dominant, nucleus-dominant, or equal distribution in cytoplasm and nucleus.
BMAL1 endogenous localization signal was predominantly cytoplasmic in 69% of cells with a smaller proportion of cells exhibiting equal levels in the nucleus and cytoplasm (18%) and nuclear-dominant staining (13%). ID2 showed an even higher incidence of cells showing a cytoplasm-dominant localization signal (89%) and with less equal cytoplasmic and nuclear staining (11%) and with no occurrence of predominantly nuclear staining. The overlay of fluorescent signals corresponding to BMAL1 and ID2 was assessed and found to be almost entirely cytoplasm dominant. i.e. of cells showing co-localization of both proteins, 85% of cells were scored as predominantly cytoplasmic. Mean percentage data were generated from three independent experiments.
HEK293 cells were then co-transfected with different expression plasmid combinations to examine whether ID2, CLOCK, and BMAL1 were localized in the same cellular compartments (Fig. 4, A and C). The subcellular distributions were classified into each of three categories (Fig. 4, B and D). Expressed individually, CLOCK-HA was found predominantly in the cytoplasm (95% of cells were cytoplasm-dominant, the remainder showing equal staining between cytoplasm and nucleus), whereas BMAL1 showed a higher level of nuclear localization with 57% of cells with equal staining in cytoplasm and nucleus, 8% nucleus-dominant, and with the remainder being 35% cytoplasm-dominant (data not shown). When overexpressed together, the CLOCK localization signal shifted dramatically to a nuclear dominant pattern, and the BMAL1-CLOCK co-staining signal occurring predominantly within the nucleus (Fig. 5, G–H). The higher incidence of nuclear localization found for BMAL1 compared with CLOCK and the shift in CLOCK localization from cytoplasm to nucleus when co-expressed with BMAL1 is consistent with published reports (30, 39). ID2, when ectopically expressed individually, was found predominantly within the cytoplasm (data not shown).
FIGURE 4.
Co-immunofluorescence reveals cytoplasmic localization of ID2 with both CLOCK and BMAL1. HEK293 cells were co-transfected with ID2 and CLOCK-HA or ID2 and BMAL1. Fluorescent dye-labeled secondary antibodies were used to visualize the cellular location of each protein. A, both the localization and co-staining signals of ID2 (green) and CLOCK (red) predominantly occur in the cytoplasm. C, both the localization and co-localization of ID2 (green) and BMAL1 (red) predominantly occurs in the cytoplasm. Cells were visualized using anti-ID2, anti-HA (for CLOCK), and anti-BMAL1 antibodies. Scale bars, 20 μm. B and D, quantification of localization data are mean ± S.E. for three independent experiments each comprising of ≥100 cell counts. The higher proportion of cells showing nuclear localization for BMAL1 compared with CLOCK is as expected from published reports (30).
FIGURE 5.
ID2 restricts CLOCK and BMAL1 nuclear localization. HEK293 cells were co-transfected with either CLOCK-HA and BMAL1 (CB state), or CLOCK-HA, BMAL1, and ID2-FLAG (ICB state). A, CLOCK and BMAL1 localization (green and red, respectively) and co-staining (yellow) when overexpressed together and in the absence of overexpressed ID2. B, CLOCK and BMAL1 localization and co-staining in the presence of overexpressed ID2. Cellular protein localizations were visualized using anti-HA (for CLOCK) and anti-BMAL1 antibodies. Scale bar, 20 μm. C–E, quantification of localization data are mean ± S.E. for three independent experiments each comprising ≥100 cell counts. CB, ectopic expression of CLOCK and BMAL1 plasmids; ICB, ectopic expression of ID2, CLOCK, and BMAL1.
We then examined the localization of proteins when ectopically expressed in pairs, specifically ID2+CLOCK-HA and ID2+BMAL1. In both instances, ID2 was primarily found within the cytoplasm, with mean cell counts for cytoplasm dominant being 90 and 93%, respectively (Fig. 4, B and D). Under these conditions, CLOCK and BMAL1 were also found primarily located in the cytoplasm, CLOCK more so than BMAL1 (Fig. 4, B and D). As a result, co-staining of ID2 with CLOCK, and ID2 with BMAL1, predominated within the cytoplasm: 92% cells showed primarily cytoplasmic staining for ID2 and CLOCK and 95% for ID2 and BMAL1. Similar overlapping patterns of primarily cytoplasmic staining were observed for ID2 with BMAL1 and ID2 with CLOCK-HA in nontransfected and transfected NIH3T3 cells (data not shown).
