Abstract
Gene duplication is a major force driving genome evolution, and examples of this mode of evolution and of the functions of duplicated genes are needed to reveal general patterns. Here, our study focuses on a particular retrogene (i.e., CG9573) that originated about 5–13 million years ago that we have named Drcd-1 related. It originated in Drosophila through retroposition of the parental gene Required for cell differentiation 1 of Drosophila (Drcd-1; CG14213), which is a known transcription cofactor. Drcd-1r is only present in D. melanogaster, D. simulans, D. sechellia, and D. mauritiana. Drcd-1r is an X to autosome retroposition event. Many retrogenes are X to autosome copies and it has been shown that positive selection underlies this bias. We sought to understand Drcd-1r mode of evolution and function to contribute to the understanding of the selective pressures acting on X to autosome retrogenes. Drcd-1r overlaps with another gene, it is within the 3′ UTR of the gene CG13102 and is encoded in the opposite orientation. We have studied the characteristics of the transcripts and quantified expression of CG13102 and Drcd-1r in wild-type flies. We found that Drcd-1r is transcribed specifically in testes. We also studied the molecular evolution of Drcd-1r and Drcd-1 and found that the parental gene has evolved under very strong purifying selection but the retrogene has evolved very rapidly (Ka/Ks ~ 1) under both positive and purifying selection, as revealed using divergence and polymorphism data. These results indicate that Drcd-1r has a novel function in the Drosophila testes. To further explore Drcd-1r function we used a strain containing a P element inserted in the region where CG13102 and Drcd-1r are located that shows recessive male sterility. Analysis of this strain reveals the difficulties that can be encountered in studying the functions of genes with overlapping transcripts. Avenues for studying of the function of this gene are proposed.
Keywords: Retrogene, Testis expression, Rcd-1, Drosophila, Positive selection
Introduction
Gene duplication provides material for the evolution of new functions (Ohno 1970). One gene duplication mechanism is retroposition (Brosius 1991). Retroposition is a molecular process that leads to the formation of intronless gene duplicates (i.e., retrogenes). Retroposition occurs when parental gene mRNA is reverse transcribed by the reverse transcriptase from a non-LTR retrotransposable element, giving rise to a cDNA that is randomly inserted into the genome (Esnault et al. 2000). Retrogenes are characterized by the absence of introns and the presence of direct repeats and a poly-A tail; however, these last two features are often not found in old retrogenes (Betran et al. 2002). These retroposed gene duplicates may either accumulate mutations, resulting in functionless retropseudogenes, or they may become functional retrogenes. Functional retrogenes must have regulatory regions to be expressed (Betran et al. 2002). Despite this, an excess of retrogenes have been described to be X to autosome retroduplicates (Bai et al. 2007; Betran et al. 2002; Emerson et al. 2004). This excess is not explained by more retrocopies been produced in this pattern (i.e., mutational biases), as retropseudogenes and retrotransposable elements do not show this bias (Emerson et al. 2004; Fontanillas et al. 2007), but positive selection (Bai et al. 2007; Betran et al. 2002; Emerson et al. 2004; Fontanillas et al. 2007). In addition, many of these retrogenes adopt male germline functions (Bai et al. 2007; Vinckenbosch et al. 2006). Understanding the selective forces that drive this pattern of duplication requires study of the mode of evolution and function of these X to autosome retrogenes.
In this work, we studied the expression and mode of evolution of CG9573, which we named Drcd-1r (Drosophila required for cell differentiation 1-related). We characterized Drcd-1r as a recently originated retrogene in Drosophila (Bai et al. 2007). The parental gene CG14213 was identified by Garces et al. (2007) as the Drosophila ortholog of Required in cell differentiation 1 (Drcd-1). Drcd-1r originated from the parental gene Drcd-1 through the process of retroposition and is located in region 29 of the 2L chromosomal arm in D. melanogaster, D. simulans, D. sechellia and D. mauritiana, whereas the parental gene is likely present in all species of Drosophila on the X chromosome. The parental gene Drcd-1 resides on the X chromosome, therefore Drcd-1r moved from the X chromosome to an autosome. From the phylogenetic distribution we infer that Drcd-1r originated about 5–13 million years ago (Bai et al. 2007; Tamura et al. 2004). In the process of retroposition, Drcd-1r lost the four introns that are present in Drcd-1 (Supplementary Figure 1). The coding region of the gene and its length have changed significantly after duplication. In D. melanogaster, the two genes show only 71% (199 residues of 279) identity at the amino acid level when removing unaligned (i.e., 25) amino acid positions (Supplementary Figure 1). Interestingly, Drcd-1r overlaps with another gene CG13102. It is within the 3′ UTR of CG13102 and encoded in the opposite orientation. Figure 1 and Supplementary Figure 2 show the location of Drcd-1r relative to CG13102. CG13102 is present in all sequenced species of Drosophila; however, its function is unknown.
Fig. 1.
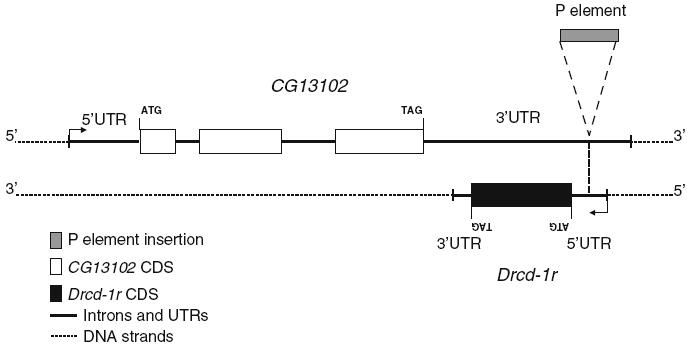
Drcd-1r genomic region. The genomic organization of the region where Drcd-1r is inserted is shown as annotated in FlyBase. The location of the P element insertion of line 11773 is also shown
Functional and structural studies of Rcd-1 (the parental gene) have been carried out in yeast and humans. Rcd-1 is responsible for nitrogen starvation-induced cell differentiation (Okazaki et al. 1998) and the homolog of Rcd-1 in mammals functions as a cell differentiation co-factor (Garces et al. 2007; Hiroi et al. 2002). Rcd-1 in humans has been shown to interact with c-myb, a transcription factor required for hematopoietic cell differentiation in humans (Haas et al. 2004). Rcd-1 contains armadillo-like repeats that are known to play a role in protein–protein interactions and believed to help in the dimerization of Rcd-1 (Garces et al. 2007). In addition, an Rcd-1 dimer has recently been described to have a DNA binding cleft formed of positively charged amino acids that provides DNA binding affinity (Garces et al. 2007). These data reveal that Rcd-1 might be a transcription factor (binding DNA) or cofactor (interacting with proteins that bind DNA).
