Abstract
Animals search for foods and decide their behaviors according to previous experience. Caenorhabditis elegans detects chemicals with a limited number of sensory neurons, allowing us to dissect roles of each neuron for innate and learned behaviors. C. elegans is attracted to salt after exposure to the salt (NaCl) with food. In contrast, it learns to avoid the salt after exposure to the salt without food. In salt-attraction behavior, it is known that the ASE taste sensory neurons (ASEL and ASER) play a major role. However, little is known about mechanisms for learned salt avoidance. Here, through dissecting contributions of ASE neurons for salt chemotaxis, we show that both ASEL and ASER generate salt chemotaxis plasticity. In ASER, we have previously shown that the insulin/PI 3-kinase signaling acts for starvation-induced salt chemotaxis plasticity. This study shows that the PI 3-kinase signaling promotes aversive drive of ASER but not of ASEL. Furthermore, the Gq signaling pathway composed of Gqα EGL-30, diacylglycerol, and nPKC (novel protein kinase C) TTX-4 promotes attractive drive of ASER but not of ASEL. A putative salt receptor GCY-22 guanylyl cyclase is required in ASER for both salt attraction and avoidance. Our results suggest that ASEL and ASER use distinct molecular mechanisms to regulate salt chemotaxis plasticity.
ANIMALS show various behaviors in response to environmental cues and modulate behaviors according to previous experience. To understand neuronal plasticity underlying learning, it is important to dissect neurons and molecules for sensing environmental stimuli, storing memory, and executing learned behaviors.
The nematode Caenorhabditis elegans has only 302 neurons and functions of sensory neurons are well characterized (White et al. 1986; Bargmann 2006). C. elegans is attracted to odorants sensed by the AWC olfactory neurons or to salts sensed by the ASE gustatory neurons (Bargmann and Horvitz 1991; Bargmann et al. 1993). The ASE neuron class consists of a bilaterally symmetrical pair, ASE-left (ASEL) and ASE-right (ASER), which sense different sets of ions including Na+ and Cl−, respectively (Pierce-Shimomura et al. 2001; Suzuki et al. 2008; Ortiz et al. 2009). ASEL is activated by an increase in salt concentration, whereas ASER is activated by a decrease in salt concentration (Suzuki et al. 2008). In the ASE gustatory neurons, a cyclic GMP (cGMP) signaling pathway mediates sensory transduction (Komatsu et al. 1996; Suzuki et al. 2008; Ortiz et al. 2009). ASEL and ASER express different sets of receptor-type guanylyl cyclases (gcys) (Ortiz et al. 2006). Of these, gcy-22, which is specifically expressed in ASER, is important for attraction to ASER-sensed ions such as Cl− (Ortiz et al. 2009).
Preference for salts changes according to previous experience (known as gustatory plasticity or salt chemotaxis learning) (Saeki et al. 2001; Jansen et al. 2002; Tomioka et al. 2006). When worms are grown on a medium that contains sodium chloride (NaCl) and food (Escherichia coli), they show attraction to NaCl by using ASE neurons (Bargmann and Horvitz 1991; Suzuki et al. 2008). In contrast, after exposure to the salt under starvation conditions, they show reduced attraction to or even avoid the salt (Saeki et al. 2001; Jansen et al. 2002; Tomioka et al. 2006). In C. elegans, it was proposed that preference for a sensory cue is defined by the sensory neuron that detects the cue (Troemel et al. 1997). ASE neurons play a major role for salt attraction (Bargmann and Horvitz 1991; Suzuki et al. 2008; Ortiz et al. 2009). However, little is known about sensory neurons that drive the learned salt avoidance; it remains unclear whether ASE neurons act as salt receptors for the learned avoidance.
We have previously shown that an insulin/PI 3-kinase signaling pathway is essential for salt chemotaxis learning (Tomioka et al. 2006). In C. elegans, the insulin-like signaling is composed of daf-2, age-1, and akt-1, which encode homologs of insulin receptor, PI 3-kinase, and protein kinase B, respectively (Morris et al. 1996; Kimura et al. 1997; Paradis and Ruvkun 1998). Mutants of daf-2, age-1, and akt-1 show attraction to salt even after starvation/NaCl conditioning (Tomioka et al. 2006).
daf-18 encodes a homolog of phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome ten), which dephosphorylates phosphatidylinositol (3,4,5)-triphosphate and counteracts the insulin/PI 3-kinase signaling (Ogg and Ruvkun 1998; Gil et al. 1999; Mihaylova et al. 1999; Rouault et al. 1999; Solari et al. 2005). Mutants of daf-18, in which the PI 3-kinase signaling is activated, show reduced attraction to NaCl even without conditioning. Since the insulin/PI 3-kinase signaling acts in ASER, we proposed that the insulin/PI 3-kinase signaling attenuates the attractive drive of ASER (Tomioka et al. 2006).