ID2 Can Sequester CLOCK and BMAL1 to Cytoplasm
As it has been confirmed that ID2 bears a cytoplasmic localization signal/nuclear export signal (NES) (26), we hypothesized that ID2 might bind to CLOCK and BMAL1 and restrict their entry into the nucleus. This would be expected to prevent CLOCK and BMAL1 transcriptional activity. To test this hypothesis, we again used immunofluorescence to stain the location of each protein. We compared a population of transfected HEK293 cells that overexpressed only CLOCK-HA and BMAL1 together (Fig. 5A, CB state) versus cells that overexpressed all three proteins, ID2-FLAG, CLOCK-HA, and BMAL1 together (Fig. 5B, ICB state). As described above, when CLOCK and BMAL1 were overexpressed together, we noted that CLOCK and BMAL1 were found in the nucleus in a majority of cells (Fig. 5E). In the ICB state, a dramatic shift in the localization of CLOCK and BMAL1 to the cytoplasm was observed; in the CB state, CLOCK was found in the nucleus in 78% of cells, dropping to 4% in the ICB state (F6,16 = 53.2, p < 0.001; Fig. 5C). BMAL1 was found predominantly in the nucleus in 36% of the cells in the CB state, compared with 0% when ID2 was introduced (F6,17 = 31.9, p < 0.01; Fig. 5D). Finally, the proportion of cells showing co-localization of CLOCK and BMAL1 in the nucleus dropped from 83% in the CB state to 0% in the ICB state (F6,17 = 274.7, p < 0.001), whereas cytoplasmic co-localization increased dramatically from 1% to 92% (p < 0.001; Fig. 5E). Similar responses were observed with increasing the concentration of ID2 plasmid (3:1:1 and 5:1:1 ratios of ID2:CLOCK:BMAL1). To further explore the mechanism of ID2 sequestration of CLOCK and BMAL1, we examined the contribution of the ID2 HLH domain and the C terminus containing putative NES. Cells were transfected with both CLOCK and BMAL1 and in combination with either full-length ID2, ID2-ΔHLH, or ID2-ΔC (Fig. 3A, supplemental Fig. S2A). Localizations of both CLOCK and BMAL1 were primarily cytoplasmic in the presence of ID2 but nuclear when co-expressed with either ID2-ΔHLH or ID2-ΔC (supplemental Fig. S1). Similar responses were observed with a 5× increased concentration of ID2 mutant plasmid.
We next tested whether NES is required for inhibition of CLOCK-BMAL1 transactivation activity on the mPer1-luc promoter as outlined above. Transfection of NIH3T3 cells with the ID2-ΔC plasmid at a matching concentration did not inhibit CLOCK-BMAL1 activity, but significant inhibition was observed at a five times higher concentration (61% ± 12% inhibition, four independent experiments; p < 0.01; supplemental Fig. S2).
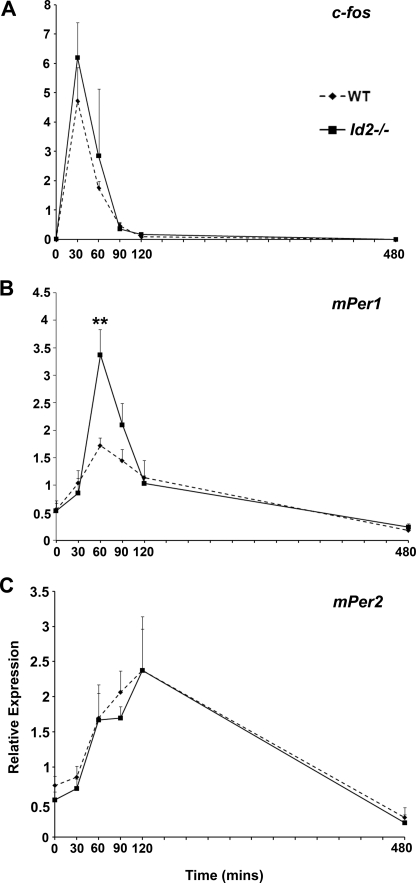
Serum Induction of mPer1 in Mouse Embryonic Fibroblasts Is Elevated in Absence of Id2
Previous data established that ID proteins have the ability to suppress CLOCK-BMAL1 transactivation of target genes including mPer1 and that Id2−/− mice exhibit larger phase shifts in response to light treatment (27). The protein interaction data outlined above illustrates that ID1, ID2, and ID3 can bind to both CLOCK and BMAL1 and that all three proteins can inhibit CLOCK-BMAL1 transactivation activity on the mPer1 promoter (27). Therefore, we sought to determine whether well characterized genes induced during clock resetting are affected by the absence of ID2. Using the in vitro model of serum-stimulated clock resetting of immortalized fibroblasts, we attempted to mimic the gene induction responses associated with phase shifting in the intact animal, such as the SCN response to photic stimuli (1, 5, 6, 16, 42–45). Total RNA was harvested from Id2+/+ and Id2−/− MEFs at 0 min, 30 min, 60 min, 90 min, 120 min and 8 h post-serum stimulation, and gene expression for c-fos, mPer1, and mPer2 determined by qRT-PCR. All three gene induction profiles in both Id2+/+ and Id2−/− cells were as predicted in terms of time of peak induction (30, 60, and 90 min, respectively) and duration of elevation (5, 6, 20, 43–45) (Fig. 6). Examination of c-fos and mPer2 gene induction did not reveal statistically significant differences between genotypes at any time point (Fig. 6, A and C). However, it is noteworthy that in four of five individual trials for c-fos expression showed higher induction in Id2−/− MEFs at 30 min (mean ± S.E. increase for four trials, 70 ± 14%). In only one trial, mPer2 expression in Id2−/− cells was higher at 120 min. In contrast, a significant 96 ± 28% increase in the induction of mPer1 was observed consistently in the Id2−/− cells; five of five individual trials showed higher mPer1 induction in Id2−/− MEFs at 60 min (t1,8 = 3.6, p < 0.01; Fig. 6B).
