Abstract
The A2A and A2B adenosine receptors (A2AR and A2BR) are implicated in many physiological processes. However, the mechanisms of their intracellular maturation and trafficking are poorly understood. In comparative studies of A2AR versus A2BR expression in transfected cells, we noticed that the levels of cell surface expression of A2BR were significantly lower than those of A2AR. A large portion of the A2BR was degraded by the proteasome. Studies of cell surface expression of A2BR chimeric molecules in transfectants suggested that A2BR does not have the dominant forward transport signal for export from the endoplasmic reticulum to the cell surface. A2BR surface expression was increased in A2BR chimeras where the A2BR carboxyl terminus (CT) was replaced or fused with the A2AR CT. Co-transfection of A2AR with A2BR enhanced surface expression of A2BR though the F(X)6LL motif in the A2AR CT. The requirements of A2AR expression for better A2BR cell surface expression was not only established in transfectants but also confirmed by observations of much lower levels of A2BR-induced intracellular cAMP accumulation in response to A2BR-activating ligand in splenocytes from A2AR−/− mice than in wild type mice. The results of mechanistic studies suggested that poor A2BR expression at the cell surface might be accounted for mainly by the lack of a dominant forward transport signal from the endoplasmic reticulum to the plasma membrane; it is likely that A2BR forms a hetero-oligomer complex for better function.
Keywords: Endoplasmic Reticulum (ER), Intracellular Trafficking, Plasma Membrane, Proteasome, Protein Degradation
Introduction
Newly synthesized proteins destined for secretory pathway have to translocate to the endoplasmic reticulum (ER)2 for correct folding, assembly, and transport to the final destination site of action. It is believed that the newly synthesized proteins are monitored by an ER quality control system to avoid accumulation of incorrectly unfolded and unassembled proteins that may disrupt proper cellular processes (1–3).
Adenosine receptors are members of G protein-coupled receptors (GPCRs). Four adenosine receptors, A1, A2A, A2B, and A3, have been cloned and characterized from several mammalian species. A2AR and A2BR interact preferably with members of the Gs family of G proteins and the A1 and A3 receptors with Gi/o proteins (4). Although adenosine receptors have been studied intensively with respect to their physiology and pharmacology (5–8), their cell biological properties such as subunit assembly, dimerization, and trafficking remain poorly understood (9–12). A better understanding of A2AR and A2BR cell biology has became especially important because their pharmacological targeting is considered in treatments of acute inflammation, wound healing, and cancer (6, 13).
Previous studies have shown that a substantial portion of glycoprotein hormone receptors in heterologous expression systems and natural tissues exist in immature forms (14–17) and contain high mannose-type N-linked glycans, which are typical for proteins located in the ER (18). This indicates that these receptors might mature inefficiently, a phenomenon that also has been found to characterize some GPCRs (19–28). For example, ∼50% of opioid receptors are retained in the ER and are targeted for ER-associated degradation (ERAD) in proteasomes (22, 23), a pathway that has subsequently been shown also to dispose other GPCRs, both the wild type and mutant forms (24–28). Is this or a similar pathway involved in the regulation of adenosine receptors? These issues have not been explored in detail in previous studies of A2BR; the data reported here have been motivated by the hypothesis that inefficient processing and maturation might be a more common property among GPCRs than thought previously. Indeed, although the A2BR was found to be retained in the intracellular compartment (29), the molecular mechanisms that govern A2BR retention have remained a mystery. Therefore, to understand the intracellular trafficking and degradation mechanisms of A2BR, we studied whether A2BR are prone to premature ERAD.
The results presented here demonstrate that a substantial portion of the newly synthesized A2BR is retained in the ER and are eventually targeted for degradation by the proteasomes. Furthermore, inhibition of proteasome activity was not sufficient to enhance maturation and cell surface expression of the receptor. Instead, it was the A2AR that was able to promote the cell surface expression of the A2BR.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
MG132 was purchased from EMD Chemicals (San Diego, CA). Lactacystin, A2AR-selective agonist CGS21680, and nonselective agonist 5-N-ethyl-carboxamidoadenosine (NECA) were purchased from Tocris (Ellisville, MO). Anti-hemagglutinin (HA) monoclonal antibody HA.11 was from Covance (Richmond, CA). Anti-Myc monoclonal antibody and Alexa Fluor 488- and 594-conjugated goat anti-rat and anti-mouse antibodies were from Invitrogen. Rat anti-HA monoclonal antibody and 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid (ABTS) tablets were purchased from Roche Applied Science. Mouse anti-calnexin monoclonal antibody was from BD Biosciences (San Jose, CA). Mouse anti-γ subunit of AP-1, mouse anti-FLAG M2 monoclonal antibodies, rabbit polyclonal anti-FLAG antibody, and rabbit polyclonal anti-Myc were purchased from Sigma. Mouse anti-sodium/potassium ATPase monoclonal antibody was purchased from Abcam (Cambridge, MA). Endoglycosidase H and PNGase F were purchased from New England Biolabs (Ipswich, MA). All other reagents were purchased from Sigma or from sources described previously (30, 32).
DNA Recombinant Procedures
Construction of Tagged A2AR, A2BR, and β2AR
A triple HA or triple Myc tag was introduced by a two-step PCR procedure (31) into the XhoI-EcoRI sites of pCI-neo vector (Promega). A triple FLAG tag was introduced by PCR using p3XFLAG-CMV-10 (Sigma) as a template into the XhoI-EcoRI sites of pCI-neo vector (Promega). Mouse cDNAs cording of full-length A2AR and A2BR were generated by pfu PCR amplification with mouse lung Marathon-Ready cDNA (Clontech) using sense (GAATTCATGGGCTCCTCGGTGTACAT) and antisense (GCGGCCGCTCAGGAAGGGGCAAACTCTG) primers for A2AR and sense (GAATTCATGCAGCTAGAGACGCAAGA) and antisense (GCGGCCGCTCATAAGCCCAGACTGAGAGTAG) primers for A2BR. Underlining of the sense and antisense primers for both A2AR and A2BR represents the EcoRI and NotI sites, respectively. The PCR products were digested with EcoRI and NotI, purified, and ligated into the triple HA- or triple Myc-tagged pCI-neo vector as described above. Human cDNA cording of full-length β2AR was generated by pfu PCR amplification with pCMV6-XL5-ADRB2 (OriGene Technologies, Rockville, MD) as a template using sense (GAATTCATGGGGCAACCCGGGAACGGCAGCGCC) and antisense (GCGGCCGCTTACAGCAGTGAGTCATTTGTACTACAATTCC) primers. Underlining of sense and antisense primers represents the EcoRI and NotI sites, respectively. The PCR products were digested with EcoRI and NotI, purified, and ligated into the triple HA-tagged pCI-neo vector as described above. Truncation mutants for A2AR, A2BR, and β2AR were created by introducing stop codon with PCR-based mutagenesis method (QuikChange site-directed mutagenesis kit, Stratagene, La Jolla, CA).
Construction of Chimeric A2AR, A2BR, and β2AR
To replace the region encoding the CT of the A2AR with A2BR, the BamHI site was introduced at Asn294 of A2BR by mutagenesis, and the BamHI-NotI fragment of A2AR (corresponding to Ile287 to terminator411) was replaced to create pCI-neoHA-A2AR-BRCT and pCI-neoHA-A2BR-ARCT. To replace the region encoding the CT of the A2AR with A2BR, the stop codon of A2BR was deleted and replaced by the BamHI site by mutagenesis to create pCI-neoHA-A2BR-ΔtgaBamHI. The BamHI-NotI fragment of A2AR was ligated to pCI-neoHA-A2BR-ΔtgaBamHI to create pCI-neoHA-A2BR-fARCT. To replace the region encoding the CT of the β2AR with A2BR, the BamHI site was introduced to β2AR cDNA at position 344, and then the BamHI-NotI fragment of β2AR was replaced to create pCI-neoHA-A2B-β2CT. To replace the region encoding the CT of the β2AR with A2BR, the BamHI-NotI fragment of β2AR was ligated into pCI-neoHA-A2BR-ΔtgaBamHI to create pCI-neoHA-A2BR-fβ2CT. The accuracy of the cDNA sequences for all constructs was verified by DNA sequencing.
Cells and DNA Transfection
The human embryonic kidney cell line, AD-293 cells (Stratagene, La Jolla, CA), was transfected with either HA-, Myc-, or FLAG-tagged adenosine receptors or HA- or FLAG-tagged β2AR constructs using the FuGENE 6 reagent (Roche Applied Science). For stable cell line, transfected cells were selected with G418 (1 mg/ml) (Invitrogen) starting 48 h after transfection in DMEM and 10% fetal bovine serum. Twenty days later, the cells surviving selection were sorted by immunofluorescence for adenosine receptor expression and subsequently maintained in G418.
Biochemical Procedures
Preparation of detergent extracts, immunoprecipitation, electrophoresis, and immunoblotting were performed as described (30, 32). Briefly, 2 × 105 AD-293 cells were transiently transfected with HA-A2BR in 6-well plates with FuGENE 6. After 72 h of transfection, the cell were washed in phosphate-buffered saline (PBS) and lysed on ice in CHAPS lysis buffer (1% CHAPS, 150 mm NaCl, 50 mm Tris, 5 mm EDTA, and Complete EDTA-free protease inhibitor mixture (Roche Applied Science)). The CHAPS-soluble supernatant was prepared by centrifugation at 21,000 × g for 20 min at 4 °C, precleared with 20 μl of protein G-Sepharose bead slurry (50%, v/v) for 1 h at 4 °C, and then immunoprecipitated with rat mAb to HA at a concentration of 0.5 μg/ml for 16 h. The immunocomplexes were pulled down with protein G-Sepharose beads and washed three times in ice-cold lysis buffer, once in ice-cold buffer containing 0.1% CHAPS, 150 mm NaCl, 50 mm Tris, and 5 mm EDTA, and once in ice-cold PBS. After the addition of gel loading buffer with 0.1 m DTT, samples were incubated at 65 °C for 10 min and loaded on 4–20% Tris-glycine SDS-PAGE followed by immunoblotting with mouse mAb to HA at 1:2000 dilution. Endoglycosidase H and N-glycanase treatments of immunoprecipitated samples were done according to the manufacturer's instructions. Densitometry was performed on scanned immunoblot and fluorography images using the ImageJ gel analysis tool (33).
