Abstract
We biochemically simulated HIV-1 DNA polymerization in physiological nucleotide pools found in two HIV-1 target cell types: terminally differentiated/non-dividing macrophages and activated/dividing CD4+ T cells. Quantitative tandem mass spectrometry shows that macrophages harbor 22–320-fold lower dNTP concentrations and a greater disparity between ribonucleoside triphosphate (rNTP) and dNTP concentrations than dividing target cells. A biochemical simulation of HIV-1 reverse transcription revealed that rNTPs are efficiently incorporated into DNA in the macrophage but not in the T cell environment. This implies that HIV-1 incorporates rNTPs during viral replication in macrophages and also predicts that rNTP chain terminators lacking a 3′-OH should inhibit HIV-1 reverse transcription in macrophages. Indeed, 3′-deoxyadenosine inhibits HIV-1 proviral DNA synthesis in human macrophages more efficiently than in CD4+ T cells. This study reveals that the biochemical landscape of HIV-1 replication in macrophages is unique and that ribonucleoside chain terminators may be a new class of anti-HIV-1 agents specifically targeting viral macrophage infection.
Keywords: HIV, Macrophage, Nucleotide, Reverse Transcription, Viral Polymerase, dNTP/rNTP
Introduction
Cellular dNTPs are mainly consumed during chromosomal replication and DNA damage repair, whereas cellular ribonucleoside triphosphates (rNTPs)3 serve not only as substrates of RNA polymerases but also as key metabolic energy carriers and substrates for numerous cell signaling pathway enzymes (e.g. ATP and GTP). Cellular dNTP biosynthesis is tightly regulated by the cell cycle in eukaryotic cells (1, 2) and is highly activated in S phase. Indeed, cells with uncontrolled cell cycles, such as cancer cells, display elevated dNTP pools compared with normal cells (3, 4).
Interestingly, cellular rNTP concentrations are much higher than dNTP concentrations (4). ATP is the most abundant rNTP, reaching up to 1–6 mm (3–5), whereas cellular dATP concentrations, even in dividing cells, are several hundred to a thousand times lower. Because transcription, metabolism, and cell signaling pathways all consume rNTPs and occur throughout the cell cycle, it is reasonable to assume that the rNTP concentrations in non-dividing cells remain similar to those in dividing cells. However, no rNTP concentration data in non-dividing cells, such as primary human macrophages, have been reported.
Chemically, the rNTP and the dNTP differ only in the presence of a 2′-OH on the sugar moiety, and the levels of rNTP in dividing cells (millimolar range) are much higher than dNTP concentrations (micromolar range). Thus, cellular replicative DNA polymerases must efficiently discriminate between dNTPs and rNTPs. In fact, many DNA polymerases have a residue or residues near their active site, which serve as a steric gate to reduce the binding affinity of any nucleotide bearing a 2′-OH (6–12). Indeed, the selectivity of the prototypical Escherichia coli DNA polymerase I for dNTPs over rNTPs is 104 to 106 (6). However, rNTP incorporation has been observed by DNA polymerases β and μ (13, 14), TdT DNA polymerase (15), and recently by replicative polymerases in yeast (16). Furthermore, the physiological and genetic consequences of rNTP incorporation by these cellular DNA polymerases remain unknown.
Human immunodeficiency virus type 1 (HIV-1) uniquely infects both activated/dividing CD4+ T cells and terminally differentiated/non-dividing macrophages (17–21). We previously reported that terminally differentiated/non-dividing macrophages harbor much lower dNTP concentrations than activated CD4+ T cells (22). In addition, the rate of proviral HIV-1 DNA synthesis in non-dividing cells, which is slower than in dividing cells (23), can be accelerated by elevating cellular dNTP concentrations (3). This suggests that HIV-1 proviral DNA synthesis in non-dividing cells is kinetically delayed compared with that in activated CD4+ T cells, due to the limited dNTP substrate pools.
In this report, we measured concentrations of both dNTP and rNTP in the two primary human HIV-1 target cell types: macrophages and activated peripheral blood mononuclear cells (PBMCs) using a quantitative tandem mass spectrometry method. This analysis allowed us to perform a series of mechanistic simulations of the proviral DNA synthesis events catalyzed by HIV-1 reverse transcriptase (RT) using physiological nucleotide pools found in the two HIV-1 target cell types. Biochemical simulations conducted in this study revealed not only the unique rNTP incorporation capability of the HIV-1 DNA polymerase in the simulated macrophage environment, but also that ribonucleoside chain terminators are a potential new class of anti-HIV-1 agents.
EXPERIMENTAL PROCEDURES
Preparation and Culture of Human Primary Macrophages and PBMCs for dNTP/NTP Assay
Human monocytes were isolated from buffy coats of HIV-1-negative, HBV/HCV-negative donors with density gradient centrifugation coupled with enrichment for CD14+ monocytes with the Rosette Sep antibody mixture (Stem Cell Technologies, Vancouver, Canada). Cells were seeded at a concentration of 1.0 × 106 cells/well (6-well plate) for 1 h at 37 °C, 5% CO2 to allow plastic adherence prior to repeated washes with 1× PBS. Monocytes were allowed to differentiate for 7 days in RPMI medium (Hyclone, Logan, UT) containing heat-inactivated 20% fetal calf serum (FCS) (Atlanta Biologicals, Lawrenceville, GA), 1% penicillin/streptomycin (Invitrogen), supplemented with 100 units/ml macrophage colony-stimulating factor (R&D Systems, Minneapolis, MN) at 37 °C, 5% CO2. For all conditions, macrophages were stained with CD11b-APC (Miltenyi Biotec, Auburn, CA) and subjected to FACS to determine purity of >99%. Human PBMCs were also isolated from buffy coats derived from healthy donors. Activated PBMCs were maintained in RPMI medium supplemented with heat-inactivated 20% FCS, 1% penicillin/streptomycin, and 2% l-glutamine (Cellgro/Mediatech, Inc., Manassas, VA); 6 μg/ml phytohemagglutinin (J-Oils Mills, Inc., Tokyo, Japan) was added to the cells 72 h prior to experiments in order to activate them.
Extraction of Intracellular Nucleotide Fraction and LC-MS/MS Analysis
For both macrophages and PBMCs, the isolated cells were washed twice with ice-cold 1× PBS to remove any residual medium. Cells were resuspended in 70% CH3OH overnight, and extracts were centrifuged at 13,000 × g for 10 min (Thermo Electron Corp., Marietta, OH). Supernatants were subsequently dried, and the resulting samples were reconstituted in HPLC mobile phase for LC-MS/MS analysis as described previously (24). The rNTP level measurements were performed using a similar approach as described above for dNTP. The stable isotopes, [13C,15N]ATP, [13C,15N]GTP, [13C,15N]CTP, and [13C,15N]TTP were used for the measurement of ATP, GTP, CTP, and UTP. The m/z parent → product MS/MS transitions 523 → 146, 539 → 162, 496 → 119, and 495 → 81 were applied for the standard stable labeled isotopes and 508 → 136, 524 → 152, 484 → 112, and 485 → 81, for the corresponding sample nucleotides, respectively.