FIGURE 6.
Serum-induced mPer1 gene expression is elevated in Id2−/− mouse embryonic fibroblasts. Serum stimulation of MEFs was used as a model system to mimic photic stimulation of the SCN. A, relative expression of c-fos in Id2−/− compared with WT (Id2+/+) MEFs shows no significant difference between genotypes. B, mPer1 expression is 2-fold higher in the peak level of gene expression in Id2−/− cells at 60 min post-initiation of serum treatment. C, relative expression of mPer2 shows no significant difference between cells. Values are mean ± S.E. fold change of expression relative to the lowest expression value from five independent experiments. c-fos, mPer1, and mPer2 expression was assessed by qRT-PCR using SYBR green and normalized to GAPDH assessed using Taqman reagents. Two-tailed Student's t test was performed at individual time points to determine statistical significance between WT and Id2−/− samples. **, p value < 0.01.
DISCUSSION
ID proteins have been implicated as repressors of transcription in multiple biological processes. They bind to bHLH proteins and prevent such complexes from contacting their DNA targets because ID proteins themselves lack the basic domain necessary for DNA binding (22–25, 41). Previous studies in our laboratory revealed that Id genes oscillate in a circadian fashion in many tissues. Specifically, Id2 oscillates with a 24-h rhythm in the SCN, fibroblasts, heart, and liver, where it also shows a distinct protein rhythm (6, 27–29). As ID2 is well established as an HLH repressor in other biological processes (22), we sought to examine whether it may play a similar role within the circadian system. The positive elements of the transcriptional-translational feedback loop of the circadian clock, CLOCK and BMAL1, are bHLH transcription factors, and our previous investigations revealed that ID proteins can repress the transactivation potential of the CLOCK-BMAL1 heterodimer (27). The simplest explanation for this repression is that ID2 binds directly to CLOCK, BMAL1, or both and prevents them from contacting the DNA and thereby reducing their transactivation potential. To test this hypothesis, we performed two complementary protein interaction assays.
We examined by co-immunoprecipitation analysis the potential interaction between ID2 and CLOCK and ID2 and BMAL1. We were successful in identifying the presence of CLOCK-HA and BMAL1-HA after the ID2-FLAG pulldown, revealing that ID2 has the capability to interact with the protein complexes that contain CLOCK and BMAL1. The two-hybrid assay demonstrated that all three ID proteins (ID1, ID2, ID3) can interact directly with both CLOCK and BMAL1. As a negative control we examined the ability of each ID protein to interact with the clock protein CRY1, which does not possess an HLH domain. As predicted, no interaction was detected between ID proteins and CRY1, but consistent with other studies, we detected an interaction between BMAL1 and CRY1 (46). It is important to note that the ID2-GAL4-CLOCK-VP16 and ID2-GAL4-BMAL1-VP16 interactions were in the range of 42–86% as robust as the established CLOCK-BMAL1 interaction. This, and the high level of specificity shown by ID proteins for their binding partners in complementary studies (32, 47, 48), lends credence to the concept that the interaction involving ID2 is an important one within the circadian system and not an artifact of the assay. This is significant because it suggests a mechanism through which ID proteins could modulate the circadian system at the level of the core oscillator and could explain the circadian phenotypes reported in the Id2−/− mouse (27).
