Abstract
Cse4 is a variant of histone H3 that is incorporated into a single nucleosome at each centromere in budding yeast. We have discovered an E3 ubiquitin ligase, called Psh1, which controls the cellular level of Cse4 via ubiquitylation and proteolysis. The activity of Psh1 is dependent on both its RING and Zinc finger domains. We demonstrate the specificity of the ubiquitylation activity of Psh1 toward Cse4 in vitro and map the sites of ubiquitylation. Mutation of key lysines prevents ubiquitylation of Cse4 by Psh1 in vitro and stabilizes Cse4 in vivo. While deletion of Psh1 stabilizes Cse4, elimination of the Cse4-specific chaperone Scm3 destabilizes Cse4 and the addition of Scm3 to the Psh1-Cse4 ubiquitylation reaction prevents Cse4 ubiquitylation, together suggesting Scm3 may protect Cse4 from ubiquitylation. Without Psh1, Cse4 overexpression is toxic and Cse4 is found at ectopic locations. Our results suggest Psh1 functions to prevent the mislocalization of Cse4.
Keywords: Cse4/CENP-A, centromere, yeast, Psh1, proteolysis, Scm3
Introduction
Every eukaryotic centromere is marked by a histone H3 variant, known as CENP-A in humans and Cse4 in budding yeast. This histone variant is incorporated into nucleosomes at centromeres where it replaces histone H3 (Meluh et al., 1998). Centromeric nucleosomes are critical to mark the site of kinetochore formation and therefore microtubule attachment and chromosome segregation. In budding yeast there is only a single centromeric nucleosome containing the histone H3 variant Cse4 (Camahort et al., 2009; Furuyama and Biggins, 2007). How Cse4 is efficiently localized to centromeres, and not other areas of the genome, is an interesting question. Factors that promote localization in budding yeast include Ndc10 and Scm3. Ndc10, along with the CBF3 complex, binds to the centromeric sequence and nucleates the formation of the inner kinetochore (Espelin et al., 1997; Espelin et al., 2003). Via the interaction between Ndc10 and Scm3, an evolutionarily conserved inner kinetochore protein that associates with Cse4, Cse4 is recruited to the centromeric nucleosome (Camahort et al., 2007; Sanchez-Pulido et al., 2009).
One process that may help control the localization of Cse4 is ubiquitin-mediated proteolysis (Collins et al., 2004). Ubiquitylation is an important post-translational lysine modification found in eukaryotes. Poly-ubiquitylation helps to control protein levels via proteasome-assisted proteolysis. However, in a limited number of cases, mono- or di-ubiquitylation controls the activity and subcellular localization of proteins (Wood et al., 2005). Ubiquitylation is a multistep process generally requiring an E1, E2, and E3. E1 is an ubiquitin-activating enzyme which activates ubiquitin, via an E1-ubiquitin intermediate. The activated ubiquitin is transferred to an E2, or ubiquitin-conjugating enzyme. The E3 generally binds to the substrate and the E2, providing the specificity for the E2 in the transfer of ubiquitin to a target. Cse4 has been shown to be poly-ubiquitylated in vivo and degraded in a proteasome-dependent manner (Collins et al., 2004). It was proposed that Cse4 may be loaded at non-centromeric sites during S phase (Pearson et al., 2004) but then ubiquitylated and proteolyzed from all regions but the centromere, where it is protected by the kinetochore (Collins et al., 2004). The proteins that might regulate Cse4 via ubiquitylation are unknown.
In the current study we present evidence that budding yeast uses an E3 ubiquitin ligase called Psh1 to specifically target Cse4 for ubiquitylation and proteolysis. Psh1 co-purifies with Cse4, as well as several other components of the kinetochore. Psh1 also localizes to centromeres by ChIP/qPCR. The interaction between Cse4 and Psh1 requires the RING domain of Psh1, but not its Zinc finger. However, both domains contribute to the control of Cse4 levels in vivo. We show that Psh1 acts as an E3 ubiquitin ligase in vitro with specificity for Cse4, and we map the level of ubiquitylation at five different lysine residues. Mutation of key lysines prevents ubiquitylation of Cse4 by Psh1 in vitro and stabilizes Cse4 in vivo, suggesting Psh1 targets these sites. Cse4-K163A provides better rescue than wild-type Cse4 when Scm3 is eliminated, consistent with Psh1 targeting this site in vivo. Interestingly, Cse4 is destabilized in the absence of Scm3 and the addition of Scm3 to an in vitro ubiquitylation reaction prevents Psh1 from ubiquitylating Cse4, suggesting Scm3 can protect Cse4 from ubiquitylation. Overexpression of Cse4 in a psh1Δ strain is toxic, and furthermore, we show that under these conditions Cse4 localizes to the ribosomal DNA repeats and a euchromatic region. Overall, our results support the idea that Psh1 normally controls the level of Cse4 to prevent its mislocalization.
Results
Identification of Psh1 as a Cse4- interacting protein
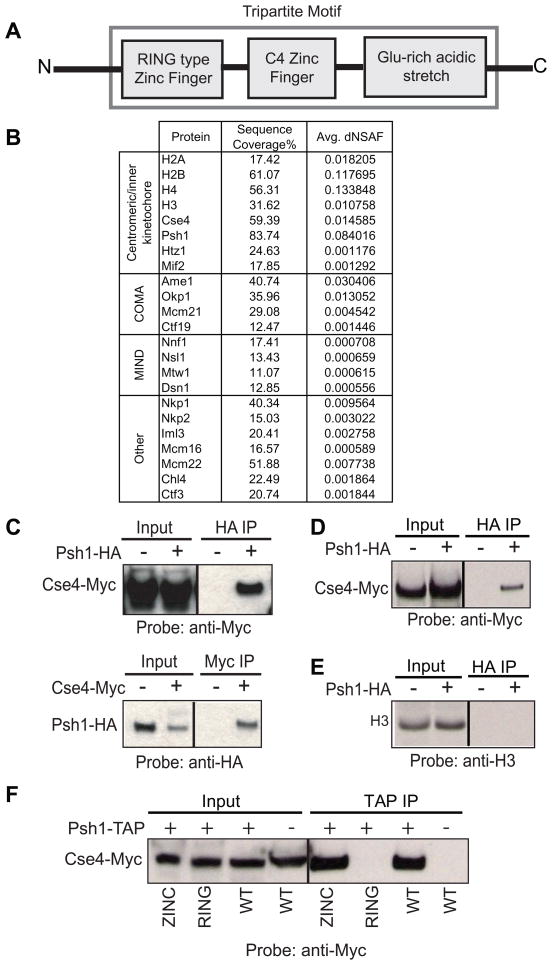
In order to discover new proteins associated with Cse4 that might be involved in the process of its localization, we immunoprecipitated Cse4 and analyzed interacting proteins using MudPIT technology. One protein that was reproducibly present was Psh1. Psh1 is a non-essential nuclear protein, named for Pob3/Spt16 Histone associated (Krogan et al., 2002). No information regarding the function of Psh1 has been published, with the exception of its behavior in large scale screens. The molecular function of Psh1 is unknown, although it contains motifs that suggest its function. Psh1 contains a RING finger, a type of zinc finger which binds two zinc cations and in some cases has been shown to possess ubiquitin ligase activity, a C4 zinc finger, and a highly acidic domain (Figure 1A). These motifs in this arrangement make it a TRIM protein (tripartite motif). This is a large family of proteins conserved from yeast to man that have been shown to have roles in various biological processes including viral pathogenesis, development, and cancer (Meroni and Diez-Roux, 2005). The conservation of the three motifs suggests that together they provide an important and common basic function.