RT Purification
The HXB2 HIV-1 RT gene was previously cloned into pET28a (Novagen), and the N-terminal hexahistidine-tagged-p66/p66 homodimer HIV-1 RT was subsequently expressed in E. coli BL21 (DE3) and purified with Ni2+-NTA chromatography followed by DEAE and SP anion exchange as described previously (25). RT of simian immunodeficiency virus (SIVagm Sab) was also previously cloned and purified (26). These RT proteins were quantified and stored in 10% glycerol dialysis buffer as described previously (27, 28).
Primer Extension Assays
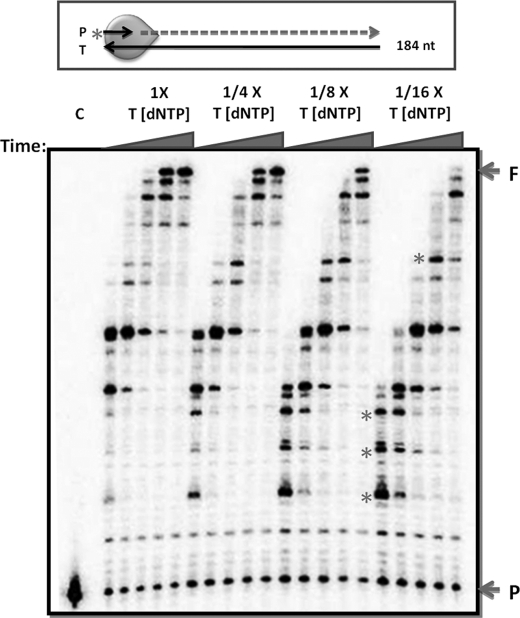
For the primer extension reaction described in Fig. 2, a 5′-end 32P-labeled 19-mer DNA primer annealed to the 184-nt region encoded in the HIV-1 NL4-3 glycosaminoglycan gene was prepared as previously described (29, 30). All DNA and RNA primers used in this study were purchased from Integrated DNA Technologies and Dharmacon Research, respectively. Assay mixtures (20 μl) contained 10 nm template-primer, the RT protein concentrations specified in the individual figure legends, four dNTPs (Amersham Biosciences) at concentrations indicated in the figure legends, and 1× Reaction Buffer (50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 5 mm MgCl2, 5 μm (dT)20). Reactions were initiated by adding the RT proteins and incubated at 37 °C for the defined times. Reactions were terminated with 10 μl of 40 mm EDTA, 99% formamide. Reaction products were immediately denatured by incubating at 95 °C for 5 min, and 4 μl of each 30-μl final reaction mixture was analyzed with 14% Urea Page gel electrophoresis (SequaGel, National Diagnostics) and scanned with a Biorad Personal Molecular Imager.
FIGURE 2.
DNA synthesis profile of HIV-1 RT at the range of the dNTP concentrations found in HIV-1 target cell types. A 5′-end 32P-labled 19-mer DNA primer (P) annealed to a 184-nt RNA template (T) encoding a portion of the HIV-1 NL4-3 glycosaminoglycan gene was extended by HIV-1 RT (40 nm; green circle) with the dNTP concentration found in human primary activated CD4 T cells (1x, dATP, 23 nm; dGTP, 20 nm; dCTP, 30 nm; TTP, 32 nm) (22) or its three serial dilutions (1:4 (1/4x), 1:8 (1/8x), and 1:16 (1/16x)) for 0. 5, 1, 2, 4, and 8 min under the reaction conditions described under “Experimental Procedures.” These reactions were analyzed by 10% polyacrylamide-urea denaturing gels. The pause sites were marked by an asterisk. F, fully extended product; P, unextended primer. C, no RT control.
For the primer reaction described in the legend to Fig. 3C, a 5′-end 32P-labeled 17-mer A primer (5′-CGCGCCGAATTCCCGCT-3′) annealed to a 40-mer RNA (5′-AAGCUUGGCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG-3′; template/primer ratio 2.5:1) was extended for various time points under the reaction conditions described above using the nucleotide pools specified in the figure legends.
FIGURE 3.
HIV-1 RT mediated incorporation of rNTPs at the intracellular concentrations found in primary human HIV-1 target cell types. A, the ratios of the rNTP and dNTP concentrations in human primary macrophages and activated PBMCs (see Table 1) are compared. The calculated -fold differences of the dNTP and rNTP concentration disparity in macrophages and activated PBMCs are marked for each of four rNTP and dNTP pairs. B, a 23-mer DNA primer (P) annealed to an RNA 40-mer template (T) (10 nm template-primer complex) was extended by HIV-1 RT (200 nm) at macrophage (M) and PBMC (PB) dNTP and rNTP concentrations (see Table 1) in the presence of [α-32P[UTP with a 1:690 ratio to the nonradioactive UTP. The reactions were terminated by 10 mm EDTA and inactivation at 95 °C for 3 min, purified with a Qiagen nucleotide removal column, and mixed with an equal amount of the 23-mer 5′-end 32P-labeled DNA primer as a loading control (LC) before application to 15% polyacrylamide denaturing gels (left). The fully extended product density (F) in the macrophage and T cell nucleotide reactions was quantified and compared after normalization by the loading control for calculating the -fold difference of the rNTP incorporation between macrophages and T cell nucleotide pools (right). C, a time course DNA synthesis by HIV-1 RT (20 nm) with the 5′-end 32P-labeled 17-mer primer annealed to the RNA 40-mer template (2 nm template-primer) at the macrophage dNTP concentration (see Table 1) for 2, 4, 8, 16, and 20 min in the presence and absence of macrophage rNTP pools measured in Table 1. C, no RT control; F, fully extended product; P, unextended primer.
Assay for rNTP Incorporation during DNA Synthesis of HIV-1 RT Using [α-32P]UTP
Ext-T DNA 23-mer primer (see below) annealed to the RNA 40-mer template (10 nm complex) was extended by 200 nm HIV-1 RT for 45 min in the 1× Reaction Buffer with the macrophage and T cell dNTP or NTP concentrations described in Table 1 using the identical non-radioactive UTP and radioactive [α-32P]UTP ratio (690:1). Following a quench with 10 mm EDTA, the reaction products were further purified with a Qiagen nucleotide removal column. The reaction products with [α-32P]UTP were normalized with a 5′-end 32P-labeled 17-mer loading control primer, which was added in an equal amount after the reactions were terminated as described (31). To monitor the entire DNA polymerization under the conditions described in this experiment, the identical reactions were conducted with the 5′-end 32P-labeled 23-mer Ext-T DNA template annealed to the 40-mer RNA template with non-radioactive dNTPs in both negative (no enzyme) and positive (250 μm dNTP substrate) controls, generating no primer extension and full primer extension, respectively (supplemental Fig. 1).
TABLE 1.