The ID2 inhibition of CLOCK-BMAL1 transactivation of the mPer1 gene is comparable with our earlier report and is similar to the level of suppression reported for mPER1, an established repressor of the clock (1, 2, 27, 33, 49). Deletion of the HLH domain rendered the ID2 protein ineffective at repressing CLOCK-BMAL1 activity, suggesting that ID2 repression is via an interaction with bHLH proteins. This is not surprising, given that the HLH domain is the area critical for the majority of reported ID2 protein interactions (22–25). In combination with our two-hybrid data, these results suggest that the inhibition of CLOCK-BMAL1 activity is in fact due to a direct action of ID2 binding to CLOCK and BMAL1 via the HLH domain and rendering them inactive.
For ID2 to interact with CLOCK and BMAL1, it is necessary for the proteins to be localized in the same cellular compartment at a particular time. Single protein overexpression revealed, as expected, CLOCK and BMAL1 in both the cytoplasm and nucleus (30, 50), although CLOCK predominantly localized to the cytoplasm. In all experimental conditions (endogenous, ectopically expressed alone, or with other proteins), ID2 was primarily localized to the cytoplasm. When CLOCK or BMAL1 were ectopically co-expressed with ID2, CLOCK and BMAL1 were found primarily in the cytoplasm. This predominantly cytoplasmic localization for ID2 is consistent with studies using GFP-tagged ID2 expressed in various cell lines (26) and immunohistochemical analysis of ID2 in mouse brain cortical and Purkinje neurons.3 In agreement with other studies (30, 50), we found co-staining of BMAL1 and CLOCK predominantly in the nucleus when the two proteins were overexpressed together. This switch, especially for CLOCK from a primarily cytoplasmic location to a primarily nuclear location, is consistent with other studies and consistent with the role of BMAL1 in facilitating entry of CLOCK into the nucleus (30, 39).
Due to this overlapping localization of ID2 with CLOCK and BMAL1 primarily in the cytoplasm, we performed a separate quantification to observe whether an overexpression of ID2 affects the localization of CLOCK and BMAL1. When ID2 was not ectopically expressed, CLOCK and BMAL1 were found together within the nucleus in the majority of the cells counted. However, when ID2 was overexpressed with CLOCK and BMAL1, their co-staining within the nucleus was considerably reduced. The simplest explanation for this finding is that ID2 binds to CLOCK and BMAL1 within the cytoplasm and restricts their nuclear entry. This, in turn, would be expected to reduce the transactivation potential of CLOCK-BMAL1. This form of sequestration has been observed in which ID2 can sequester the bHLH proteins OLIG1 and OLIG2 to the cytoplasm in a mechanism that suppresses the formation of OLIG-E2A complexes (51).
The proposal that cytoplasmic localization by ID2 as a method of restricting CLOCK and BMAL1 nuclear activity is supported by the results of our mutant ID2 experiments. The deletion of the HLH region would be expected to render the ID2-ΔHLH protein incapable of binding to CLOCK or BMAL1, thereby permitting their nuclear accumulation and normal transactivation activity. A small section of the ID2 C-terminal region (between amino acids 103–119) is required for cytoplasmic localization and contains a putative NES (amino acids 106–115) (26). Therefore, the ID2-ΔC protein would be predicted to have limited capability in restricting CLOCK and BMAL1 to the cytoplasm but might still inhibit CLOCK-BMAL1 transactivation. Conversely, the N terminus appears to have limited function in regulating localization. The ID2-ΔN mutant, even though it is additionally missing the C terminus at amino acids 121–134, would still be expected to harbor its NES and could thus remain functional in sequestering CLOCK and BMAL1 to the cytoplasm and inhibit their transactivation activity.
Consistent with the mPer1 promoter reporter results, the nuclear location of both CLOCK and BMAL1 in the presence of ID2-ΔHLH supports the notion that the HLH domain is necessary for interaction with CLOCK and BMAL1. The nuclear location of both CLOCK and BMAL1 in the presence of ID2-ΔC also suggests that the putative ID2 C terminus NES is necessary for sequestration of CLOCK and BMAL1 to the cytoplasm. The mPer1 promoter reporter results that reveal no inhibition of CLOCK-BMAL1 activity at matching doses of ID2-ΔC but inhibition at high doses also indicate that cytoplasmic sequestration contributes to the inhibition of CLOCK-BMAL1 transcriptional activity by ID2.