Our results reveal that Drcd-1r is a novel gene with testis-specific function that has evolved under both positive and purifying selection, consistent with the way many new testis-specific genes have been seen to evolve (Betran and Long 2003; Ting et al. 1998; Long and Langley 1993; Tracy et al. 2010; Dorus et al. 2008; Arguello et al. 2006). We also analyzed a mutant stock (11773) that has a P element inserted in the 5′UTR (annotated using a single cDNA from testis, AT13107) of Drcd-1r. This region is also annotated as the 3′UTR of CG13102 using a single cDNA RE57454 from embryos (see Fig. 1 and Supplementary Figure 2). While males homozygous for the chromosome containing this P element insertion are sterile and have been described as having a post-meiotic differentiation defect by Castrillon et al. (1993), our analyses reveal the difficulties of studying the function of a gene whose transcript overlaps with another gene.
Materials and methods
Strains
The expression of CG13102 and Drcd-1r was first studied in wild-type D. melanogaster flies from strains EC-180 (Ecuador-180) and EC-175 (Ecuador-175). Expression was also studied using the stock 11773 containing a P element insertion that was obtained from Bloomington Drosophila Stock Center. This stock is balanced using CyO, a balancer for the 2nd chromosome. Because the P element insertion leads to recessive sterility in males, the stock is maintained with heterozygous males against this balancer (P{PZ}ms(2)29F07717 CG957307717 CG1310207717 cn1/CyO; ry506). Curly (i.e., males heterozygous for the P element insertion) and non-Curly (i.e., males homozygous for the P element insertion) males and females are produced every generation while only the non-Curly males are sterile. After receiving the stock, it was checked to confirm the position of the P element insertion. DNA was extracted and PCR performed with flanking primers and primers in the P element and flanking region. The infertility of this stock was also confirmed (data not shown). A complementation test was performed because it is the standard way to confirm if the recessive male-sterile phenotype of a mutation (P element insertion in stock 11773) is induced by the mutation. We utilized two stocks (Df(2L)M22-14/CyO and Df(2L)exel6021/CyO) that have chromosome deficiencies with breakpoints from 29C1-2 to 30C8-9 and 29F7 to 30A2, respectively, both of which span the P element insertion site 29F. The two stocks are balanced using CyO and were also obtained from Bloomington Drosophila Center. We crossed fertile Curly males from two stocks (i.e., Df(2L)M22-14/CyO and Df(2L)exel6021/CyO) with fertile homozygous females from the P element insertion stock 11773 (i.e., non-Curly females). We monitored the fertility of non-Curly male descendants. Several vials with at least five of those males were mass-mated with fertile Curly females fromstock 11773.
Isolines from the wild were used for our polymorphism analyses. Twelve isolines for D. melanogaster from a single population in Zimbabwe (Hollocher et al. 1997) and 10 for D. simulans from a single population in Madagascar were used in these analyses. The Zimbabwe and Madagascar lines were kindly provided by the Chug-I. Wu and Chip Aquadro laboratories, respectively. DNA was extracted from a single fly for each of the D. melanogaster and D. simulans isolines. DNA extractions were carried out using the Puregene kit (QIAGEN, Valencia, CA) with single-fly modifications.
Expression analyses
RNA was extracted from whole adult males, testis and male carcasses (gonadectomized males) of EC180 and EC175 (Strains from Ecuador) and the mutant stock 11773. One hundred testes were dissected and kept in RNA-later buffer (Applied Biosystems/Ambion, Austin, TX). RNA was extracted using an RNeasy kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions and quantified using a Nano-drop spectrophotometer. Analyzing the expression of intronless genes (such as Drcd-1r) is challenging because genomic contamination can produce a band of the same size as that expected from cDNA. Therefore, we digested possible contaminating DNA in the total RNA (DNase I amplification grade; Invitrogen, Carlsbad, CA) and ran controls including DNA-digested total RNA without reverse transcriptase. Oligo-dT (Promega, Madison, WI) was used for first strand cDNA synthesis. RT-PCRs were then carried out using the primers shown in Supplementary Table 1. Forward and reverse primers were designed in the coding region of CG13102 and another set of primers was designed in the coding region of Drcd-1r that is also the 3’UTR of CG13102. A fifth primer was designed in the 3′UTR of Drcd-1r and is referred to as the specific primer. This primer was used instead of oligo-dT for first strand cDNA synthesis in the Drcd-1r-specific RT-PCR. PCR products were purified using a QIAGEN PCR purification kit (QIAGEN, Valencia, CA) and sequenced using an ABI sequencer with fluorescent BigDye terminator nucleotides (Applied Bio-systems, Foster City, CA). 5′ and 3′ Rapid Amplification of cDNA Ends (RACE) were performed using RNA from testes of wild-type males and sterile males (i.e., non-Curly) from stock 11773 to reveal if the P element insertion affects transcriptional start and end sites. 5′ RACE was performed to identify the Transcription Start Site (TSS) of Drcd-1r and 3′ RACE was performed to identify the Transcription End Site (TES) of CG13102. We used the Ambion kit 1700 (Applied Biosystems/Ambion, Austin, TX) for both 5′ and 3′ RACE. This protocol allows RACE only on mRNA that has a cap. RACE PCR products were either directly cleaned or extracted from the gel and then sequenced. Primer sequences used in different surveys are provided in Supplementary Table 1.
Quantitative real-time RT-PCR (QRT-PCR) was used to measure the amount of RNA produced by both CG13102 and Drcd-1r from sterile and wild-type males in testis and gonadectomized bodies (i.e., carcasses). RNA was extracted from wild-type males of stock EC-175 and sterile males homozygous for the P element insertion of stock 11773. The same two sets of primers as above were used, one set in the coding region of CG13102 and the other in the overlapping region shared by both CG13102 and Drcd-1r. Gapdh2 was used to normalize the QRT-PCR (see Supplementary Table 1). QRT-PCR was performed using the ABI 7300 Real Time PCR system and the SYBR Green PCR Core reagents from Applied Biosystems (Foster City, CA). The RT reaction was performed as described above using oligo-dT as the primer. The PCR efficiency for each pair of primers was determined according to Schmittgen and Livak (Schmittgen and Livak 2008) utilizing the equation m = −(1/Log E), where m is the slope of the line and E is the efficiency. The results of this assessment are shown in Supplementary Table 2. The efficiencies were comparable for all PCR primer pairs.
The estimates of transcript abundance for CG13102 and Drcd-1r were normalized to estimates from the control gene Gapdh2 on a per-plate basis to account for variation in absolute amounts of RNA. This was accomplished by subtracting the control gene value CT (per each replicate) from the region-gene-of-interest value CT (per each replicate) and obtaining ΔCT. Average ΔCTs for different genes and strains were compared using ANOVA. Threshold cycle numbers (i.e., CT values) were obtained with default ABI software parameters. QRT-PCR products were run in a gel to control for spurious amplification.