In C. elegans, diacylglycerol (DAG) regulates functions of motor neurons and sensory neurons. egl-30, which encodes the α-subunit of heterotrimeric G-protein Gq, facilitates production of DAG and enhances locomotory movements (Brundage et al. 1996; Lackner et al. 1999). In the AWC olfactory neurons, a novel protein kinase C-ɛ/η (nPKC-ɛ/η) ortholog TTX-4 (also known as PKC-1), which is one of DAG targets, plays an essential role in attraction behavior to AWC-sensed odors (Okochi et al. 2005; Tsunozaki et al. 2008). GOA-1 Goα regulates olfactory adaptation by antagonizing Gqα–DAG signaling (Matsuki et al. 2006).
This study investigated the involvement of the ASE taste receptor neurons in the starvation-induced salt avoidance. We show that both ASEL and ASER contribute to salt chemotaxis learning. Activation of the PI 3-kinase signaling and the Gq/DAG/PKC signaling acted antagonistically in reversal of ASER function, whereas these signaling pathways did not have prominent effects on ASEL function. In ASER, GCY-22 was required for both salt attraction and avoidance. These results suggest that ASE neurons are important for bidirectional chemotaxis and also suggest that distinct molecular mechanisms regulate functions of ASEL and ASER in salt chemotaxis learning.
MATERIALS AND METHODS
Strains and culture:
C. elegans strains were cultivated at 20° under standard conditions (Brenner 1974), except that the E. coli strain NA22 was used as a food source. Strain used were wild-type N2, che-1(p674) I, age-1(hx546), age-1(m333) II, daf-18(e1375), daf-18(mg198) IV, akt-1(ok525), gcy-22(pe902), gcy-22(pe905), gcy-22(pe917), gcy-22(pe922), gcy-22(tm2364), lsy-6(ot71), ttx-4(nj3) V, dyf-11(pe554) X, OH7621 otIs204[ceh-36∷lsy-6, elt-2∷gfp], OH8585 otIs4[gcy-7∷gfp]; otEx3822[ceh-36∷CZ-caspase3(p17); gcy-7∷caspase3(p12)-NZ; myo-3∷mCherry], and OH8593 ntIs1[gcy-5∷gfp] V; otEx3830[ceh-36∷CZ-caspase3(p17); gcy-5∷caspase3(p12)-NZ; myo-3∷mCherry].
Salt chemotaxis assays:
Experiments for Figures 1A, 6A, and 6B were performed on a 9-cm assay plate using a 100 mm NaCl agar plug as a source of chemoattractant (5 mm in diameter and 7 mm thick) (supporting information, Figure S1A), which was previously described (Tomioka et al. 2006). In experiments for Figure 1B, a 100 mm ammonium chloride agar plug (100 mm ammonium chloride adjusted to pH 6.0 with ammonium hydroxide, 5 mm potassium phosphate, pH 6.0, 1 mm CaCl2, 1 mm MgSO4, 2% agar) was placed on a plate (Figure S1A) and animals were preexposed to a mock-conditioning liquid (5 mm potassium phosphate, pH 6.0, 1 mm CaCl2, 1 mm MgSO4) for 1 hr at room temperature (23°). In other experiments, the chemotaxis assay was performed using a 9-cm assay plate (5 mm potassium phosphate, pH 6.0, 1 mm CaCl2, 1 mm MgSO4, 2% agar, 10 ml), on which a salt gradient had been formed for 18–24 hr by placing two agar plugs containing 100 mm of NaCl (100 mm NaCl, 5 mm potassium phosphate, pH 6.0, 1 mm CaCl2, 1 mm MgSO4, 2% agar) close to the edge of the plate (Figure S1B). Just before placing the animals, the plugs were removed, and 1 μl each of 0.5 m sodium azide was spotted at the gradient peak(s) and at point(s) on the opposite end of the plate (Figure S1).