Our previous data explored the role of ID2 in vivo using a genetic deletion of Id2 and probing of the circadian system (27). Id2−/− mice exhibit enhanced phase shifts and an increased rate of entrainment when subjected to a large delay of the photoschedule. We wanted to explore the molecular basis for this phenotype. Because our current and previous data also demonstrate that overexpression of ID2 with CLOCK and BMAL1 leads to repression of CLOCK-BMAL1 transactivation potential, we hypothesized that the absence of ID2 would result in an increase in CLOCK-BMAL1 activity. We predict that a Zeitgeber induction of mPer1 in the Id2−/− mouse would be elevated. Using MEFs derived from Id2−/− mice, we mimicked the synchronization/phase shift stimulation on these cells. The addition of high levels of serum to fibroblasts is considered to be comparable with photic stimulation of the SCN by activating signal transduction cascades culminating in pCREB activation, induction of the period genes, and generation of phase shifts (5, 6, 17). In serum-stimulated cells, induction profiles of the immediate early genes c-fos, mPer1, and mPer2 in both Id2+/+ and Id2−/− cells were as predicted in terms of time of peak induction and duration of elevation (5, 6, 20, 43–45). However, a significant increase in the induction of mPer1 was observed in the Id2−/− cells. As mPer1 is a state variable of the circadian pacemaker required for normal phase resetting (1, 2, 10, 52), it is possible that an increased induction of PER1 is responsible for the entrainment phenotype described in the intact animal.
These results suggest an important role for ID2 in the circadian clockwork in limiting the magnitude of phase responses and also suggest that under appropriate circumstances, reduction or absence of ID2 could result in higher activity of CLOCK-BMAL1. In turn, this would facilitate a stronger response to photic stimuli via transactivation of the mPer1 gene promoter. A proposed model for the role of ID2 in entrainment of the circadian clock is described in Fig. 7 and is based on an indirect effect of ID2 upon transcriptional activation by the CRE.
FIGURE 7.
Proposed model for the role of ID2 in clock resetting. A, serum stimulation of MEF or light activation of retinal photoreceptors, retinohypothalamic tract (RHT), and in turn, SCN neuron (1, 5, 6, 16, 42). Intracellular signaling pathways are activated culminating in the phosphorylation of CREB on Ser-133 and Ser-142 and permitting CREB binding to its response element, CRE, in the promoter region of the immediate early gene/clock gene mPer1. This promotes transcription of the target gene (12, 13, 61). The CRE and E-box elements are located in the promoter regions of the mPer1 gene. CLOCK-BMAL1 heterodimer binding to the E-box element within the gene promoter is considered responsible for rhythmic transcriptional activity in various clock and clock-controlled genes (12, 13). Studies in the Clock Δ19 homozygous mutant mouse suggest an additional role of this binding: that CLOCK-BMAL1 binding to the E-box element is permissive to, or gates, transcriptional activation by the CRE within the same promoter region (19, 20). Target gene (mPer1 and mPer2) induction by activation of the CRE is reduced in the Clock (Clk/Clk) mutant mouse SCN and MEFs following light and serum stimulation, respectively. mPer1 is a state variable of the clock and its induction or suppression is deemed critical to phase shifting of the clock (1, 2, 10, 42). B, in the wild type cell, the presence of ID2 interferes with the dimerization of CLOCK-BMAL1 by binding to both canonical clock proteins. ID2-CLOCK and ID2-BMAL1 cannot bind to the E-box element because ID2 lacks a basic DNA binding domain. The quantity of the available CLOCK-BMAL1 heterodimer is reduced, leading to reduced transcription of the target gene mPer1. C, in the Id2−/− cell, the number of CLOCK-BMAL1 heterodimers is increased due to the absence of ID2, thereby increasing the occurrence of binding of CLOCK-BMAL1 to the E-box element and permitting maximal activation of the promoter when there is coincidental binding of pCREB to the CRE. This results in enhanced transcription of the target gene mPer1. As mPer1 is a state variable of the clock, a larger phase shift of the clock is produced (27). Additionally, a direct role for CLOCK in signaling to the mPer1 gene has also been proposed independent of the pCREB pathway, in which Ca2+ -dependent protein kinase C phosphorylation of CLOCK can regulate mPer1 induction (62). Sequestration of CLOCK to the cytoplasm and reduction in the quantities of available CLOCK-BMAL1 heterodimer by ID2 could modulate this proposed signaling pathway.
The action of ID2 upon CLOCK-BMAL1 may even explain some aspects of clock output regulation observed in the Id2−/− liver (29). In the liver, both Id2 gene and protein are rhythmic with a protein peak phase of approximately circadian time 12 (29), coincident with the proposed peak of BMAL1-CLOCK heterodimer activity (53, 54). Although a majority of CCGs that are abnormally regulated in the Id2−/− liver are different from those identified in the Clk/Clk mutant mouse (29, 53, 55, 56), there is some overlap in the CCGs misregulated by both (29). This suggests that an ID2 interaction with CLOCK and BMAL1 could contribute to some of the regulation of clock output driven specifically by CLOCK-BMAL1.
