Abstract
CD151, a transmembrane protein of the tetraspanin family, is implicated in the regulation of cell-substrate adhesion and cell migration through physical and functional interactions with integrin receptors. In contrast, little is known about the potential role of CD151 in controlling cell proliferation and survival. We have previously shown that β4 integrin, a major CD151 partner, not only acts as an adhesive receptor for laminins but also as an intracellular signaling platform promoting cell proliferation and invasive growth upon interaction with Met, the tyrosine kinase receptor for hepatocyte growth factor (HGF). Here we show that RNAi-mediated silencing of CD151 expression in cancer cells impairs HGF-driven proliferation, anchorage-independent growth, protection from anoikis, and tumor progression in xenograft models in vivo. Mechanistically, we found that CD151 is crucially implicated in the formation of signaling complexes between Met and β4 integrin, a known amplifier of HGF-induced tumor cell growth and survival. CD151 depletion hampered HGF-induced phosphorylation of β4 integrin and the ensuing Grb2-Gab1 association, a signaling pathway leading to MAPK stimulation and cell growth. Accordingly, CD151 knockdown reduced HGF-triggered activation of MAPK but not AKT signaling cascade. These results indicate that CD151 controls Met-dependent neoplastic growth by enhancing receptor signaling through β4 integrin-mediated pathways, independent of cell-substrate adhesion.
Keywords: Cell Surface Receptor, Integrin, Receptor tyrosine Kinase, Signal Transduction, Tumor, CD151, HGF, MET, Tetraspanin, Tumor Growth
Introduction
CD151 is a cell surface protein of the tetraspanin family. These proteins contain four transmembrane domains, two extracellular loops, and two short cytoplasmic tails (1, 2). Tetraspanins are major partners of integrins that regulate cell adhesion to extracellular matrix components as well as cell-cell contacts, thereby modulating cell migration. In particular, CD151 is typically associated with α3β1, α6β1, and α6β4 integrin receptor complexes for laminins (3, 4), and its expression is increased during epithelial-mesenchymal transition (5). Not surprisingly, the activity of tetraspanins as pro-migratory molecules has been implicated in tumor invasion and metastasis (6, 7). For example, elevated CD151 expression has been correlated with poor prognosis and increased metastasis in colon, lung, and prostate cancers (8–10). In line with this, the association between integrin α6β4 and CD151 has been linked to tumor cell motility, increased metastasis, and poor prognosis in colon cancer (11). More recently, it has been shown that a CD151-specific antibody can prevent tumor cell invasion and metastasis by modulating cell-substrate adhesion (12). Notably, although the function of CD151 in the regulation of integrin-dependent adhesion and migration is well established, its role in cell growth and survival signaling is poorly understood.
Tetraspanins can interact with several partners in addition to integrins and form intricate protein networks in tetraspanin-enriched microdomains at the cell surface, which may function as platforms regulating receptor signaling (13). In this context, previous studies have demonstrated that CD151, CD9, and CD82 can modulate cancer cell motility and morphogenesis elicited by the activation of Met and EGF receptor tyrosine kinase receptors (RTKs)4 (14–21). Because of its role in promoting integrin-dependent adhesion, CD151 has been envisaged so far mostly as a linking molecule in the functional cross-talk between integrin-mediated signals, RTK activation, and cancer cell migration. Intriguingly, activated RTKs are known to cross-talk with integrin receptors not only to regulate migration but also to promote cell growth and survival (22, 23). One paradigmatic example is integrin α6β4, a major CD151 partner (24), which cooperates with RTKs to promote anchorage-independent growth, cell transformation, and tumor progression. Accordingly, we have shown previously that the long cytosolic tail of β4 integrin acts as an adhesion-independent signaling substrate that amplifies tumor cell growth, survival, and invasive phenotypes mediated by RTK Met (25, 26). In this scenario, our working hypothesis was that the tetraspanin CD151 might participate in the control of cell proliferation and tumor growth by acting as molecular bridge between oncogenic Met receptor and integrins. Therefore, in this work, we specifically focused on the growth and survival responses elicited by hepatocyte growth factor (HGF)/Met signaling in tumor cells and demonstrated that CD151 positively controls Met-dependent signaling leading to cell proliferation, protection from anoikis, and in vivo tumorigenesis. Mechanistic studies revealed that CD151 is required for the phosphorylation of β4 integrin elicited by Met and for the ensuing adhesion-independent pathway promoting tumor cell growth.
MATERIALS AND METHODS
Cell Culture and Cell Transfection
Tumor cell lines were derived from ATCC. GTL16 cells, derived from a human gastric carcinoma, were previously described by Giordano et al. (47) (see also Trusolino et al. (23)). Cells were grown in standard culture medium supplemented with 10% fetal bovine serum. The expression constructs encoding β4 integrin, Grb2, Gab1, and human HGF (poly-His-tagged) have been described previously (25–27). The shRNA expression vector targeting β4 has been previously described (25). For ectopic expression experiments, human CD151 cDNA was subcloned into a lentiviral expression construct (pRRLsinPPThCMV-MCSpre). Lentiviral particles were produced as described (28) and used to transduce target cells in the presence of 8 μg/ml Polybrene (Sigma-Aldrich). The K-RASG12V vector was from F. d'Adda di Fagagna (The FIRC Institute of Molecular Oncology, Milan, Italy). cDNA transfection of A549 cells was performed using Lipofectamine2000 (Invitrogen).
Antibodies and Other Reagents
Primary antibodies were as follows: anti-phosphotyrosine and a-Gab1 were from Upstate Biotech Millipore (Charlottesville, VA); anti-actin was from Santa Cruz Biotechnology (Santa Cruz, CA); anti-AKT, anti-p42/44 MAPK(Erk1/2), and anti-phospho-p42/44 MAPK were from Cell Signaling (Danvers, MA); and anti-Met monoclonal antibodies (DO24 and DL21 clones) have been previously described (29). Anti-β4 integrin (clone 450-11A) was from BD Biosciences; anti-β1 integrin (clone 18) and anti-Grb2 were from BD Transduction Laboratories. Anti-human CD151 (clone 11G5a) from Serotec (Raleigh, NC) was used for immunoprecipitation; anti-CD151 (clone 11B1), kindly provided by Prof. Ashman (University of Newcastle, Australia), was used for immunoblotting. Secondary antibodies were purchased from Amersham Biosciences. Purified recombinant HGF was kindly provided by Genentech Inc. (South San Francisco, CA). Methyl-β-cyclodextrin was purchased from Sigma-Aldrich.