Figure 1.—
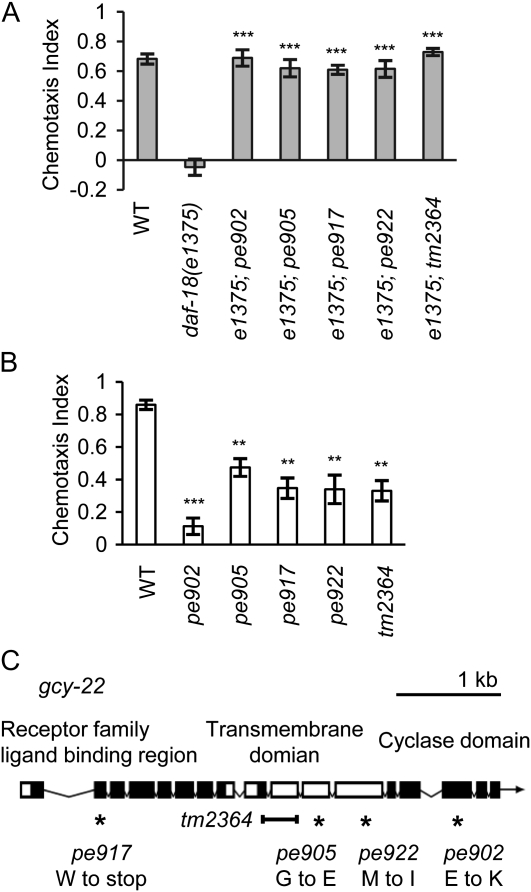
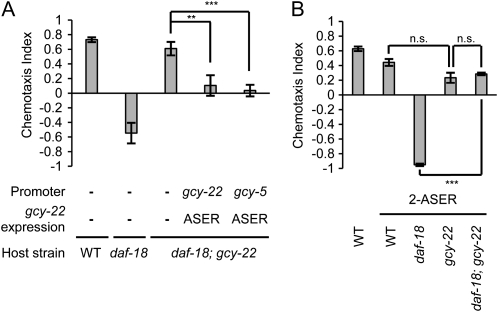
Loss of a receptor-type guanylyl cyclase GCY-22 suppresses the chemotaxis defect of daf-18 mutants. (A) pe902, pe905, pe917, and pe922 were obtained in a suppressor screen for mutations that rescue the chemotaxis defect of daf-18(e1375) mutants. Naive animals were tested for NaCl chemotaxis. A deletion mutation gcy-22(tm2364) also suppresses the chemotaxis defect of the daf-18 mutants. (B) pe902, pe905, pe917, pe922, and tm2364 cause defects in attraction behavior to the ASER-sensed chemical ammonium chloride. (C) Genomic structure of gcy-22 receptor-type guanylyl cyclase. Solid boxes indicate predicted protein domains. Locations of the four suppressor mutations in the gcy-22 gene are depicted. Error bars represent standard error of the mean (SEM). (***) P < 0.001, (**) P < 0.01, Bonferroni t-test.
Figure 6.—
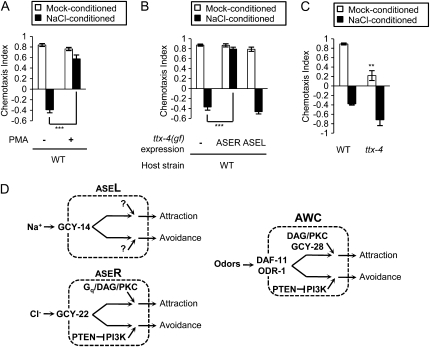
Constitutive activation of the Gq/diacylglycerol/PKC signaling fixes ASER to promote attraction behavior. (A) Animals were treated in a mock-conditioning liquid or a NaCl-conditioning liquid with or without the diacylglycerol analog PMA. Treatment with PMA disrupts starvation/NaCl learning and causes attraction behavior after the NaCl conditioning. (B) TTX-4(A160E) was expressed in ASER under the gcy-5 promoter or in ASEL under the gcy-7 promoter. Expression of TTX-4(A160E) in ASER causes attraction behavior even after NaCl conditioning. (C) ttx-4 mutants show defects in salt attraction but not in salt avoidance. (D) Schematic of molecular mechanisms in ASE neurons and AWC neurons that regulate gustatory and olfactory plasticity, respectively (see text). Guanylyl cyclases GCY-22, GCY-14, DAF-11, and ODR-1 are required for chemosensation. GCY-22 acts in ASER and GCY-14 acts in ASEL for sensing ions (Ortiz et al. 2009). In AWC, ODR-1 and DAF-11 are required for sensing odors (Birnby et al. 2000; L'etoile and Bargmann 2000). Error bars represent SEM. (***) P < 0.001, (**) P < 0.01, Bonferroni t-test.
In experiments for Figures 1–4 (except 1B), and 5E, naive animals were used. In experiments for Figure 1B, Figure 5, A–D, and Figure 6, conditioned animals were used. For naive assays, young adults grown on NGM plates seeded with E. coli NA22 at 20° were washed three times with wash buffer (5 mm potassium phosphate, pH 6.0, 1 mm CaCl2, 1 mm MgSO4, 0.5g/liter gelatin) and directly placed at the center of the assay plates, and then incubated at room temperature (23°) for 30 min. For learning assays, washed young adults were transferred to a conditioning buffer with 20 mm NaCl (NaCl conditioning) or without (mock conditioning), and incubated at room temperature (23°) for 1 hr. After the incubation, animals were directly placed at the center of the assay plates, and then incubated at room temperature (23°) for 30 min. The chemotaxis index was calculated as described in Figure S1, A was the number of animals within the area including peak(s) of the salt gradient, B was the number of animals within the area including the control spot(s), N was the number of all animals on an assay plate, and C was the number of animals that did not move in the central region (Figure S1). In all experiments, 100–200 animals were put on an assay plate.