Interference between transcripts of CG13102 and Drcd-1r in the overlapping region was evaluated by looking for double-stranded RNA (dsRNA) following previously described methodology (Puig et al. 2004; Aravin et al. 2001). Total RNA was obtained from forty males from stocks EC175 and 11773. This methodology involves treating RNA with RNase to degrade single-stranded RNA, removing the RNase using ethanol precipitation, heating the sample at 95°C for 5 min to denature existing dsRNA and finally reverse transcribing the RNA. Two regions of Drcd-1r covering the overlap with CG13102 were evaluated for the presence of dsRNA using PCR and nested PCR. The primers used are listed in Supplementary Table 1. The negative control for these reactions involves PCR and nested PCR of a sample treated with RNase but not denatured.
Sequence analyses
Genomic DNA extractions were performed on single flies using the Puregene kit. Drcd-1r was PCR amplified from genomic DNA of ten D. melanogaster flies from different Zimbabwe strains [ZH13, ZH19, ZH20, ZH21, ZH23, ZH24, ZH27, ZH28, ZH32, and ZH40; (Hollocher et al. 1997)] and ten D. simulans flies from different Madagascar strains (M1, M2, M4, M5, M24, M37, M50, M242, M252, and M258). The primers used for this amplification are provided in Supplementary Table 1. PCR products were sequenced from both strands using the above and internal primers and as described above. PCR products from heterozygotes were cloned using a TOPO cloning kit and one insert was sequenced to resolve haplotypes. GenBank accession numbers for the sequences obtained in this work are HM358897-HM358925.
The sequences were aligned using Clustal W (Thompson et al. 1994) and manually adjusted. The KA/KS ratio (ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site) for both the parental gene Drcd-1 and its retrogene Drcd-1r were compared using PAML4 software (Yang 2007). For this analysis, the Drcd-1 sequences from eleven Drosophila species and sequences of the retrogene (Drcd-1r) from D. melanogaster, D. simulans and D. sechellia (Clark et al. 2007) were used. A phylogeny was also provided (see below). This tree was constructed taking into account the phylogenetic information and the time of the gene duplication event (Ting et al. 2000; Bai et al. 2007). Several likelihood ratio models were compared and this allowed us to examine differing hypotheses regarding evolutionary pressures acting on Drcd-1 and Drcd-1r that made sense a priori. We tested if the retrogene and parental gene have evolved with the same Ka/Ks ratio. We also wanted to know if the retrogene protein changed faster after duplication and if it has been evolving under positive or purifying selection. This was done by performing several comparisons: a model with single ratio versus a model with two ratios (parental vs. retrogene), and a model with two ratios versus a model with three ratios (parental vs. retrogene vs. lineage after duplication). We also compared the best of these models (i.e., the two ratio model; see below) with a model in which the ratio of the retrogene lineages is fixed to having the value of 1 to test for purifying or positive selection. In our case given that the retrogene is evolving with a ratio greater than one (see below), this comparison tests for positive selection. The comparisons were tested considering that twice the difference of likelihood between the models that we want to compare should distribute as a χ2 distribution with as many degree of freedom as the difference in parameters between the models.
The coding sequences used for Drcd-1 and Drcd-1r in the above divergence analyses were the annotated coding regions in FB2008_06 released July 3, 2008 from FlyBase (Wilson et al. 2008). For Drcd-1r, GD22367, GM12572 and CG9573 sequences for D. simulans, D. sechellia and D. melanogaster were retrieved. For Drcd-1, we used the reported orthologs: GM22976 in D. sechellia, GE15860 in D. yakuba, GG19241 in D. erecta, GF20275 in D. ananassae, GA12828 in D. pseudoobscura, GK16372 in D. willistoni, GJ15574 in D. virilis, and GH24151 in D. grimshawi. We added GI21711 of D. mojavensis and GL27101 of D. persimilis as the Drcd-1 orthologs in these species based on sequence and synteny conservation. We were not able to use the D. simulans Drcd-1 sequence because the D. simulans genome has not been completely sequenced in the Drcd-1 region. Drcd-1 and Drcd-1r alignments of these sequences revealed indels and therefore ambiguity in the alignment of the 3’ region (See Supplementary Figure 1). We performed PAML analyses removing this region to be conservative (i.e., not to spuriously inflate protein divergence between paralogs).
Next, site models (NSsites) of CODEML software implemented in PAML were used to assess the possibility of positive selection acting on a few sites in Drcd-1r. Site-specific likelihood models M7 and M8 were applied to the sequences with the appropriate tree topology (Nielsen and Yang 1998; Yang et al. 2000). Model M7, which does not allow for sites under positive selection was compared to model M8, which allows for sites under positive selection. M7 assumes a beta distribution for ω (i.e., Ka/Ks ratio or dn/ds ratio as designated by the authors) between 0 and 1 over all sites while M8 adds an additional site class (ω ≥ 1) with ω estimated from the data. A likelihood ratio test was performed by calculating two times the log likelihood values and comparing this value to a χ2 distribution with two degrees of freedom (see PAML manual for details on conservative number of the degrees of freedom in this comparison; http://abacus.gene.ucl.ac.uk/software/pamlDOC.pdf). Posterior probabilities of codons under positive selection were computed in model M8 using Bayes Empirical Bayes (BEB) approach (Yang et al. 2005) and implemented in PAML when the LRT was significant. The tree provided for the site analyses was ((Drcd-1r_D. simulans, Drcd-1r_D. sechellia), Drcd-1r_D. melanogaster). Again, taking a conservative approach we performed the site models (above and below) removing 18 codons in D. simulans sequences and 11 codons in D. melanogaster sequences that did not align unambiguously.
Drcd-1r sequences were also analyzed using the HyPhy package and the above tree topology. Sequences were uploaded to the HyPhy package available at http://www.datamonkey.org (Pond and Frost 2005). Random effects likelihood (REL) analyses were performed in an attempt to detect positively selected codons in Drcd-1r. A Bayes factor threshold of 50 that corresponds to very high posterior probabilities (P ~ 1/Bayes factor) was used for REL analysis (Kosakovsky Pond and Frost 2005). This codon analysis is supposed to be more realistic than the PAML site models because it allows for synonymous rate variation across sites (Kosakovsky Pond and Frost 2005).