Knockdown of Gene Expression by shRNA
CD151 expression was stably suppressed in tumor cells by lentiviral-mediated expression of shRNA specifically targeting the CD151 transcript, using short hairpin RNA (shRNA) cloned into lentivirus expression vector pLKO.1-puro control vector (Sigma-Aldrich). For most experiments, the targeted sequence was 5′-CTCAAGTACCTGCTGTTTA-3′, whereas in selected experiments, a second sequence was used: 5′-TGGAGATCATCGCTGGTAT-3′ (indicated as “shCD151_2”). The sequences were BLAST-searched against all human sequences and were not found to have significant homology to genes other than CD151. Following incubation with lentiviral particles, infected cells were selected in culture in the presence of 1 μg/ml puromycin (Sigma-Aldrich).
Cell Viability Assays
Cells were seeded in 24-well culture dishes (Costar) at an initial density of 2 × 104 cells/well. Either the next morning (0 h) or 72 h later, upon culturing in medium supplemented with 0.5% FBS, the cells were incubated for 1 h with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) diluted in culture medium. Eventually, 300 μl of dimethyl sulfoxide (DMSO) were added to each well to dissolve the colored product of the mitochondrial-based reaction, and the optical density was measured at 570 nm in a microtiter plate reader.
Cell Growth Curves
Tumor cells were seeded in multiple 96-well dishes (Costar) at an initial density of 3 × 103 cells/well. The cells were cultured in medium supplemented with 0.5% FBS, with or without HGF (20 ng/ml). Every 24 h, a fraction of the wells were fixed with 11% glutaraldehyde and stained with crystal violet, and the absorbance was read using a standard colorimetric system at 562 nm.
Adhesion-independent Cell Growth in Soft Agar
For the analysis of colony formation in soft agar, 2 × 103 cells were resuspended in medium containing 10% FBS plus 0.5% SeaPlaque agar (Lonza Group Ltd., Basel, Switzerland). Cells were seeded in 24-well plates containing a 1% agar underlay and maintained in the absence or presence of HGF (20 ng/ml). Foci of cellular growth were visualized 3 weeks after seeding by staining with tetrazolium salt 0.2 mg/ml. Spectrometric measurements at 570 nm were recorded using a DTX 880 multimode plate reader (Beckman-Coulter).
Adhesion-independent Cell Survival (Anoikis Assay)
24-well cell culture dishes were coated with 200 μl of polyhydroxyethylmethacrylate (poly-HEMA) (12 mg/ml in 95% ethanol) according to standard protocols (30, 31). The solution was left to dry for 48 h at room temperature. Cells were then seeded in the poly-HEMA-coated wells (at an initial concentration of 2 × 104) and cultured in medium supplemented with 1% serum, with or without HGF (10 ng/ml), for 72 h. Then the surviving cells were collected in a new multiwell dish, and cell survival was assessed by ATP content using a luminescence assay (ATPlite 1 step kit, PerkinElmer Life Sciences). Measurements were recorded using a DTX 880 multimode plate reader (Beckman-Coulter).
RNA Isolation and Real-time PCR
Total RNA from tumor cell lines was isolated by using the RNeasy Protect mini kit (Qiagen, Inc, Chatsworth, CA) according to the manufacturer's instructions. cDNA preparation was performed according to standard procedures using M-MLV RT (Promega, Madison, WI) and oligo(dT) (Sigma-Aldrich) primers. Gene expression was determined by real-time quantitative PCR with ABI PRISM 7900HT (Applied Biosystems, Milan, Italy) using the following SYBR Green primer pairs: h-CD151 (NM_004357) forward, 5′-GCACCGTTTGCCTCAAGT-3′, reverse, 5′-ACCACCAGGATGTAGGCTGT-3′; h-HGF (NM_000601.4) forward, 5′-CGAGGGAAGGTGACTCTGAA-3′, reverse, 5′-CCACGACCAGGAACAATGAC-3′; and h-GAPDH (NM_002046) forward, 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
Protein Analysis, Immunoprecipitation, and Western Blotting
In most experiments, cellular proteins were solubilized using a 1% Triton X-100-containing buffer (Extraction Buffer), as described previously (27) and quantified using the Bio-Rad protein assay (Bio-Rad Laboratories GmbH, Munich, Germany). Cell extracts were electrophoresed on SDS-polyacrylamide gel, transferred onto nitrocellulose membranes (Hybond, GE Healthcare Europe GmbH, Munich, Germany), and blocked in phosphate-buffered saline, 0.1% Tween 20, 5% BSA. The membrane was incubated with appropriate dilutions of primary antibody followed by the appropriate peroxidase-conjugated secondary antibody. Detection was performed with the ECL system (Amersham Biosciences). For CD151 co-immunoprecipitation experiments, cellular proteins were solubilized in a buffer containing 1% Brij 97 or 0.5% Nonidet P-40, plus 1 m Tris, pH 8, 5 m NaCl, 1 m MgCl2, and a mixture of protease inhibitors (32). To detect CD151 in Western blotting, SDS-PAGE in non-reducing conditions was applied.
For experiments with cells in suspension, A549 cells were cultured for 48 h without fetal serum and then detached by incubating with 5 mm EDTA in PBS. After centrifugation, cell pellets were resuspended in serum-free medium, with or without HGF, and incubated for 10 min at 37°.