Figure 4.—
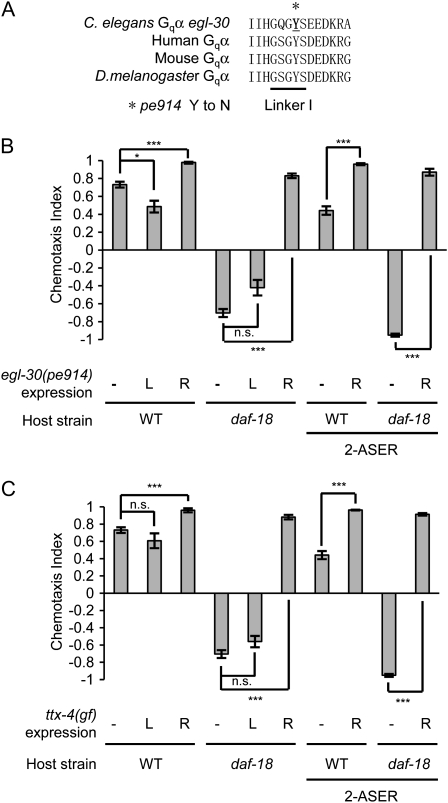
Gq/PKC signaling promotes salt attraction antagonistic to the insulin/PI 3-kinase signaling. (A) pe914 mutation causes an amino acid substitution in the linker I domain of the EGL-30 Gqα protein. (B) egl-30(pe914) was expressed under the ASER-specific gcy-5 promoter or under the ASEL-specific gcy-7 promoter. Expression of egl-30(pe914) in ASER switches the avoidance behaviors of daf-18(mg198) mutants and daf-18(mg198); lsy-6(ot71) mutants. (C) A gain-of-function form of nPKC TTX-4, TTX-4(A160E), was expressed in ASER under the gcy-5 promoter or in ASEL under the gcy-7 promoter. Expression of TTX-4(A160E) in ASER switches the avoidance behaviors of daf-18(mg198) mutants and daf-18(mg198); lsy-6(ot71) mutants. Error bars represent SEM. (***) P < 0.001, (*) P < 0.05, Bonferroni t-test. n.s., not significant.
Figure 2.—
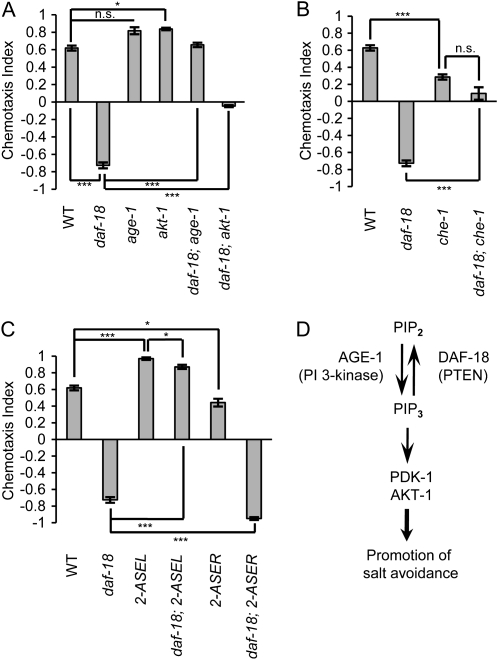
ASER promotes salt avoidance and ASEL promotes salt attraction in daf-18 mutants. (A) NaCl chemotaxis of wild-type animals, daf-18(mg198null), age-1(hx546rf), akt-1(ok525null), daf-18(mg198); age-1(m333null), and daf-18(mg198); akt-1(ok525null) mutants under naive conditions. On the new assay format (see Figure S1B), daf-18(mg198) mutants show salt avoidance. Mutations of age-1 and akt-1 suppress the salt avoidance of the daf-18 mutants. (B) Effect of the dysfunction of ASE neurons due to the che-1(p674) mutation. The che-1(p674) mutation suppresses the salt avoidance of daf-18(mg198) mutants. (C) Chemotaxis of daf-18(mg198) mutants with 2-ASEL (otIs204[ceh-36p∷lsy-6]) or 2-ASER (lsy-6(ot71)). ASER promotes aversion and ASEL promotes attraction in daf-18 mutants. (D) A model deduced from the analysis of the role of ASER in daf-18 mutants. ASER promotes salt avoidance when the PI 3-kinase signaling is activated. Error bars represent SEM. (***) P < 0.001, (*) P < 0.05, Bonferroni t-test. n.s., not significant.