Intracellular dNTP and rNTP concentrations of primary human monocyte-derived macrophage and activated PBMCs as determined by LC-MS/MS
| Concentration |
||||
|---|---|---|---|---|
| dCTP | dGTP | dATP | TTP | |
| μm | ||||
| Activated PBMC | 3.67 ± 2.65 | 1.52 ± 1.01 | 9.22 ± 4.5 | 16.0 ± 5.25 |
| Macrophages | 0.07 ± 0.05 | 0.07 ± 0.05 | 0.04 ± 0.03 | 0.05 ± 0.04 |
| -Fold difference | ||||
| Amount |
||||
|---|---|---|---|---|
| dCTP | dGTP | dATP | TTP | |
| pmol/106 cells | ||||
| Activated PBMC | 1.17 ± 0.85 | 0.49 ± 0.32 | 2.95 ± 1.46 | 4.85 ± 1.56 |
| Macrophages | 0.19 ± 0.13 | 0.18 ± 0.13 | 0.10 ± 0.07 | 0.13 ± 0.10 |
| Concentration |
||||
|---|---|---|---|---|
| CTP | GTP | ATP | UTP | |
| μm | ||||
| Activated PBMC | 182 ± 24 | 1,745 ± 128 | 6,719 ± 560 | 690 ± 100 |
| Macrophages | 25 ± 8 | 323 ± 95 | 1,124 ± 339 | 173 ± 47 |
| -Fold difference | ⅕ | ⅙ | 1/4 | |
| Amount |
||||
|---|---|---|---|---|
| CTP | GTP | ATP | UTP | |
| pmol/106 cells | ||||
| Activated PBMC | 58 ± 8 | 558 ± 41 | 2,150 ± 179 | 221 ± 32 |
| Macrophages | 66 ± 21 | 859 ± 253 | 2,990 ± 902 | 460 ± 124 |
Single Nucleotide Incorporation Rates of HIV-1 RT in Cellular Nucleotide Pools
For analyzing the impact of rNTPs on the dNTP incorporation rate of HIV-1 RT in Fig. 4, four different 5′-end 32P-labeled primers (Primer A, 5′-TCGCCCTTAAGCCGCGC-3′; Ext-T primer, 5′-CTTATAACGATCGCCCTTAAGCC; Ext-G primer, 5′-GAATTCCCGCTAGCAATATTCT-3′; Ext-C primer, 5′-TATAACGATCGCCCTTAAGCCG-3′) were individually annealed to the 40-mer RNA template, and the reactions were conducted under the conditions described in the legend to Fig. 2 except using the nucleotide concentrations defined in the figure legends.
FIGURE 4.
Comparison of dNTP and rNTP incorporation rates of HIV-1 RT at the nucleotide concentrations found in human macrophages. Incorporation kinetics of HIV-1 RT (green circle) for individual dNTPs and rNTPs at their concentrations found in macrophages (see Table 1) were compared by single nucleotide extension reactions described under “Experimental Procedures.” A, a representative time course reaction set for dATP alone, ATP alone, or both dATP and ATP at their macrophage concentrations. P, unextended 17-mer primer; E, extended 18-mer product. C, no RT control. B, the percentages of the primer extension at early time points were plotted to compare the incorporation rate for each of dNTPs or rNTPs alone as well as both dNTPs and rNTPs at their macrophage concentrations. C, identical reactions were conducted with SIVagm Sab-1 RT (SIV). D, steady state Km values of HIV-1 RT to rNTPs determined by single nucleotide incorporation assay. E, comparison of rNTP concentrations (black bars) of macrophages and PBMC and the Km values (red zone) of HIV-1 RT to rNTPs. Gray bars, average concentrations of dNTPs of the two target cell types obtained from Table 1.
Steady State Kinetics of HIV-1 RT with rNTPs
The single nucleotide incorporation reactions were conducted using the four template-primers (20 nm) used in Fig. 4 and 20 nm RT in the 1× reaction buffer for 5 min at least in triplicate at the ranges of NTP concentrations described in the figure legend. The Km values were determined as described below.
Primer Extension with 3′-End rNMP Primer
5′-End 32P-labeled 23-mer Ext-T primer with either 3′-end dCMP or rCMP was annealed to the 40-mer RNA template, and 10 nm template-primer was extended by 40 nm HIV-1 RT at the dNTP concentration found in macrophages under the 1× reaction buffer condition for the time course described.
Visualization, Quantification, and Kinetic Analysis of the Primer Extension Reactions
All primer extension reactions in this study were analyzed with urea-denaturing 10–16% polyacrylamide gels and scanned with a phosphor imager (Bio-Rad) and quantified with the Bio-Rad Quantity One software. Nonlinear regression analysis was done with Kaleida Graph, rates were determined with a single exponential, and Michaelis-Menten fits were completed as described previously (32).
Test for Cytotoxicity of 3′-Deoxyadenosine (Adenosine Chain Terminator; rACT)
Human primary activated CD4 T cells, a human microglia cell line, CHME5 (33), and a human monocytic cell line, U937 (National Institutes of Health AIDS Research and Reference Reagent Program), were cultured as described previously. For isolating CD4 T cells, human PBMCs were harvested from Ficoll density gradients (Lymphoprep, Axis-Shield PoC AS, Oslo, Norway) from buffy coat purchased from the New York Blood Center, and monocytes were then purified using immunomagnetic selection with anti-CD14 antibody-conjugated magnetic beads, following the manufacturer's recommendations (Miltenyi Biotech). CD4+ T lymphocytes were isolated from the monocyte-depleted buffy coats after the immunomagnetic selection as described above and cultured with IL-2 (National Institutes of Health AIDS Research and Reference Reagent Program) for activation. CHME5, U937, and human primary activated CD4+ T cell were cultured in the presence and absence of varying concentrations of rACT for 48 h, and the percentages of live and dead cells were analyzed by both FACS analysis and trypan blue staining.
Assays for Inhibition of HIV-1 Infection and Proviral DNA Synthesis by rACT in Macrophages and T Cells
The purified human monocytes from three different donors were differentiated to macrophages as described previously, and the D3HIV-GFP vector system expressing GFP was also prepared as described previously (3, 22, 33, 34). Equal p24 level of the produced vector was used for infecting human macrophages preincubated with different concentrations of rACT (RI Chemicals) for 12 h. One-half of the harvested cells were used for determining the percentages of the GFP-expressing and/or propidium iodide-stained cells by FACS at 7 days postinfection. All cells used to assess transduction efficiency were gated or stained with propidium iodide to ensure that live cells were measured. The remaining cells were used for the extraction of genomic DNA. A quantitative 2-LTR circle PCR assay using real-time PCR was normalized by total genomic DNA (Bio-Rad). The primers and amplification protocol have been used in our previous studies (3, 35). rACT was tested in T cells as follows. Human primary activated CD4+T cells (2.5 × 105 cells) isolated from two donors were pretreated with 0, 0.1, and 1 mm rACT for 4 h, and then the cells were transduced with PD3-HIV-GFP. The transduced cells were fixed at 48 h post-transduction and analyzed by FACS for determining the percentage of GFP-positive cells. The analysis was conducted in duplicate per donor, and the means and S.D. values were derived from all data obtained.
RESULTS
Quantitative Mass Spectrometry Assay for dNTP and rNTP Concentrations in Primary HIV-1 Target Cell Types
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was employed to measure dNTP and rNTP pools in human primary macrophages and activated human PBMCs, which are routinely used for culturing macrophage-tropic and T cell-tropic HIV-1, respectively (Table 1). These two cell types were isolated from buffy coats of healthy donors as described under “Experimental Procedures.” Known amounts of 13C/15N-labeled individual standards were used to determine dNTP and rNTP quantities in the samples with LC-MS/MS, and the cellular nucleotide concentrations were calculated using their cell volumes, which were previously reported (22).