The McDonald–Kreitman test (McDonald and Kreitman 1991) was used to compare the within-species polymorphism and between-species divergence in order to understand further the mode of evolution of Drcd-1r. This test, which contrasts the ratio of fixed synonymous differences to nonsynonymous differences with the ratio of the polymorphic synonymous to nonsynonymous sites, was performed using D. melanogaster and D. simulans data and DNAsp software (Rozas et al. 2003). We also performed a modified McDonald–Kreitman test in which synonymous unpreferred changes were ignored because they might potentially contribute more to polymorphism than divergence due solely to codon usage (Schlenke and Begun 2003).
Again, alignments of Drcd-1r sequences from D. simulans and D. melanogaster revealed indels and therefore ambiguity in the alignment. Once more, taking a conservative approach we performed the McDonald–Kreitman test removing 18 codons in D. simulans sequences and 11 codons in D. melanogaster sequences.
Results
Expression of CG13102 and Drcd-1r in wild-type males
Unexpected biases in the patterns of duplication of retrogenes (i.e., excess of X to autosome retroduplicates) have been described (Bai et al. 2007; Betran et al. 2002; Emerson et al. 2004). In addition, these genes show testis-specific transcription (Bai et al. 2007; Betran et al. 2002; Emerson et al. 2004). Becasue Drcd-1r is an X to autosome retroposition event that was inserted within the 3′ UTR of the gene CG13102, we studied the pattern of expression and transcript length of Drcd-1r and CG13102 as a first approach to understand Drcd-1r function and the possible effects of a mutant that carries a P element insertion in the region.
Some information was available about the expression and transcript length of CG13102 and Drcd-1r in males prior to this work. The annotation of the CG13102 transcript in FlyBase shown in Fig. 1 was supported by a single-embryo cDNA and the one for Drcd-1r by a transcript from testis. In addition, microarray data has recently become available in FlyAtlas (Chintapalli et al. 2007) revealing that Drcd-1r is upregulated 6.2 times compared to whole body in testis and is either expressed at the same level or downregulated in the rest of the tissues studied. CG13102 is upregulated 11.2 times in midgut and is either expressed at the same level or downregulated in the rest of the tissues studied. In particular, it is downregulated in testis. We carried out expression analyses of both genes in testis from additional strains (i.e., wild-type strains EC180 and EC175 and the P element mutant strain 11773) to characterize the transcripts in this tissue and the level of expression of both genes.
RT-PCR was carried out for CG13102 and Drcd-1r in testis and the rest of the body (i.e., carcasses). A specific primer instead of oligo-dT was used to make the cDNA for Drcd-1r to ensure amplification occurred only when this gene was expressed (see “Materials and methods”). Figure 2 depicts expression of CG13102 and Drcd-1r in testis and in carcasses of wild-type flies. We observed expression of both genes in testis but only CG13102 was expressed in carcasses. We conclude that Drcd-1r is the only testis-specific gene of the pair. This is in agreement with the data reviewed above.
Fig. 2.
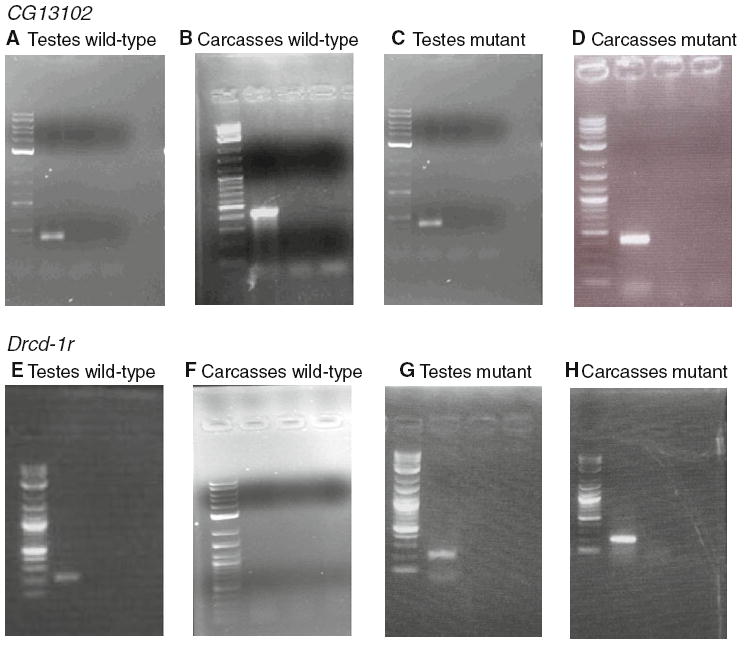
RT-PCR results are shown. The first lane is always the ladder, the second lane is RT+, the third lane is an RT- control and the fourth lane is a PCR negative control. a, b, c and d correspond to CG13102 results using RNA from testis of wild-type males, carcasses from wild-type males, testis from sterile males and carcasses from sterile males, respectively. e, f, g and h correspond to Drcd-1r results using RNA from testis of wild-type males, carcasses from wild-type males, testis from sterile males and carcasses from sterile males, respectively
The 3′RACE product of CG13102 and the 5′RACE product of Drcd-1r from wild-type chromosomes of EC-180 male testis were obtained and sequenced. The 5′RACE product of Drcd-1r coincides with that annotated in FlyBase (Supplementary Figure 2). Interestingly, we observed a 3′UTR of CG13102 of the same length as described in the embryo shown in Supplementary Figure 2 and reported to be 1,308 bp for some transcripts, however the 3′UTR is much shorter (just 515 bp) in some other transcripts. Only the short transcripts would not be affected by the P element insertion.
Expression of CG13102 and Drcd-1r in males of the mutant stock (11773)
Studies in wild-type (EC-180) males showed expression of Drcd-1r and CG13102 in testis. CG13102 was expressed ubiquitously in males whereas Drcd-1r was expressed only in testis. We studied the expression of both genes in testis of individuals from stock 11773 homozygous for the P element insertion (non-Curly males of the stock that is maintain with the CyO balancer) to confirm the effects on transcription of the P element insertion. We use the same procedure as before to study the expression pattern in wild-type to make sure that we detected both transcripts independently. Both genes were transcribed in testis of non-Curly males of stock 11773 (Fig. 2). We expected that there would be a lack of expression of Drcd-1r in the non-Curly flies due to the P element insertion in its 5′UTR. However, we found transcription of both genes in the testis of mutant males and we observed abnormal transcription of Drcd-1r in carcasses (Fig. 2). Thus Drcd-1r, which was not transcribed in wild-type carcasses, is transcribed in carcasses from the strain with the P element insertion (Fig. 2). We conclude that the P element insertion is driving broad expression of Drcd-1r possibly due the presence of known or cryptic regulatory regions in the P element, because the transcript begins within the P element (see below). This broad expression if also occurring in testis could be responsible for the sterility phenotype.