Xenograft Transplantation Experiments
Lentivirus-transduced A549 cells (7 × 106) were injected subcutaneously into both of the flanks of immunodeficient CD1 nu−/− female mice (Charles River Laboratories, Lecco, Italy). Tumor growth was monitored by external measuring with a caliper every 4 days, and volume estimation was obtained applying a formula described previously (28). All the experiments were performed on groups of six animals per experimental point. All animal procedures were approved by the Ethical Commission of the University of Torino, Italy and by the Italian Ministry of Health.
Statistics
Results are means of at least two or three different experiments + S.E. Comparisons were made using two-tailed Student's t test (or one-way analysis of variance test, when more than two experimental groups were compared). p values < 0.05 were considered to be statistically significant.
RESULTS
CD151 Is Required to Mediate HGF-induced Cell Proliferation, Adhesion-independent Growth, and Survival
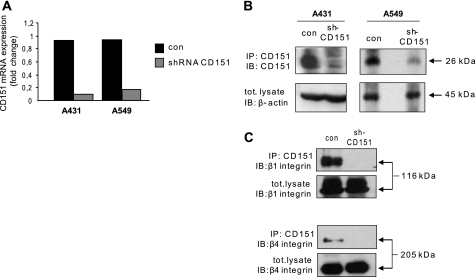
To elucidate the functional relevance of CD151 in cancer cell behavior, we transduced A431 (human epidermoid carcinoma) and A549 (non-small cell lung carcinoma) cells with lentiviral vectors carrying either shRNAs directed against CD151 (sh-CD151) or an empty vector control (con). The constructs contained a puromycin resistance cassette, and over 80% of the cells survived drug selection, indicating a very high efficiency of gene transfer. We therefore obtained pools of shRNA-expressing cells that were analyzed in subsequent experiments. The silencing of CD151 in both A431 and A549 tumor cell lines was demonstrated by quantitative PCR analysis, which indicated a down-regulation of mRNA expression levels to 10% of controls (Fig. 1A). This was further confirmed by Western blotting analysis (Fig. 1B). Moreover, although β1 and β4 integrins are tightly associated with CD151 in control cells, we could not co-purify these integrins with the residual tetraspanin expressed in silenced cells (Fig. 1C), indicating the achievement of a virtually complete functional knock-out of CD151.
FIGURE 1.
Generation of CD151-deficient cells. A431 and A549 carcinoma cells were transduced with lentiviral vectors to express shRNA sequences targeting CD151 transcript (shCD151) or with empty lentiviral vectors (con). CD151 silencing was verified by real-time quantitative PCR (A) as well as by immunoblotting (IB) (B). The expression of β-actin was checked by Western blotting on total lysates to provide a loading control. IP, immunoprecipitation. C, the co-immunoprecipitation of β1 and β4 integrins with CD151 was assayed by Western blotting in control and CD151-deficient A431 cells. tot. lysate, total lysate.
Consistent with previous reports (21, 33–35), CD151 down-regulation partly impaired integrin-dependent functions, especially cell adhesion and haptotactic migration on laminin substrates (data not shown and supplemental Fig. 1A). However, CD151-deficient cells could efficiently migrate through fibronectin-coated filters, both spontaneously and in response to HGF, a pleiotropic factor inducing proliferation and directional cell migration (supplemental Fig. 1B).
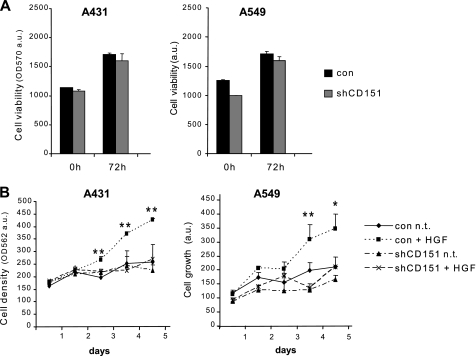
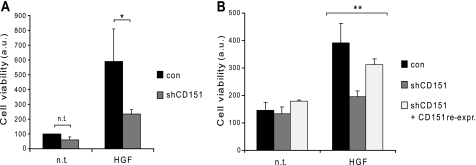
Importantly, although the viability of CD151-deficient A549 and A431 cancer cells was not basally affected (Fig. 2A), we found that their proliferative response to HGF was impaired (Fig. 2B). This finding suggested that CD151 might have a role in signaling pathways controlling tumor cell growth in addition to its well known activities as a pro-adhesive and a pro-migratory molecule. It has been shown that cell-substrate adhesion can support growth factor signaling in certain cells (36, 37). Therefore, to address this issue, we investigated the functional relevance of CD151 in the ability of cells to grow and survive in the absence of anchorage to the extracellular matrix (a major hallmark of the transformed phenotype induced by Met oncogene activation). In soft agar assays, which evaluate the clonal expansion of non-adherent cancer cells, CD151-deficient A549 cells formed less than 40% foci upon HGF stimulation when compared with control cells (Fig. 3A). In addition, HGF-induced cell survival was assessed by culturing tumor cells in wells coated with poly-HEMA. This treatment prevents cell-substrate adhesion and rapidly induces cell death (by a process known as anoikis), representing a powerful experimental tool to assay anti-apoptotic survival responses in tumor cells. Again, similar to that observed in soft agar assays, HGF stimulation efficiently protected control cells from anoikis but not CD151-deficient cells (Fig. 3B). To rule out potential off-target effects of the shRNA used for CD151 knockdown, we exogenously re-established the expression of this tetraspanin in gene-deficient A549 cells and found that this partially restored HGF-induced protection from anoikis (Fig. 3B). Together, these results indicate that CD151 is a positive effector of Met-dependent tumorigenesis in vitro.
FIGURE 2.
CD151 is required for cell growth response induced by HGF. A, cell viability was assessed in control (con) and CD151-deficient cells by MTT assays at two distinct time points (see “Materials and Methods” for details). Values in the graphs indicate the amount of reaction product, revealed by absorbance at 570 nm (average of two independent experiments performed in duplicate +S.E.). a.u., arbitrary units. B, HGF-induced cell proliferation was assessed by growth curves (see “Materials and Methods” for details). Cell staining with crystal violet was eventually revealed by absorbance at 562 nm. Three independent experiments were performed in triplicate with consistent results (**, p < 0.001; *, p < 0.05). con n.t., control untreated.