Figure 1.
Psh1 interacts with Cse4 via its RING domain and is part of the kinetochore. A. Psh1 is a member of the TRIM protein family, containing a RING finger motif, and C4 Zinc finger motif, and a glutamine-rich acidic stretch. B. TAP purification of Psh1 brings down Cse4 as well as several other proteins in the kinetochore as detected by MudPIT. Sequence coverage indicates the percentage of the amino acid sequence detected by MS. Normalized spectral counts were used to estimate relative protein levels, as described in (Zybailov et al., 2006), with spectral counts for peptides shared between multiple proteins being distributed (dNSAF) based on spectral counts unique to each protein (Zhang et al., 2010). C. In whole cell extract from a strain carrying Cse4-Myc and Psh1-HA, Cse4 pulls down Psh1 and Psh1 pulls down Cse4. Control IPs were performed from strains lacking the tag on Psh1 or Cse4. D. Psh1 pulls down Cse4 from MNase-solubilized chromatin preparation. A control IP was performed on a strain lacking the HA tag on Psh1. E. Psh1 does not pull down H3 from the same MNase-solubilized chromatin preparation. F. Strains were constructed bearing Cse4-Myc and TAP-Psh1, TAP-Psh1-Zinc finger mutant, TAP-Psh1-RING finger mutant, or no TAP tag (−). TAP-Psh1-RING does not pull down Cse4-Myc. For information regarding all of the strains used in this work please see Supplemental Table 1.
In a Psh1-TAP preparation that was analyzed by mass spectrometry, we find all of the canonical histones, Htz1, Cse4, the inner kinetochore protein Mif2, and several subcomplexes of the kinetochore including all the subunits of the middle kinetochore COMA and MIND complexes, and several components of the outer kinetochore (Figure 1B), indicating that Psh1 can associate with the kinetochore.
To further confirm the physical interaction between Cse4 and Psh1, we performed co-immunoprecipitation (co-IP) and western blotting using an epitope-tagged strain (Cse4-Myc/Psh1-HA). Both whole-cell extracts and micrococcal nuclease(MNase)-solubilized chromatin preparations were used for co-IP. MNase-solubilized chromatin was prepared as previously described (Camahort et al., 2009). We were able to detect the interaction between Psh1 and Cse4 in whole-cell extracts (Figure 1C) as well as in MNase-solubilized chromatin preparations (Figure 1D). Psh1 does not pull down histone-H3 in the MNase-solubilized chromatin preparation (Figure 1E) despite the sequence similarity between Cse4 and H3, suggesting that the interaction is specific for Cse4.
In order to identify the domains of Psh1 important for the interaction with Cse4, we utilized RING and zinc finger mutants (Hereafter refer to as RING and ZINC mutants). We have introduced double mutations, H47A/C50A in the RING finger and C150A/C153A in the zinc finger respectively, disrupting the two fingers. Co-IP using whole-cell extracts show that the ZINC mutant but not RING mutant can immunoprecipitate Cse4 (Figure 1F). This result demonstrates that the RING finger domain of Psh1 is critical for the physical interaction with Cse4.
Psh1 localizes to centromeres
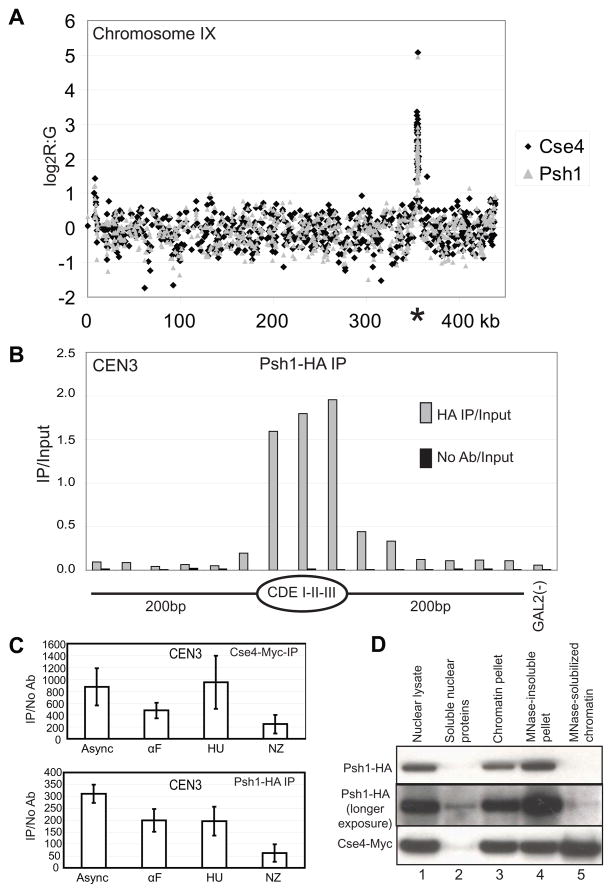
The MudPIT data combined with the Western blotting analysis strongly suggested that Psh1 specifically associates with Cse4. We examined whether Psh1 localizes to centromeres using ChIP chip. Using the strain with Cse4-Myc and Psh1-HA, we performed ChIP for each protein. The material was then used for hybridization to a custom in-house DNA microarray containing probes spanning the entire yeast genome. The sequences from all 16 centromeres were highly enriched in both the Psh1 and Cse4 ChIP; chromosome IX is shown as a representative chromosome (Figure 2A). These results are consistent with Psh1 localizing at centromeres. We next tested the localization of Psh1 at high resolution using chromatin treated with micrococcal nuclease followed by ChIP. The DNA was examined by qPCR using primer pairs spanning centromere 3 (Figure 2B). This analysis indicates that Psh1 localizes precisely at the centromeric DNA sequence, similar to the localization pattern of Cse4 (Camahort et al., 2009). We further tested whether there was a cell cycle dependent pattern to the localization of Psh1 at CEN3 using ChIP/qPCR. Psh1 appears to localize to CEN3 in G1, S and G2/M arrested cultures (Figure 2C), suggesting Psh1 localizes to the centromere throughout the cell cycle. Although the signal for Psh1 decreases in the nocodazole arrested culture, so does the signal for Cse4, and both are still present at levels much higher than background. We have previously reported difficulty with ChIP at the centromeric nucleosome, especially when the kinetochore is assembled (Camahort et al., 2009), so this reduction may either reflect less localization or simply a lower accessibility due to steric hindrance from other kinetochore components.
Figure 2.
Psh1 localizes to centromeres. A. ChIP was performed on a strain expressing Cse4-Myc and Psh1-HA. The eluate from the HA ChIP (grey triangles, Psh1) and Myc ChIP (black diamonds, Cse4) was competitively hybridized to DNA microarrays. The log2 of the red (eluate) to green (total chromatin) ratio is shown on the y axis plotted against the SGD coordinates (kb) for Chromosome IX. The location of the centromere is indicated with an asterisk. Data for all probes are available in Supplemental Table 2. The values for the y axis are the average of biological replicates. B. ChIP was performed on MNase-solubilized chromatin for Psh1-HA (HA IP) followed by qPCR for CEN3 (CDEI-II-III) and the surrounding region. “No Ab” indicates the signal for an IP performed in which the HA antibody was omitted. GAL2 serves as a negative control locus. C. ChIP was performed on asynchronous (asynch) cultures, and cultures arrested with alpha factor (αF), hydroxyurea (HU), and nocodazole (NZ) followed by qPCR for CEN3. D. A strain expressing Psh1-HA and Cse4-Myc was fractionated to collect nuclei (nuclear lysate). The nuclear lysate was further subfractionated into soluble nuclear proteins, chromatin pellet, MNase-solubilized chromatin, and the MNase insoluble pellet. Each fraction was probed for Psh1 and Cse4. While a substantial fraction of Cse4 is solubilized with MNase, the majority of Psh1 remains in the insoluble chromatin pellet. For a similar experiment performed in synchronized cultures, see Supplemental Figure 1.