Next, we asked if the P element insertion modifies the TSS of Drcd-1r and the TES of CG13102. We performed 5′RACE for the gene Drcd-1r and 3′RACE for the gene CG13102 using RNA from non-Curly males (homozygous for the P element insertion). Our results showed that both genes are affected by the P element insertion in testis. Transcripts of CG13102 in males homozygous for the P element insertion are affected; the 3′UTR of the most abundant transcript extends approximately 700 bases into the P element with a spliced cryptic intron. Transcripts of Drcd1-r are also affected; 5′RACE from males homozygous for the P element insertion showed that the most abundant transcript starts with 16 bases of the P element. Thus, most often the TSS of Drcd-1r and the TES of CG13102 include part of the P element (data not shown). Shorter TSSs for Drcd-1r (i.e., 116 bp shorter than in wild-type) and TESs for CG13102 that do not extend into the region where the P element is inserted were also sometimes obtained.
While it is likely that CG13102 is still transcribed in the same cell types because its 5′ region is not perturbed, Drcd-1r is expressed in a different pattern. We observed expression in somatic cells that is likely driven by a regulatory region within the P element. Additional analyses also indirectly (by detecting dsRNAs that are not produced in wild-type flies) revealed a different pattern of transcription in testes (see below).
Are CG13102 and Drcd-1r transcribed at the same level in wild-type and mutant males?
To understand the effects of the P element insertion on the expression level of the genes, we carried out a quantitative real-time RT-PCR (QRT-PCR) experiment with RNA from sterile male testis (males homozygous for the P element insertion) and compared it to the RNA of wild-type male testis from stock EC175. We ran and analyzed three different PCR reactions for each: (1) using PCR primers in the Drcd-1r region that overlap CG13102; (2) using primers in the coding region of CG13102; and (3) using primers for Gapdh2 (normalizer). We obtained and compared the ΔCT (the difference between the CT of the gene of interest and the CT of Gapdh2) mean values. The mean and the standard error of the ΔCT values using the PCR primers in the Drcd-1r region for fertile males (EC175 line) were −0.785 ± 0.5185 and for sterile males (stock 11773) they were 1.4767 ± 0.8833, showing a statistically significant difference (F1,5 = 18.524; P = 0.008). We infer that in sterile males the quantity of transcripts from the overlap region is lower than in fertile males (wild-type line). This could be explained by the presence of RNA interference (dsRNA degradation) between Drcd-1r and CG13102 transcripts in the mutants due to the broader expression of Drcd-1r in infertile males (i.e., expression in carcasses of the males homozygous for the P element insertion; Fig. 2). It is likely due to Drcd-1r and CG13102 being transcribed in the same cell types in the testis of infertile males as discussed below. The mean and the standard errors of the ΔCT values for the coding region of CG13102 in fertile males were 3.6075 ± 1.2572 and in sterile males they were −0.6166 ± 0.3079, revealing a significant difference (F1,5 = 30.910; P = 0.003) with an increase of the amount of transcript in testes of sterile males. These data reveal effects that are different depending on the region of the gene, likely revealing a variety of transcripts (as described above) and their non-overlapping expression in wild-type testis but overlapping expression in the testis of mutant individuals. Interestingly, the observed mean and standard error ΔCT values for the mRNA (cDNA) from carcasses of fertile males for the coding region of CG13102 were 3.59 ± 0.5003, which is not significantly different from that obtained for wild-type male testis above (3.6075 ± 1.2572; P > 0.05). This suggests that the alterations (i.e., changes possibly related to the phenotype) are restricted to testis cells.
Here we only look at the level of transcript and it is possible that, because transcript lengths and sequences of 5′UTR of Drcd-1r or the 3′UTR of CG13102 have changed, translation or other features encoded in the UTRs could be altered.
Is there interference between transcripts of CG13102 and Drcd-1r in wild-type or mutant males?
Given that these two genes are overlapping and Drcd-1r is mis-expressed in mutant males (i.e., transcribed in tissues where it was not transcribed in wild-type males), we studied the presence of double stranded RNA in wild-type and mutant males. RNA obtained from whole males of stocks EC175 and 11773 was assessed for the presence of dsRNA in two regions of Drcd-1r. In sterile males from stock 11773, we detected double-stranded RNA (dsRNA) within the overlapping region at the 5′ end of Drcd-1r but not at the 3′ end (Fig. 3 and data not shown). However, we did not observe dsRNA in either region in the wild-type stock (Fig. 3 and data not shown). We infer that the occurrence of interference between transcripts of the overlapping region is a result of an alteration of the pattern of expression of Drcd-1r in sterile males (i.e., the P element insertion is driving broad, somatic and testes expression of Drcd-1r) and is most likely due to new expression in testis of Drcd-1r (see QRT-PCR results above). This RNA interference could be responsible for or contribute to the sterility phenotype, as could the Drcd-1r mis-expression described above (see additional discussion below).
Fig. 3.
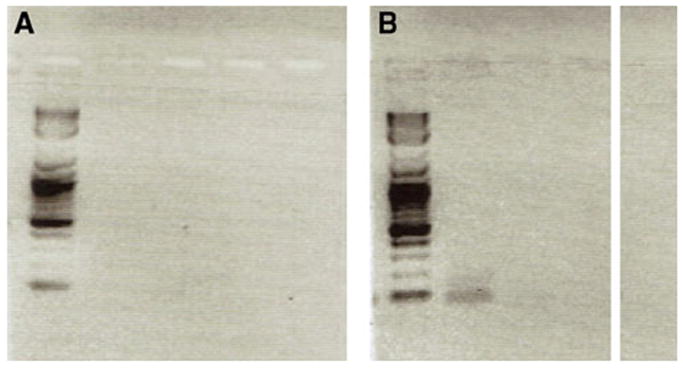
dsRNA detection results are shown. a and b correspond to wildj-type and infertile male assays in the 5′ region of overlap. Lane 1 corresponds to the ladder. Lane 2 corresponds to RT+ treated with RNase and denatured. Lane 3 corresponds to RT− treated with RNase and denatured. Lane 4 corresponds to RT+ treated with RNase and not denatured. Lane 5 is the negative control for PCR
Complementation test of the mutant stock 11773
Curly males from two stocks (i.e., Df(2L)M22-14/CyO and Df(2L)exel6021/CyO) were crossed with fertile homozygous females for the P element insertion from stock 11773 (i.e., non-Curly females). Non-Curly males derived from these crosses were mass-mated with fertile Curly females from stock 11773. These non-Curly males are heterozygous for the P element insertion over a deficiency and are predicted to be sterile, given that the sterility effects described for this stock are recessive. To our surprise, abundant offspring were produced from those crosses, revealing that the insertion might not be causing the observed sterility unless it is caused by the level of interference between transcripts or level of mis-expression of the transcript (as observed above). If the phenotype is caused by RNA interference or mis-expression in testis, it remains possible that there is a dosage effect and that heterozygotes for deficiencies that have only one copy of the P element insertion do not display enough interference or mis-expression (see “Discussion”).