FIGURE 3.
CD151 mediates anchorage-independent growth and cell survival induced by HGF. A, A549 cells were grown in soft agar, in the presence or absence of HGF, to test their ability to survive and proliferate without anchorage to the extracellular matrix. After 3 weeks, living cells were stained with tetrazolium salts and scored by evaluating absorbance at 570 nm (see “Materials and Methods” for details). Values shown in graphs are means +S.E. of three independent experiments performed in triplicate, normalized to non-stimulated control cells; *, p < 0.05. a.u., arbitrary units; n.t., untreated; n.s., not significant; con, empty lentiviral vectors. B, A549 cells were grown in suspension in poly-HEMA-coated dishes to induce anoikis. After 72 h, cell viability was assessed by the MTT assay (see “Materials and Methods”). HGF induced cell survival in control cells, whereas this response was lost in CD151-deficient cells and partially rescued by CD151 re-expression (re-expr.) (quantitative PCR analysis of CD151 expression in the different cell lines is shown in supplemental Fig. 2). Data shown are the means +S.E. of three independent experiments performed in triplicate. Statistical significance was established by globally comparing the three experimental conditions by one-way analysis of variance test; **, p < 0.0001. Moreover, Student's t test analysis of paired samples indicated statistical significance of shCD151 and CD151-re-expression condition versus control, with p < 0.001 for both cases (not shown).
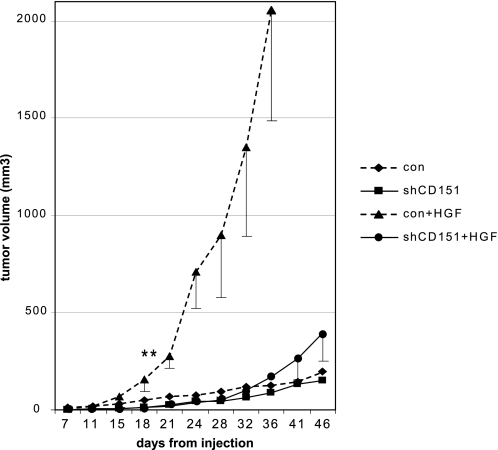
CD151 Is Required to Mediate HGF-induced Tumor Growth in Vivo
To further investigate the role of CD151 in HGF-mediated enhancement of tumor cell growth and survival, we exploited a previously established experimental model of HGF-driven tumorigenesis in mice. Thus, A549 tumor cells carrying an autocrine loop of HGF (28) were transduced with shRNA knocking down CD151 levels (expression analysis in supplemental Fig. 3) and then subcutaneously injected into nude mice; tumor size was monitored over a period of 2 months. CD151 knockdown did not significantly affect the growth of parental tumor cells in vivo (Fig. 4). On the other hand, consistent with previous findings, autocrine HGF overexpression remarkably accelerated tumor growth. Strikingly, this hyperproliferative response was almost totally abrogated in CD151-deficient cells (Fig. 4). These data confirm, in an in vivo setting, the critical role of CD151 in mediating Met-dependent tumor growth.
FIGURE 4.
CD151 is required for HGF-dependent tumorigenesis in vivo. A549 cells, either control or carrying a HGF autocrine loop, were transduced with shRNA-expressing constructs to knock down CD151 levels (see expression analysis in supplemental Fig. 3) and then injected subcutaneously in nude mice to score their tumorigenic potential. Tumor volume was periodically measured, indicating that although HGF autocrine signaling induced striking tumor growth, this effect was almost completely blunted in CD151-deficient cells. Two independent experiments were performed, including five mice per each experimental group, showing consistent results. Significance was assessed by Student's t test analysis on the complete series of values of the four experimental groups, revealing that only control (con) tumors expressing HGF were statistically different from any other condition; **, p < 0.005.
CD151 Is Required to Mediate HGF-induced Activation of the MAPK Pathway, Independently of Cell-Substrate Adhesion
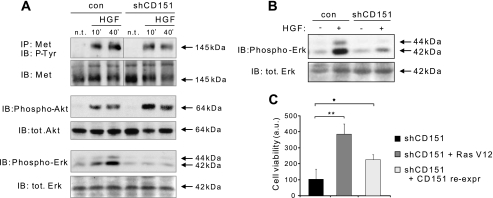
We furthermore analyzed the signaling consequences of CD151 depletion in response to HGF stimulation. CD151 knockdown did not affect tyrosine phosphorylation of Met in A549 tumor cells, indicating that the catalytic activity of the receptor was not impaired (Fig. 5A, top panels). Similarly, HGF-induced AKT phosphorylation was not impaired by CD151 down-regulation, suggesting that this major Met-driven signaling pathway is not dependent on CD151 (Fig. 5A, middle panels). On the other hand, silencing CD151 expression curbed HGF-induced MAPK-Erk1/2 activation (Fig. 5A, bottom panels); therefore, this tetraspanin seems to be required for stimulation of selected pathways downstream from Met activation.
FIGURE 5.
CD151 knockdown subverts HGF-induced signaling in cancer cell lines. A, control (con) and CD151-deficient A549 cells were starved 48 h and treated with 50 ng/ml HGF for the indicated times (or left untreated). Tyrosine-phosphorylated (P-Tyr) Met was detected by immunoprecipitation (IP) followed by immunoblotting (IB) (top row). Moreover, total protein cell lysates were probed to detect activated Akt (Phospho-Akt), total Akt (tot. Akt), activated MAPK/Erk (Phospho-Erk), and total MAPK/Erk (total Erk). CD151 knockdown impaired HGF-induced MAPK activation but was without significant effect on Met and Akt activation. n.s., not significant. B, A549 cells were serum-starved for 48 h, put in suspension, and stimulated or not with HGF (50 ng/ml for 15 min). Erk activation and total Erk were revealed with appropriate antibodies. CD151 knockdown hampers Erk activation in HGF-stimulated cells, in the absence of cell-substrate adhesion. C, hyperactivation of RAS by ectopic overexpression of K-RASG12V or re-expression of CD151 enhanced HGF-dependent soft agar growth of CD151-deficient cells. A549 cells were grown in soft agar in the presence of 50 ng/ml HGF. Foci of viable cells were eventually scored as in Fig. 3A. Data shown are the means +S.E. of two independent experiments performed in triplicate; **, p < 0.001; *, p < 0.01. a.u., arbitrary units.