Macrophages were found to harbor a much lower dNTP concentration (40–70 nm) than activated PBMCs (1.52–16.0 μm) (Table 1 and Fig. 1). We previously determined the dNTP concentrations of human primary macrophages and activated CD4+ T cells using an enzyme-based assay (22); the results are also presented in Fig. 1 for comparative purposes. The dNTP concentrations of macrophages and activated PBMCs determined by LC-MS/MS were very similar to those of macrophages and activated CD4+ T cells previously determined by our enzyme assay and other studies (36) except for the macrophage dGTP concentration, which was 3-fold higher in the MS/MS assay than in the enzyme-based assay. Basically, in the LC-MS/MS assay, the dNTP concentration in macrophages is 22–320-fold lower than in activated T cells. In contrast, as shown in Table 1, the rNTP concentration in macrophages was only 4–7-fold lower than that in activated PBM cells, supporting the idea that non-dividing cells still maintain high rNTP concentrations, presumably due to multiple roles of rNTPs in various cellular events, such as transcription, cell signaling, and cellular metabolism, which also occur in non-dividing cells.
FIGURE 1.
Comparison of dNTP concentrations of human primary macrophages and activated PBMCs with steady state Km and pre-steady state Kd values of HIV-1 RT to dNTP substrates. The dNTP concentrations of macrophages and activated PBMC, which were determined by LC-MS/MS and summarized in Table 1, are plotted with black bars. The gray bars represent dNTP concentrations of human primary macrophages and activated CD4+ T cells previously determined by the enzyme-based dNTP assay (22). The red and blue zones represent the ranges of the previously determined HIV-1 Km (22, 37–39) and Kd (34, 40, 41) values of HIV-1 RT to dNTP substrates, respectively.
Also indicated in Fig. 1 are the steady state and pre-steady state HIV-1 RT dNTP kinetic parameters: the Km values (red zone) (22, 37–39), which are the dNTP concentrations giving 50% of the maximum reaction rate, and the Kd values (blue zone) (34, 40, 41), which indicate the dNTP binding affinity. The T cell dNTP concentrations are generally higher than the published Km and Kd values of HIV-1 RT. In contrast, the macrophage dNTP pools measured are considerably lower than the Km and Kd values of HIV-1 RT (22, 38, 39). This suggests that viral DNA synthesis is relatively restricted by dNTP availability in macrophages, most likely due to suboptimal substrate binding.
DNA Synthesis Profiles of HIV-1 RT in Cellular dNTP Pools
The dNTP concentration-dependent DNA synthesis profile of HIV-1 RT was investigated in the presence of dNTP pools intended to simulate the cellular microenvironments described in the legend to Fig. 1 (22). We employed a time course primer extension reaction with HIV-1 RT (Fig. 2) using a 19-mer 5′ 32P-labeled DNA primer (P) annealed to an RNA template encoding a 184-nt portion of the HIV-1 gag gene in the simulated T cell dNTP pool (1×) and three substrate dilutions 1:4, 1:8, and 1:16 (1/4x, 1/8x, and 1/16x) (the dNTP concentration in the 1:16 dilution is still higher than that of macrophages). As assessed by the amount of the 184-bp full-length products (F in Fig. 2), DNA synthesis proceeded more slowly in the diluted/low dNTP pools as compared with the simulated T cell dNTP pool. This kinetic delay resulted in increasing early pausing by RT (see the asterisk in Fig. 2). Because the macrophage dNTP pool is an additional 2–20 times lower in concentration than even the 1:16 dilution of the simulated T cell dNTP pool, it is likely that viral DNA synthesis in the macrophage environment will be kinetically slower and more distributive, with substantially more pausing than that seen in the 1/16x lanes in Fig. 2. Collectively, Figs. 1 and 2 suggest that limited dNTP pools contribute to the delayed proviral HIV-1 DNA synthesis previously observed in macrophages (22, 23). This is consistent with the observation that HIV-1 proviral synthesis is accelerated by elevating cellular dNTP pools in non-dividing cells but not in dividing cells, which already have dNTP levels well above the Kd and Km of HIV-1 RT (3, 42).
HIV-1 RT Incorporates rNTP from Cellular dNTP/rNTP Pools Found in HIV-1 Target Cell Types
One key observation revealed by quantification of dNTP and rNTP concentrations in the two HIV-1 target cell types, summarized in Table 1, is that rNTP and dNTP concentration disparities exist in macrophages and activated PBMCs. As illustrated in Fig. 3A, due to the relatively high rNTP concentrations and extremely low dNTP concentrations, macrophages display 4–80 times greater disparity between rNTP and dNTP concentrations than those in activated PBMCs. This unexpected greater relative difference of rNTP and dNTP pools in macrophages enabled us to hypothesize that HIV-1 RT might opportunistically incorporate rNTPs in the macrophage environment, particularly when DNA synthesis kinetics are largely compromised due to the limited dNTP pools as shown in Fig. 2.
To test this hypothesis, we performed primer extension/DNA synthesis reactions of HIV-1 RT (Fig. 3B) with a non-radioactive 23-mer primer annealed to a 40-mer RNA template in the presence of dNTP and rNTP concentrations that simulated those measured in macrophages and activated PBMCs (Table 1 and Fig. 3A). To visualize the incorporation of rNTPs during DNA synthesis, low levels of [α-32P[UTP ( of non-radioactive UTP concentrations) were included in the simulated nucleotide pools. As shown in Fig. 3B (left), the polymerase reaction of HIV-1 RT with dNTPs alone did not generate any visible bands in the absence of rNTP, which was expected. However, fully extended, radiolabeled products (F) were detected in reactions performed using simulated macrophage dNTP/rNTP pools (Fig. 3B). In contrast, no radiolabeled extension products were detected in reactions performed using simulated PBMC dNTP/rNTP pools (Fig. 3B). Control experiments with a 5′-end 32P-labeled primer confirmed the efficient extension of the primer and the production of full-length products in both the simulated PBMCs and simulated macrophage environments (supplemental data 1). Thus, the absence of a radiolabeled extension product under the PBMC condition in Fig. 3B reflects a lack of rNTP incorporation and not a failure to complete DNA synthesis. Quantification of the radioactively labeled extended products in Fig. 3B (right) revealed that rNTP incorporation in the simulated macrophage environment was 22 times more efficient than in the simulated T cell environment.