Sequence analyses
Divergence data was analyzed using the PAML software (see “Materials and methods”) to understand the mode of evolution for Drcd-1r and the parental gene Drcd-1. Ka/Ks ratios for different branches were calculated under different models and their likelihoods compared to infer if the functions and constraints of the parental gene and retrogene have remained the same or not and to explore if there has been positive selection acting on the Drcd-1r. See Table 1 for results. Figure 4 shows a tree with labeled nodes to help interpret the results.
Table 1.
PAML analyses for Drcd-1 and Drcd-1r
| Branch | Free-ratio model | One-ratio model | Two-ratio model Drcd-1 vs. Drcd-1r | Three-ratio model Drcd-1 vs. Drcd-1r vs. dup. lineage | Two-ratio fixed model Drcd-1 vs. Drcd-1r = 1 |
|---|---|---|---|---|---|
| 15–16 | 999.000 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 16–17 | 0.0054 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 17–18 | 0.0061 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 18–19 | 0.0045 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 19–20 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 20–21 | 0.0315 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 21–1 | 0.0210 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 21–2 | 0.0092 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 20–22 | 1.5565 | 0.0563 | 1.0649 | 1.6341 | 1.0000 |
| 22–3 | 0.5326 | 0.0563 | 1.0649 | 0.9060 | 1.0000 |
| 22–23 | 1.8069 | 0.0563 | 1.0649 | 0.9060 | 1.0000 |
| 23–4 | 1.2459 | 0.0563 | 1.0649 | 0.9060 | 1.0000 |
| 23–5 | 0.5157 | 0.0563 | 1.0649 | 0.9060 | 1.0000 |
| 19–24 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 24–6 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 24–7 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 18–8 | 0.0047 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 17–25 | 0.0057 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 25–9 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 25–10 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 16–11 | 0.0120 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 15–26 | 0.0001 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 26–27 | 0.1584 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 27–12 | 0.0048 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 27–13 | 0.0063 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| 26–14 | 0.0079 | 0.0563 | 0.0083 | 0.0083 | 0.0083 |
| p | 53 | 28 | 29 | 30 | 28 |
| l | −4561.76 | −4825.46 | −4573.28 | −4572.44 | −4573.34 |
See Fig. 4 for branch definition. p is the number of parameters in the model, and l is log likelihood of the model. ω̂, the estimated KA/KS ratio for the branch, is shown in a given model. See text for comparisons of the likelihoods of the models. The alignment of the sequences compared is 936 bp
Fig. 4.
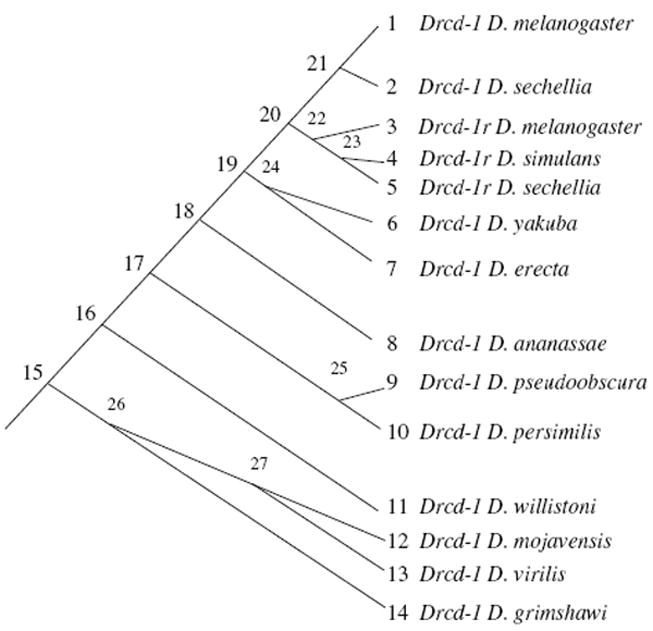
For Drcd-1r and its parental gene (Drcd-1), species and gene phylogeny used by PAML is shown. Nodes and tips are labeled to help interpret Table 1
In particular, a free-ratio model that assumes that all branches evolve at different ratios was run. The log likelihood value for this model was −4561.76. The number of parameters for this model was 53. This model was then compared to a one-ratio model that assumes all branches evolve at a single ratio. The log likelihood value of the one-ratio model was −4825.46. A comparison revealed that the free-ratio model was significantly more likely than the one one-ratio model (X2 = 527.41, df = 25, P < 10−6), indicating that the Ka/Ks ratios are indeed different among the lineages. The two-ratio model allows for the estimation of a different Ka/Ks ratio in the parent gene Drcd-1 and the retrogene Drcd-1r. The two-ratio model was significantly more likely than the one-ratio model (X2 = 504.36, df = 1, P < 10−6). The estimated Ka/Ks ratio of the retrogene was 1.0649. This is a very large ratio and close to 1. The Ka/Ks ratio for the parental gene (Drcd-1) is very small 0.0083. We then compared the two-ratio model with a three-ratio model that allows for the estimation of a different evolutionary ratio right after the duplication (internal branch 20–22; Fig. 4). The three-ratio model was not significantly more likely than the two-ratio model (X2 = 0.12, df = 1, P = 0.7290). We next tested if the single retrogene ratio is different from 1 to uncover the action of positive selection. The two-ratio model where the retrogene lineage was fixed to the ratio of 1 was compared to the two-ratio model. The comparison revealed that these two models do not significantly differ (X2 = 1.68, df = 1, P = 0.1949). This revealed that the retrogene is evolving at a Ka/Ks ratio of ~ 1 (i.e., the mode of evolution predicted for pseudogenes). However, this does not imply that Drcd-1r is a pseudogene because in all lineages the open reading frame is intact. This is further explored below using site models and polymorphism data. These additional approaches revealed the action of positive selection in addition to purifying selection.
PAML site model analysis did not reveal significant differences between models M7 and M8 (2Δl = 3.2516, df = 2; P > 0.05). However, random effects likelihood (REL) analyses using the Hyphy package detected seven codons with a Bayes factor greater than 50 (P < 0.02) that have likely been under positive selection (180, 204, 240, 252, 261 and 281; shown in Supplementary Table 3 and Supplementary Figure 3). Only one of these codons (180) corresponds to a previously conserved aromatic amino acid (F184 in humans (Garces et al. 2007)). Amino acids known to form the dimerization surface and a positively charged cleft that possibly binds DNA (Garces et al. 2007) are often conserved, but many Rcd-1 amino acids conserved from humans to flies have changed in Drcd-1r (Supplementary Figure 4). This leads us to conclude that Drcd-1r is likely to have a different function from the parental gene. It is possible that its function would be related to transcription (i.e., be similar to the parental gene) because many amino acids involved in dimerization and possibly DNA binding are conserved (see additional discussion below).