Figure 3.—
The receptor-type guanylyl cyclase GCY-22 in ASER promotes salt avoidance. (A) Rescue experiments with expression of gcy-22 under its own promoter or ASER-specific gcy-5 promoter in the daf-18(e1375); gcy-22(pe917) mutant. (B) Suppression of the avoidance behavior of daf-18; 2-ASER mutants by the gcy-22(pe917) mutation. NaCl chemotaxis of wild-type animals, lsy-6(ot71), daf-18(mg198); lsy-6(ot71), lsy-6(ot71) gcy-22(pe917), and daf-18(mg198); lsy-6(ot71) gcy-22(pe917) mutants under naive conditions. Error bars represent SEM. (***) P < 0.001, (**) P < 0.01, Bonferroni t-test. n.s., not significant.
Figure 5.—
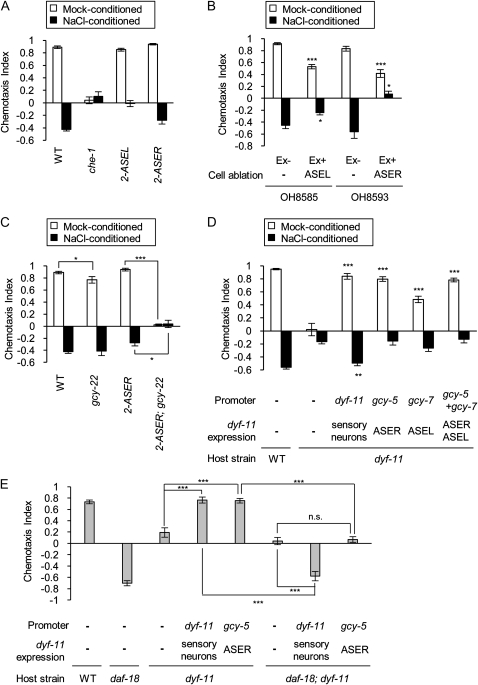
Taste receptor neurons ASE promote salt avoidance and other sensory neurons are also required for salt avoidance. (A) Learning assays were performed with wild-type animals, che-1(p674) mutants, 2-ASEL animals (otIs204[ceh-36p∷lsy-6]), and 2-ASER animals (lsy-6(ot71)). che-1 mutants do not show salt chemotaxis plasticity, whereas 2-ASEL animals and 2-ASER animals show plasticity. (B) Learning assays were performed with the OH8585 strain (ASEL ablated) and OH8593 strain (ASER ablated). Both strains show impaired learned avoidance but still show plasticity. (C) Learning assays were performed with wild-type animals, gcy-22(pe917) mutants, lsy-6(ot71) animals (2-ASER), and lsy-6(ot71) gcy-22(pe917) mutants. ASER promotes learned salt avoidance depending on gcy-22. (D) dyf-11 was expressed under the dyf-11 promoter, in ASER under the gcy-5 promoter, or in ASEL under the gcy-7 promoter in the dyf-11(pe554) mutant background. The dyf-11 mutation causes dysfunction of sensory neurons. Selective rescue of ASER, ASEL, or both restore salt attraction after mock conditioning, but do not rescue the learned avoidance. (E) dyf-11 was expressed under the dyf-11 promoter or in ASER under the gcy-5 promoter in the dyf-11(pe554) mutant background or in the daf-18(mg198); dyf-11(pe554) mutant background. Expression of DYF-11 in ASER restores the attractive response in dyf-11 mutants. The daf-18 mutation downregulates the response of ASER but does not cause salt avoidance. Error bars represent SEM. (***) P < 0.001, (**)P < 0.01, (*) P < 0.05, Bonferroni t-test. n.s., not significant.
Genetic screens for suppressor mutations that improve salt chemotaxis of daf-18 mutants:
Mutagenesis with EMS (ethyl methanesulfonate) was performed as described (Brenner 1974). F1 progeny (15,000) of mutagenized daf-18(e1375) mutants were divided into 46 independent groups and cultured separately. F2 animals were tested for naive chemotaxis; animals that were attracted to salt were collected. These suppressor candidates were cultured and the progeny were further screened with the same procedure for additional five generations to concentrate suppressor mutants. Finally, single worms were picked from each group and the progeny were tested for suppression.