Next, we investigated the impact of macrophage nucleotide pools on the entire ensemble of RT-mediated DNA synthesis. To do this, we conducted a time course primer extension reaction using a 5′-end 32P-labeled 17-mer annealed to an RNA 40-mer. Unlike in Fig. 3B (which shows only extension products that have incorporated a radiolabeled rNTP), this analysis reveals all extended primer products because the primer itself is labeled. In Fig. 3C, we compared the efficiency of DNA synthesis by HIV-1 RT in the simulated macrophage environment that contained either dNTPs alone or dNTPs plus rNTPs. The results show that RT-mediated DNA synthesis was enhanced by the presence of rNTPs in the simulated macrophage environment, as reflected by an increase in fully extended synthesis products (F) and a reduction in paused products (*). This suggests that rNTPs may serve as substrate for HIV-1 RT in the macrophage environment. Importantly, a similar kinetic enhancement by rNTPs was also observed during DNA synthesis of HIV-1 RT with DNA template in the macrophage nucleotide pools (supplemental data 2), suggesting that HIV-1 RT can incorporate rNTPs during both (+)- and (−)-strand proviral DNA synthesis in the macrophage nucleotide environment. In addition, as predicted by the lack of rNTP incorporation (Fig. 3B), no kinetic enhancement by rNTPs was observed in the T cell nucleotide pools (data not shown).
Comparison of dNTP and rNTP Incorporation Rates of HIV-1 RT with the Nucleotide Pools Found in Macrophages
We next analyzed the incorporation kinetics for individual dNTPs and rNTPs by HIV-1 RT at their macrophage concentrations. For this, we first employed a similar primer extension reaction as that used in Fig. 3C, except we used a specific primer and a single dNTP, rNTP, or both to estimate individual nucleotide incorporation rates (see “Experimental Procedures”). As shown in Fig. 4A, HIV-1 RT almost equally incorporates dATP and ATP at macrophage concentrations. As summarized in Fig. 4B (blue dotted lines), HIV-1 efficiently incorporates ATP and CTP but less efficiently incorporates GTP and UTP. Importantly, however, upon combining dNTPs and rNTPs in a reaction (Fig. 4, A and B, purple dotted lines), the total single nucleotide extension rate appeared to be elevated compared with the rates of either dNTP (red line) or rNTP alone. Basically, when primer extension products were plotted by time, primer extension was accelerated in the presence of both dNTPs and rNTPs at their macrophage concentrations compared with the reactions containing either dNTPs or rNTPs alone. The results of this single nucleotide assay are consistent with the data from our multiple nucleotide incorporation assay (Fig. 3C). These data suggest that rNTPs, which are normally the substrates of RNA polymerases, not only are incorporated into DNA by HIV-1 RT but also positively affect the DNA synthesis rate in a simulated macrophage environment.
Next, we tested if RT of SIVagm Sab, which is another lentivirus capable of infecting macrophages, also incorporates rNTPs as well as dNTPs under conditions that simulate those present in macrophages. As shown in Fig. 4C, SIV RT also utilizes ATP at the simulated macrophage concentration, and the addition of ATP to dATP resulted in more efficient DNA synthesis. This shows that the effective utilization of rNTPs during DNA synthesis is probably common among RTs of lentiviruses that replicate in macrophages.
Steady-state Kinetic Analysis of HIV-1 RT rNTP Incorporation
Next, we investigated the enzymatic efficiency of rNTP incorporation by HIV-1 RT. For this, we determined the Km values of HIV-1 RT for rNTPs using a single nucleotide extension assay (22, 32) and compared these values with measured cellular rNTP concentrations found in macrophages and PBMCs. As summarized in Fig. 4, D and E, the Km values for ATP and GTP are well within the range of cellular ATP and GTP concentrations found in macrophages, whereas the Km values of UTP and CTP were higher than their macrophage concentrations. The data in Fig. 4, D and E, also suggest that HIV-1 RT should be capable of utilizing rNTPs when present at the concentrations found in activated T cells. However, rNTP incorporation is presumably prevented in the activated T cell environment due to the high levels of dNTPs, which compete much more effectively for binding to RT and are preferred substrate (see Fig. 3B and supplemental data 1).
Effect of rNMP at the 3′-End of the Primer on HIV-1 RT Activity
We next tested if HIV-1 RT can extend an incorporated rNMP at the 3′-end of a primer. For this, we compared the incorporation of a single dNTP into DNA primers containing either dCMP or rCMP at the 3′-ends. These two types of primers were 5′-end 32P-labeled and annealed to a 40-mer RNA template (Fig. 5). These primers were extended by HIV-1 RT (20 nm) in the presence of the TTP concentration found in macrophages (40 nm). As shown in Fig. 5A, HIV-1 RT displayed an equal extension capability with both the 3′-end rCMP and dCMP primers at the macrophage TTP concentration. Next, we conducted the same experiments except with both dATP and dGTP, which are the first and second nucleotides to be incorporated, respectively. As shown in Fig. 5B, HIV-1 RT was also equally capable of incorporating the second nucleotide (dGTP) from both 3′-end dCMP and rCMP primers. In fact, the efficient extension of 3′-end rNMP is not surprising because HIV-1 RT initiates both (−)- and (+)-proviral DNA synthesis using RNA primers (e.g. tRNA3Lys and polypurine tract RNA primer) containing 3′-end rNMPs during viral replication. This result, together with the data in Figs. 3 and 4, clearly demonstrates that rNTPs are utilized by HIV-1 RT, including their incorporation into newly synthesized DNA and their subsequent extension. Furthermore, this readily occurs and does not negatively impact overall DNA polymerization kinetics in the simulated macrophage environment.
FIGURE 5.
Extension of DNA primers containing 3′-end dCMP and rCMP by HIV-1 RT in macrophage dNTP concentrations. A, diagram of the template (T) and primers (P) used in this experiment. Two 5′-end 32P-labeled DNA primers containing dCMP or CMP at their 3′ ends were individually annealed to the RNA 40-mer template. These two template-primers were extended by HIV-1 RT (40 nm; green circle), and single nucleotide (dATP) extension time courses were preformed with dTTP for the first nucleotide incorporation. B, the two template-primers were also extended by HIV-1 RT with both dATP and dGTP for the first and second dNTP incorporations at their macrophage concentrations (see Table 1), respectively, from the 3′-end dCMP or rCMP primers. The reactions were conducted for 30, 60, 90, 120, 240, 480, and 720 s. P, unextended primer; E1, one nucleotide extended product; E2, two nucleotide extended products; C, no RT control.
Inhibition of HIV-1 Proviral DNA Synthesis in Macrophages by 3′-Deoxyadenosine
We hypothesized that because HIV-1 RT can incorporate rNTPs given macrophage nucleotide pools, rNTP analogs lacking 3′-OH, termed ribonucleoside chain terminators (rNCTs), can be incorporated during HIV-1 replication and may inhibit viral reverse transcription in human macrophages. To test this, we employed a ribonucleoside chain terminator, 3′-deoxyadenosine (rACT).