To further explore the mode of evolution of Drcd-1r and understand if the fast evolution is due to relaxation of constraints or positive selection, we performed the McDonald–Kreitman test. This test compares the ratio of within-species polymorphism for synonymous and nonsynonymous sites to between-species divergence. We performed the test with the sequences for Drcd-1r from D. simulans and D. melanogaster (Table 2). The ratio of the fixed nonsynonymous to synonymous substitutions was 3.0769 (80/26) and the ratio of the nonsynonymous to synonymous polymorphisms was 0.2692 (7/26), which is a highly significant difference (G (with Williams correction) = 31.026; P < 10−4). The modified McDonald–Kreitman test also showed significant differences in the same direction (G (with Williams correction) = 10.508; P = 0.0012). The ratio of fixed nonsynonymous to synonymous substitutions was 13.33 (80/6) and the ratio of nonsynonymous to synonymous polymorphisms was 1.1667 (7/6). These ratios clearly show an excess of nonsynonymous fixed substitutions as compared to nonsynonymous polymorphic sites, which indicates that the protein has been under recurrent positive selection between species. The number of polymorphic sites shows an excess of synonymous sites as compared to nonsynonymous sites, which reveals that the retrogene Drcd-1r has also been under purifying selection within species. Differences between the species do not seem to have accumulated in any particular region (Supplementary Figure 3).
Table 2.
McDonald–Kreitman and modified McDonald–Kreitman tests for Drcd-1r in the D. melanogaster and D. simulans lineages
| Fixed | Polymorphic | |
|---|---|---|
| Replacement | 80 | 7 |
| Synonymous | 26 (6) | 26 (6) |
G (with Williams correction) = 31.026; P < 0.0001. Values in brackets correspond to the changes to preferred codons used for the modified McDonald–Kreitman test: G (with Williams correction) = 10.508; P = 0.0012
Discussion
Examples are beginning to accumulate in Drosophila of newly duplicated genes (often retrogenes) acquiring male germline-specific expression (Bai et al. 2007; Betran and Long 2003; Betran et al. 2002; Long and Langley 1993; Yuan et al. 1996; Hwa et al. 2004; Tripoli et al. 2005; Arguello et al. 2006; Sturgill et al. 2007). Some of these duplicates have been proven to have important functions in the male germline (Kalamegham et al. 2007; Zhong and Belote 2007; Timakov and Zhang 2001). Some of them are transcription factors specific to the male germline that are required for the progression of meiosis (Chen et al. 2005; Hiller et al. 2001, 2004). The data we provide in this work leads us to propose that Drcd-1r is another example of this type of duplication that has been recruited for male germline function in Drosophila. As outlined above, Drcd-1r originated from Drcd-1 and could have a function related to the parental gene (i.e., be a novel transcription factor or cofactor). However, it could also have acquired a related but different function as exemplified by the case of OdsH. OdsH was believed to be a fast evolving male germline transcription factor (Sun et al. 2004; Ting et al. 1998) but recent data revealed that OdsH binds satellite DNA and likely has a function in its decondensation (Bayes and Malik 2009). This means that the interactions of Drcd-1r with other proteins and DNA need to be studied to learn about its function (see below).
Sequence analyses of polymorphism and divergence revealed that Drcd-1r has been evolving under strong positive and purifying selection. A McDonald–Kreitman test revealed that many amino acid substitutions have fixed not only to change the function of this gene after duplication, but subsequent to duplication the protein has been changing rapidly under positive selection in every lineage. This pattern of evolution is often seen in male germline genes and has been attributed to sexual selection, sexual antagonistic coevolution or genomic conflicts (Pröschel et al. 2006; Swanson et al. 2001; Swanson and Vacquier 2002; Presgraves 2007; Presgraves and Stephan 2007).
We also studied expression in testis for wild-type males and mutants of Drcd-1r. The P element inserted in its 5′UTR affects the transcription and expression pattern of Drcd-1r. In males homozygous for the P element insertion, Drcd-1r transcription initiates within the P element, which leads to somatic transcription and likely changes the testis tissue expression as interpreted from the occurrence of dsRNA only in infertile males. The P element also changes the 5′UTR in the mutant line. Translation control elements (TCE) in the 5′UTR of Drosophila testis-specific genes have been described in several instances, and their loss often leads to a sterility phenotype when the protein is produced before it is needed (Blumer et al. 2002; Kempe et al. 1993; Schaäfer et al. 1990, 1995). However, we did not find the previously described TCE. In any case, mis-expression accompanied or not by de-repression might be occurring in homozygous mutant individuals. Mis-expression of a gene with transcriptional regulation properties (Drcd-1r) could lead to a phenotype, but it would often be a dominant phenotype unless there is dosage effect (i.e., slight mis-expression does not cause a phenotype). We surmise that the recessive phenotype could be caused by production of sufficient dsRNA due to the presence of both Drcd-1r and CG13102 transcripts in testes cells where this is not expected to occur.
However, complementation tests revealed that a single copy of the mutant allele even in the absence of the wild-type allele produces fertile individuals. While our hypothesis could still explain this outcome (i.e., threshold levels of dsRNA or mis-expression are needed to cause the phenotype), it is also possible that this P element is not responsible for the phenotype. Additional Drcd-1r mutants are needed to clarify the fertility effects. The direct study of Drcd-1r protein–protein interactions and interactions with DNA need to be studied to find out more about Drcd-1r function/s. Another approach that would also help to separate the function of the two overlapping genes (Drcd-1r and CG13102) would be to make a deletion of both genes, provide them in separate constructs and study the phenotype when either gene is missing.
Drcd-1r is an X to autosome duplicated gene. It has been proposed that this type of duplication might be beneficial due either to male germline X inactivation or sexual antagonism (Betran et al. 2002, 2004; Emerson et al. 2004; Bai et al. 2007; Ranz et al. 2003; Wu and Xu 2003). In the case of X inactivation in the male germline (Hense et al. 2007; Lifschytz and Lindsley 1972), it is thought to be beneficial to have genes copy from the X onto autosomes where transcription can occur for a longer time (Bai et al. 2007; Betran et al. 2002; Emerson et al. 2004). Under the standard sexual antagonistic ontogenetic conflict hypothesis, males and females need the same gene but prefer different alleles (i.e., homologous traits are selected in different directions in the different sexes; (Chippindale et al. 2001)). In this case, if either sex-biased or sexual antagonistic mutations are partially dominant, it might be beneficial to have the male-biased genes located on the autosomes (Ranz et al. 2003; Rice 1992; Wu and Xu 2003). Interestingly, recent expression data (Vibranovski et al. 2009) have revealed that the parental gene transcript (Drcd-1) is present during spermatogenesis at similar times and at a higher levels than the retrogene in most stages. Detailed cellular expression patterns need to be described for Drcd-1 and Drcd-1r proteins to provide more data about their spermatogenesis functions.