Quantification of Transfected Adenosine Receptors by Enzyme-linked Immunoassays
5 × 105 Cells were transiently transfected in 6-well plates with FuGENE 6 with DNA at a ratio of 3:2. Half of the cells were placed onto 6-well plates, and the rest were further divided into six aliquots and placed onto 96-well poly-d-lysine-coated plates (BD Biosciences) after 48 h of transfection. The cells were further cultured for 24 h to allow their attachment to the plates. To quantify HA- or FLAG-tagged A2AR, A2BR, and β2AR expression at the cell surface, cells were fixed with 4% paraformaldehyde for 15 min at room temperature, washed three times with PBS, and incubated with PBS containing 1% bovine serum albumin for 1 h and then with the same buffer containing mouse mAb to HA or FLAG at a 1:1000 dilution for 1 h as described previously (34). Empty vector-transfected cells were used as a control. Cells were washed four times with PBS, and peroxidase activity was measured by incubating the cells for 1 h at 37 °C with water-soluble substrate (ABTS, 1 mg/ml) following the manufacturer's instructions. The oxidized product was detected by reading its absorbance at 405 nm using Varioskan (Thermo Scientific). To verify the total expression of HA- or FLAG-tagged A2AR, A2BR, and β2AR, cells on the 6-well plates were washed, lysed, and immunoprecipitated with rat mAb to HA followed by immunoblotting as described above. β-Actin from 5% of each of the total cell lysates was monitored simultaneously by immunoblotting using mouse anti β-actin antibody (Sigma) as a loading control. In transient expression assays (except for Fig. 1), the band intensity of total receptor expression was measured by ImageJ software, and cell surface expression level was adjusted based on the total level of individual receptor expression.
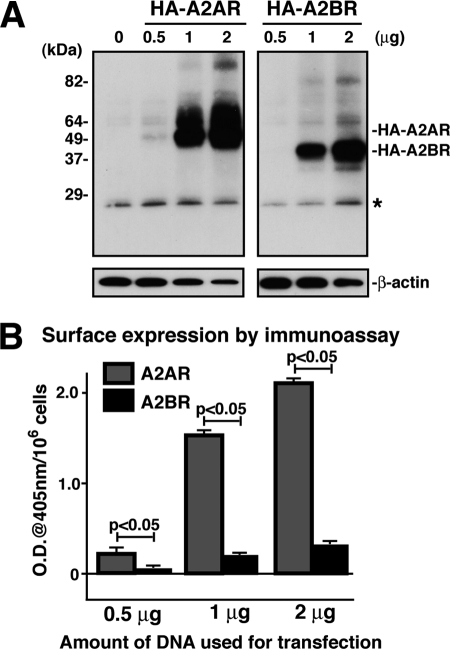
FIGURE 1.
Both the total and cell surface expression levels of A2BR are lower than those of A2AR. A, AD-293 cells were transfected with HA-A2AR or HA-A2BR with different amounts of DNA (0.5–2 μg), lysed in 1% CHAPS, and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with β-actin monoclonal antibody as a loading control. Shown is a representative of three independent experiments. The asterisk indicates IgG light chain. B, AD-293 cells were transfected with HA-A2AR or HA-A2BR as in A and plated onto 96-well plates. After a 72-h transfection, cells were fixed with 4% paraformaldehyde and incubated with mouse mAb to HA followed by goat anti-mouse HRP-conjugated antibody. Cells were washed extensively with PBS, and receptors at the cell surface were determined by enzyme-linked immunoassay with ABTS solution (1 mg/ml). OD at 405 nm was measured and expressed as means (OD/106 cells) ± S.D. (n = 6).
Metabolic Labeling
1 × 106 AD-293 cells expressing HA-A2AR or HA-A2BR were starved for 30 min in methionine/cysteine-free DMEM (Invitrogen) supplemented with 1% l-glutamine and pulsed for 15 min with 100 μCi/ml EasyTagTM EXPRESS35S protein labeling mix (PerkinElmer Life Sciences). The culture medium was then replaced with complete medium and chased for the indicated times. At each time point, cells were washed with PBS and lysed in ice-cold lysis buffer (1% CHAPS, 150 mm NaCl, 50 mm Tris, 5 mm EDTA, and Complete EDTA-free protease inhibitor mixture (Roche Applied Science)). Cell debris was removed by centrifugation, and the lysates were precleared with 20 μl of protein G-Sepharose bead slurry (50%, v/v) for 1 h at 4 °C. The supernatants were incubated overnight at 4 °C with mouse mAb to HA (0.5 μg/ml). Immunocomplexes were pulled down with protein G-Sepharose beads and washed three times in ice-cold lysis buffer, once in ice-cold buffer containing 0.1% CHAPS, 150 mm NaCl, 50 mm Tris, and 5 mm EDTA, and once in ice-cold PBS. After the addition of gel loading buffer with 0.1 m DTT, samples were incubated at 65 °C for 10 min and loaded onto 4–20% Tris-glycine SDS-PAGE followed by fluorography.
Immunofluorescence
AD-293 cells expressing HA-A2AR or HA-A2BR were grown on poly-d-lysine-coated coverslips (BD Biosciences), fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, and permeabilized with 0.1% saponin or 0.2% Triton X-100. Adenosine receptors and organelle marker proteins were visualized using rat mAb to HA and mouse mAbs against organelle markers followed by incubation with secondary antibody coupled to Alexa-488 or Alexa-594 (Molecular Probes). Images were taken with a Keck three-dimensional fusion microscope, developed in the CenSSIS Laboratory at Northeastern University, or a Leica TCS SP2 AOBS confocal microscope at the Tufts University/New England Medical Center Imaging Facility.
Studies of Functional A2AR and A2BR Expression ex Vivo Splenocytes
As the naive, nonactivated immune cells express only a little A2BR, the splenocytes were activated under conditions that induce activation of H-2b anti-H-2d allogeneic T cells, with the highest levels of A2AR and A2BR expression determined to be after 3–4 days of mixed lymphocyte culture. This allowed us to observe differences between A2BR-induced cAMP expression in cells from wild type and A2AR-deficient mice. cAMP assay was performed as described previously (13). In these assays the splenocytes from wild type mice were compared with splenocytes from A2AR or A2BR knock-out mice after incubation with the A2AR-selective agonist CGS21680 (1 μm) or with nonselective agonist NECA (1 or 10 μm), which triggered cAMP production by both A2AR and A2BR.
Animals
C57BL/6 background A2AR−/− mice were described previously (13). C57BL/6 background A2BR−/− and age-matched control (A2BR+/+) mice were obtained from parallel breeding in a colony of A2BR−/− mice. A2BR−/− mice were backcrossed 11 times to C57BL/6 mice (13). In the control experiment, no mixed lymphocyte response was detected after 6 days of cell culture with A2BR−/− spleen cells as responders and wild type C57BL/6 spleen cells as stimulators or with C57BL/6 as responders and A2BR−/− as stimulators. Wild type C57BL/6 and BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA). The experimental protocol was approved by the animal experimental committee of Northeastern University.
Statistical Analysis
Data are expressed as means ± S.D. (n = 6) (with n = number of cell culture). Differences between groups were evaluated using Student's t test. A p value of <0.05 was considered statistically significant.
RESULTS
A2BR Is Retained in the ER and Degraded
First, we compared the cell surface expression of A2BR with A2AR, the closest family member of GPCR for A2BR (4). For this purpose, we constructed mouse A2BR and A2AR in a CMV promoter-based expression vector by tagging HA at the amino terminus to take advantage of labeling the cell surface receptor without detergent. AD-293 cells were then transiently transfected with different amount of plasmid. The total protein expression levels of A2BR and A2AR were studied by immunoprecipitation followed by immunoblotting, whereas cell surface expression was determined by enzyme-linked immunoassay as described (34). To ensure that an equal amount of lysate was used for immunoprecipitation, we always monitored the β-actin level by immunoblotting in 5% of total lysate. As shown in Fig. 1A, total expression of both CHAPS-solibilized A2BR and A2AR depends on the amount of DNA (0.5–2 μg), but the A2BR total expression level was consistently lower than that of A2AR (∼40%). Cell surface expression of A2BR depends on its total expression level; however, the cell surface expression level of A2BR was significantly lower than that of A2AR (Fig. 1B). We were not able to further increase cell surface expression of A2BR by simply increasing the amount of DNA used for transfection (up to 6 μg), because it actually always resulted in even lower A2BR expression than when using 2 μg of DNA, as judged by immunoprecipitation followed by immunoblotting (data not shown). This is probably because of the toxicity of the transfection reagent and/or because the “overexpression” could promote more aggressive degradation of transfected protein. Observations of lower levels of A2BR expression have been made not only using AD-293 cells or the HA tag, as the same outcomes were also observed in experiments with transfected COS-7 cells or transfected Myc-tagged A2BR and A2AR (data not shown). Thus, although cell surface expression of A2BR depends on its total cellular expression, both the total expression and surface expression of A2BR were consistently and significantly lower than those of A2AR.
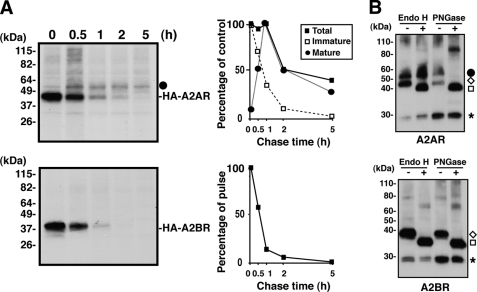
To test whether a lower expression level of A2BR in steady state reflects its degradation, we established stable AD-293 cell lines that express HA-A2BR or HA-A2AR and performed pulse-chase analysis. The cells were starved for 30 min in DMEM without methionine and cysteine and then pulse-labeled with [35S]methionine/cysteine for 15 min and chased up to 5 h in complete medium. At different time points, cells were washed in PBS, lysed in buffer containing 1% CHAPS and protease inhibitors mixture, and immunoprecipitated with mouse anti-HA antibody followed by fluorography. As shown in Fig. 2A, A2BR was initially synthesized as a ∼39-kDa band, which was sensitive to both endoglycosidase H and N-glycanase treatment (Fig. 2B), indicating that this band remained in an immature form. This band virtually disappeared within 2 h. In contrast, the A2AR was initially synthesized as a ∼42-kDa band and then processed to a mature ∼54-kDa band, because this form was resistant to endoglycosidase H treatment (Fig. 2, A and B). The ∼42-kDa immature form gradually disappeared along with the chase time. On the contrary, the ∼54-kDa mature form peaked at 1 h of chase and declined at later times. Although ∼40% of the total A2AR was degraded in 5 h, A2BR was degraded much faster than A2AR, and no mature form was observed. In agreement with these results, the subcellular localization of A2BR was confirmed by immunofluorescence using known markers of intracellular location, predominantly in the ER, as A2BR co-localized well with the ER protein calnexin (Fig. 3). A2BR was poorly co-localized with the γ subunit of AP-1, the trans-Golgi network marker, and with another plasma membrane protein, sodium/potassium ATPase (35), suggesting that a majority of the A2BR was retained in the ER. These observations of A2BR localization were in marked contrast with the cell surface expression of A2AR, which co-localized well with sodium/potassium ATPase (Fig. 3), providing another evidence of inefficient transport of A2BR to the plasma membrane. Interestingly, A2AR was not only localized in the plasma membrane but was also localized intracellularly, as reflected by the co-localization of A2AR with calnexin (Fig. 3), indicating that some A2AR molecules were also localized in the ER. Data from the pulse-chase analysis (Fig. 2A) suggest that the ∼40% degradation of A2AR after a 5-h chase may reflect the ER staining of A2AR in the steady state.