We compared Psh1 with Cse4 for their presence in different subcellular fractions using the strain with Cse4-Myc and Psh1-HA. The fractions tested by western blotting were nuclear lysate, soluble nuclear proteins, chromatin pellet, MNase-insoluble pellet and MNase-solubilized chromatin. We found that both proteins were highly enriched in nuclear lysate, the chromatin pellet and in the MNase-insoluble pellet. However, only Cse4, but not Psh1, was strongly detected in the MNase-solubilized chromatin fraction, which corresponds to mononucleosomes (Figure 2D). This suggests that even though Psh1 localizes to centromeres and physically interacts with Cse4, it is not as effectively solubilized as Cse4 upon MNase treatment and may remain associated with the kinetochore, the massive protein complex assembled at the centromere. Longer exposure of the Psh1 Western blot reveals a low level of Psh1 in the MNase-solubilized fraction. The signal in Figure 1D presumably derives from the concentration of this low amount by immunoprecipitation. We used the same subcellular fractionation procedure to determine whether there were any differences in Psh1 behavior in asynchronous, G1, S, and G2/M arrested cultures. The level of Psh1 in the various fractions is constant (Supplemental Figure 1), consistent with the ChIP/qPCR results suggesting that localization of Psh1 does not change drastically over the course of the cell cycle.
Psh1 specifically ubiquitylates Cse4
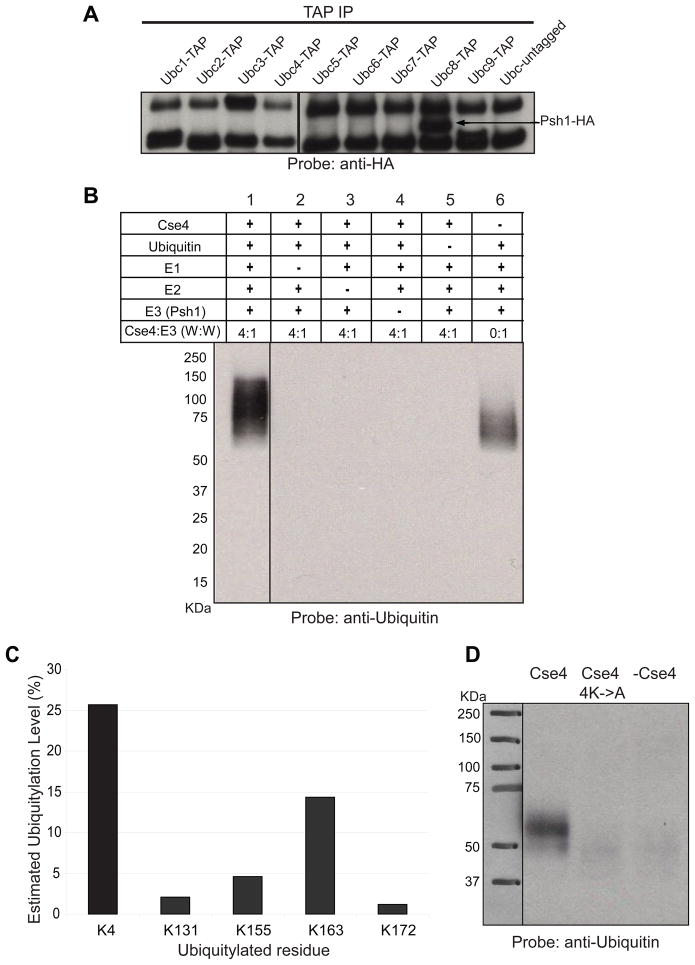
Psh1 contains motifs which suggest it could act as an E3 ubiquitin ligase. Since we did not know which E2 might function with Psh1, we used TAP tag strains to determine which E2 interacts with Psh1. By testing the co-immunoprecipitation of all nine yeast E2s (Ubc1-9) with Psh1, we determined that only Ubc8 interacts with Psh1 (Figure 3A).
Figure 3.
Psh1 acts as an E3 for Cse4 in vitro. A. Budding yeast has nine different E2 genes. Strains were constructed that expressed each E2 with a TAP tag (or an untagged control) and Psh1-HA. An antibody to the TAP tag was used for immunoprecipitation and then Western blotting was used to detect Psh1-HA. The two bands present in every lane represent non-specific hybridization. B. Recombinant proteins were used to carry out ubiquitylation in vitro. Following SDS-PAGE, Western blotting was carried out with anti-ubiquitin antibody. Molecular weight markers are indicated. C. Following the ubiquitylation reaction, the products were subjected to MS analysis. Five lysines (4, 131, 155, 163, 172) in Cse4 were detected as ubiquitylated. The level of ubiquitylation on Cse4 was calculated by subtracting the level observed in the absence of Psh1 from the ubiquitylation level following treatment with Psh1. D. Cse4 with K131, 155, 163, and 172 mutated to alanine (Cse4 4K->A) was included in a Psh1 ubiquitylation reaction. Psh1 does not efficiently ubiquitylate this mutant Cse4 protein. Supplemental Figure 2 shows the ubiquitylation activity of Psh1 on H2A, H2B, H3, H4, and Scm3.
Next we wanted to test the E3 activity of Psh1 in vitro using recombinant proteins. UbcH2, the human homolog of Ubc8, was used as the E2. The reaction components included in each lane are indicated (Figure 3B). Ubiquitylated proteins were detected using western blotting with anti-ubiquitin antibody. The first sample, containing all the components required for the reaction, shows more ubiquitylated proteins migrating above 50 KDa compared to the background signal detected in the absence of Cse4 (lane 6), indicating Cse4 is ubiquitylated. The products are dependent on the presence of the E1, E2, ubiquitin, and Psh1. The size of the products is consistent with poly-ubiquitylation. We tested the specificity of ubiquitylation by performing the same assay with histones H2A, H2B, H3, H4 and also Scm3 in place of Cse4. None of these proteins appear to be ubiquitylated (Supplemental Figure 2). These results demonstrate that the ubiquitin ligase (E3) activity of Psh1 is specific for Cse4.
We analyzed the Cse4 sample by mass spectrometry to determine the sites of ubiquitylation. We found that K4, K131, K155, K163 and K172 of Cse4 are ubiquitylated. Our recombinant Cse4 shows some ubiquitylation on K4 without treatment with Psh1. K4 is in the N terminal tail of Cse4 and may be more exposed than the residues in the histone fold domain. Ubiquitylation of K131, 155, 163, and 172 is only observed following treatment with Psh1. K163 shows the highest level of ubiquitylation for the residues in the histone fold domain (Figure 3C). K131, 163, and 172 are not conserved in the histone fold domain of H3. These lysine residues, especially K163, might therefore be important for stability of Cse4 in vivo. However, we note that single-residue alanine-scanning mutagenesis of Cse4 shows that these lysines, when mutated individually, do not show any growth or ploidy defects (Camahort et al., 2009)(S. Nakanishi and A. Shilatifard, personal communication). MS data also reveals that Psh1 is auto-ubiquitylated at K303 at a level of 33%; this explains the background signal detected with anti-ubiquitin antibody for the sample without Cse4 (Figure 3B, lane 6) and for the reactions performed with other targets (Supplemental Figure 2).