If we consider that all four Id genes are rhythmically expressed in the SCN and share peak phases in the late subjective night (27), it is possible that a combinatorial effect of multiple ID proteins could confer a strong repressive effect upon CLOCK-BMAL1 heterodimer availability. A knockdown of all four Id genes might therefore be predicted to have a more profound effect upon the circadian system than what has been observed in the Id2−/− mouse.
These data implicate ID2 as a modulator of the circadian system. We propose that the ID proteins might also act in a similar manner as the bHLH orange proteins, other transcriptional repressors of bHLH activity that include BMAL1 as a target (7–9, 57). The action of ID2 describes an autoregulatory feedback loop that closes outside the core oscillator and that affects aspects of circadian timing in a similar manner as described for Neurospora VIVID (58). This includes photic entrainment and clock output, acting as a repressor on the positive elements of the clock, and yet is not required for circadian rhythmicity. There are other examples of additional autoregulatory loops that contribute to the circadian system but are not essential for the persistence of the oscillator (CREB, cAMP responsive element modulator, cAMP, Dec1, Dec2, clockwork orange, and frequency antisense transcript) (2, 7–9, 57, 59). It is hypothesized that the occurrence of multiple interlocked feedback loops provides stability and precision for the clock and to which ID2 could additionally contribute.
Our previous studies implicated ID2 in clock resetting and entrainment and in the regulation of clock output. In summary of our current study, the HLH transcriptional repressor ID2 can interact directly with the canonical clock proteins CLOCK and BMAL1 and can sequester CLOCK and BMAL1 to the cytoplasm; and the absence of Id2 results in an increase in serum-induced mPer1, leading to a hypothesized molecular correlate for the enhanced phase shift response observed in vivo. This suggests that ID2 may be an important modulator of the mammalian circadian clock, regulating aspects of both clock input and output and that this could be achieved in part through interaction with canonical components of the oscillator.
Supplementary Material
Acknowledgments
We thank M. P. Antoch, H. Axelson, M. A. Israel, R. W. Lim, J. B. Hogenesch, and J. E. Baggs for generous provision of plasmids, N. P. Watson and M. A. Israel for help in generating MEF cell lines, and members of the K.T. Vaughan lab for assistance with immunofluorescence analysis.
This work was supported, in whole or in part, by NIGMS, National Institutes of Health Grant GM087508 (to G. E. D.). This work was also supported by a grant from the University of Notre Dame (to G. E. D.), a University Research fellowship and grant from the Royal Society (to G. E. D.), a University of Notre Dame Fisher Research fellowship (to S. M. W.), and National Science Foundation Research Experience for Undergraduate (REU) Summer Research Fellowship Award 0453325 (to S. J. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
G. E. Duffield, unpublished data.
- SCN
- suprachiasmatic nucleus
- ANOVA
- analysis of variance
- bHLH
- basic helix-loop-helix
- NES
- nuclear export signal
- CREB
- cAMP response element binding protein
- CCG
- clock-controlled gene
- CRE
- cAMP-response element
- qRT-PCR
- quantitative RT-PCR
- MEF
- mouse embryonic fibroblast
- CB
- ectopic expression of CLOCK and BMAL1
- CBI
- ectopic expression of CLOCK, BMAL1 and ID2.