FIGURE 2.
Impaired maturation of A2BR. A, stable cell lines expressing HA-A2AR or HA-A2BR were starved, labeled with [35S]methionine/cysteine for 15 min, and then chased in complete medium for up to 5 h. The cells were lysed with buffer containing 1% CHAPS at each time point and subjected to immunoprecipitation using mouse mAb to HA. The immunoprecipitates were subjected to SDS-PAGE followed by fluorography. Molecular makers are shown on the left. The black circle represents the mature form. The intensity of each band was measured and plotted (on the right). Data are shown as a percentage of intensity of the pulse band (0 h) for the total (mature form plus immature form) and immature forms. For the mature form, data are shown as a percentage of intensity of the maximum labeling of the mature receptor at 1 h of chase. Shown is a representative of three independent experiments. The black squares, black circles, and white squares represent the total (mature plus immature form), mature form, and immature form, respectively, as revealed in B. B, HA-A2AR and HA-A2BR were isolated by immunoprecipitation from stably transfected AD-293 cells with rat mAb to HA. The immunoprecipitates were then either mock-treated (−) or treated with endoglycosidase H (endo H) or N-glycanase (PNGase) before analysis by SDS-PAGE and immunoblotting. The black circle, white diamond, and white square represent the mature form, immature form, and deglycosylated form, respectively. The asterisk indicates the IgG light chain.
FIGURE 3.
Poor expression of A2BR at the cell surface. Immunofluorescence localization of HA-A2BR and HA-A2AR in transfected AD-293 cells is shown. Cells were co-immunostained with rat mAb to HA and mouse mAbs against organelle markers. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. CaNX, calnexin; γ-1, gamma subunit of AP-1; Na+/K+ ATPase, sodium/potassium ATPase; white bar, 10 μm.
Stabilization of A2BR by Inhibiting Proteasome Activity and Low Temperature Is Not Sufficient for Transport of A2BR from the ER to the Cell Surface
Next, we sought to determine the mechanism of proteolytic degradation of A2BR. For this purpose, a stable cell line expressing HA-A2BR was pretreated with lysosomal inhibitors (leupeptin, chloroquine, and NH4Cl) or proteasomal inhibitors (MG132 and lactacystin) for 2 h and then starved, labeled with [35S]methionine/cysteine for 15 min, and chased in complete medium for 2 h. Protease inhibitors were included throughout the experiments. Cell extracts were then immunoprecipitated with anti-HA antibody followed by fluorography as described above. As shown in Fig. 4A, only proteasomal inhibitors (MG132 and lactacystin) inhibited A2BR degradation, indicating that A2BR is efficiently degraded by the proteasome. This proteasomal degradation of A2BR was also confirmed at the steady state level by immunoprecipitation followed by immunoblotting after cells were treated with proteasomal inhibitors for 16 h (Fig. 4B). In contrast to results with proteasomal inhibitors MG132 and lactacystin, which strongly stabilized A2BR, treatment with lysosomal inhibitors somewhat destabilized total A2BR levels for an unknown reason. We often observed a ∼33-kDa band in addition to the a ∼39-kDa band of A2BR in lysates from transiently transfected cells (Fig. 1A) and proteasome inhibitor-treated cells (Fig. 4, A and B, black squares). To test whether this ∼33-kDa band represented a form deglycosylated by the cytosolic N-glycanase or a major degradation product from the CT, we treated HA-A2BR-expressing cells with the proteasomal inhibitor MG132 for 16 h. Then HA-A2BR was immunoprecipitated and treated with either endoglycosidase H or N-glycanase (Fig. 4C). We found that the migration of the ∼33-kDa band was not affected by any of these treatments, indicating that this molecule was already deglycosylated.
FIGURE 4.
A2BR is degraded mainly by the proteasome. A, a stable cell line expressing HA-A2BR was pretreated with lysosomal protease inhibitors leupeptin (200 μg/ml), choloroquin (200 μm), and NH4Cl (50 mm) or with proteasome inhibitors MG132 (10 μm) and lactacystin (10 μm) for 2 h at 37 °C and then starved, labeled with [35S]methionine/cysteine for 15 min, and chased in complete medium containing protease inhibitors for 2 h. The cells were lysed with buffer containing 1% CHAPS at each time point and subjected to immunoprecipitation using mouse mAb to HA. The immunoprecipitates were subjected to SDS-PAGE followed by fluorography. Molecular makers are shown on the left. The arrow indicated the position of monomeric HA-A2BR. The black square represents the faster migrating band of HA-A2BR. B, a stable cell line expressing HA-A2BR was treated with lysosomal protease inhibitors leupeptin (200 μg/ml), choloroquin (200 μm), and NH4Cl (50 mm) or with proteasome inhibitors MG132 (10 μm) and lactacystin (10 μm) for 16 h at 37 °C. Then the cells were lysed and subjected to immunoprecipitation using rat mAb to HA. The immunoprecipitates were then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. The arrow indicates the position of monomeric HA-A2BR. The black square represents the faster migrating band of HA-A2BR. 5% of the total cell lysate was analyzed by SDS-PAGE and immunoblotting using mAb to β-actin as a loading control. Cell surface expression of HA-A2BR with protease inhibitors was determined by enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR in each treatment was expressed as percent means ± S.D. of nontreated cells (n = 6). C, a stable cell line expressing HA-A2BR was treated with MG132 (10 μm) for 16 h at 37 °C. HA-A2BR was isolated by immunoprecipitation from stably transfected AD-293 cells with antibody to HA. The immunoprecipitates were then either mock-treated (−) or treated with endoglycosidase H (endo H) or N-glycanase (PNGase) before analysis by SDS-PAGE and immunoblotting. The arrow indicates the position of monomeric HA-A2BR, and the black square represents the deglycosylated form. D, a stable cell line expressing HA-A2BR was incubated for 24 h at 37 °C, 24 h at 30 °C, or 24 h at 30 °C plus an additional hour at 37 °C. Then cells were lysed, subjected to immunoprecipitation, and analyzed by SDS-PAGE and immunoblotting. 5% of the total cell lysate was analyzed by SDS-PAGE and immunoblotting using mAb to β-actin as a loading control. The cell surface expression of HA-A2BR was determined by enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR was expressed as percent means ± S.D. of cells incubated at 37 °C (n = 6). E, immunofluorescence localization of HA-A2BR in transfected AD-293 cells after incubation of the cells at 30 °C for 24 h. Cells were co-immunostained with rat mAb to HA and mouse mAb to Na+/K+ ATPase. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm.
We reasoned that if the A2BR degradation was blocked in the secretory pathway, we could expect enhanced A2BR expression on the cell surface in the presence of proteasome inhibitors. Therefore, we tested whether inhibitors of proteasomal activity, which prevent degradation of A2BR, could enhance the cell surface expression of A2BR. As shown in Fig. 4B, the surface expression of A2BR was not increased even when its degradation by the proteasome was inhibited. The data in Fig. 4B suggest that A2BR was already destined to be on the degradation pathway rather than the secretory pathway.
This interpretation was supported by studies of the effects of low temperature (shown in Fig. 4, D and E). It has been reported that low temperature incubation blocks cystic fibrosis transmembrane conductance regulator (CFTR) degradation and promotes its cell surface expression (36, 37). Therefore, we tested whether this was also the case for A2BR by culturing HA-A2BR stably expressing cells at 30 °C for 24 h and then determining the total protein level and cell surface expression by immunoprecipitation followed by immunoblotting and enzyme-linked immunoassay, respectively. We show that lowering the culture temperature from 37 to 30 °C for 24 h resulted in stabilization of A2BR by 2-fold, as assessed by immunoprecipitation followed by immunoblotting, but did not enhance its surface expression, as assessed by an enzyme-linked immunoassay (Fig. 4D) and immunofluorescence (Fig. 4E). Because it has been shown that low temperature incubation can affect protein trafficking from the ER to the plasma membrane (38), we incubated the HA-A2BR-expressing cells at 30 °C for 24 h to stabilize A2BR and then further incubated them for 1 h at 37 °C to resume normal protein transport machinery and check whether additional incubation was able to enhance A2BR expression at the cell surface. The additional 1 h of incubation time at 37 °C after a 24-h incubation at 30 °C showed minimum enhancement of A2BR expression at the plasma membrane (Fig. 4D), demonstrating that inhibition of A2BR degradation by low temperature does not promote A2BR surface expression. This is consistent with the previous results, where inhibition of proteasomal activity showed no effect on its surface localization (Fig. 4B).