We purified recombinant Cse4 that had K131, 155, 163, and 172 mutated to alanine and tested the ability of Psh1 to ubiquitylate this protein. Psh1 was not able to ubiquitylate this protein in vitro, consistent with these lysine residues being the primary targets of recombinant Psh1 (Figure 3D). The difference in migration of ubiquitylated Cse4 in part B and D likely reflects the lower efficiency of the ubiquitylation reaction in part D due to the lower concentration of Cse4 protein used as a result of the solubility of the Cse4-4K->A mutant.
Psh1 controls the levels of Cse4
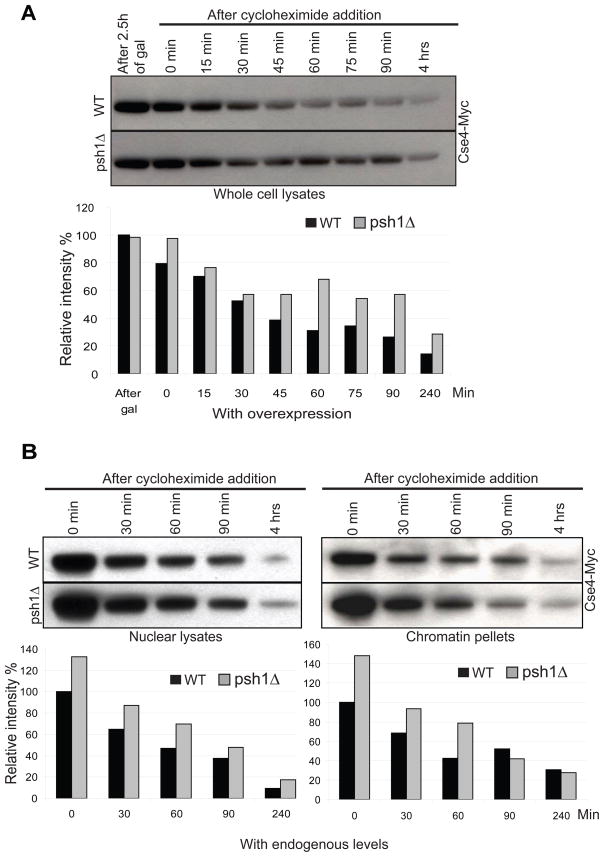
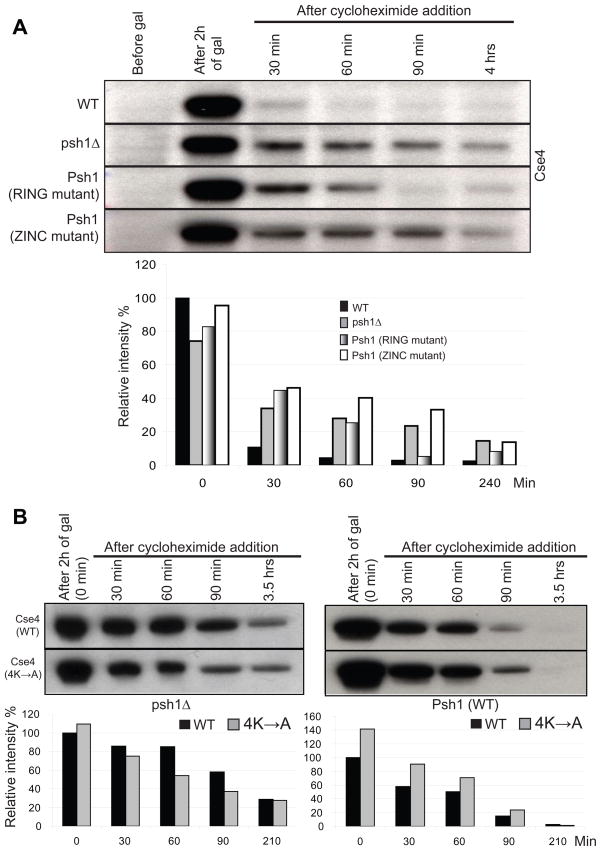
It has been previously reported that the cellular levels of Cse4 are controlled by ubiquitin-mediated proteolysis, thus ensuring its specific localization to the centromere (Collins et al., 2004). If Psh1 is an E3 that ubiquitylates Cse4 in vivo and targets it for proteolysis, then deletion of Psh1 might be expected to stabilize the level of Cse4. We analyzed the stability of Cse4 by monitoring the level of Cse4-Myc, integrated at the URA3 locus and expressed from the GAL promoter, in wild type and psh1Δ strains (Figure 4A)or in strains expressing Cse4-Myc at endogenous levels (Figure 4B). Cycloheximide was added to repress protein translation and the level of Cse4 was determined by Western blotting at different time points. As would be expected if Psh1 targets Cse4 for ubiquitin-mediated proteolysis, Cse4 is more stable in a psh1Δ strain, confirming the role of Psh1 in controlling cellular Cse4 levels. Deletion of Psh1 stabilizes Cse4 only partially and this indicates the presence of other factors/mechanisms responsible for the degradation of Cse4 in vivo. A similar observation has been reported by Collins et al.; a mutant version of Cse4 lacking all possible ubiquitylation sites (lysines) was still degraded (Collins et al., 2004).
Figure 4.
Deletion of PSH1 stabilizes Cse4 protein level in vivo. Histograms indicate intensity of Cse4-Myc. A. Cse4-Myc was integrated at the URA3 locus under the control of the gal promoter. Cse4-Myc was induced by exposure to galactose for 2.5 hours in a wild-type (WT) strain or a strain in which PSH1 was deleted (psh1Δ). Cycloheximide was added and cells were collected at the timepoints indicated for Western blot analysis using an anti-Myc antibody. 70 μg of total protein was loaded per lane. B. Endogenous levels of Cse4-Myc were measured in nuclear lysates and chromatin pellets at various times following cycloheximide treatment in WT and psh1Δ strains. 30 μg of total protein was loaded per lane.
To examine which domains of Psh1 were required in vivo to target Cse4, a pulse-chase assay was performed in four different W303 backgrounds: wild type, psh1Δ, RING mutant and ZINC mutant. Cse4 is stabilized in both RING and ZINC mutants compared to wild type cells (Figure 5A). Even though the mutation in the zinc finger does not affect the physical interaction between the two proteins (Figure 1D), it appears to abolish the ability of Psh1 to target Cse4 for degradation and stabilizes Cse4 to a similar extent as a complete deletion of Psh1 (Figure 5A). Both domains are required for Psh1-mediated degradation of Cse4.
Figure 5.
Features of Psh1 and Cse4 determine Cse4 stability. Histograms indicate intensity of Cse4. A. Strains with the genotype indicated (WT, psh1Δ, Psh1-RING, Psh1-ZINC) were transformed with a 2 micron plasmid containing Cse4 under control of the gal promoter. Cse4 was induced by exposure to galactose for 2 hours, cycloheximide was added, and cells were collected at the timepoints indicated for analysis by Western blotting using an anti-Cse4 antibody. 35 μg of total protein was loaded per lane. B. The stability of the Cse4-4K->A mutant was measured as in part A. The strain background in part A is W303 while the background in part B is S288C, which may account for the difference in Cse4 stability.
Given the identification of key lysine residues in Cse4 that are targeted by Psh1, we asked whether expression of Cse4 with lysines 131, 155, 163, and 172 mutated to alanine would stabilize Cse4. Using a pulse-chase experiment, we found that this Cse4 mutant protein was more stable than the wild-type Cse4 protein when Psh1 was present, but the mutant was actually less stable in the absence of Psh1 (Figure 5B). This result is consistent with these lysines being targeted by Psh1 in vivo.