Mapping and cloning of pe902, pe905, pe917, and pe922:
For mapping of pe902, pe905, pe917, and pe922, a tester strain that carries the daf-18(e1375) mutation in the CB4856 background was generated by repeated outcrosses. Mapping was performed using the SNPs (single nucleotide polymorphisms) between this strain and the suppressor mutants. All of these alleles were mapped to the left arm of chromosome V (see results). After sequencing the gcy-22 genomic region of each suppressor mutant, the mutations were identified.
pe902 was identified as a G to A substitution, whose 5′ and 3′ flanking sequences are CCGGCAAATTTTTCCAGGTA and AACAATTGGTGATGGATATT, respectively. pe905 was identified as a G to A substitution, whose 5′ and 3′ flanking sequences are CAACTTGTGTAAATTCATCG and TTATCTCTTGACTCACCAAC, respectively. pe917 was identified as a G to A substitution, whose 5′ and 3′ flanking sequences are TTAAATTTCAGCCTAACATG and GTGTTAACTAACTGTCGAGA, respectively. pe922 was identified as a G to A substitution, whose 5′ and 3′ flanking sequences are TTACCTATCACAAAAGAAAT and GATATCTACTCATTCGCGAT, respectively.
Mapping and cloning of pe914:
Before mapping of pe914, the original pe914; e1375 mutant was outcrossed with N2 strain and the e1375 mutation was removed. The pe914 single mutant showed hyperactive locomotion and egg-laying constitutive phenotype, which is a typical phenotype caused by activation of egl-30 Gq signaling. pe914 was mapped to the right arm of chromosome I using SNPs between CB4856 and the pe914 strain. Genomic region of egl-30 was sequenced and a missense mutation was identified. pe914 was identified as a T to A substitution, whose 5′ and 3′ flanking sequences are GAATTATCCACGGTCAGGGA and ATTCGGAAGAGGACAAGCGA, respectively.
PMA treatment:
Young adult animals were washed and soaked in a mock-conditioning liquid or a NaCl-conditioning liquid with 1.0 μg/ml phorbol 12-myristate 13-acetate (PMA) for 1 hr. After the exposure, animals were directly placed at the center of the assay plates and then incubated at room temperature (23°) for 30 min.
Cloning of gcy-22 cDNA:
yk1153.b08 (a generous gift from Y. Kohara) corresponds to 3′-terminal half of a predicted gcy-22 cDNA. To obtain predicted 5′-terminal half of gcy-22 cDNA, reverse transcription reaction was performed using total RNA as a template and a gene-specific primer 5′-TAGATCTTCTGTTATCTGCATCAC-3′ and SuperScriptIII kit (Invitrogen). 5′-terminal gcy-22 cDNA was amplified by RT-PCR using primers 5′-GCTAGCATGAGTTTCATATCAAAATGTTTTATTTGC-3′ and 5′-TAATGGTCCAACTTGTGACACCGTTGGATC-3′. Then, the obtained 5′-terminal fragment of gcy-22 cDNA was sequenced. The 5′-terminal gcy-22 cDNA and yk1153.b08 were fused by PCR using primers 5′-GCTAGCATGAGTTTCATATCAAAATGTTTTATTTGC-3′ and 5′-GGTACCTTAGATAGATTCTCCATTCTCCTTCGCCTC-3′, which yielded a full-length gcy-22 cDNA. The full-length gcy-22 cDNA was found to be identical to the gene model T03D8.5. The full-length gcy-22 cDNA was inserted into the pPD-DEST vector to generate a gcy-22 destination vector.
Plasmid construction and germline transformation:
venus∷dyf-11, egl-30(pe914), ttx-4(A160E), and gcy-22 expression vectors were constructed using the GATEWAY system (Invitrogen). Procedures for creating these original cDNA constructs were described previously (Okochi et al. 2005; Tomioka et al. 2006; Kunitomo and Iino 2008). Expression constructs composed of particular promoters and the cDNAs were created by LR reactions (site-specific recombination) between the entry plasmids and the destination plasmids. To generate entry vectors carrying gcy-22 promoter sequences, the 2-kb promoter region of gcy-22 were amplified by PCR from C. elegans genomic DNA and then inserted into the pDONR201 vector by site-specific recombination. The ttx-4(A160E) cDNA was inserted into the pPD-DEST vector to generate a ttx-4(A160E) destination plasmid.
Plasmids were injected into the gonad arms as previously described (Mello et al. 1991). Expression constructs for egl-30(pe914), ttx-4(A160E), and gcy-22 were injected at 20 ng/μl and dyf-11∷venus expression constructs were injected at 5 ng/μl, along with myo-3p∷venus (20 ng/μl) as a transformation marker and pPD49.26 as carrier DNA. In each case, the final concentration of injected DNA was 100 ng/μl.
RESULTS
The receptor-type guanylyl cyclase GCY-22 in ASER promotes avoidance in daf-18 PTEN mutants:
Previously, we have reported that the insulin/PI 3-kinase signaling is required in the ASER neuron to attenuate salt attraction after starvation/salt conditioning (Tomioka et al. 2006). daf-18 encodes a homolog of phosphatase PTEN, which dephosphorylates PIP3 to negatively regulate the PI 3-kinase signaling (Ogg and Ruvkun 1998; Gil et al. 1999; Mihaylova et al. 1999; Rouault et al. 1999; Solari et al. 2005). Mutants of daf-18 show reduced attraction even without conditioning (naive conditions), suggesting that the PI 3-kinase signaling is important for the regulation of ASER function (Tomioka et al. 2006).