Chemical Chaperones Failed to Stabilize A2BR
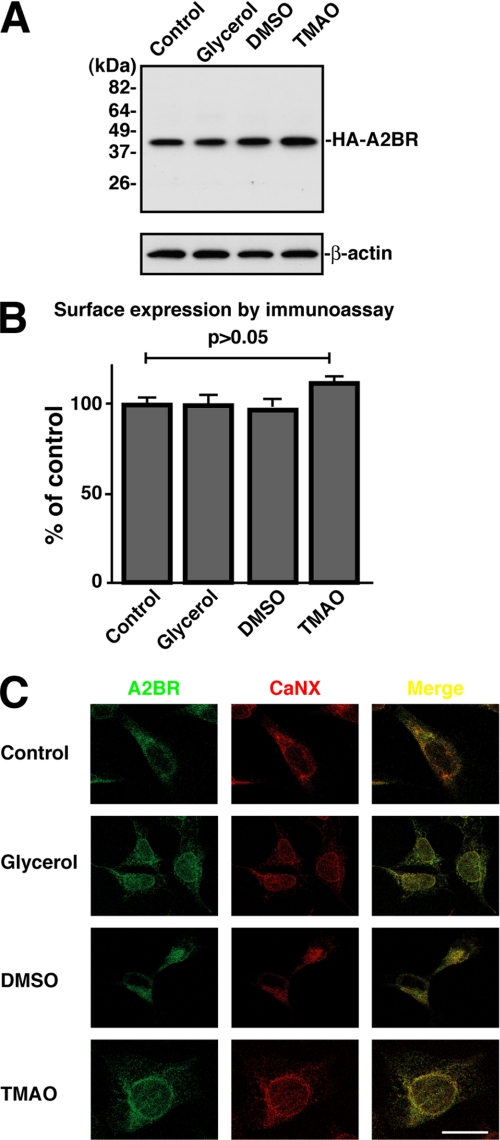
We next sought to determine whether the A2BR was degraded by the proteasome because of (i) the unfolding status of the A2BR structure and/or (ii) restricted transport from the ER due to certain signal(s) within A2BR structure. Chemical chaperons are used to nonspecifically help the folding of proteins during protein synthesis and intracellular processing. If it is the unfolding that triggers A2BR degradation, then treatment of HA-A2BR-expressing cells with nonspecific chemical chaperones (glycerol, dimethyl sulfoxide (DMSO), and osmolyte trimethylamine N-oxide (TMAO)) was expected to help its folding and promote cell surface expression (39–42). To test this hypothesis, we treated the HA-A2BR-expressing cells with 4% glycerol, 2% DMSO, or 75 mm TMAO for 24 h, and then the total A2BR and cell surface A2BR expression were determined by immunoprecipitation followed by immunoblotting and enzyme-linked immunoassay, respectively. We show that treatment of the HA-A2BR-expressing cells with 4% glycerol or 2% DMSO for 24 h neither stabilized A2BR nor promoted cell surface expression (Fig. 5, A and B). Only treatment with 75 mm TMAO for 24 h slightly stabilized A2BR by up to 15% (Fig. 5A) and enhanced its cell surface expression (Fig. 5B). If chemical chaperons had really helped stabilize A2BR, we expected to see an altered proteinase digestion pattern when the membrane fraction was isolated and treated with exogenous proteinase K, which is used to indicate the folding status of proteins (43). Therefore, after HA-A2BR-expressing cells were treated with 4% glycerol, 2% DMSO, or 75 mm TMAO for 24 h, crude membrane fractions were prepared by centrifugation and incubated with different concentrations of proteinase K. The digested A2BR was recovered by immunoprecipitation with anti-HA antibody and immunoblotting. As shown in supplemental Fig. 1, the digested fragments in membrane to the chemical chaperon-treated cells and the nontreated cells were almost identical. These results also suggested that the chemical chaperon did not affect the conformation of A2BR. Furthermore, in agreement with these biochemical data, chemical chaperon treatments did not significantly alter A2BR localization as assessed by immunofluorescence, as the majority of the A2BR was still well co-localized with the ER marker protein calnexin (Fig. 5C). In addition, after these chemical chaperon treatments, only a very small portion of the A2BR was co-localized with sodium/potassium ATPase, which is quite similar to findings with untreated cells (supplemental Fig. 2). We also tested whether a longer incubation with these chemical chaperons could help the folding of A2BR. To this end we treated the HA-A2BR-expressing cells with 2% glycerol, 1% DMSO, or 75 mm TMAO for up to 3 days. No apparent stabilization and enhancement of surface expression of A2BR were observed (data not shown). Thus, nonselective chemical chaperone treatments failed to stabilize and enhance the cell surface localization of A2BR.
FIGURE 5.
Chemical chaperons failed to enhance cell surface expression of A2BR. A, AD-293 cells expressing HA-A2BR were incubated without (Control) or with chemical chaperons (4% glycerol, 2% DMSO, or 75 mm TMAO) for 24 h at 37 °C. The steady state levels of HA-A2BR under these conditions were analyzed by immunoprecipitation, SDS-PAGE, and immunoblotting. 5% of the total cell lysate was analyzed by SDS-PAGE and immunoblotting using mAb to β-actin as a loading control. Shown is a representative of three independent experiments. B, AD-293 cells expressing HA-A2BR were treated as in A, and the cell surface expression of HA-A2BR was determined by enzyme-linked immunoassay. Relative surface expression of HA-A2BR with chemical chaperons, as described in A, was expressed as percent means ± S.D. of control cells (n = 6). Shown is a representative of three independent experiments. C, immunofluorescence localization of HA-A2BR in transfected AD-293 cells after incubating the cells with chemical chaperons as described in A. Cells were co-immunostained with rat mAb to HA and mouse mAb to calnexin (CaNX). The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm.
Studies of A2BR Truncation Mutants Suggested That A2BR CT Does Not Have ER Retention Signals
Next, we checked the possibility of whether A2BR was degraded because of restricted transport from the ER due to a certain signal(s) within the A2BR structure. It is known that the CTs of different GPCRs differ in length and structure, thus recruiting important molecules that regulate transport machinery and/or signaling. Indeed, A2AR, the closest family member to A2BR, possesses a CT much longer in length than that of A2BR (125 versus 41 residues in mouse), suggesting that A2AR and A2BR may interact with different molecules for transport from the ER to the plasma membrane and signaling. Because the role of the CT for transport machinery in A2BR molecules is not well understood, we determined the role of the CT of A2BR in its transport from the ER to the cell surface by creating the truncation mutants. Sequences of specific ER retention signals have been reported previously in the primary sequences of other receptors (44), but we were not able to find them in sequences of A2BR CT. The one possible candidate could be Arg293-Asn294-Arg295, which is considered an arginine-based ER-sorting motif (45). It is highly unlikely that this motif in the A2BR CT is a dominant ER retention signal, because A2AR CT also possesses Arg286-Ile287-Arg288 at identical positions to A2BR, and A2AR shows much better expression on the cell surface. However, it was still possible that A2BR CT does possess some yet to be discovered ER retention signal(s) that prevents A2BR surface expression. If this is the case, then one can expect to see enhanced A2BR surface expression due to the CT truncation in A2BR. If, however, the A2BR CT is required for its efficient transport to the cell surface, then the deletion of CT will significantly decrease its surface expression. To test this hypothesis, we created truncation mutant constructs of A2BR (see Fig. 6A). These constructs were transiently expressed in AD-293 cells, and total protein expression and cell surface expression were determined. The truncation of A2BR CT did indeed further reduce its already low level of surface expression by ∼50% for 306 and 292 stop mutants (Fig. 6B). In contrast, deletion of the CTs of A2AR and β2AR almost completely abolished their surface expression (Fig. 6, C and D). The report that deletion of the CTs of the α2B adrenergic receptor, the angiotensin II type 1A receptor (46), and the A1 adenosine receptor (47) results in markedly reduced (>95%) cell surface expression suggests that the relatively less severe (i.e. only 50%) reduction of surface expression due to CT deletion of A2BR could be unique. Because ∼50% of the A2BR was still expressed at the cell surface even without CT, these results suggest that the CT of A2BR is not strictly required for its surface transport. However, the CT of A2BR is capable of improving its surface expression. Furthermore, these results also suggest that the CT of A2BR has neither an apparent ER retention motif nor dominant forward transport signal(s) from the ER to the cell surface. Because the immunoassay used for CT truncation of A2BR could represent the lower limit for the assay and observations of A2BR wild type and CT truncation mutants could represent the relatively same numbers of receptors, we tested whether replacement or fusion of the A2BR CT with the CT of either A2AR or β2AR would affect their surface expression to further confirm whether this was actually the case.
FIGURE 6.
CT of A2BR has no dominant forward transport signal from the ER to the cell surface. A, amino acid sequence of mouse A2BR CT. The mutant constructs are indicated by arrows. 7TM, represents the seventh transmembrane domain of A2BR. B, HA-A2BR wild type and 325, 305, and 292 stop truncation mutants were transiently transfected to AD-293 cells. The cells were lysed and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of HA-A2BR was determined by enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR truncation mutants was adjusted by total expression levels and expressed as percent means ± S.D. of wild type (n = 6). Shown is a representative of three independent experiments. C, HA-A2AR wild type, CT deletion mutant (ΔCT), and chimeric mutant, where CT of A2AR was replaced by CT of A2BR (BRCT) and transiently transfected to AD-293 cells. The cells were lysed and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each construct was determined by enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR mutants was adjusted by total expression levels and expressed as percent means ± S.D. of wild type (n = 6). Shown is a representative of three independent experiments. D, HA-β2AR wild type, CT deletion mutant (ΔCT), and chimeric mutant where CT of β2AR was replaced by CT of A2BR and transiently transfected to AD-293 cells. The cells were lysed and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each construct was determined by enzyme-linked immunoassay, as shown on the right. The relative surface expression of HA-β2AR mutants was adjusted by total expression levels and expressed as percent means ± S.D. of wild type (n = 6). Shown is a representative of three independent experiments.