Supplementary Material
Acknowledgments
We thank Dr. Chitra Chandrasekaran, Sidi Chen and Dr. Manyuan Long for comments on this work. We also thank Mao-Lien Wu, Chung-I Wu, Tessa Bauer DuMont and Chip Aquadro for providing stocks. Research was supported by UTA startup funds and grant R01 GM071813 from the National Institutes of Health, USA to EB.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10709-010-9474-8) contains supplementary material, which is available to authorized users.
References
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Arguello JR, Chen Y, Yang S, Wang W, Long M. Origination of an X-linked testes chimeric gene by illegitimate recombination in drosophila. PLoS Genet. 2006;2:e77. doi: 10.1371/journal.pgen.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Feschotte C, Betran E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326:1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Long M. Dntf-2r: a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics. 2003;164:977–988. doi: 10.1093/genetics/164.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Emerson JJ, Kaessmann H, Long M. Sex chromosomes and male functions: where do new genes go? Cell Cycle. 2004;3:873–875. [PubMed] [Google Scholar]
- Blumer N, Schreiter K, Hempel L, Santel A, Hollmann M, Schafer MA, Renkawitz-Pohl R. A new translational repression element and unusual transcriptional control regulate expression of don juan during Drosophila spermatogenesis. Mech Dev. 2002;110:97–112. doi: 10.1016/s0925-4773(01)00577-9. [DOI] [PubMed] [Google Scholar]
- Brosius J. Retroposons—seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, Dinardo S, Wasserman SA. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller M. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Gibson JR, Rice WR. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci USA. 2001;98:1671–1675. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo De Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Dorus S, Freeman ZN, Parker ER, Heath BD, Karr TL. Recent origins of sperm genes in Drosophila. Mol Biol Evol. 2008;25:2157–2166. doi: 10.1093/molbev/msn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Fontanillas P, Hartl DL, Reuter M. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLoS Genet. 2007;3:2256–2267. doi: 10.1371/journal.pgen.0030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces RG, Gillon W, Pai EF. Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 2007;16:176–188. doi: 10.1110/ps.062600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Siegert M, Schurmann A, Sodeik B, Wolfes H. c-Myb protein interacts with Rcd-1, a component of the CCR4 transcription mediator complex. Biochemistry. 2004;43:8152–8159. doi: 10.1021/bi035857y. [DOI] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hiroi N, Ito T, Yamamoto H, Ochiya T, Jinno S, Okayama H. Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J. 2002;21:5235–5244. doi: 10.1093/emboj/cdf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollocher H, Ting CT, Wu ML, Wu CI. Incipient speciation by sexual isolation in Drosophila melanogaster: variation in mating preference and correlation between sexes. Evolution. 1997;51:1175–1181. doi: 10.1111/j.1558-5646.1997.tb03965.x. [DOI] [PubMed] [Google Scholar]
- Hwa JJ, Zhu AJ, Hiller MA, Kon CY, Fuller MT, Santel A. Germ-line specific variants of components of the mitochondrial outer membrane import machinery in Drosophila. FEBS Lett. 2004;572:141–146. doi: 10.1016/j.febslet.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Kalamegham R, Sturgill D, Siegfried E, Oliver B. Drosophila mojoless, a retroposed GSK-3, has functionally diverged to acquire an essential role in male fertility. Mol Biol Evol. 2007;24:732–742. doi: 10.1093/molbev/msl201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe E, Muhs B, Schafer M. Gene regulation in Drosophila spermatogenesis: analysis of protein binding at the translational control element TCE. Dev Genet. 1993;14:449–459. doi: 10.1002/dvg.1020140606. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis. Proc Natl Acad Sci USA. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- Mcdonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin: Springer; 1970. [Google Scholar]
- Okazaki N, Okazaki K, Watanabe Y, Kato-Hayashi M, Yamamoto M, Okayama H. Novel factor highly conserved among eukaryotes controls sexual development in fission yeast. Mol Cell Biol. 1998;18:887–895. doi: 10.1128/mcb.18.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays. 2007;29:386–391. doi: 10.1002/bies.20555. [DOI] [PubMed] [Google Scholar]
- Presgraves DC, Stephan W. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol Biol Evol. 2007;24:306–314. doi: 10.1093/molbev/msl157. [DOI] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Caceres M, Ruiz A. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon- induced antisense RNA. Proc Natl Acad Sci USA. 2004;101:9013–9018. doi: 10.1073/pnas.0403090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sexually antagonistic genes: experimental evidence. Science. 1992;256:1436–1439. doi: 10.1126/science.1604317. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schäfer M, Kuhn R, Bosse F, Schäfer U. A conserved element in the leader mediates post-meiotic translation as well as cytoplasmic polyadenylation of a Drosophila spermatocyte mRNA. EMBO J. 1990;9:4519–4525. doi: 10.1002/j.1460-2075.1990.tb07903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Nayernia K, Engel W, Schäfer U. Translational control in spermatogenesis. Dev Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Ting CT, Wu CI. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science. 2004;305:81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timakov B, Zhang P. The hsp60B gene of Drosophila melanogaster is essential for the spermatid individualization process. Cell Stress Chaperones. 2001;6:71–77. doi: 10.1379/1466-1268(2001)006<0071:thgodm>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu CI. The phylogeny of closely related species as revealed by the genealogy of a speciation gene, Odysseus. Proc Natl Acad Sci USA. 2000;97:5313–5316. doi: 10.1073/pnas.090541597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy C, Rio J, Motiwale M, Christensen SM, Betran E. Convergently recruited nuclear transport retrogenes are male biased in expression and evolving under positive selection in Drosophila. Genetics. 2010;184:1067–1076. doi: 10.1534/genetics.109.113522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripoli G, D’elia D, Barsanti P, Caggese C. Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Genome Biol. 2005;6:R11. doi: 10.1186/gb-2005-6-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB, Consortium F. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I, Xu EY. Sexual antagonism and X inactivation—the SAXI hypothesis. TIGS. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yuan X, Miller M, Belote JM. Duplicated proteasome subunit genes in Drosophila melanogaster encoding testes-specific isoforms. Genetics. 1996;144:147–157. doi: 10.1093/genetics/144.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Belote JM. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007;134:3517–3525. doi: 10.1242/dev.004770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