To further investigate how the insulin/PI 3-kinase signaling modulates ASER function to regulate salt chemotaxis, we screened for suppressors of the chemotaxis defect in daf-18 mutants. Suppressor mutants were expected to show salt attraction similar to wild-type animals in the daf-18 genetic background. About 30,000 EMS-mutagenized haploid genomes were screened in the daf-18(e1375) genetic background by salt chemotaxis assay (see materials and methods). Through the screening, we obtained 23 suppressor mutants (data not shown). Of these, four suppressor mutations, pe902, pe905, pe917, and pe922, which clearly suppressed the chemotaxis defect of the daf-18 mutant, were picked up for further analysis (Figure 1A).
The suppressor mutations pe902, pe905, pe917, and pe922, were mapped by using the SNPs between the N2 strain and the CB4856 strain (Wicks et al. 2001). All of these suppressor mutations were mapped to the left arm of chromosome V and resided between two SNPs, pkP5135 and cE5-269. This region contains the gcy-22 gene, which is required for attraction to ASER-sensed ions including chloride ion (Ortiz et al. 2009). The four alleles caused reduced attraction to ammonium chloride in the wild-type background, similar to the deletion mutant, gcy-22(tm2364) (Figure 1B). The tm2364 mutation suppressed chemotaxis defect of the daf-18(e1375) mutant (Figure 1A), indicating that loss of gcy-22 improves chemotaxis of daf-18 mutants. By sequencing the gcy-22 genomic region of each suppressor mutant, nonsense and missense mutations were identified (Figure 1C; Figure S2; materials and methods).
gcy-22 encodes a receptor-type guanylyl cyclase, which is specifically expressed in the ASER neuron (Ortiz et al. 2006; Ortiz et al. 2009). gcy-22 mutants show diminished calcium responses of ASER to changes of salt concentration (Ortiz et al. 2009), suggesting that GCY-22 mediates sensory transduction. One possible explanation is that ASER promotes both salt attraction and aversion depending on the activity of the insulin/PI 3-kinase signaling.
ASER promotes salt avoidance and ASEL promotes salt attraction in daf-18 mutants:
Next, we analyzed salt chemotaxis of daf-18 mutants in a modified chemotaxis assay (Figure S1B). The new assay format is suitable for characterizing salt avoidance because all animals that avoid salt contribute negatively to the chemotaxis index, which was not the case in the former assay format (Figure S1A).
The new assay format revealed that daf-18 mutants showed a remarkable salt avoidance, whereas wild-type animals were attracted under naive conditions (Figure 2A). The avoidance in the daf-18 mutants was suppressed by mutations of age-1 PI3K and akt-1 Akt/PKB (Figure 2A), indicating that the behavioral switching is dependent on the activity of the PI 3-kinase signaling. The mutation of akt-1 did not restore wild-type chemotaxis in the daf-18 mutants, suggesting that other molecules act in parallel with AKT-1. These results suggest that activation of the PI 3-kinase signaling promotes salt avoidance rather than downregulates salt attraction under naive conditions.
To identify neurons that promote salt avoidance in daf-18 mutants, we tested whether ASE neurons are involved in the avoidance behavior of daf-18 mutants. che-1 encodes a zinc finger transcription factor, which is required for specification of ASE neurons, and mutants of che-1 show defects in salt-attraction behavior (Uchida et al. 2003). A che-1 mutation suppressed the salt-avoidance behavior, suggesting that ASE mediates the salt aversion in daf-18 mutants (Figure 2B).
The ASE neurons consist of a pair of neurons, ASEL and ASER, which are bilaterally symmetrical in morphology but functionally distinct (Pierce-Shimomura et al. 2001; Suzuki et al. 2008; Ortiz et al. 2009). lsy-6 encodes a microRNA that regulates determination of the asymmetrical cell fates (Johnston and Hobert 2003). Loss of lsy-6 leads to a “2-ASER” mutant phenotype, whereas ectopic expression of lsy-6 causes a “2-ASEL” phenotype (Johnston and Hobert 2003; Ortiz et al. 2009). Calcium responses of the transformed ASEL (or ASER) to changes in concentration of various ions are comparable to that of authentic ASEL (or ASER) (Ortiz et al. 2009).