Studies of A2AR and β2AR Chimeras with A2BR Used as “Diagnostic” Tools Suggested That A2BR Does Not Have Dominant Transport Signals from the ER to the Plasma Membrane
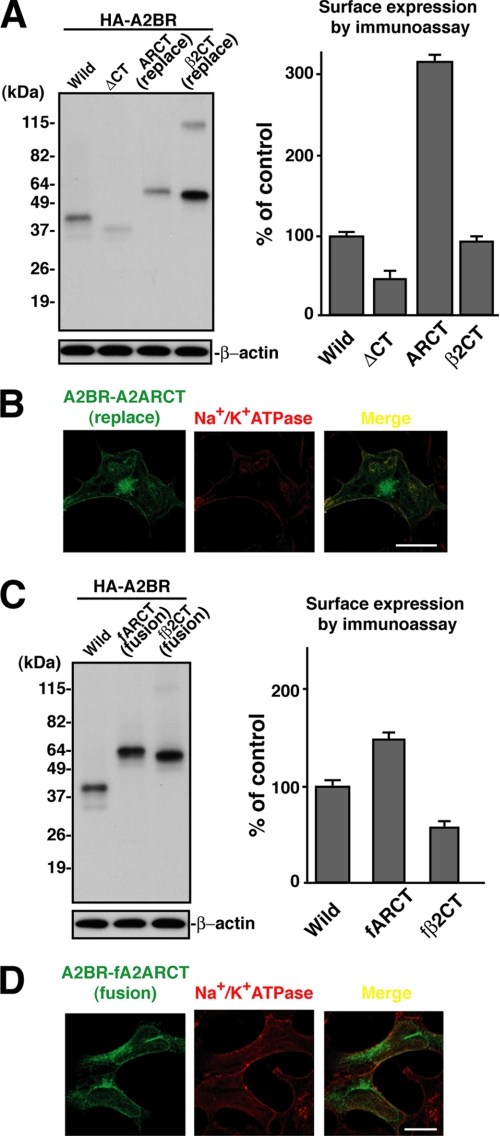
We constructed these chimeras to determine the presence of ER retention signal(s) in CT of A2BR and/or lack of dominant forward transport signal(s) from the ER to the cell surface. We chose A2AR and β2AR for the following reasons. A2AR possesses a primary structure very similar to that of A2BR except for the secondary extracellular domain, and their CTs share similar functions. Although the role of the A2AR CT has been the subject of interesting studies (48), β2AR is one of the most extensively studied GPCRs, and it has been shown to be well expressed at the plasma membrane (49). Furthermore, it is reported that replacement of the CT of β2AR with that of the GABABR1, which contains a well characterized arginine-based ER retention signal, results in its complete retention inside the cells (50). These data suggest that β2AR without CT has no dominant forward transport signal(s) to overcome the ER retention motif and export it to the plasma membrane. Indeed, as described above, the CT deletion mutant of β2AR resulted in markedly reduced (>95%) cell surface expression as compared with the wild type (Fig. 6D). For these reasons, we created the following chimeras: HA-A2AR-A2BRCT (where CT of A2AR was replaced by CT of A2BR) and HA-β2AR-A2BRCT (where CT of β2AR was replaced by CT of A2BR). These chimeric constructs were transiently transfected into AD-293 cells, and total protein expression and cell surface expression were determined. We expected that these chimeras could show better cell surface expression than CT truncation mutants of A2AR and β2AR (less than 5% of wild type) but not better than wild type. As shown in Fig. 6, C and D, transfected cells exhibited ∼34- and ∼45-kDa bands corresponding to HA-A2AR-A2BRCT and HA-β2AR-A2BRCT, respectively. We found that the surface expression of both the HA-A2AR-A2BRCT and HA-β2AR-A2BRCT chimeras improved to the levels of 20 and 30% of wild type as compared with CT deletion mutants (less than 5% of wild type). These results suggested that the CT of A2BR has neither an apparent ER retention motif nor a dominant forward transport signal(s) from the ER to the cell surface. If the A2BR possesses no dominant forward transport signal(s) from the ER to the cell surface, one can expect to see improved cell surface expression of A2BR, once the appropriate signal(s) is obtained. To test this, we created the chimeras HA-A2BR-A2ARCT (CT of A2BR was replaced by CT of A2AR) and HA-A2BR-β2CT (CT of A2BR was replaced by CT of β2AR) and transiently transfected them into AD-293 cells; then total protein expression and cell surface expression were determined. As shown in Fig. 7A, the transfected constructs exhibited ∼63- and ∼61-kDa bands corresponding to HA-A2BR-A2ARCT and HA-A2BR-β2CT, respectively. HA-A2BR-A2ARCT dramatically improved (more than 300% of wild type) its surface expression as assessed by enzyme-linked immunoassay (Fig. 7A). On the contrary, HA-A2BR-β2CT showed almost identical surface expression with wild type (Fig. 7A). In agreement with the enzyme-linked immunoassay results, immunofluorescence microscopy revealed that the HA-A2BR-A2ARCT chimera, but not the HA-A2BR-β2CT, showed more co-localization with sodium/potassium ATPase, a plasma membrane marker (Fig. 7B) (data not shown). The above results suggested that once A2BR acquires the appropriate forward transport signal(s) from the ER to plasma membrane at the CT, it could be transported to the plasma membrane even more efficiently than the wild type. To avoid eliminating any of the possible, but as yet unknown transport signal(s) and any interacting partners in the A2BR CT, and to further confirm these results given above, we created the following chimeras: HA-A2BR-fA2ARCT (where the CT of A2AR was fused to the very end of CT of A2BR) and HA-A2BR-fβ2CT (where CT of β2AR was fused to the very end of CT of A2BR). These constructs were transiently transfected into AD-293 cells, and total protein expression and cell surface expression were determined. As shown in Fig. 7C, the transfected cells exhibited ∼65- and ∼62-kDa bands corresponding to HA-A2BR-fA2ARCT and HA-A2BR-fβ2CT. As expected, HA-A2BR-fA2ARCT dramatically improved (∼150% of wild type) their surface expression as assessed by enzyme-linked immunoassay (Fig. 7C). In agreement with the enzyme-linked immunoassay results, immunofluorescence microscopy revealed that the HA-A2BR-fA2ARCT chimera showed more co-localization with sodium/potassium ATPase, a plasma membrane marker (Fig. 7D), but its co-localization is less clear than the HA-A2BR-A2ARCT chimera (Fig. 7B). Although the CT of β2AR unexpectedly failed to promote cell surface expression of A2BR chimeras, results from HA-A2BR with A2AR chimeras strongly suggested that inefficient transport of A2BR might be due to the absence of a dominant transport signal(s) from the ER to plasma membrane. However, it could be delivered more efficiently to the cell surface once it acquired a dominant signal(s).
FIGURE 7.
CT of A2AR is able to transport A2BR more efficiently to the cell surface. A, HA-A2BR wild type, deletion of CT truncation mutant (ΔCT), the chimeric mutant in which CT of A2BR was replaced by CT of A2AR (ARCT (replace)), or the chimeric mutant in which CT of A2BR was replaced by CT of β2AR (β2CT (replace)) was transiently transfected to AD-293 cells. The cells were lysed and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each construct was determined by enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR mutants was adjusted by total expression levels and expressed as percent means ± S.D. of wild type (n = 6). Shown is a representative of three independent experiments. B, immunofluorescence localization of HA-A2BR-A2ARCT (CT of A2BR was replaced by CT of A2AR) in transfected AD-293 cells. Cells were co-immunostained with rat mAb to HA and mouse mAb against Na+/K+ ATPase. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm. C, HA-A2BR wild type, the chimeric mutant in which CT of A2BR was fused by CT of A2AR (fARCT), or the chimeric mutant in which CT of A2BR was fused by CT of β2AR (fβ2CT) was transiently transfected to AD-293 cells. The cells were lysed and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each construct was determined by enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR mutants was adjusted by total expression levels and expressed as percent means ± S.D. of wild type (n = 6). Shown is a representative of three independent experiments. D, immunofluorescence localization of HA-A2BR-fA2ARCT (CT of A2AR was fused to CT of A2BR) in transfected AD-293 cells. Cells were co-immunostained with rat mAb to HA and mouse mAb to Na+/K+ ATPase. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm.
The A2AR-A2BR Interaction Enhanced A2BR Surface Expression
Next, we checked the possibility that A2AR helps A2BR cell surface expression by forming a heterodimer when transfected together. In order to test this, the HA-A2BR and FLAG-A2AR or FLAG-β2AR were transiently co-transfected into AD-293 cells; then the total expression levels of A2BR, as well as FLAG-A2AR and FLAG-β2AR, and cell surface expression of A2BR were determined by immunoprecipitation followed by immunoblotting and enzyme-linked immunoassay, respectively. We found that the total expression level of HA-A2BR at steady state was slightly improved when FLAG-A2AR was co-transfected (Fig. 8A). Remarkably, the cell surface expression of HA-A2BR was dramatically improved by FLAG-A2AR co-transfection but not by FLAG-β2AR (Fig. 8B). These observations prompted the more detailed studies of interactions between A2AR and A2BR in Myc-A2BR and HA-A2AR immunoprecipitates from extracts of co-transfected AD-293 cells.
FIGURE 8.
Efficient cell surface expression of A2BR requires A2AR co-expression. A, HA-A2BR and empty vector (Vec), FLAG-A2AR (FA2AR), or FLAG-β2AR (Fβ2AR) were transiently co-transfected to AD-293 cells. The cells were lysed in 1% CHAPS-containing buffer and subjected to immunoprecipitation using rat mAb to HA or rabbit polyclonal antibody to FLAG after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA or FLAG. Not, represents nontransfected cells. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The asterisk indicates IgG heavy chain. B, the cell surface expression of each co-transfectant was determined by enzyme-linked immunoassay. Relative surface expression of HA-A2BR were adjusted by total expression levels and expressed as percent means ± S.D. of HA-A2BR plus empty vector-transfected cells (n = 6). Shown is a representative of three independent experiments. C, HA-A2AR and Myc-A2BR were transiently co-transfected to AD-293 cells. The cells were lysed in 1% CHAPS-containing buffer and subjected to immunoprecipitation (IP) using rat mAb to HA (A2AR) or rabbit polyclonal antibody to Myc (A2BR) after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting (Wb) using mouse mAb to HA (A2AR) or Myc (A2BR). D, immunofluorescence localization of HA-A2AR and Myc-A2BR in stably co-transfected AD-293 cells. Cells were immunostained with rat mAb to HA and mouse mAb to Myc. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm.
The physical association between Myc-A2BR and HA-A2AR in co-transfectants is demonstrated in Fig. 8C by detection of: (i) HA-A2AR molecules by immunoblotting of anti-Myc antibody “pulldowns” in immunoprecipitates with Myc-A2BR (lane 2) and (ii) Myc-A2BR molecules by immunoblotting of anti-HA antibody pulldowns in HA-A2AR immunoprecipitates (lane 4). In both cases, we observed co-precipitated HA-A2AR and Myc-A2BR, suggesting that A2AR and A2BR form the heterodimer. Confocal microscopy also shows clearly increased A2BR surface expression if co-transfected with A2AR (Fig. 8D).
The F(X)6LL Motif in A2AR CT Is Responsible for Rescuing A2BR to the Cell Surface
Next, we sought to determine which domain or motif of A2AR is responsible for rescuing A2BR cell surface expression. We first tested whether A2AR-A2BRCT and/or A2BR-A2ARCT could rescue A2BR cell surface expression when transfected together. The cell surface expression of HA-A2BR was determined by immunoassay, and total expression levels were determined by immunoprecipitation followed by immunoblotting. As expected, FLAG-A2BR-A2ARCT, as compared with FLAG-A2AR-A2BRCT, brings more HA-A2BR (∼250%) when co-transfected together (Fig. 9A). This result was also confirmed by immunofluorescence study. HA-A2BR and FLAG-A2AR-A2BRCT co-localized well only in the intracellular compartment but not at the plasma membrane (Fig. 9B). On the contrary, HA-A2BR co-localized well with FLAG-A2BR-A2ARCT not only in the intracellular compartment but also at the plasma membrane (Fig. 9B). These results suggested that the A2AR CT might be necessary to rescue A2BR. To confirm this and further investigate the molecular mechanism in detail, we made various truncation mutants from the CT of A2AR (Fig. 9C). HA-A2AR truncation mutants were transiently transfected into AD-293 cells, cell surface expression of mutants was determined by immunoassay, and total expression levels were determined by immunoprecipitation followed by immunoblotting. This experiment showed us which sequence was involved in the cell surface expression of A2AR itself. As shown in Fig. 9B, the cell surface expression of HA-A2AR 372, 305, and 301 stop mutants was identical with wild type. However, the HA-A2AR 298 stop mutant showed ∼60% of the surface expression of wild type and further truncation of the CT (295, 291, and 285 stop mutants) showed 5–30% of the surface expression of wild type. The immunofluorescence of each mutant construct indicated that plasma membrane localizations of HA-A2AR 372, 305, and 301 stop mutants were well retained, which was similar to the wild type. However, HA-A2AR 295, 291, and 285 stop mutants localized mainly to the intracellular compartment (Fig. 9E), indicating that the RIREFRQTFRKIIRTH sequence in the CT is essential for efficient cell surface expression of A2AR itself. Among these 16 amino acids, we focused on F(X)6LL (where X can be any residue and L is leucine or isoleucine), which is proposed to be an ER export motif for several GPCRs (46). For this reason, we made a mutant in which all Phe290, Ile297, and Ile298 sequences are replaced with Ala (Fig. 10A, A2AR FII/AAA) and transiently transfected into AD-293 cells; cell surface expression of mutants was determined by immunoassay, and total expression levels were determined by immunoprecipitation followed by immunoblotting. As expected, the FLAG-A2AR FII/AAA mutant showed ∼5% of the cell surface expression of wild type (Fig. 10B). This was further confirmed by immunofluorescence study, which showed that the majority of this mutant co-localized with the ER marker, calnexin, but was poorly expressed at the plasma membrane (Fig. 10C). Therefore, we concluded that the F(X)6LL motif in A2AR CT is primarily responsible for its cell surface expression. From these results, it is reasonable to expect that the F(X)6LL motif in the A2AR CT sequence is the minimal sequence required for A2BR cell surface expression when transfected together. Thus, we co-transfected HA-A2BR with FLAG-A2AR wild type or FLAG-A2AR FII/AAA into AD-293 cells, and the cell surface and total expression of the receptors were measured. As shown in Fig. 10D, co-transfection of FLAG-A2AR FII/AAA with HA-A2BR resulted in almost completely blocking the cell surface expression of HA-A2BR, suggesting that FLAG-A2AR FII/AAA may form a heterodimer with HA-A2BR and interfere with its export from the ER. This result was further confirmed by an immunofluorescence study, which showed only intercellular staining and co-localization (Fig. 10D) representing ER but not plasma membrane staining (data not shown). Collectively, we concluded that A2AR rescued A2BR cell surface expression through the F(X)6LL motif in A2AR CT.