Scm3 may protect Cse4 from proteolysis by Psh1
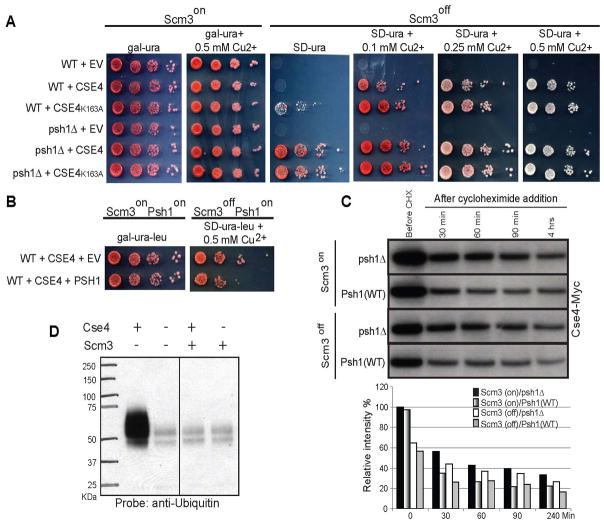
Scm3 is an essential protein in budding yeast that is required for Cse4 deposition; shut off normally results in a metaphase arrest (Camahort et al., 2007). We have previously shown that the growth of a strain in which Scm3 is either deleted or turned off (Scm3off) can be rescued by overexpression of Cse4 (Camahort et al., 2009). We reasoned that if Psh1 was responsible for proteolysis of Cse4, then deletion of PSH1 might improve the rescue. Strains were constructed that had the sole endogenous copy of Scm3 under control of the gal promoter and Cse4 was expressed from a 2 micron plasmid under the control of a copper inducible promoter whose activity can be titrated with copper. Increasing levels of copper result in higher levels of induction, but some expression occurs even without the addition of copper. When Scm3 is on, there is no difference in growth when PSH1 is deleted. However, when we delete PSH1 and turn off Scm3, we find that the rescue of growth is more efficient than when PSH1 is present (Figure 6A, compare WT+CSE4 and psh1Δ+CSE4). The previously reported mitotic delay associated with Scm3off+CSE4 (Camahort et al., 2009) is largely alleviated by deletion of PSH1 (Supplemental Figure 3A). Furthermore, the rescue is more efficient when CSE4 carries the K163A mutation, but only when PSH1 is still present (Figure 6A, compare the difference between WT+CSE4 and WT+CSE4K163A and psh1Δ+CSE4 and psh1Δ+CSE4K163A), and this also reduces the mitotic delay (Supplemental Figure 3A). Using ChIP followed by qPCR, we find that Psh1 still localizes to CEN3 in the absence of Scm3 to levels very similar as those reported in Figure 2C (Supplemental Figure 4).
Figure 6.
Deletion or overexpression of PSH1 alters rescue of Scm3 off strains. Strains were constructed in which the only copy of Scm3 was under the control of the gal promoter, allowing the expression of Scm3 to be controlled with glucose/galactose. A. Cse4 was under the control of a copper-inducible promoter on a 2 micron vector. “EV” indicates empty vector. When Scm3 is expressed (Scm3on), no growth differences are observed in 10-fold serial dilutions. Lower levels of induction of Cse4 (0, 0.1 mM Cu2+) rescue Scm3 off when PSH1 is deleted (psh1Δ) as compared to WT. Cse4K163A rescues Scm3off better than Cse4 when PSH1 is present (WT). For cytometric analysis of DNA content in these samples see Supplemental Figure 3A. For ChIP analysis showing Psh1 is present at CEN3 in the absence of Scm3, see Supplemental Figure 4. B. When Psh1 is overexpressed from its own promoter on a 2 micron vector, the rescue of Scm3off by overexpression of Cse4 is less efficient. For cytometric analysis of DNA content in these samples see Supplemental Figure 3B. C. The stability of an integrated copy of Cse4-Myc under the control of its endogenous promoter was measured +/− Scm3 and +/− Psh1. Scm3 was shut off for 1 hour in YPD for 1 hour prior to the addition of cycloheximide, which we have previously reported is sufficient to eliminate any detectable Scm3 (Camahort et al., 2007). Histograms indicate intensity of Cse4-Myc. D. When Scm3 is included in a Psh1-Cse4 ubiquitylation reaction, Cse4 is not ubiquitylated.
In a complimentary experiment, we tested whether overexpression of PSH1 from its own promoter on a 2 micron vector would reduce the rescue observed in the Scm3off strain when CSE4 is overexpressed. Indeed, overexpression of PSH1 reduces the rescue (Figure 6B) and the mitotic delay becomes more pronounced (Supplemental Figure 3B). Collectively these results strongly suggest that Psh1 can control the cellular levels of Cse4 by targeting Cse4 for degradation via ubiquitylation and proteolysis.
Since elimination of Scm3 seemed to sensitize cells to the deletion of PSH1, we tested the idea that Scm3 might protect Cse4 from the action of Psh1. In the absence of Scm3, the steady state level of Cse4-Myc (expressed at endogenous level) is lower (before CHX, Figure 6C). As expected, Cse4 levels are stabilized when Psh1 is deleted. In addition, we performed an ubiquitylation reaction in the presence of Scm3, and found that under these conditions Cse4 is protected from ubiquitylation by Psh1 (Figure 6D). Together these data suggest Scm3 might protect Cse4 from ubiquitylation by Psh1.
Deletion of PSH1 results in increased mislocalization of Cse4
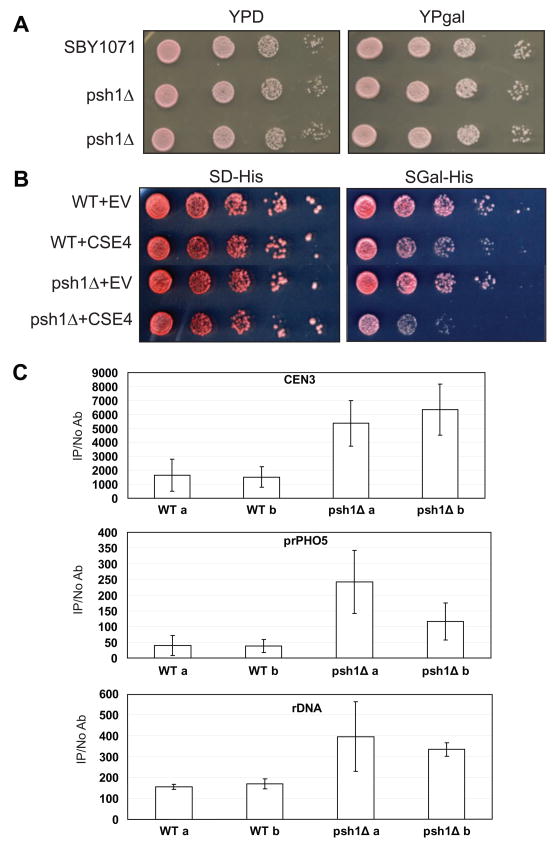
PSH1 is not an essential gene and deletion strains do not show an obvious growth phenotype (Figure 7A). A simple deletion does not cause an elevation in loss of a non-essential linear chromosome (data not shown). We decided to examine what happens when an integrated copy of Cse4 is overexpressed from the gal promoter and PSH1 is deleted. Under these conditions we still do not observe a growth phenotype (Figure 7A). We had previously examined the localization of Cse4 under these conditions and we did not observe any significant mislocalization (Camahort et al., 2009). However, when we delete PSH1 and overexpress Cse4 from the gal promoter on a two micron vector, we observe slow growth compared to a strain in which Psh1 is present (Figure 7B, compare WT+CSE4 and psh1Δ+CSE4). Thus, very high levels of Cse4 cause a growth defect in the absence of PSH1. Since there appears to be mechanisms for regulating Cse4 that are independent of Psh1, high levels of expression of Cse4 may be necessary to reveal the action of Psh1.