First, we tested the cytotoxicity of rACT in human primary CD4+ T cells, human primary macrophages, human microglial CHME5, and human premonocytic U937 cell lines. Cells were incubated with different concentrations of rACT up to 1 mm for 2 days, and cell survival was determined as described under “Experimental Procedures.” As shown in Fig. 6A, no significant cytotoxicity of rACT was observed in primary human cells or transformed cell lines at concentrations up to 1 mm. Next, we tested if rACT could inhibit the replication of HIV-1 in macrophages. For this, we measured the transduction efficiency of an HIV-1 vector in primary human macrophages in the presence and absence of rACT. Human macrophages, which were prepared from multiple donors, were preincubated with various concentrations of rACT and then transduced with an HIV vector (D3-GFP), which expresses all HIV-1 proteins except Env and Nef (replaced with enhanced GFP) (22). Virus infectivity was determined by FACS analysis for GFP expression at 7 days postinfection. As shown in Fig. 6, B and C, treatment with rACT (0.1 and 1 mm) reduced the percentage of macrophages expressing GFP compared with no treatment. However, the reduction of HIV-1 vector infectivity by rACT might conceivably result from the inhibition of cellular RNA polymerase-mediated transcription of the GFP gene or some other off-target effects. To verify that this was not the case, we measured the copy number of HIV-1 2-LTR DNA circles that are formed in the nucleus after the completion of reverse transcription and are commonly used to monitor HIV-1 proviral DNA synthesis kinetics (43). As shown in Fig. 6D, rACT (0.1 and 1 mm) substantially reduced the 2-LTR circle copy number in macrophages. We also examined the effect of rACT on HIV-1 infection in activated CD4+ T cells. As shown in Fig. 6E, rACT resulted in less of an inhibitory effect in T cells at both rACT concentrations compared with macrophages (Fig. 6C). This suggests that this ribonucleoside analog is preferentially incorporated by HIV-1 RT in macrophages where the disparity between dNTP and NTP is vastly larger compared with T cells (Fig. 3A).
FIGURE 6.
Inhibition of HIV-1 reverse transcription by 3′-deoxyadenosine chain terminator (rACT) in human primary macrophage and its cytotoxicity. A, cytotoxicity of rACT in human primary CD4+ T cell, U937, and CHME5. These three types of cells were cultured and treated with different concentrations of rACT up to 1 mm for 2 days, and the percentages of live and dead cells were determined by FACS and/or trypan blue staining. The percentage of the live cells in the absence of rACT was used for normalization (100%), and the base line cell death of these cell types in the absence of rACT was less than 5%. The cytotoxicity of rACT is shown in the FACS described in B by the propidium iodide staining. B, human primary macrophages were preincubated with different concentrations of rACT (0, 10, 100, and 100 μm) for 24 h and then transduced with HIV-GFP vector (equal p24 amounts). The percentages of GFP-positive macrophages and propidium iodide-positive macrophages in a representative donor, which were determined by FACS at 7 days post-transduction, are shown. C, the percentages of the GFP-positive macrophages in three donors are summarized. D, the genomic DNAs were extracted from the transduced macrophages in B and were applied for the quantitative 2-LTR circle PCR as described under “Experimental Procedures.” The viral production was determined by p24 ELISA with the collected media. These experiments were performed in triplicate on cells from three different blood donors. E, the percentages of the GFP-positive activated CD4+ T cells from two donors. The transduction and drug treatment were conducted identically as in D except the infection duration was 2 days. Error bars, S.D.
Thus, the data presented in Fig. 6 using primary macrophages from multiple donors show that rACT effectively inhibits HIV-1 reverse transcription in macrophages, suggesting that rNCTs are potentially a new class of HIV-1 RT inhibitor specifically targeting the unique ability of HIV-1 to infect terminally differentiated/non-dividing macrophages.
DISCUSSION
Whereas dNTPs exclusively serve as building blocks of DNA during chromosomal DNA replication and DNA repair, rNTPs are more versatile and are used for not only RNA synthesis but also cellular energy transfer (e.g. ATP) and as substrates of various cell signaling pathway enzymes (e.g. ATP and GTP). All nucleotides are clearly not compartmentalized in either the nucleus or the cytoplasm (4), so both cellular and viral DNA and RNA polymerases have evolved to efficiently differentiate dNTPs and rNTPs. Specifically, this chemical difference between these two nucleotides is the 2′-OH of their sugar moiety. This can be attributed in part to the fact that the dNTP binding sites of cellular DNA polymerases possess a highly conserved gate control mechanism using a bulky amino acid (e.g. tyrosine) that sterically hinders entrance of the 2′-OH of rNTPs and effectively eliminates their binding to the active site (6–9).
In dividing cells, the rNTP concentration is much higher than that of dNTPs. Our data show that this disparity is even greater in terminally differentiated macrophages, simply because the dNTP concentration difference (22–320-fold) between these two cell types is much larger than the rNTP concentration difference (4–7-fold). Viruses, such as HIV-1, that need to synthesize DNA in macrophages where DNA synthesis is terminally absent encounter not only very low dNTP concentrations that can restrict viral genomic DNA synthesis but also a very large dNTP/rNTP disparity. Our data (Fig. 2) clearly demonstrate that DNA synthesis by HIV-1 RT is significantly delayed at dNTP concentrations that approximate those found in macrophages. These findings prompted us to hypothesize that HIV-1 RT may incorporate rNTPs during DNA synthesis, particularly in the macrophage environment, where dNTP concentrations are limiting (well below the Km and Kd of the enzyme) and rNTP concentrations remain very high. This delayed DNA synthesis generates frequent and extended pausing (Fig. 2), which probably increases the opportunity for HIV-1 RT to incorporate rNTPs, especially when the disparity between dNTP and rNTP concentrations is very large (as is the case in macrophages). Additionally, in our previous study (22) reporting the HIV-1 RT based-dNTP assay, we did not observe the incorporation of rNTPs by HIV-1 RT, and this must be due to the use of much lower rNTP concentrations (12.5 μm each rNTP) compared with the rNTP concentrations found in macrophages and used in our current study (∼1,124 μm; Table 1).
What are the consequences of rNTP incorporation into proviral HIV-1 DNA in the macrophage? Recent work by the Kunkel laboratory (16) suggests that rNTP incorporation during chromosomal DNA synthesis may be more common than previously thought. This is further supported by the existence of multiple rNTP DNA repair mechanisms (44–46), and their existence predicts a deleterious effect of rNTP misincorporation at high levels. Interestingly, biochemical evidence that host DNA repair polymerases β and μ, TdT, and yeast replicative polymerases are able to incorporate rNTPs has been reported (14–16, 47), although it remains to be determined if rNTP incorporation by these polymerases during DNA repair actually happens in live cells. The biological impact of rNMP incorporation during DNA synthesis by both viruses and host cells requires further study.
In general, there are two ways to investigate the cellular incorporation of nucleotides during DNA and RNA synthesis. One way is to monitor the incorporation of the radiolabeled nucleotides into cellular nucleic acids (DNA and RNA) after the treatment with radioactively labeled nucleosides that are converted to their triphosphate forms. In fact, we attempted to quantify amounts of rNMP incorporation in newly synthesized provirus DNAs with 3H-labeled ribonucleosides in the infected macrophages, but this has proved to be technically difficult because a majority of the radioactivity (rNTP incorporation) was detected in cellular RNA even after their removal in the isolated cellular DNA with excessive treatment with RNases. Furthermore, the use of the radioactive ribonucleosides makes it more difficult to explain the predicted data because it is also possible that these radioactive ribonucleotides (i.e. rNDPs) can be converted to corresponding deoxyribonucleotides (i.e. dNDPs) by host ribonucleotide reductase and then eventually phosphorylated to dNTPs, which will be incorporated into the proviral DNA. This unwanted incorporation of the radioactive dNTPs is possible in macrophages because it is also well known that non-dividing cells, such as macrophages, utilize the p53R2 subunit for dNTP biosynthesis, especially when the cells are exposed to DNA damage (48). However, it remains to be tested if the HIV-1 infection induces DNA damage and the p53R2 expression in macrophages.