FIGURE 9.
A2AR CT is required for efficient cell surface expression of A2AR itself as well as of A2BR. A, HA-A2BR and empty vector (Vec), FLAG-A2AR-A2BRCT (AR-BRCT), or FLAG-A2BR-A2ARCT (BR-ARCT) were transiently co-transfected to AD-293 cells. The cells were lysed in 1% CHAPS-containing buffer and subjected to immunoprecipitation using rat mAb to HA or rabbit polyclonal antibody to FLAG after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA or FLAG. Not, represents nontransfected cells. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each co-transfectant was determined by an enzyme-linked immunoassay, as shown on the right. Relative surface expression of HA-A2BR was adjusted by total expression levels and expressed as percent means ± S.D. of HA-A2BR plus empty vector-transfected cells (n = 6). Shown is a representative of three independent experiments. B, immunofluorescence localization of HA-A2BR and FLAG-A2AR-A2BRCT or FLAG-A2BR-A2ARCT in transiently co-transfected AD-293 cells. Cells were immunostained with rat mAb to HA and mouse mAb to FLAG. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm. C, schematic representation of amino acid sequence of mouse A2AR CT. The mutant constructs used in this study are also indicated at the bottom. 7TM, represents the seventh transmembrane domain of A2AR. D, HA-A2AR wild type or truncation mutants shown in C were transiently transfected to AD-293 cells. The cells were lysed in 1% CHAPS-containing buffer and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. The cell surface expression of each transfectant was determined by an enzyme-linked immunoassay, as shown on the right. Relative surface expression of each mutant construct was adjusted by total expression levels and expressed as percent means ± S.D. of wild type HA-A2AR-transfected cells (n = 6). Shown is a representative of three independent experiments. E, immunofluorescence localization of HA-A2AR wild type and truncation mutants in transiently transfected AD-293 cells. Cells were immunostained with rat mAb to HA. The bound primary antibodies were detected by using Alexa Fluor 594 (red)-conjugated goat anti-rat IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. White bar, 10 μm.
FIGURE 10.
The F(X)6LL motif in A2AR CT is responsible for efficient cell surface expression of A2BR when transfected together. A, schematic representation of amino acid sequence of mouse A2AR F(X)6LL motif in CT and its mutant, FII/AAA, where all Phe290, Ile297, and Ile298 sequences in this motif were replaced by Ala. The actual position of the motif is indicated by the bold vertical line. 7TM, represents the seventh transmembrane domain of A2AR. B, FLAG-A2AR wild type or mutant FLAG-A2AR FII/AAA was transiently transfected to AD-293 cells. The cells were lysed in 1% CHAPS-containing buffer and subjected to immunoprecipitation using rat mAb to HA after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA. Not, represents not transfected cells. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each transfectant was determined by an enzyme-linked immunoassay, as shown on the right. The relative surface expression of each mutant construct was adjusted by the total expression level and expressed as percent means ± S.D. of wild type HA-A2AR-transfected cells (n = 6). Shown is a representative of three independent experiments. C, immunofluorescence localization of HA-A2AR FII/AAA in transiently transfected AD-293 cells. Cells were immunostained with rat mAb to HA and mouse mAb to calnexin (CaNX). The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm. D, HA-A2BR and either empty vector (Vec), FLAG-A2AR wild type, or FLAG-A2AR FII/AAA mutant was transiently co-transfected to AD-293 cells. The cells were lysed in 1% CHAPS-containing buffer and subjected to immunoprecipitation using rat mAb to HA (A2BR) or rabbit polyclonal antibody to FLAG (A2AR) after 72 h of transfection. The immunoprecipitates were washed and then analyzed by SDS-PAGE and immunoblotting using mouse mAb to HA or FLAG. Not, represents nontransfected cells. 5% of the total cell lysate was subjected to SDS-PAGE and blotted with mAb to β-actin as a loading control. The cell surface expression of each transfectant was determined by an enzyme-linked immunoassay, as shown on the right. The relative surface expression of HA-A2BR was adjusted by total expression levels and expressed as percent means ± S.D. of HA-A2BR plus empty vector-transfected cells (n = 6). Shown is a representative of three independent experiments. E, immunofluorescence localization of HA-A2BR and FLAG-A2AR FII/AAA mutant in transiently co-transfected AD-293 cells. Cells were immunostained with rat mAb to HA and mouse mAb to FLAG. The bound primary antibodies were detected by using either Alexa Fluor 488 (green)- or Alexa Fluor 594 (red)-conjugated goat anti-rat or mouse IgG antibody. Cells were analyzed by immunofluorescence with confocal microscopy. Merged images are shown in yellow. White bar, 10 μm.
Genetic in Vivo Evidence That A2AR Chaperones A2BR in Splenocytes in Vivo
The GPCR heterodimerization described above could be at least in part due to a potentially nonphysiological association, an artifact of studies of GPCR overexpressing transfectants. To address this common caveat of GPCR studies, we compared the expression levels of functional A2AR and A2BR in splenocytes from wild type or A2AR knock-out or A2BR knock-out mice by taking advantage of available unique in vivo genetic controls. We reasoned that if A2AR is indeed required in vivo for A2BR expression and function on the cell surface (as suggested by data in Fig. 8), then we would expect much decreased levels of functional A2BR on cells from A2AR gene-deficient mice. As described in Fig. 11 we compared the levels of A2AR- or A2BR-triggered intracellular cAMP accumulation in parallel assays of the cells from wild type or A2AR or A2BR knock-out mice (13). To determine and compare the levels of surface expression of functional A2BR in the presence or absence of A2AR, we incubated the splenocytes from wild type and knock-out mice with the A2AR-selective agonist CGS21680, which triggers cAMP accumulation in the assay conditions used only due to A2AR signaling, or with the nonselective adenosine receptor agonist NECA, which triggers cAMP accumulation by both A2AR and A2BR (4). The difference between NECA and CGS21680 represents the contribution of cell surface-expressed and functional A2BR, which is determined by subtracting the CGS21680-A2AR-induced cAMP accumulation from that induced by NECA (the sum of (A2AR + A2BR)-triggered cAMP accumulation). Fig. 11A represents a summary of different independent assays (see Fig. 11, B–D). We show that very low A2BR-induced cAMP accumulation was detected in cells from A2AR-deficient mice (Fig. 11A). The analysis of Fig. 11, B and D, estimates the range of contribution of A2AR signaling in the cAMP increases detected after incubation of cells with NECA or CGS21680. In the key experiment with activated splenocytes from A2AR knock-out mice (Fig. 11C), the cAMP accumulation upon A2AR-selective CGS21680 agonist treatment was very low, as expected, as there is no A2AR expression. In support of our previous observations (Figs. 9 and 10) with transfectants, little cAMP accumulation was detected in response to the A2BR-activating ligand (Fig. 11, A and D). One caveat with regard to interpreting the results of Fig. 11 is that deleting A2AR or A2BR may influence the admixture of various cell types in the spleen or the number of different receptors on individual types of splenocytes. Such changes could contribute to the apparent differences observed in A2BR signaling.
FIGURE 11.
Impaired expression of functional A2BR in splenocytes from A2AR knock-out mice. A, summary of A2BR-triggered cAMP assays in B–D. A2BR-triggered cAMP accumulation was estimated by subtracting cAMP accumulation with A2AR-selective agonist (1 μm CGS21680) treatment from nonselective agonist (10 μm NECA) treatment. Data represent the mean ± S.E. (n = 8). *, p < 0.05. B–D, in a pharmacological control for A2AR and A2AR + A2BR selectivity of cAMP-elevating CGS21680 and NECA, respectively, the A2AR + A2BR antagonist ZM241385 (ZM) inhibited both NECA- and CGS21680-induced cAMP in lymphoid cells following a mixed lymphocyte culture from wild type (B), A2BR knock-out (C), and A2AR knock-out (D) mice spleen. Cells were treated in vitro with 1 or 10 μm NECA, 1 μm CGS21680, and/or 1 μm ZM241385 for 15 min at 37 °C. *, p < 0.05.
DISCUSSION
Studies of GPCR maturation and trafficking to the cell surface are important because many receptor mutants leading to intracellular retention have been identified that cause disease via impaired signaling (51). Proper trafficking control of the receptors is therefore essential for accurate signal propagation. In this report we show that the majority of A2BR was degraded by the proteasome, and only a small portion was forwarded to the cell surface due to the lack of an effective forward transport signal from the ER. The A2BR is not the only known membrane protein with ineffective transport to the cell surface. The most representative example is CFTR. It was shown that the ΔF508 mutation in the CFTR gene causes severe cystic fibrosis. This mutant is reported to be degraded by the proteasome due to its unfolding status, and this is accepted as an explanation as to why mutant CFTR never reaches the plasma membrane. In mammalian cells, however, only ∼20% of newly synthesized wild type CFTR folds correctly and is transported from the ER to the plasma membrane through the secretory pathway, whereas the remaining ∼80% of CFTR is degraded from the ER through the ubiquitin-proteasome pathway (52). It the importance of the ERAD mechanisms in avoiding any unfolded or unassembled proteins accumulation in the cell is now well recognized. Thus, there are precedents that not only mutated proteins but also even wild type membrane proteins can be degraded by the proteasome. Indeed, many wild type GPCRs have been shown to be retained in the ER, and only a small portion of synthesized GPCR molecules is transported to the plasma membrane for their function. Among such GPCRs are the δ-opioid receptors (22, 23), luteinizing hormone receptors (24), olfactory receptors (26), and the thyrotropin-releasing hormone receptor (28).