Figure 7.
Ectopic localization of Cse4 increases when PSH1 is deleted. A. Cse4-Myc was integrated under the control of the gal promoter. Cse4 was integrated at the URA3 locus and overexpressed from the gal promoter (YPgal) in a WT or psh1Δ strain. No growth defects are observed in 10-fold serial dilutions. B. Cse4 was overexpressed from the gal promoter (gal-his) on a 2 micron plasmid in either a WT or psh1Δ strain. “EV” indicates an empty vector control as a point of comparison. Under these circumstances, overexpression of Cse4 is toxic in a psh1Δ strain as demonstrated by 10-fold serial dilutions. C. From the strains shown in (B) overexpressing Cse4, ChIP was performed for Cse4 followed by qPCR for CEN3, the PHO5 promoter, and the rDNA. The values on the y axis are arbitrary units representing the enrichment for each sequence from ChIP performed with and without antibody with respect for total chromatin for each sample. Biological replicates for the WT and psh1Δ strains are shown (a and b). The error bars represent the standard deviation for qPCR performed in triplicate.
We previously reported that Cse4 localizes to the rDNA in a wild-type strain background (Camahort et al., 2009). The level of Cse4 at the rDNA is above background, but below the levels observed at the centromere. One possible explanation for the localization at the rDNA is that positive supercoils accumulate due to high levels of transcription and since Cse4 may assemble into right-handed nucleosomes (Furuyama and Henikoff, 2009), this region of the genome may be permissive for the assembly of Cse4 nucleosomes. We reasoned that when Cse4 was overexpressed and PSH1 was deleted, the mislocalization of Cse4 to the rDNA might increase. We examined Cse4 levels at the rDNA, CEN3, and a negative control site, the PHO5 promoter, using ChIP followed by qPCR. We found that when PSH1 was deleted, the level of Cse4 at CEN3 was elevated an average of 3.7-fold over that observed in a wild-type strain, and the level at the rDNA was elevated an average of 2.2-fold (Figure 7C). In addition, we observed an average of 4.6-fold elevation of Cse4 at the promoter of the PHO5 gene, a site where Cse4 is not normally found. These results suggest that Psh1 can help prevent Cse4 localization at non-centromeric sites.
Discussion
Several proteins have been shown to contribute positively to the targeting of Cse4 to centromeres, including Scm3 and Ndc10. Psh1 appears to be a negative regulator of Cse4, contributing to its ubiquitylation and proteolysis. Overexpression of CID, the fly homolog of CSE4, has been shown to result in non-centromeric localization and partial ectopic kinetochore formation (Van Hooser et al., 2001). Overexpression of CENP-A in human cells is associated with genome-instability (Amato et al., 2009; Tomonaga et al., 2003). Mechanisms that control the levels of the centromeric histone variant may help to prevent lethal events associated with misincorporation. We have previously shown that Cse4 normally shows a low level of localization to the rDNA (Camahort et al., 2009). Here we show that Psh1 helps to prevent higher levels of localization of Cse4 to this locus as well as another locus, the promoter of the PHO5 gene, which does not normally contain Cse4. When levels of Cse4 are very high and Psh1 is deleted, Cse4 may be promiscuously assembled into nucleosomes. Because Cse4 nucleosomes can nucleate kinetochore formation, Psh1 may help prevent titration of kinetochore components by non-centromeric sites. Mislocalization of Cse4 could also affect transcription.
It is curious that Psh1 targets Cse4 for proteolysis and also co-purifies with the kinetochore. How then is Cse4 in the centromeric nucleosome protected from ubiquitylation? One possibility is that although the localization of Psh1 to the kinetochore is not cell cycle regulated, its activity may be regulated. Psh1 was heavily post-translationally modified (unpublished observation from Psh1-TAP experiment), which could contribute to its regulation. A second possibility that is not mutually exclusive is that Cse4 that is associated with Scm3 may be protected from the action of Psh1. These Cse4-Scm3 complexes may be either soluble or chromatin associated. We observed that deletion of Psh1 stabilized both the total nuclear pool and chromatin associated Cse4 (Figure 4B), but we do not believe this experiment allows us to distinguish whether Psh1 can target soluble or chromatin associated Cse4 since the chromatin pool may be influenced by the soluble pool. The soluble pool of Cse4 is very difficult to measure since the levels are so low.
Scm3 is required for the deposition of Cse4 at centromeres and is intimately associated with the centromeric nucleosome (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007). Scm3 appears to be a Cse4-specific nucleosome assembly factor (Shivaraju et al., manuscript in preparation). Our experiments suggest that the protection of Cse4 from Psh1 is dependent in part on Scm3. A simple deletion of PSH1 does not show any obvious growth phenotype. However, when Scm3 is not present, growth defects due to the deletion or overexpression of Psh1 become more obvious, suggesting Cse4 is more vulnerable to the action of Psh1 under these conditions (Figure 6). Furthermore, the levels of Cse4 are lower in the absence of Scm3 and the addition of Scm3 to the in vitro ubiquitylation assay prevents the ubiquitylation of Cse4. Many of our experiments rely on overexpression of Cse4 to reveal the function of Psh1. Because Scm3 is present at such low levels in cells (Ghaemmaghami et al., 2003), overexpression of Cse4 is likely to increase the level of Scm3-free Cse4, which then exposes the activity of Psh1. Under normal growth conditions, this pool of Cse4 may be very low, making it difficult to measure the activity of Psh1. We speculate that Cse4 that is associated with Scm3 is “on track” for appropriate targeting and incorporation into centromeric DNA. However, Scm3-free Cse4, either soluble or chromatin associated, may be a key target for Psh1 since this Cse4 may have a higher probability of being mislocalized.
While Psh1 provides one avenue for regulation of Cse4, there are clearly additional mechanisms since a deletion of Psh1 does not completely stabilize Cse4. This finding is consistent with a previous report that concluded that the additional degradation mechanism(s) was independent of ubiquitylation since a Cse4 mutant lacking all lysine residues was still degraded (Collins et al., 2004). These additional means of regulating Cse4 remain to be discovered.
Materials and methods
Yeast strains
All the strains used are listed in Supplemental Table 1.