We have demonstrated above that CD151 expression is required for HGF-dependent functional responses independent from cell-substrate adhesion, such as growth in soft agar and protection from anoikis. Accordingly, we tested whether HGF could induce MAPK activation in non-adherent tumor cells and whether this pathway would require CD151 expression. Indeed, we found that HGF elicited MAPK-Erk1/2 phosphorylation in suspended A549 cells, whereas this function was strongly impaired upon CD151 knockdown (Fig. 5B), consistent with that seen in adherent cells. Thus, the role of CD151 to enhance HGF-dependent activation of MAPK is independent from cell adhesion to the extracellular matrix.
Notably, the hyperactivation of MAPK cascade by ectopic expression of an active form of K-RAS (K-RASG12V) could increase the anchorage-independent growth of CD151-deficient A549 cells (Fig. 5C); consistent with Fig. 3B, increased growth was also observed by re-expression of CD151 in knocked down cells (Fig. 5C). Together, these findings substantiate the notion that CD151 sustains HGF-dependent and adhesion-independent responses through stimulation of the MAPK signaling axis.
CD151 Is Required for Functional Coupling between Met and β4 Integrin
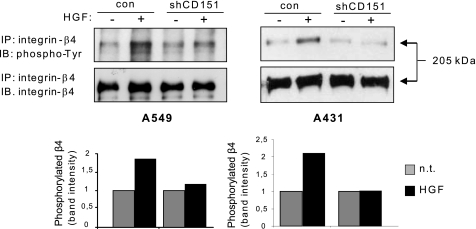
We finally asked about the molecular mechanism mediating HGF-dependent activation of MAPK signaling in the presence of CD151. We have previously demonstrated that HGF-induced proliferation is promoted by the interaction of Met with β4 integrin in multiple tumor cells, including A549 and A431 carcinomas. Following HGF-induced tyrosine phosphorylation, β4 amplifies Met signaling by acting as an associated docking platform for the recruitment of intracellular signal transducers (24, 26, 27). This β4 integrin-based signaling pathway mediates adhesion-independent biological outcomes, including anchorage-independent growth (26), and leads to dedicated stimulation of the MAPK cascade in response to HGF (27). Intriguingly, CD151 is a major partner of β4 integrin (25). We therefore asked whether the ability of CD151 to promote HGF-dependent tumor growth may implicate β4 integrin as a signaling intermediate. Actually, we found that Met-dependent phosphorylation of β4 integrin was lost in both A549 and A431 CD151-deficient cells (Fig. 6), suggesting the requirement of this tetraspanin in Met-β4 signaling complexes.
FIGURE 6.
CD151 mediates β4 integrin phosphorylation by Met. A549 and A431 cells (control (con) and CD151-depleted) were serum-starved for 48 h and treated or not with HGF 50 ng/ml for 20 min. CD151 knockdown inhibits HGF-induced β4 integrin tyrosine phosphorylation. IP, immunoprecipitation; IB, immunoblotting. n.s., not significant.
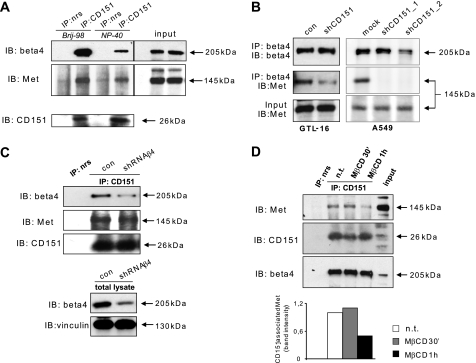
Because CD151 is typically associated with β4, and CD151 and Met can form physical complexes in certain tumor cells (15), we asked whether this tetraspanin could actually be an intermediate player required for the formation of Met-β4 complexes on the cell surface. Currently available antibodies do not allow the efficient purification CD151 immunocomplexes in A549 cells; therefore, to reveal a potential association of the three endogenous proteins, we analyzed a tumor model found to contain high amounts of CD151, together with Met and β4 integrin, namely GTL16 carcinoma cells. Indeed, we found that CD151 immunocomplexes contained both Met and β4 integrin, suggesting the formation of a ternary complex (Fig. 7A). Notably, tetraspanin-based receptor complexes are localized into specific compartments on the cell surface, known as tetraspanin-enriched microdomains (44), which can only be isolated using mild membrane detergents, such as Brij 97. Consistently, we found that CD151-Met-β4 complexes could be recovered by using mild detergents that preserve tetraspanin-enriched microdomains, whereas they were disrupted in the presence of Triton X-100 (Fig. 7A and data not shown). Of note, it was shown previously that Met-β4 complexes also should be recovered in the absence of Triton X-100 (23).
FIGURE 7.
CD151 mediates the formation of a ternary complex coupling β4 integrin with Met. A, CD151 was immunopurified from different lysates of GTL16 tumor cells (upon protein solubilization with either Brij 97 or Nonidet P-40 detergent (NP-40)), and immunocomplexes were analyzed by Western blotting with specific antibodies to detect associated receptor molecules. Both β4 integrin and Met were selectively found in association with CD151. Specificity controls were provided by protein immunoprecipitation (IP) with a non-related antibody (nrs). IB, immunoblotting. B, β4 integrin and Met were efficiently co-immunoprecipitated in control GTL16 and A549 cells, whereas this association was strikingly reduced in CD151-deficient cells. Comparable expression levels of Met and β4 integrin in both cell lines were confirmed by immunoblotting. Two independent shRNA sequences were used to knock down CD151 expression in A549 cells (see ”Materials and Methods“ for details and supplemental Fig. 4 for validation). C, the expression of β4 integrin was silenced in GTL-16 cells by using shRNA targeting β4 (bottom panels). CD151 was immunopurified from different cell lysates, and immunocomplexes were analyzed by Western blotting with antibodies specific for Met and β4 integrin (upper panels), as in panel A. CD151-Met interaction takes place even in the absence of β4 expression. nrAb, non-related antibody. D, GTL-16 cells were treated or not with 10 mm methyl-β-cyclodextrin (MβCD) for the indicated times. CD151 was immunoprecipitated from cell lysates, and immunocomplexes were analyzed by Western blotting with antibodies specific for the β4 integrin and the Met receptor (as above). Methyl-β-cyclodextrin treatment disrupted Met-CD151-β4 complexes. n.t., untreated.