Throughout this work, we measured the cell surface expression of the receptors by immunoassay, which enabled us to show the exact percentage expression of control. This method could represent the lower limit of the A2BR CT deletion mutant (Fig. 6B). However, we believe that this assay is the most reliable and convenient method of checking the expression of cell surface receptors, especially that of A2BR. Using a radiolabeled ligand, the sensitivity might be better than with an immunoassay in some respects, but this has its own disqualifying negatives. Indeed, the radiolabeled ligand assay usually requires the isolation of membrane, and this process could disturb A2BR expressed in low numbers at the cell surface. In this sense, this method might yield misleading results far removed from the situation with nearly intact cells. FACS analysis to check cell surface expression may not be suitable for A2BR, because the procedure of harvesting cells would also destroy low number, surface-expressed A2BR. For these reasons, the immunoassay is the best remaining option, which only requires fixation of the cells (as we put HA, FLAG, and Myc tags at the N terminus of the receptor, which was already exposed at the cell surface) before incubating them with the antibodies and substrate to measure cell surface expression of receptors.
In the absence of selective and membrane-permeable pharmacological chaperones, we used nonselective chemical chaperones to check A2BR folding status. Because it has been shown that even nonselective chemical chaperones are able to stabilize proteins and prevent their degradation (39–42), we tested the three most common chemical chaperones for their effect on the folding status of A2BR. In contrast to observations with other proteins (39–42), we found that these chaperones did not stabilize A2BR. Therefore, we were not completely able to exclude the possibility that A2BR degradation is due to its unfolded conformation. However, if unfolding of A2BR is a main factor that triggered degradation, then the question is why we observed improved surface expression of A2BR-A2ARCT and HA-A2BR-fA2ARCT chimeric proteins (300 and 150% of wild type, respectively). Thus, it appears that the unfolding status of A2BR could be only a small factor in determining the A2BR degradation.
In general, a simple explanation of GPCR retention within intracellular compartments in the absence of appropriate signals for forward export or in the case of a lack of private chaperones or escort proteins might be due to the persistent interaction with proteins of the general quality control machinery. Although this hypothesis remains somewhat plausible, some observations argue for the existence of specific ER export mechanisms. The export of many proteins from the ER is mandatory, to ensure their proper destinations, and it requires signal-mediated sorting into ER-derived transport vesicles (53). For instance, transport of the vesicular stomatitis virus glycoprotein (VSV-G) has served as a model for studying protein folding and export from the ER. VSV-G, a type I transmembrane protein that traffics to the cell surface and possesses a cytoplasmically exposed CT sequence of 29 residues, is required for transport from the ER. Within this tail sequence, a conserved YTDIEM motif is necessary for efficient export of VSV-G from the ER (54, 55). For GPCR, the F(X)6LL motif is highly conserved in the membrane-proximal CT of GPCRs (46), and thus this F(X)6LL motif may provide a common mechanism for ER export of GPCRs. Interestingly, β2AR has an F(X)6LL motif in its CT (46), and both A2AR and A2BR also have an F(X)6LL motif in their CTs. Considering our present results, the F(X)6LL motif does not seem to be an efficient ER export signal for A2BR. As with A2BR, although α1D-adrenergic receptors possesses an F(X)6LL motif (46), it is also reported that heteromerizaion with α1B-adrenergic receptors is required for efficient cell surface expression (56). However, there are many other membrane proteins that are efficiently exported from the ER but do not contain apparent motifs, suggesting that divergent ER export systems may exist. Therefore, it will be interesting to determine in future studies why the F(X)6LL motif in A2BR is not sufficient to deliver the receptor to the cell surface. One of the possibilities is that not only FLL but also amino acid residues between Phe and Leu-Leu and/or nearby sequences could be important for ER export. Now we are investigating the precise molecular mechanisms of why the F(X)6LL motif in A2BR is not efficient for ER export.
Figs. 6 and 7 show the results of studies of A2BR chimeras where the CT of A2BR was replaced or fused with the CT of A2AR or β2AR CT. It is instructive that the β2AR CT had less effect on cell surface localization of A2BR than the A2AR CT, suggesting that not all export signals in the CT of different GPCR molecules are similarly effective in enabling the A2BR surface expression. Because the HA-A2BR-A2ARCT (replaced) chimera showed more efficient transport than HA-A2BR-fA2ARCT (fused) to the cell surface, it suggests that the conformation of the chimera could be also important. Therefore, we were not able to exclude the possibility that it is the unfavorable conformation of both the HA-A2BR-β2CT (replaced) and HA-A2BR-fβ2CT (fused) chimeras that prevents their export from the ER, so that these chimeras are not expressed at levels exceeding those for wild type molecule. Another possibility could be that the kinetics of internalization, recycling, and/or degradation at the lysosome for A2BR-A2ARCT and A2BR-β2CT chimeras might be different, which could reflect the different cell surface expressions of these chimeras at the steady state.
There are several A2AR-interacting proteins that may regulate the trafficking (48, 57–59), but to our knowledge only E3KARP and Ezrin are reported to interact with A2BR (29). However, the precise molecular mechanisms that regulate A2BR trafficking and function are still uncertain. Unlike GABABR2, which is not expressed at the cell surface due to the ER retention motif in CT (60), a small portion of A2BR can be transported from the ER to the cell surface, probably by constitutive flow. Our results suggested that the majority of A2BR is degraded by the proteasome; however, once it acquires the appropriate transport signal, A2BR can be transported to the cell surface more efficiently.
Indeed, this was the case. Co-expression of A2AR together with A2BR enhanced A2BR surface expression (Fig. 8). Model-building for A2AR-A2BR interactions was limited by the absence of the whole crystal structure data for both A2AR and A2BR. Although there has been an important advance in the crystal structure of human A2AR with antagonist, unfortunately the analysis was done without CT (61). Nevertheless, the existing data and interpretations of studies of human β2AR (62) do allow the suggestion that that the CT of both A2AR and A2BR could be relatively unstructured. This is, in turn, supported by the absence of any known structures in their CT by Web-based software (Protein sequence analysis in Pôle Bioinformatique Lyonnais). Therefore, because both A2AR and A2BR are expected to have relatively unstructured CT, it is highly unlikely that the CT of both A2AR and A2BR are involved in their dimerization. We expect that dimerization may be mediated by other domains, probably transmembrane domains. The next interesting question is how the A2AR-A2BR dimer may use the F(X)6LL motif in A2AR CT as a dominant signal for efficient transport from the ER, because A2BR has no dominant forward transport signal from the ER on its own. It is likely that the A2AR-A2BR dimer can exit from the ER more efficiently than an A2BR monomer and/or homodimer because the coat protein complex II may recognize its F(X)6LL motif in the A2AR CT. This hypothesis is supported by the results shown in Fig. 10D, where FLAG-A2AR FII/AAA almost completely blocked the cell surface expression of HA-A2BR when transfected together. In this case, A2AR FII/AAA and A2BR heterodimer were not able to exit from the ER, because coat protein complex II no longer recognized the mutant motif. This mechanism greatly contrasts with that for the GABAB receptor, because the CT of GABABR2 interacts with that of GABABR1 through a coiled-coil structure to mask its ER retention motif, RXR(R) (63). Therefore it is likely that A2BR function as a hetero-oligomeric complex for better functioning. Like the T cell receptor assembly (64), if A2BR does not meet the A2AR and/or other components (possibly at the ER) and fails to assemble into a complex, it will be recognized as an unassembled protein, and thus A2BR will be degraded by the proteasome.
We conducted our studies (except for Fig. 11) through exogenously overexpressed proteins in a model cell line. Overexpression could affect their folding status, degradation, and trafficking. We also encountered technical limitations in studies of cAMP response with agonist and/or antagonist in transfected cells, where we obtained less clear results than those from the ex vivo data (Fig. 11). This could be explained by the high background of nontreated cells, due in part to cell damage by the transfection reagent, leading to a higher concentration of adenosine levels in the media. To overcome this limitation due to the transfection reagent, we considered using stable A2AR and/or A2BR transfectants, but the limitation here was that it was hard to accomplish an equal expression level of transfected molecules per cells. Controlling the expression levels is critical for these cases, based on our results in Fig. 1 (i.e. cell surface expression of A2AR and A2BR depends on total expression levels). However, and more importantly, these results in transfected cells described above were confirmed by a cAMP accumulation assay using functional A2AR and A2BR in splenocytes from wild type, A2AR knock-out, and A2BR knock-out mice (Fig. 11).
In summary, we have provided possible mechanisms as to the inefficient transport of A2BR to the cell surface. We showed that poor A2BR expression at the cell surface might be accounted for mainly by the lack of dominant forward transport signals from the ER to the plasma membrane. As it is now well recognized that many GPCRs form hetero-oligomer complexes for trafficking and function (60), it is likely that A2BR could assemble into a hetero-oligomer complex with A2AR and other interacting partners and then be transported together to the cell surface more efficiently. It will be important in future studies to identify whether A2BR has other interacting partners, including other GPCRs, and what their possible role may be in enabling the function and cell surface expression of A2BR.
Supplementary Material
Acknowledgments
We thank Dr. Rob Jackson, Tufts Center for Neuroscience Research, for help with confocal microscopy, and Dr. Alenka Lovy-Wheeler for technical assistance with the Leica confocal microscope.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA111985 and R01 CA112561 (to M. V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. 1 and 2.
- ER
- endoplasmic reticulum
- AR
- adenosine receptor
- CT
- carboxyl terminus
- β2AR
- β2 adrenergic receptor
- GPCR
- G protein-coupled receptor
- ERAD
- ER-associated degradation
- NECA
- 5-N-ethyl-carboxamidoadenosine
- ABTS
- 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)
- DMSO
- dimethyl sulfoxide
- TMAO
- osmolyte trimethylamine N-oxide
- CFTR
- cystic fibrosis transmembrane conductance regulator
- VSV-G
- vesicular stomatitis virus glycoprotein.