The other way is to employ competitive nucleoside inhibitors, and our biochemical analyses predicted that rNCTs lacking 3′-OH should inhibit HIV-1 proviral DNA synthesis in macrophages if HIV-1 RT incorporates rNTPs during proviral DNA synthesis. Indeed, this prediction was confirmed in the experiment described in the legend to Fig. 6, using the rNCT, 3′-deoxyadenosine (rACT) inhibitor. Importantly, a much lower rACT-inhibitory effect was observed in T cells (Fig. 6E) compared with macrophages. Clearly, this supports the series of biochemical observations presented in this study, where HIV-1 RT incorporates rNTPs more efficiently under macrophage nucleotide pools relative to those found in T cells.
The macrophage data presented in Fig. 6 imply that rACT can be efficiently converted to the triphosphate form by cellular nucleoside and nucleotide phosphorylases. Terminally differentiated macrophages still need to carry out transcription, protein synthesis, and intracellular signaling and so presumably have the machinery necessary to convert rACT to its active form. Moreover, the 2′,3′-dideoxynucleoside analogs, such as azidothymidine and dideoxy Inosine, are known to be efficiently phosphorylated to their 5′-triphosphate forms in macrophages (49). Thus, we conclude that rACT is probably phosphorylated in macrophages. In addition, the fact that rACT is not toxic to primary human cells (activated CD4+ T cells and macrophages) and several human cell lines at concentrations of up to 1 mm suggests that host transcription machinery, as well as metabolic systems and cell signaling enzymes, are not significantly affected by rACT-triphosphate (3′-deoxy-ATP) at these concentrations.
Considering how rACT acts to inhibit HIV-1 replication, it is important to note the possibility that rACT-triphosphate (3′-deoxy-ATP) could be converted by cellular ribonucleotide reductases to 2′,3′-dideoxyadenosine triphosphate (ddATP), which can also inhibit HIV-1 replication. However, the 3′-OH of rNTPs, which is absent in rACT, is essential to the highly conserved catalysis and chemistry of ribonucleotide reductases for 2′-OH removal (50), suggesting that rACT-triphosphate cannot be converted to ddATP by ribonucleotide reductases. In general, adenosine analogs have not been well conceived as antiviral agents due to the metabolic conversion of adenine to inosine by host adenosine deaminase and then to hypoxanthine. In our proof of concept experiment, we employed only rACT because it is currently the only kind of rNCT commercially available. However, we envision that other kinds of rNCTs, such as rCCT, could be more practical for future in vivo tests.
Finally, our data (Fig. 6E) indicate that rNCTs are much less effective in inhibiting HIV-1 replication in activated CD4+ T cells than in macrophages, which is consistent with the biochemical observation that there is no significant incorporation of rNTP by HIV-1 RT at dNTP/rNTP concentrations that simulate those present in activated T cells (Fig. 3B). How then can this approach to HIV-1 replication inhibition be an effective anti-HIV therapy? We believe that rNCTs may be very useful in terms of preventing the establishment and persistence of long lived productive reservoirs of virus infection in vivo. This approach may have utility not only for macrophages but also for other non-dividing cell types that HIV-1 can infect, such as naive resting T cells and memory T cells. In addition, the use of Nucleoside Reverse Transcriptase Inhibitors and less toxic (Fig. 6A) rNCTs in combination may offer the potential for synergy and offer opportunities for greater therapeutic effectiveness and/or dose reduction, which would lead to lowered cost and/or reduced NRTI-associated side effects, such as neuropathy. Finally, it is worth adding that targeted approaches to HIV-1 inhibition in macrophages are likely to be a useful antiviral strategy because macrophage infection is generally accepted as essential during HIV-1 pathogenesis (19–21, 51–55).
In summary, we characterized proviral DNA synthesis catalyzed by HIV-1 RT under conditions that recapitulated the dNTP and rNTP concentrations found in the two major virus target cell types: activated T cells and terminally differentiated macrophages. Our data show that rNTPs are used as substrates by HIV-1 reverse transcriptase under conditions that simulate those found in macrophages but not under conditions that simulate the environment of in a dividing HIV-1 target cell type. Finally, we show that ribonucleoside chain terminators (rNCTs) have potential as antiviral drugs and that one such molecule (3′-deoxyadenosine) can efficiently inhibit HIV-1 replication in macrophages. This suggests a new category of anti-HIV-1 agents, with potential activity against virus infection of non-dividing host cells.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Dewhurst, Dr. Tracy Baas, and Erin Noble for critically reading the manuscript. We also thank Dr. Joseph Hollenbaugh for his assistance with CD4 T-cell experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants AI049781 (to B. K.), T32 AI007362 (to E. M. K.), TL1 RR024135 (to A. L.), 2P30-AI-050409, R01-AI-076535, 5R37-AI-041980, and 5R37-AI-025899. This work was also supported by the Department of Veterans Affairs (to R. F. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- rNTP
- ribonucleoside triphosphate
- rNCT
- ribonucleoside chain terminator
- ddNTP
- 2′,3′-dideoxyribonucleoside triphosphate
- PBMC
- peripheral blood mononuclear cell
- RT
- reverse transcriptase
- nt
- nucleotide.