Importantly, we found that CD151 depletion in both GTL16 and A549 carcinoma cells disrupted the physical association between Met and β4 (Fig. 7B), suggesting that this tetraspanin forms a molecular bridge between the two receptors. These results were further confirmed by expressing an alternative shRNA targeting a different sequence of CD151 (Fig. 7B).
Although the physical association between CD151 and β4 has been characterized previously (3), not much is known about CD151-Met interaction. Interestingly, we found that this complex is formed independently of β4 integrin, as demonstrated by expression knockdown in GTL16 cells (Fig. 7C). The association was further confirmed by expressing ectopic CD151 in COS cells, which contain endogenous Met but not β4 (supplemental Fig. 5). The findings above strongly suggest a direct interaction between CD151 and Met, irrespective of the known function of this tetraspanin in integrin-regulated cell-substrate adhesion.
Furthermore, although tetraspanin-enriched microdomains are thought to be distinct from prototypical lipid rafts, it has been shown that cholesterol depletion can affect the distribution of certain tetraspanin-associated molecules, suggesting a further degree of complexity in cell surface organization (45). Interestingly, upon treatment with the cholesterol-depleting drug methyl-β-cyclodextrin (MβCD), β4 integrin was still in association with CD151 as expected (46), whereas Met could not be found in the complex any further (Fig. 7D), suggesting that the formation of the triple complex is restricted to cholesterol-enriched tetraspanin-enriched microdomains. Altogether, these findings indicate that CD151 interacts with both Met and β4, mediates their association in a signaling complex found in specific membrane compartments, and promotes the ensuing Met-dependent phosphorylation of β4 cytoplasmic domain.
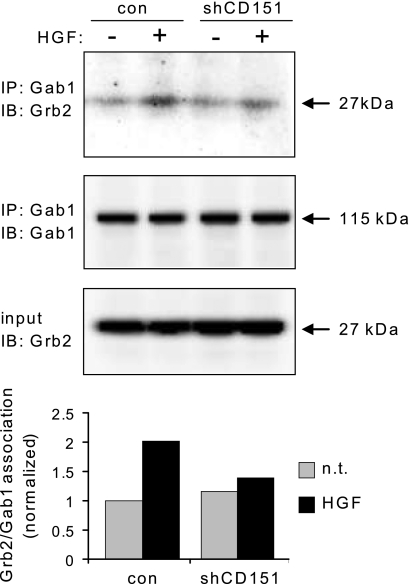
The subsidiary role of tyrosine phosphorylated β4 in enhancing HGF-dependent activation of MAPK relies, at least in part, on a preferential recruitment of Grb2 to the multiadaptor Gab1, with consequent privileged stimulation of Ras-mediated signals (27). Indeed, upon CD151 silencing, the HGF-induced association between Gab1 and Grb2 was hampered (Fig. 8). In summary, CD151 sustains the physical and functional cooperation between Met and β4, and it is required for efficient transduction of Met/β4-dependent signals leading to selective activation of the Grb2-Sos-Ras-MAPK cascade.
FIGURE 8.
CD151 is required for efficient Gab1-Grb2 coupling in the Met signaling cascade. CD151-deficient and CD151-proficient A549 cells were transiently transfected with Gab1 and stimulated with 50 ng/ml HGF for 15 min. Gab1 was precipitated from cell lysates, and the immunocomplexes were probed with the indicated antibodies by Western blotting. CD151 down-modulation impairs HGF-induced association of Gab1 with Grb2 in A549 cells. IP, immunoprecipitation; IB, immunoblotting; Con, empty lentiviral vectors; n.s., not significant.
DISCUSSION
Tetraspanin molecules, such as CD9, CD82, and CD151, are expressed in tumor cells and have been reported to regulate neoplastic progression via mechanisms that, although only partly understood, are thought to implicate cancer cell adhesion, migration, and invasion (7). In the case of CD151, its role in neoplastic progression is controversial. On the one side, several reports have documented increased expression of CD151 in tumor samples and, in some cases, have correlated such overexpression with increased metastatic propensity (8–10). On the other side, an inhibitory function for this tetraspanin in invasion and metastasis has also been suggested, possibly as a consequence of sustained adhesion to extracellular matrix, which in turn reduces tumor cell locomotion (38–40). In line with this, a CD151 antibody capable of blocking tumor cell migration, invasion, and metastasis was shown to exert agonistic functions facilitating CD151-mediated stabilization of integrin complexes, with consequent production of firm cell-matrix adhesion and reduced tumor cell motility (12). The functional role of CD151 in cell-substrate adhesion is further substantiated by the observation that this tetraspanin can physically interact with α3β1, α6β1, and α6β4 integrin receptors (4).