REFERENCES
- 1.Wojcikiewicz R. J. (2004) Trends Pharmacol. Sci. 25, 35–41 [DOI] [PubMed] [Google Scholar]
- 2.Hanson M. A., Stevens R. C. (2009) Structure 17, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mustafi D., Palczewski K. (2009) Mol. Pharmacol. 75, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fredholm B. B., Ijzerman A. P., Jacobson K. A., Klotz K. N., Linden J. (2001) Pharmacol. Rev. 53, 527–552 [PMC free article] [PubMed] [Google Scholar]
- 5.Fredholm B. B., Hokfelt T., Milligan G. (2007) Acta Physiol. (Oxf.) 190, 3–7 [DOI] [PubMed] [Google Scholar]
- 6.Linden J. (2005) Mol. Pharmacol. 67, 1385–1387 [DOI] [PubMed] [Google Scholar]
- 7.Sitkovsky M. V., Lukashev D., Apasov S., Kojima H., Koshiba M., Caldwell C., Ohta A., Thiel M. (2004) Annu. Rev. Immunol. 22, 2101–2126 [DOI] [PubMed] [Google Scholar]
- 8.Ohta A., Sitkovsky M. (2001) Nature 414, 916–920 [DOI] [PubMed] [Google Scholar]
- 9.Milligan G., Ramsay D., Pascal G., Carrillo J. J. (2003) Life Sci. 74, 181–188 [DOI] [PubMed] [Google Scholar]
- 10.Yang D., Koupenova M., McCrann D. J., Kopeikina K. J., Kagan H. M., Schreiber B. M., Ravid K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 792–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montesinos M. C., Shaw J. P., Yee H., Shamamian P., Cronstein B. N. (2004) Am. J. Pathol. 164, 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun C. X., Zhong H., Mohsenin A., Morschl E., Chunn J. L., Molina J. G., Belardinelli L., Zeng D., Blackburn M. R. (2006) J. Clin. Invest. 116, 2173–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta A., Gorelik E., Prasad S. J., Ronchese F., Lukashev D., Wong M. K., Huang X., Caldwell S., Liu K., Smith P., Chen J. F., Jackson E. K., Apasov S., Abrams S., Sitkovsky M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13132–13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hipkin R. W., Sánchez-Yagüe J., Ascoli M. (1992) Mol. Endocrinol. 6, 2210–2218 [DOI] [PubMed] [Google Scholar]
- 15.VuHai-LuuThi M. T., Misrahi M., Houllier A., Jolivet A., Milgrom E. (1992) Biochemistry 31, 8377–8383 [DOI] [PubMed] [Google Scholar]
- 16.Vannier B., Loosfelt H., Meduri G., Pichon C., Milgrom E. (1996) Biochemistry 35, 1358–1366 [DOI] [PubMed] [Google Scholar]
- 17.Apaja P. M., Aatsinki J. T., Rajaniemi H. J., Petäjä-Repo U. E. (2005) Endocrinology 146, 3224–3232 [DOI] [PubMed] [Google Scholar]
- 18.Parodi A. J. (2000) Annu. Rev. Biochem. 69, 69–93 [DOI] [PubMed] [Google Scholar]
- 19.Fishburn C. S., Elazar Z., Fuchs S. (1995) J. Biol. Chem. 270, 29819–29824 [DOI] [PubMed] [Google Scholar]
- 20.Imai Y., Soda M., Inoue H., Hattori N., Mizuno Y., Takahashi R. (2001) Cell 105, 891–902 [DOI] [PubMed] [Google Scholar]
- 21.Wüller S., Wiesner B., Löffler A., Furkert J., Krause G., Hermosilla R., Schaefer M., Schülein R., Rosenthal W., Oksche A. (2004) J. Biol. Chem. 279, 47254–47263 [DOI] [PubMed] [Google Scholar]
- 22.Petaja-Repo U. E., Hogue M., Laperriere A., Walker P., Bouvier M. (2000) J. Biol. Chem. 275, 13727–13736 [DOI] [PubMed] [Google Scholar]
- 23.Petaja-Repo U. E., Hogue M., Laperriere A., Bhalla S., Walker P., Bouvier M. (2001) J. Biol. Chem. 276, 4416–4423 [DOI] [PubMed] [Google Scholar]
- 24.Pietilä E. M., Tuusa J. T., Apaja P. M., Aatsinki J. T., Hakalahti A. E., Rajaniemi H. J., Petäjä-Repo U. E. (2005) J. Biol. Chem. 280, 26622–26629 [DOI] [PubMed] [Google Scholar]
- 25.Apaja P. M., Tuusa J. T., Pietilä E. M., Rajaniemi H. J., Petäjä-Repo U. E. (2006) Mol. Biol. Cell 17, 2243–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu M., Echeverri F., Moyer B. D. (2003) Traffic 4, 416–433 [DOI] [PubMed] [Google Scholar]
- 27.Andersson H., D'Antona A. M., Kendall D. A., Von Heijne G., Chin C. N. (2003) Mol. Pharmacol. 64, 570–577 [DOI] [PubMed] [Google Scholar]
- 28.Cook L. B., Zhu C. C., Hinkle P. M. (2003) Mol. Endocrinol. 17, 1777–1791 [DOI] [PubMed] [Google Scholar]
- 29.Sitaraman S. V., Wang L., Wong M., Bruewer M., Hobert M., Yun C. H., Merlin D., Madara J. L. (2002) J. Biol. Chem. 277, 33188–33195 [DOI] [PubMed] [Google Scholar]
- 30.Moriyama K., Bonifacino J. S. (2002) Traffic 3, 666–677 [DOI] [PubMed] [Google Scholar]
- 31.Higuchi R. (1990) in PCR Protocols: A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. eds) pp. 177–183, Academic Press, San Diego, CA [Google Scholar]
- 32.Dell'Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S. (1999) Mol. Cell 3, 11–21 [DOI] [PubMed] [Google Scholar]
- 33.Lee C. M., Hickey M. M., Sanford C. A., McGuire C. G., Cowey C. L., Simon M. C., Rathmell W. K. (2009) Oncogene 28, 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammad S., Baldini G., Granell S., Narducci P., Martelli A. M., Baldini G. (2007) J. Biol. Chem. 282, 4963–4974 [DOI] [PubMed] [Google Scholar]
- 35.Göransson O., Deak M., Wullschleger S., Morrice N. A., Prescott A. R., Alessi D. R. (2006) J. Cell Sci. 119, 4059–4070 [DOI] [PubMed] [Google Scholar]
- 36.Denning G. M., Anderson M. P., Amara J. F., Marshall J., Smith A. E., Welsh M. J. (1992) Nature 358, 761–764 [DOI] [PubMed] [Google Scholar]
- 37.Gelman M. S., Kopito R. R. (2002) J. Clin. Invest. 110, 1591–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Morchoe C. C., Jones W. R., 3rd, Jarosz H. M., O'Morchoe P. J., Fox L. M. (1984) J. Cell Biol. 98, 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamarappoo B. K., Verkman A. S. (1998) J. Clin. Invest. 101, 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato S., Ward C. L., Krouse M. E., Wine J. J., Kopito R. R. (1996) J. Biol. Chem. 271, 635–638 [DOI] [PubMed] [Google Scholar]
- 41.Burrows J. A., Willis L. K., Perlmutter D. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1796–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan C. M., Nickols H. H., Limbird L. E. (2003) J. Biol. Chem. 278, 35678–35686 [DOI] [PubMed] [Google Scholar]
- 43.Wada I., Ou W. J., Liu M. C., Scheele G. (1994) J. Biol. Chem. 269, 7464–7472 [PubMed] [Google Scholar]
- 44.Teasdale R. D., Jackson M. R. (1996) Annu. Rev. Cell Dev. Biol. 12, 27–54 [DOI] [PubMed] [Google Scholar]
- 45.Michelsen K., Yuan H., Schwappach B. (2005) EMBO Rep. 6, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duvernay M. T., Zhou F., Wu G. (2004) J. Biol. Chem. 279, 30741–30750 [DOI] [PubMed] [Google Scholar]
- 47.Pankevych H., Korkhov V., Freissmuth M., Nanoff C. (2003) J. Biol. Chem. 278, 30283–30293 [DOI] [PubMed] [Google Scholar]
- 48.Milojevic T., Reiterer V., Stefan E., Korkhov V. M., Dorostkar M. M., Ducza E., Ogris E., Boehm S., Freissmuth M., Nanoff C. (2006) Mol. Pharmacol. 69, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 49.Lefkowitz R. J. (2007) Acta Physiol. (Oxf.) 190, 9–19 [DOI] [PubMed] [Google Scholar]
- 50.Salahpour A., Angers S., Mercier J. F., Lagacé M., Marullo S., Bouvier M. (2004) J. Biol. Chem. 279, 33390–33397 [DOI] [PubMed] [Google Scholar]
- 51.Ulloa-Aguirre A., Janovick J. A., Miranda A. L., Conn P. M. (2006) ACS Chem. Biol. 1, 631–638 [DOI] [PubMed] [Google Scholar]
- 52.Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. (1998) EMBO J. 17, 6879–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barlowe C. (2003) Trends Cell Biol. 13, 295–300 [DOI] [PubMed] [Google Scholar]
- 54.Nishimura N., Bannykh S., Slabough S., Matteson J., Altschuler Y., Hahn K., Balch W. E. (1999) J. Biol. Chem. 274, 15937–15946 [DOI] [PubMed] [Google Scholar]
- 55.Sevier C. S., Weisz O. A., Davis M., Machamer C. E. (2000) Mol. Biol. Cell 11, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hague C., Uberti M. A., Chen Z., Hall R. A., Minneman K. P. (2004) J. Biol. Chem. 279, 16641–16649 [DOI] [PubMed] [Google Scholar]
- 57.Burgueño J., Blake D. J., Benson M. A., Tinsley C. L., Esapa C. T., Canela E. I., Penela P., Mallol J., Mayor F., Jr., Lluis C., Franco R., Ciruela F. (2003) J. Biol. Chem. 278, 37545–37552 [DOI] [PubMed] [Google Scholar]
- 58.Sun C. N., Cheng H. C., Chou J. L., Lee S. Y., Lin Y. W., Lai H. L., Chen H. M., Chern Y. (2006) Mol. Pharmacol. 70, 454–466 [DOI] [PubMed] [Google Scholar]
- 59.Gsandtner I., Charalambous C., Stefan E., Ogris E., Freissmuth M., Zezula J. (2005) J. Biol. Chem. 280, 31898–31905 [DOI] [PubMed] [Google Scholar]
- 60.Prinster S. C., Hague C., Hall R. A. (2005) Pharmacol. Rev. 57, 289–298 [DOI] [PubMed] [Google Scholar]
- 61.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science. 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granier S., Kim S., Shafer A. M., Ratnala V. R., Fung J. J., Zare R. N., Kobilka B. (2007) J. Biol. Chem. 282, 13895–13905 [DOI] [PubMed] [Google Scholar]
- 63.Margeta-Mitrovic M., Jan Y. N., Jan L. Y. (2000) Neuron 27, 97–106 [DOI] [PubMed] [Google Scholar]
- 64.Yang M., Omura S., Bonifacino J. S., Weissman A. M. (1998) J. Exp. Med. 187, 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.