TAP purification
Tandem Affinity Purification (TAP) method was adopted from (Puig et al., 2001). Nine liters of yeast cultures were grown to late log phase (OD600 ~ 1.5) and the cell pellet was washed first with water and then with TAP extract buffer (40mM Hepes-KOH (pH 7.5), 10% glycerol, 350mM NaCl, 0.1% Tween-20, 0.5mM DTT and protease inhibitors). The pellet was resuspended in 20mL of TAP extract buffer and the cells were lysed by bead beating. The extract was incubated with 0.1U of Benzonase (Novagen, 70664-3) for 20min and centrifuged to remove cell debris. Cell lysate was incubated for 2h at 4°C with 350uL (50% slurry) of IgG sepharose beads (GE Healthcare, 17-0969-01), prewashed with TAP extract buffer. Beads were recovered by centrifugation, transferred to a 10mL Bio-Rad Poly-Prep column and washed first with TAP extract buffer (10mL x 4), then with 10mL of TEV cleavage buffer (10mM Tris (pH 8.0), 150mM NaCl, 0.1% NP-40, 0.5mM EDTA, 10% glycerol, 0.5mM DTT and protease inhibitors) by gravity flow. Beads were resuspended in 1mL of TEV cleavage buffer, transferred to a 1.5mL microfuge tube and incubated with 10uL of AcTEV protease (Invitrogen, 12575-015), overnight at 4°C. Beads suspension was transferred to the column and TEV cleaved products were eluted by gravity flow. Washed the beads with 3mL of Calmodulin binding buffer (CBB) (10mM Tris (pH 8.0), 1mM MgOAc, 1mM imidazole, 2mM CaCl2, 0.1% NP-40, 10% glycerol, 0.5mM DTT and protease inhibitors) containing 300mM NaCl and pooled the washing with TEV cleavage elution. Three microliters of 1M CaCl2 and 200uL (50% slurry) of Calmodulin sepharose (GE Healthcare, 17-0529-01), prewashed with CBB containing 300mM NaCl, were added and incubated for 2h at 4°C. Washed the beads with CBB containing NaCl and transferred to a 1mL spin column (Bio-Rad). Proteins were eluted with 1mL (200μL x 5) of Calmodulin elution buffer (10mM Tris (pH 8.0), 0.15M NaCl, 1mM MgOAc, 1mM imidazole, 2mM EGTA, 0.1% NP-40, 10% glycerol, 0.5mM DTT and protease inhibitors). Proteins were TCA precipitated and subjected to MudPIT analysis.
Chromatin fractionation and MNase-solubilized chromatin Co-IP
Chromatin fractionation procedure has been previously reported (Zhang et al., 2005). Cells were grown to mid log phase (OD600 ~0.8), harvested and washed first with cold water then with SB (20mM Tris (pH 7.4), 1M sorbitol). The cells were resuspended in PSB (200mM Tris (pH 7.4), 20mM EDTA, 1M NaCl and 100mM β-mercaptoethanol), incubated for 10min at room temperature and centrifuged. Washed cells with wash buffer (20mM Tris (pH 7.4), 1M NaCl) and with SB. Resuspended in SB and incubated with glusulase (PerkinElmer, NEE154001EA) for 1h at 30oC. Washed twise with SB, resuspended in EBX-0.1 (20mM Tris (pH 7.4), 0.1M NaCl, 0.25% Triton X-100, 15mM β-mercaptoethanol and protease inhibitors) and added Triton X-100 to a final concentration of 0.5%. Gently swirled on ice for 10min, layered the cell lysate on a cushion of NIB (20mM Tris (pH 7.4), 0.1M NaCl, 1.2M sucrose, 15mM β-mercaptoethanol) and centrifuged. Nuclear pellet was resuspended in EBX-0.1. Triton X-100 was added to a final concentration of 1% and swirled on ice for 15min to lyse nuclei. Resulting nuclear lysate was centrifuged to separate the soluble nuclear proteins from chromatin pellet. Resuspended the pellet in EBX-0.1 containing 0.5mM β-mercaptoethanol and CaCl2 was added to a final concentration of 3mM. Chromatin suspension was digested with 100–250 units of MNase (Worthington Biochemicals Corp., LS004798) for 15–30min at 37°C. The digestion was stopped by adding EGTA and EDTA to a final concentration of 25mM. MNase-solubilized chromatin was separated from the insoluble chromatin by centrifugation and the pellet was resuspended in EBX-0.1. For Co-IP, MNase-solubilized chromatin was incubated with 4μL of anti-HA (Covence PRB.101P) overnight at 4°C. Thirty microliters (50% slurry) of protein G beads (GE Healthcare, 17-0618-01), pre-blocked with 1% BSA, were added and incubated for 2h at 4°C. The beads were washed three times with EBX-0.1 and proteins were eluted with SDS buffer (10mM Tris pH 7.5, 1mM EDTA and 1% SDS). Immunoprecipitates were subjected to SDS-PAGE and western blotting.
Whole cell extract Co-IP
Cells were grown to mid log phase (OD600 ~0.8) and harvested. The cell pellet was washed with PBS (137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, 1mM KH2PO4, pH 7.4) and resuspended in lysis buffer (50mM Tris (pH 7.5), 150mM NaCl, 0.1% NP-40, 1mM DTT, 10% glycerol and protease inhibitors). Cells were bead beated and the debris was removed by centrifugation. Dilution/wash buffer (50mM Tris (pH 7.5), 150mM NaCl, 0.1% NP-40) was added to the cell lysate and incubated with the antibody over night at 4°C. The antibodies used are as follows: anti-HA and anti-Myc (Covence, PRB.101P and MMS-150P respectively), anti-TAP (Open Biosystems, CAB1001). Thirty microliters (50% slurry) of protein G beads, pre-blocked with 1% BSA, were added and incubated for 2h at 4°C. The beads were washed three times with dilution/wash buffer and proteins were eluted with SDS buffer (10mM Tris pH 7.5, 1mM EDTA and 1% SDS). Immunoprecipitates were subjected to SDS-PAGE and western blotting.
Pulse-chase assay
Cultures were grown to OD600 ~0.6 in appropriate media with 2% raffinose. Galactose was added to a final concentration of 4% and incubated for another 2h. Cycloheximide (Sigma) was added to a final concentration of 10μg/mL and 100mL culture fractions were collected at different time points. Whole cell extracts were prepared by bead beating and protein concentrations were determined using Bradford assay. Proteins were analyzed by SDS-PAGE and western blotting and the same amount of total proteins were used from each fraction.
SDS-PAGE and Western Blotting
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using NuPAGE Novex 4–12% Bis-Tris pre-cast gels (Invitrogen). Western blotting was performed according to standard protocol using a nitrocellulose (Whatman, Protran) or PVDF (Millipore, Immobilon-P) membrane. Primary antibodies used are as follows: anti-Myc (Covance, 9E10, MMS-150P, 1:2500), anti-HA (Covance, PRB-101P, 1:2500 and Roche, 3F10, 1:1000), anti-H3 (abcam, ab1791, 1:2500), anti-ubiquitin (Invitrogen, 13–1600, 1:1000), anti-Cse4 (polyclonal rabbit antibody against recombinant Cse4, 1:25000). Secondary antibodies were HRP linked, anti-rabbit IgG (from donkey), anti-mouse IgG (from sheep) and anti-rat IgG (from goat) (GE Healthcare, NA934V, NA931V and NA935V respectively).
ChIP chip
The ChIP chip methods and analysis have been previously published (Camahort et al., 2009).
MNase-ChIP-qPCR
The procedure was adopted from Camahort et al., 2009. Cultures were grown to mid log phase and fixed with formaldehyde (1% final) for 20min at 30°C. Cell pellet was resuspended in buffer-Z (1M sorbitol, 50mM Tris (pH 7.4), 10mM β-mercaptoethanol), zymolyase (USBiological, Z1005) was added and incubated for 30min at 30°C for spheroplasting. CaCl2 (3mM final) was added to the harvested chromatin and subjected to MNase digestion with 100U/mL (Worthington). The digestion was stopped by adding EGTA and EDTA to a final concentration of 25mM and diluted with 3X buffer-L (150mM HEPES-KOH (pH 7.5), 420mM NaCl, 3mM EDTA, 3% Triton X-100, 0.3% deoxycholate). ChIP was performed using anti-HA (Covance, PRB-101P, 1:150) and for the control samples antibody was omitted. Quantitative PCR was performed as described in Camahort et al., 2009.