Less is known about the function of CD151 in the regulation of tumorigenic properties, such as deregulated proliferation and evasion from apoptosis. It was reported that the growth of certain carcinoma cells and/or their ability to form tumors in mice may basally decrease upon CD151 knockdown (14, 21, 41). We have not seen the same thing in A549 and A431 carcinoma cells, possibly indicating a distinctive requirement for CD151 expression in different cells. In this work, we specifically focused on the growth and survival responses elicited by HGF/Met signaling in tumor cells and demonstrated that CD151 expression is critically implicated, both in vitro and in vivo. Unexpectedly, this specific function of CD151 does not proceed from its regulation of integrin-mediated adhesion because the effects are observed independently of cell attachment to the extracellular matrix. In fact, here we demonstrated for the first time that CD151 sustains adhesion-independent functions, such as tumor cell growth in soft agar and protection from anoikis induced by HGF-Met signaling. Moreover, we found that CD151 is necessary to direct Met activity toward tyrosine phosphorylation of β4 integrin, which triggers a signaling pathway leading to dedicated stimulation of MAPK-regulated proliferative signals (27). Tetraspanins are known for their ability to organize laterally into tetraspanin-enriched microdomains and promote the formation of multimolecular complexes including plasma membrane receptors and associated molecules (2). In line with this assumption, independent studies have shown that CD151 can associate with Met (14) as well as with β4 integrin (25). Our data suggest the existence of triple Met-CD151-β4 complexes on the cell surface within cholesterol-enriched microdomains, and importantly, demonstrate that a valid Met-β4 association depends on the presence of CD151.
The function of CD151 as a membrane-associated scaffold for optimization of RTK-integrin cross-talk may well extend beyond the HGF/Met case. For instance, β4 integrin has also been implicated as an intermediate effector of EGF-dependent signaling (42). However, in contrast to that seen for Met, we could not reveal any physical association between CD151 and EGF receptor in cancer cells (not shown), suggesting a specificity of this tetraspanin in regulating complex formation between β4 integrin and Met.
Of note, this dual role of CD151 as a modifier of adhesion-related responses (cross-talk with integrins, regulation of cell migration, and invasion) and as an adhesion-independent mediator of tumor growth and survival recalls a similar behavior of β4 integrin. In fact, on the one hand, β4 acts as a mechanical adhesive device by incorporating into hemidesmosomes and participating in the architecture of stable cell-matrix interactions (43); on the other hand, it acts as a servo-signaling apparatus to amplify RTK-triggered transduction pathways, irrespective of integrin engagement by matrix ligands (23, 24). This dichotomy of CD151 is reflected in some apparent inconsistencies. For example, the impairment of cell-substrate adhesion caused by CD151 knockdown can be reverted by adding an activating anti-β1 integrin antibody; however, this treatment cannot rescue the loss of phosphorylation of multiple focal adhesion proteins (such as focal adhesion kinase, Src, p130Cas, and paxillin) normally modulated by cell adhesion and integrin activation (35). One could speculate that CD151 hijacks these signal transducers to convey adhesion-independent growth signals. Consistently, we observed that HGF-induced phosphorylation of focal adhesion kinase at the Src-specific phosphorylation sites Tyr-861 and Tyr-925 is reduced in CD151-deficient cells.5
In conclusion, we demonstrate here that CD151 is crucially implicated in the association and the signaling cross-talk between Met and β4 integrin, as well as for proper implementation of Met/β4-dependent MAPK activation and tumor growth. This unveils a novel player in RTK-regulated oncogenic networks and puts forward the notion that targeting CD151 might be beneficial to interfere with Met-driven malignancy.
Supplementary Material
Acknowledgments
We are grateful to Leonie K. Ashman (University of Newcastle, Australia) for anti-CD151 antibodies. Thanks go to all the members of the Tamagnone and Giordano laboratories at IRCC for helpful discussion and advice.
This work was supported by grants from the Italian Association for Cancer Research (AIRC) and Regione Piemonte (grants to L. Trusolino, L. Tamagnone, and P. M. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods and Figs. 1–5.
M. Franco and L. Tamagnone, unpublished observations.
- RTK
- receptor tyrosine kinase receptor
- HGF
- hepatocyte growth factor
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- poly-HEMA
- polyhydroxyethylmethacrylate
- h
- human.
REFERENCES
- 1.Boucheix C., Rubinstein E. (2001) Cell Mol. Life Sci. 58, 1189–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemler M. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 3.Sincock P. M., Fitter S., Parton R. G., Berndt M. C., Gamble J. R., Ashman L. K. (1999) J. Cell Sci. 112, 833–844 [DOI] [PubMed] [Google Scholar]
- 4.Sterk L. M., Geuijen C. A., van den Berg J. G., Claessen N., Weening J. J., Sonnenberg A. (2002) J. Cell Sci. 115, 1161–1173 [DOI] [PubMed] [Google Scholar]
- 5.Zavadil J., Bitzer M., Liang D., Yang Y. C., Massimi A., Kneitz S., Piek E., Bottinger E. P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6686–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazo P. A. (2007) Cancer Sci. 98, 1666–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zöller M. (2009) Nat. Rev. Cancer 9, 40–55 [DOI] [PubMed] [Google Scholar]