REFERENCES
- 1.Reppert S. M., Weaver D. R. (2001) Annu. Rev. Physiol. 63, 647–676 [DOI] [PubMed] [Google Scholar]
- 2.Dunlap J. C., Loros J. J., DeCoursey P. J. (2004) Chronobiology: Biological Timekeeping, Sinauer Associates, Sunderland, MA [Google Scholar]
- 3.Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peirson S. N., Butler J. N., Duffield G. E., Takher S., Sharma P., Foster R. G. (2006) Biochem. Biophys. Res. Commun. 351, 800–807 [DOI] [PubMed] [Google Scholar]
- 5.Balsalobre A., Damiola F., Schibler U. (1998) Cell 93, 929–937 [DOI] [PubMed] [Google Scholar]
- 6.Duffield G. E., Best J. D., Meurers B. H., Bittner A., Loros J. J., Dunlap J. C. (2002) Curr. Biol. 12, 551–557 [DOI] [PubMed] [Google Scholar]
- 7.Grechez-Cassiau A., Panda S., Lacoche S., Teboul M., Azmi S., Laudet V., Hogenesch J. B., Taneja R., Delaunay F. (2004) J. Biol. Chem. 279, 1141–1150 [DOI] [PubMed] [Google Scholar]
- 8.Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. (2002) Nature 419, 841–844 [DOI] [PubMed] [Google Scholar]
- 9.Rossner M. J., Oster H., Wichert S. P., Reinecke L., Wehr M. C., Reinecke J., Eichele G., Taneja R., Nave K. A. (2008) PLoS One 3, e2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama M., Kouzu Y., Takahashi S., Wakamatsu H., Moriya T., Maetani M., Watanabe S., Tei H., Sakaki Y., Shibata S. (1999) J. Neurosci. 19, 1115–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maywood E. S., Mrosovsky N., Field M. D., Hastings M. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 15211–15216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7728–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tischkau S. A., Mitchell J. W., Tyan S. H., Buchanan G. F., Gillette M. U. (2003) J. Biol. Chem. 278, 718–723 [DOI] [PubMed] [Google Scholar]
- 14.von Gall C., Duffield G. E., Hastings M. H., Kopp M. D., Dehghani F., Korf H. W., Stehle J. H. (1998) J. Neurosci. 18, 10389–10397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNulty S., Schurov I. L., Sloper P. J., Hastings M. H. (1998) Eur. J. Neurosci. 10, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 16.Balsalobre A., Marcacci L., Schibler U. (2000) Curr. Biol. 10, 1291–1294 [DOI] [PubMed] [Google Scholar]
- 17.Balsalobre A., Brown S. A., Marcacci L., Tronche F., Kellendonk C., Reichardt H. M., Schütz G., Schibler U. (2000) Science 289, 2344–2347 [DOI] [PubMed] [Google Scholar]
- 18.Hirota T., Okano T., Kokame K., Shirotani-Ikejima H., Miyata T., Fukada Y. (2002) J. Biol. Chem. 277, 44244–44251 [DOI] [PubMed] [Google Scholar]
- 19.Jung H., Choe Y., Kim H., Park N., Son G. H., Khang I., Kim K. (2003) Neuroreport 14, 15–19 [DOI] [PubMed] [Google Scholar]
- 20.Shearman L. P., Weaver D. R. (1999) Neuroreport 10, 613–618 [DOI] [PubMed] [Google Scholar]
- 21.Duffield G. E. (2003) J. Neuroendocrinol. 15, 991–1002 [DOI] [PubMed] [Google Scholar]
- 22.Ruzinova M. B., Benezra R. (2003) Trends Cell Biol. 13, 410–418 [DOI] [PubMed] [Google Scholar]
- 23.Andres-Barquin P. J., Hernandez M. C., Israel M. A. (2000) Histol. Histopathol. 15, 603–618 [DOI] [PubMed] [Google Scholar]
- 24.Norton J. D. (2000) J. Cell Sci. 113, 3897–3905 [DOI] [PubMed] [Google Scholar]
- 25.Yokota Y. (2001) Oncogene 20, 8290–8298 [DOI] [PubMed] [Google Scholar]
- 26.Kurooka H., Yokota Y. (2005) J. Biol. Chem. 280, 4313–4320 [DOI] [PubMed] [Google Scholar]
- 27.Duffield G. E., Watson N. P., Mantani A., Peirson S. N., Robles-Murguia M., Loros J. J., Israel M. A., Dunlap J. C. (2009) Curr. Biol. 19, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Iino M., Shigeyoshi Y., Hashimoto S. (2002) Nature 418, 534–539 [DOI] [PubMed] [Google Scholar]
- 29.Hou T. Y., Ward S. M., Murad J. M., Watson N. P., Israel M. A., Duffield G. E. (2009) J. Biol. Chem. 284, 31735–31745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondratov R. V., Chernov M. V., Kondratova A. A., Gorbacheva V. Y., Gudkov A. V., Antoch M. P. (2003) Genes Dev. 17, 1921–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amelio A. L., Miraglia L. J., Conkright J. J., Mercer B. A., Batalov S., Cavett V., Orth A. P., Busby J., Hogenesch J. B., Conkright M. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20314–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jögi A., Persson P., Grynfeld A., Påhlman S., Axelson H. (2002) J. Biol. Chem. 277, 9118–9126 [DOI] [PubMed] [Google Scholar]