In vitro ubiquitylation assay
Human E1 (UBE1), E2 (UbcH2-human homolog of budding yeast Ubc8) and ubiquitin from Boston Biochem (E-305, E2-607 and U-100H respectively) were used for the assay. E1, E2 (0.1μg each), 0.5ug of FLAG-His6-Psh1 and 4ug of ubiquitin were mixed in the reaction buffer (4mM HEPES-NaOH (pH 7.9), 6mM KOAc, 5mM MgCl2, 1mM DTT). Energy regeneration solution-ERS (Boston Biochem, B-10) was used as the energy source. Two micrograms of Cse4 was added as the substrate so that Cse4:E3 ratio was 4:1 (w/w). Total reaction volume was 30μL. Control samples were prepared as shown in the table in Figure 3B. Samples were incubated for 1h at 30°C and the reaction was stopped by adding LDS sample buffer (Invitrogen, NP0007) and boiling for 5min. Ubiquitylated proteins were analyzed by SDS-PAGE and western blotting. For the determination of ubiquitylation sites, after incubation, samples were TCA precipitated and subjected to mass spectrometric analysis.
Psh1 mutagenesis
Site-directed mutagenesis was performed using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene).
Recombinant protein expression and purification
Methods for histone expression and purification have been published (Camahort et al., 2009). Full length Psh1 was cloned between the XhoI and NotI sites of pBacpak8-Flag-6xHis vector after being amplified from yeast genomic DNA using specific primers (Fwd: GGGCTCGAGATGGGCGACGAATTACACAA, Rev: GGGGCGGCCGCTTATTCATCGTCACTGTCTC). pBacpak8-Flag-6xHis-Psh1 was transfected with Cellfectin into Sf21 cells along with purified baculovirus genome and incubated for 3 days at 27°C. The entire primary culture (P1) was amplified by adding it to a fresh batch of Sf21 cells. The virus was harvested at the end of 7 days. The viral culture was used to infect 50 million cells and cultured for 48hrs at 27°C. Cells were harvested by centrifugation and washed twice in 1x PBS containing protease inhibitors. Cells were resuspended in lysis buffer (50mM HEPES pH 7.9, 500mM NaCl, 2mM MgCl2, 0.2% Triton X-100, 10% Glycerol, 0.5mM EDTA and protease inhibitors) and kept on ice for 30 min. The cell debris was pelleted by ultracentrifugation (40,000 rpm for 30 min using a fixed angle rotor 70Ti, Beckman). The expressed protein was bound to FLAG M2 agarose beads for 4hrs. at 4°C. The beads were then washed 4 times with the lysis buffer and the bound protein eluted in the Elution buffer (50 mM HEPES pH 7.9, 100mM NaCl, 2mM Mgcl2, 0.02% NP40, 10% glycerol and protease inhibitors) containing 0.5mg/ml of 3x FLAG peptide.
Supplementary Material
Supplemental Figure 1, related to Figure 2. The subcellular fractionation pattern of Psh1 does not vary over the course of the cell cycle.
Supplemental Figure 2, related to Figure 3. Psh1 does not efficiently ubiquitylate other histones or Scm3. Cse4 was used as a positive control. The reactions were analyzed by Western blotting with anti-ubiquitin antibody. The strong band at the bottom of the Western blots is ubiquitin. A. Psh1 was incubated with recombinant H2A, H2B, H3, H4, or Cse4. B. Psh1 was incubated with recombinant Scm3 or Cse4.
Supplemental Figure 3, related to Figure 6. Analysis of DNA content for strains shown in Figure 6. An aliquot of each culture was collected and fixed with 70% ethanol. Following treatment with RNAse in 50 mM sodium citrate, DNA was stained with Sytox green and cells were briefly sonicated. DNA content was analyzed using a BD FACSCalibur flow cytometer and FlowJo 8.8.4 software. A. Strains grown in gal-ura + 0.1 mM Cu2+ (Scm3on) show similar profiles. When cells are transferred to SD-ura + 0.1 mM Cu2+ and allowed to grow 2–3 generations (Scm3off), differences in DNA profiles become apparent. B. Strains grown in gal-ura-leu (Scm3on) show similar profiles. When cells are transferred to SD-ura-leu + 0.5 mM Cu2+ and allowed to grow 2–3 generations (Scm3off), differences in DNA profiles become apparent.
Supplemental Figure 4, related to Figure 6. Psh1 localizes to CEN3 in Scm3off strains by ChIP followed by qPCR. α-Cse4 antibody was used to pull down Cse4 and α-HA antibody was used to pull down Psh1; qPCR was used to examine their localization at CEN3. A. ChIP for Cse4 was carried out on biological replicate cultures (a and b) grown in SD-ura + 0.5 mM Cu2+ (Scm3 off). The level of each is shown compared to a ChIP performed without antibody. Each qPCR was performed in triplicate to generate the standard deviation shown with the error bars. B. ChIP-qPCR data for Psh1-HA under Scm3on (from Figure 2C- Async.) and Scm3off conditions plotted side by side shows that Psh1 is present at CEN3 in both conditions.
Acknowledgments
We thank Steven Tan, Lili Pan, Raymond Camahort, and Bethany Harris for technical assistance. We thank Ed Choi and Kausik Si for reagents, and Sue Biggins for strains and plasmids. GH, MS, MM, and JLG were supported in part by R01GM080477.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 Is Essential to Recruit the Histone H3 Variant Cse4 to Centromeres and to Maintain a Functional Kinetochore. Mol Cell. 2007 doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the Architecture of a Simple Kinetochore Using DNA–Protein Crosslinking. The Journal of Cell Biology. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the Essential Saccharomyces cerevisiae Kinetochore Protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and Histones CenH3-H4 Assemble the Core of Centromere-Specific Nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1, related to Figure 2. The subcellular fractionation pattern of Psh1 does not vary over the course of the cell cycle.
Supplemental Figure 2, related to Figure 3. Psh1 does not efficiently ubiquitylate other histones or Scm3. Cse4 was used as a positive control. The reactions were analyzed by Western blotting with anti-ubiquitin antibody. The strong band at the bottom of the Western blots is ubiquitin. A. Psh1 was incubated with recombinant H2A, H2B, H3, H4, or Cse4. B. Psh1 was incubated with recombinant Scm3 or Cse4.
Supplemental Figure 3, related to Figure 6. Analysis of DNA content for strains shown in Figure 6. An aliquot of each culture was collected and fixed with 70% ethanol. Following treatment with RNAse in 50 mM sodium citrate, DNA was stained with Sytox green and cells were briefly sonicated. DNA content was analyzed using a BD FACSCalibur flow cytometer and FlowJo 8.8.4 software. A. Strains grown in gal-ura + 0.1 mM Cu2+ (Scm3on) show similar profiles. When cells are transferred to SD-ura + 0.1 mM Cu2+ and allowed to grow 2–3 generations (Scm3off), differences in DNA profiles become apparent. B. Strains grown in gal-ura-leu (Scm3on) show similar profiles. When cells are transferred to SD-ura-leu + 0.5 mM Cu2+ and allowed to grow 2–3 generations (Scm3off), differences in DNA profiles become apparent.
Supplemental Figure 4, related to Figure 6. Psh1 localizes to CEN3 in Scm3off strains by ChIP followed by qPCR. α-Cse4 antibody was used to pull down Cse4 and α-HA antibody was used to pull down Psh1; qPCR was used to examine their localization at CEN3. A. ChIP for Cse4 was carried out on biological replicate cultures (a and b) grown in SD-ura + 0.5 mM Cu2+ (Scm3 off). The level of each is shown compared to a ChIP performed without antibody. Each qPCR was performed in triplicate to generate the standard deviation shown with the error bars. B. ChIP-qPCR data for Psh1-HA under Scm3on (from Figure 2C- Async.) and Scm3off conditions plotted side by side shows that Psh1 is present at CEN3 in both conditions.