REFERENCES
- 1.Björklund S., Skog S., Tribukait B., Thelander L. (1990) Biochemistry 29, 5452–5458 [DOI] [PubMed] [Google Scholar]
- 2.Chabes A. L., Björklund S., Thelander L. (2004) J. Biol. Chem. 279, 10796–10807 [DOI] [PubMed] [Google Scholar]
- 3.Jamburuthugoda V. K., Chugh P., Kim B. (2006) J. Biol. Chem. 281, 13388–13395 [DOI] [PubMed] [Google Scholar]
- 4.Traut T. (1994) Mol. Cell. Biochem. 140, 1573–4919 [DOI] [PubMed] [Google Scholar]
- 5.Stridh S. (1983) Arch. Virol. 77, 223–229 [DOI] [PubMed] [Google Scholar]
- 6.Astatke M., Ng K., Grindley N. D., Joyce C. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3402–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnin A., Lázaro J. M., Blanco L., Salas M. (1999) J. Mol. Biol. 290, 241–251 [DOI] [PubMed] [Google Scholar]
- 8.Cases-Gonzalez C. E., Gutierrez-Rivas M., Ménendez-Arias L. (2000) J. Biol. Chem. 275, 19759–19767 [DOI] [PubMed] [Google Scholar]
- 9.Gao G., Orlova M., Georgiadis M. M., Hendrickson W. A., Goff S. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner A. F., Jack W. E. (1999) Nucleic Acids Res. 27, 2545–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun G., O'Neil P. K., Yu H., Ron Y., Preston B. D., Dougherty J. P. (2001) J. Virol. 75, 11902–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang G., Franklin M., Li J., Lin T. C., Konigsberg W. (2002) Biochemistry 41, 10256–10261 [DOI] [PubMed] [Google Scholar]
- 13.Bergoglio V., Ferrari E., Hübscher U., Cazaux C., Hoffmann J. S. (2003) J. Mol. Biol. 331, 1017–1023 [DOI] [PubMed] [Google Scholar]
- 14.Nick McElhinny S. A., Ramsden D. A. (2003) Mol. Cell. Biol. 23, 2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulé J. B., Rougeon F., Papanicolaou C. (2001) J. Biol. Chem. 276, 31388–31393 [DOI] [PubMed] [Google Scholar]
- 16.Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letvin N. L., Eaton K. A., Aldrich W. R., Sehgal P. K., Blake B. J., Schlossman S. F., King N. W., Hunt R. D. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2718–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuitemaker H., Kootstra N. A., Fouchier R. A., Hooibrink B., Miedema F. (1994) EMBO J. 13, 5929–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van't Wout A. B., Kootstra N. A., Mulder-Kampinga G. A., Albrecht-van Lent N., Scherpbier H. J., Veenstra J., Boer K., Coutinho R. A., Miedema F., Schuitemaker H. (1994) J. Clin. Invest. 94, 2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westervelt P., Trowbridge D. B., Epstein L. G., Blumberg B. M., Li Y., Hahn B. H., Shaw G. M., Price R. W., Ratner L. (1992) J. Virol. 66, 2577–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu T., Mo H., Wang N., Nam D. S., Cao Y., Koup R. A., Ho D. D. (1993) Science 261, 1179–1181 [DOI] [PubMed] [Google Scholar]
- 22.Diamond T. L., Roshal M., Jamburuthugoda V. K., Reynolds H. M., Merriam A. R., Lee K. Y., Balakrishnan M., Bambara R. A., Planelles V., Dewhurst S., Kim B. (2004) J. Biol. Chem. 279, 51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien W. A., Namazi A., Kalhor H., Mao S. H., Zack J. A., Chen I. S. (1994) J. Virol. 68, 1258–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromentin E., Gavegnano C., Obikhod A., Schinazi R. F. (2010) Anal. Chem. 82, 1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. (1991) EMBO J. 10, 3905–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skasko M., Diamond T. L., Kim B. (2009) Biochemistry 48, 5389–5395 [DOI] [PubMed] [Google Scholar]
- 27.Weiss K. K., Bambara R. A., Kim B. (2002) J. Biol. Chem. 277, 22662–22669 [DOI] [PubMed] [Google Scholar]
- 28.Weiss K. K., Isaacs S. J., Tran N. H., Adman E. T., Kim B. (2000) Biochemistry 39, 10684–10694 [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan M., Fay P. J., Bambara R. A. (2001) J. Biol. Chem. 276, 36482–36492 [DOI] [PubMed] [Google Scholar]
- 30.Balakrishnan M., Roques B. P., Fay P. J., Bambara R. A. (2003) J. Virol. 77, 4710–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aggarwal S., Bradel-Tretheway B., Takimoto T., Dewhurst S., Kim B.PLoS One 5, e10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy E. M., Hergott C., Dewhurst S., Kim B. (2009) Biochemistry 48, 11161–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamburuthugoda V. K., Santos-Velazquez J. M., Skasko M., Operario D. J., Purohit V., Chugh P., Szymanski E. A., Wedekind J. E., Bambara R. A., Kim B. (2008) J. Biol. Chem. 283, 9206–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss K. K., Chen R., Skasko M., Reynolds H. M., Lee K., Bambara R. A., Mansky L. M., Kim B. (2004) Biochemistry 43, 4490–4500 [DOI] [PubMed] [Google Scholar]
- 35.Skasko M., Kim B. (2008) J. Virol. 82, 7716–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Bercoff D., Wurtzer S., Compain S., Benech H., Clavel F. (2007) J. Virol. 81, 4540–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp E. B., Miglietta J. J., Shrutkowski A. G., Shih C. K., Grob P. M., Skoog M. T. (1991) Nucleic Acids Res. 19, 3035–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston B. D., Poiesz B. J., Loeb L. A. (1988) Science 242, 1168–1171 [DOI] [PubMed] [Google Scholar]
- 39.Wainberg M. A., Drosopoulos W. C., Salomon H., Hsu M., Borkow G., Parniak M., Gu Z., Song Q., Manne J., Islam S., Castriota G., Prasad V. R. (1996) Science 271, 1282–1285 [DOI] [PubMed] [Google Scholar]
- 40.Kati W. M., Johnson K. A., Jerva L. F., Anderson K. S. (1992) J. Biol. Chem. 267, 25988–25997 [PubMed] [Google Scholar]
- 41.Reardon J. E. (1992) Biochemistry 31, 4473–4479 [DOI] [PubMed] [Google Scholar]
- 42.Piccirilli J. A., Vyle J. S., Caruthers M. H., Cech T. R. (1993) Nature 361, 85–88 [DOI] [PubMed] [Google Scholar]
- 43.Butler S. L., Hansen M. S., Bushman F. D. (2001) Nat. Med. 7, 631–634 [DOI] [PubMed] [Google Scholar]
- 44.Eder P. S., Walder R. Y., Walder J. A. (1993) Biochimie 75, 123–126 [DOI] [PubMed] [Google Scholar]
- 45.Rydberg B., Game J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16654–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekiguchi J., Shuman S. (1997) Mol. Cell 1, 89–97 [DOI] [PubMed] [Google Scholar]
- 47.Bergoglio V., Canitrot Y., Hogarth L., Minto L., Howell S. B., Cazaux C., Hoffmann J. S. (2001) Oncogene 20, 6181–6187 [DOI] [PubMed] [Google Scholar]
- 48.Håkansson P., Hofer A., Thelander L. (2006) J. Biol. Chem. 281, 7834–7841 [DOI] [PubMed] [Google Scholar]
- 49.Arts E. J., Marois J. P., Gu Z., Le Grice S. F., Wainberg M. A. (1996) J. Virol. 70, 712–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandes P. A., Ramos M. J. (2003) Chemistry 9, 5916–5925 [DOI] [PubMed] [Google Scholar]
- 51.Boyer J. C., Bebenek K., Kunkel T. A. (1996) Methods Enzymol. 275, 523–537 [DOI] [PubMed] [Google Scholar]
- 52.Dorr P., Westby M., Dobbs S., Griffin P., Irvine B., Macartney M., Mori J., Rickett G., Smith-Burchnell C., Napier C., Webster R., Armour D., Price D., Stammen B., Wood A., Perros M. (2005) Antimicrob. Agents Chemother. 49, 4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulick R. M., Lalezari J., Goodrich J., Clumeck N., DeJesus E., Horban A., Nadler J., Clotet B., Karlsson A., Wohlfeiler M., Montana J. B., McHale M., Sullivan J., Ridgway C., Felstead S., Dunne M. W., van der Ryst E., Mayer H. (2008) N. Engl. J. Med. 359, 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson D. A., Operskalski E. A., Busch M. P., Mosley J. W., Koup R. A. (1998) J. Infect. Dis. 178, 1163–1166 [DOI] [PubMed] [Google Scholar]
- 55.Willey S., Peters P. J., Sullivan W. M., Dorr P., Perros M., Clapham P. R. (2005) Antiviral. Res. 68, 96–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.