- 8.Ang J., Lijovic M., Ashman L. K., Kan K., Frauman A. G. (2004) Cancer Epidemiol. Biomarkers Prev. 13, 1717–1721 [PubMed] [Google Scholar]
- 9.Hashida H., Takabayashi A., Tokuhara T., Hattori N., Taki T., Hasegawa H., Satoh S., Kobayashi N., Yamaoka Y., Miyake M. (2003) Br. J. Cancer 89, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tokuhara T., Hasegawa H., Hattori N., Ishida H., Taki T., Tachibana S., Sasaki S., Miyake M. (2001) Clin. Cancer Res. 7, 4109–4114 [PubMed] [Google Scholar]
- 11.Gesierich S., Paret C., Hildebrand D., Weitz J., Zgraggen K., Schmitz-Winnenthal F. H., Horejsi V., Yoshie O., Herlyn D., Ashman L. K., Zöller M. (2005) Clin. Cancer Res. 11, 2840–2852 [DOI] [PubMed] [Google Scholar]
- 12.Zijlstra A., Lewis J., Degryse B., Stuhlmann H., Quigley J. P. (2008) Cancer Cell 13, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemler M. E. (2003) Annu. Rev. Cell Dev. Biol. 19, 397–422 [DOI] [PubMed] [Google Scholar]
- 14.Klosek S. K., Nakashiro K., Hara S., Goda H., Hasegawa H., Hamakawa H. (2009) Biochem. Biophys. Res. Commun. 379, 1097–1100 [DOI] [PubMed] [Google Scholar]
- 15.Klosek S. K., Nakashiro K., Hara S., Shintani S., Hasegawa H., Hamakawa H. (2005) Biochem. Biophys. Res. Commun. 336, 408–416 [DOI] [PubMed] [Google Scholar]
- 16.Murayama Y., Shinomura Y., Oritani K., Miyagawa J., Yoshida H., Nishida M., Katsube F., Shiraga M., Miyazaki T., Nakamoto T., Tsutsui S., Tamura S., Higashiyama S., Shimomura I., Hayashi N. (2008) J. Cell. Physiol. 216, 135–143 [DOI] [PubMed] [Google Scholar]
- 17.Odintsova E., Voortman J., Gilbert E., Berditchevski F. (2003) J. Cell Sci. 116, 4557–4566 [DOI] [PubMed] [Google Scholar]
- 18.Sridhar S. C., Miranti C. K. (2006) Oncogene 25, 2367–2378 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi M., Sugiura T., Abe M., Ishii K., Shirasuna K. (2007) Int. J. Cancer 121, 1919–1929 [DOI] [PubMed] [Google Scholar]
- 20.Todeschini A. R., Dos Santos J. N., Handa K., Hakomori S. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X. H., Richardson A. L., Torres-Arzayus M. I., Zhou P., Sharma C., Kazarov A. R., Andzelm M. M., Strominger J. L., Brown M., Hemler M. E. (2008) Cancer Res. 68, 3204–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo W., Pylayeva Y., Pepe A., Yoshioka T., Muller W. J., Inghirami G., Giancotti F. G. (2006) Cell 126, 489–502 [DOI] [PubMed] [Google Scholar]
- 23.Trusolino L., Bertotti A., Comoglio P. M. (2001) Cell 107, 643–654 [DOI] [PubMed] [Google Scholar]
- 24.Sterk L. M., Geuijen C. A., Oomen L. C., Calafat J., Janssen H., Sonnenberg A. (2000) J. Cell Biol. 149, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertotti A., Comoglio P. M., Trusolino L. (2005) Cancer Res. 65, 10674–10679 [DOI] [PubMed] [Google Scholar]
- 26.Bertotti A., Comoglio P. M., Trusolino L. (2006) J. Cell Biol. 175, 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michieli P., Mazzone M., Basilico C., Cavassa S., Sottile A., Naldini L., Comoglio P. M. (2004) Cancer Cell 6, 61–73 [DOI] [PubMed] [Google Scholar]
- 28.Follenzi A., Naldini L. (2002) Methods Enzymol. 346, 454–465 [DOI] [PubMed] [Google Scholar]
- 29.Prat M., Crepaldi T., Gandino L., Giordano S., Longati P., Comoglio P. (1991) Mol. Cell. Biol. 11, 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiucci G., Ravid D., Reich R., Liscovitch M. (2002) Oncogene 21, 2365–2375 [DOI] [PubMed] [Google Scholar]
- 31.Xu L. H., Yang X., Bradham C. A., Brenner D. A., Baldwin A. S., Jr., Craven R. J., Cance W. G. (2000) J. Biol. Chem. 275, 30597–30604 [DOI] [PubMed] [Google Scholar]
- 32.Geary S. M., Cambareri A. C., Sincock P. M., Fitter S., Ashman L. K. (2001) Tissue Antigens 58, 141–153 [DOI] [PubMed] [Google Scholar]
- 33.Takeda Y., Kazarov A. R., Butterfield C. E., Hopkins B. D., Benjamin L. E., Kaipainen A., Hemler M. E. (2007) Blood 109, 1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winterwood N. E., Varzavand A., Meland M. N., Ashman L. K., Stipp C. S. (2006) Mol. Biol. Cell 17, 2707–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada M., Sumida Y., Fujibayashi A., Fukaguchi K., Sanzen N., Nishiuchi R., Sekiguchi K. (2008) FEBS J. 275, 3335–3351 [DOI] [PubMed] [Google Scholar]
- 36.Yamada K. M., Even-Ram S. (2002) Nat. Cell Biol. 4, E75–E76 [DOI] [PubMed] [Google Scholar]
- 37.Comoglio P. M., Boccaccio C., Trusolino L. (2003) Curr. Opin. Cell Biol. 15, 565–571 [DOI] [PubMed] [Google Scholar]
- 38.Chometon G., Zhang Z. G., Rubinstein E., Boucheix C., Mauch C., Aumailley M. (2006) Exp. Cell Res. 312, 983–995 [DOI] [PubMed] [Google Scholar]
- 39.García-López M. A., Barreiro O., García-Díez A., Sánchez-Madrid F., Peñas P. F. (2005) J. Invest. Dermatol. 125, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 40.Peñas P. F., García-Díez A., Sánchez-Madrid F., Yáñez-Mó M. (2000) J. Invest Dermatol. 114, 1126–1135 [DOI] [PubMed] [Google Scholar]
- 41.Sadej R., Romanska H., Baldwin G., Gkirtzimanaki K., Novitskaya V., Filer A. D., Krcova Z., Kusinska R., Ehrmann J., Buckley C. D., Kordek R., Potemski P., Eliopoulos A. G., Lalani el-N., Berditchevski F. (2009) Mol. Cancer Res. 7, 787–798 [DOI] [PubMed] [Google Scholar]
- 42.Mainiero F., Pepe A., Yeon M., Ren Y., Giancotti F. G. (1996) J. Cell Biol. 134, 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margadant C., Frijns E., Wilhelmsen K., Sonnenberg A. (2008) Curr. Opin. Cell Biol. 20, 589–596 [DOI] [PubMed] [Google Scholar]
- 44.Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Trends Cell Bio.l 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 45.Israels S. J., McMillan-Ward E. M. (2007) Thromb. Haemost. 98, 1081–1087 [PubMed] [Google Scholar]
- 46.Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giordano S., Ponzetto C., Di Renzo M. F., Cooper C. S., Comoglio P. M. (1989) Nature 339, 155–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.