- 33.Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. (1999) Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 34.Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 35.Florio M., Hernandez M. C., Yang H., Shu H. K., Cleveland J. L., Israel M. A. (1998) Mol. Cell. Biol. 18, 5435–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iavarone A., Garg P., Lasorella A., Hsu J., Israel M. A. (1994) Genes Dev. 8, 1270–1284 [DOI] [PubMed] [Google Scholar]
- 37.Chen B., Han B. H., Sun X. H., Lim R. W. (1997) Nucleic Acids Res. 25, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirayama J., Nakamura H., Ishikawa T., Kobayashi Y., Todo T. (2003) J. Biol. Chem. 278, 35620–35628 [DOI] [PubMed] [Google Scholar]
- 39.Kwon I., Lee J., Chang S. H., Jung N. C., Lee B. J., Son G. H., Kim K., Lee K. H. (2006) Mol. Cell. Biol. 26, 7318–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peirson S. N., Butler J. N., Foster R. G. (2003) Nucleic Acids Res. 31, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benezra R., Davis R. L., Lassar A., Tapscott S., Thayer M., Lockshon D., Weintraub H. (1990) Ann. N.Y. Acad. Sci. 599, 1–11 [DOI] [PubMed] [Google Scholar]
- 42.Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 43.Albrecht U., Sun Z. S., Eichele G., Lee C. C. (1997) Cell 91, 1055–1064 [DOI] [PubMed] [Google Scholar]
- 44.Shigeyoshi Y., Taguchi K., Yamamoto S., Takekida S., Yan L., Tei H., Moriya T., Shibata S., Loros J. J., Dunlap J. C., Okamura H. (1997) Cell 91, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 45.Takumi T., Matsubara C., Shigeyoshi Y., Taguchi K., Yagita K., Maebayashi Y., Sakakida Y., Okumura K., Takashima N., Okamura H. (1998) Genes Cells 3, 167–176 [DOI] [PubMed] [Google Scholar]
- 46.Langmesser S., Tallone T., Bordon A., Rusconi S., Albrecht U. (2008) BMC Mol. Biol. 9, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun X. H., Copeland N. G., Jenkins N. A., Baltimore D. (1991) Mol. Cell. Biol. 11, 5603–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langlands K., Yin X., Anand G., Prochownik E. V. (1997) J. Biol. Chem. 272, 19785–19793 [DOI] [PubMed] [Google Scholar]
- 49.Sangoram A. M., Saez L., Antoch M. P., Gekakis N., Staknis D., Whiteley A., Fruechte E. M., Vitaterna M. H., Shimomura K., King D. P., Young M. W., Weitz C. J., Takahashi J. S. (1998) Neuron 21, 1101–1113 [DOI] [PubMed] [Google Scholar]
- 50.Tamaru T., Isojima Y., van der Horst G. T., Takei K., Nagai K., Takamatsu K. (2003) Genes Cells 8, 973–983 [DOI] [PubMed] [Google Scholar]
- 51.Samanta J., Kessler J. A. (2004) Development 131, 4131–4142 [DOI] [PubMed] [Google Scholar]
- 52.Masubuchi S., Kataoka N., Sassone-Corsi P., Okamura H. (2005) Journal of Neuroscience 25, 4719–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller B. H., McDearmon E. L., Panda S., Hayes K. R., Zhang J., Andrews J. L., Antoch M. P., Walker J. R., Esser K. A., Hogenesch J. B., Takahashi J. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M. (2001) Cell 107, 855–867 [DOI] [PubMed] [Google Scholar]
- 55.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. (2002) Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 56.Oishi K., Miyazaki K., Kadota K., Kikuno R., Nagase T., Atsumi G., Ohkura N., Azama T., Mesaki M., Yukimasa S., Kobayashi H., Iitaka C., Umehara T., Horikoshi M., Kudo T., Shimizu Y., Yano M., Monden M., Machida K., Matsuda J., Horie S., Todo T., Ishida N. (2003) J. Biol. Chem. 278, 41519–41527 [DOI] [PubMed] [Google Scholar]
- 57.Lim C., Chung B. Y., Pitman J. L., McGill J. J., Pradhan S., Lee J., Keegan K. P., Choe J., Allada R. (2007) Curr. Biol. 17, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heintzen C., Loros J. J., Dunlap J. C. (2001) Cell 104, 453–464 [DOI] [PubMed] [Google Scholar]
- 59.O'Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H. (2008) Science 320, 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finkel T., Duc J., Fearon E. R., Dang C. V., Tomaselli G. F. (1993) J. Biol. Chem. 268, 5–8 [PubMed] [Google Scholar]
- 61.Gau D., Lemberger T., von Gall C., Kretz O., Le Minh N., Gass P., Schmid W., Schibler U., Korf H. W., Schütz G. (2002) Neuron 34, 245–253 [DOI] [PubMed] [Google Scholar]
- 62.Shim H. S., Kim H., Lee J., Son G. H., Cho S., Oh T. H., Kang S. H., Seen D. S., Lee K. H., Kim K. (2007) EMBO Rep. 8, 366–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.