To dissect contributions of ASEL and ASER to the aversive response of daf-18 mutants, 2-ASEL and 2-ASER strains that carry the daf-18 mutation were tested for salt chemotaxis. lsy-6; daf-18 double mutants with 2-ASER exhibited stronger salt avoidance compared to the daf-18 mutants, whereas daf-18 mutants with 2-ASEL exhibited attraction to NaCl (Figure 2C). These results suggest that ASER promotes salt avoidance and ASEL promotes salt attraction in daf-18 mutants and also suggest that activation of the insulin/PI 3-kinase signaling switches the function of ASER (Figure 2D). They also complement our previous cell-specific rescue experiments showing that the DAF-18 acts in ASER to promote salt attraction (Tomioka et al. 2006).
The attraction of daf-18; gcy-22 mutants was reduced by expression of gcy-22 under the gcy-5 promoter, which drives specific expression in ASER (Figure 3A). On the other hand, the strong avoidance of lsy-6; daf-18 mutants with 2-ASER was suppressed by the gcy-22 mutation (Figure 3B). Taken together, these results suggest that GCY-22 in ASER promotes salt avoidance in daf-18 mutants. In the daf-18; gcy-22 mutant, ASER is dysfunctional and ASEL promotes salt attraction, which leads to the suppression of the avoidance behavior of daf-18.
A Gq/diacylglycerol/PKC signaling promotes salt attraction antagonistic to the insulin/PI 3-kinase signaling:
Another suppressor allele, pe914, was identified as a missense mutation in egl-30, which encodes the α-subunit of the heterotrimeric G-protein Gq (Tomioka et al. 2006). egl-30(pe914) is a presumptive gain-of-function mutation because it caused hyperactive locomotion and constitutive egg-laying phenotypes (data not shown), which are typical phenotypes of egl-30(gf) alleles (Schade et al. 2005; Williams et al. 2007). pe914 is a missense mutation in linker I, a well-conserved domain of Gqα across phylogeny (Figure 4A). Expression of egl-30(pe914) in ASER of daf-18 mutants suppressed the avoidance behavior and switched it to strong attraction. (Figure 4B).
In C. elegans motoneurons, egl-30 Gqα facilitates production of DAG (Brundage et al. 1996; Lackner et al. 1999). An nPKC-ɛ/η ortholog, ttx-4, which is a target of DAG, regulates locomotion behavior (Sieburth et al. 2007). TTX-4 also plays an essential role for the attractive response to odors (Okochi et al. 2005; Tsunozaki et al. 2008). Similar to the effects of egl-30(pe914), expressing TTX-4(A160E), a gain-of-function form of TTX-4 (Dekker et al. 1993; Okochi et al. 2005), in ASER switched the avoidance behavior of daf-18 mutants to attraction (Figure 4C). Therefore, the Gq/PKC signaling acts antagonistically to the PI 3-kinase signaling.
Taste-receptor ASE neurons promote the learned avoidance:
Wild-type animals show salt attraction after exposure to a salt-free liquid without food (mock conditioning). In contrast, they show salt avoidance after exposure to a salt-containing liquid without food (NaCl conditioning) (hereafter referred to as “learned avoidance”). In salt-attraction behaviors, ASE neurons function as main salt receptors (Bargmann and Horvitz 1991; Suzuki et al. 2008). To ask whether ASE neurons are also involved in salt avoidance after NaCl conditioning, che-1 mutants were subjected to chemotaxis assay after NaCl conditioning. Consistent with the previous report (Hukema et al. 2006), che-1 mutants did not show either attraction or avoidance after conditioning (Figure 5A). This suggests that ASE neurons are required for the learned salt avoidance.
Next, we tested the learning abilities of animals with genetically ablated ASEL or ASER neuron (Ortiz et al. 2009). In these strains, the apoptosis-inducing protein caspase is expressed in ASEL or ASER (Ortiz et al. 2009). Both ablated animals showed defects in the aversive response after NaCl conditioning compared to control animals (Figure 5B), suggesting that both ASEs are required for the learned avoidance. Both ablated animals and animals with 2-ASER or 2-ASEL showed chemotaxis plasticity following the conditioning (Figure 5, A and B), suggesting that both ASEs contribute to salt chemotaxis plasticity. The learned avoidance of animals with 2-ASER was fully dependent on gcy-22, indicating that ASER promotes learned salt avoidance depending on GCY-22 function (Figure 5C). The gcy-22 mutant also showed learned avoidance, suggesting that GCY-22-independent plasticity, probably mediated by ASEL, also contributes to salt chemotaxis learning (Figure 5C).
Next, we asked whether ASE neurons are sufficient to direct the learned avoidance. We tested salt chemotaxis of animals with disrupted chemosensory neurons except ASER or ASEL. dyf-11 encodes a protein that is required for cilium biogenesis of sensory neurons. Loss of dyf-11 causes truncation of sensory endings and defects in responses to water-soluble compounds (Kunitomo and Iino 2008). Animals with functional ASER or ASEL, or both, by cell-specific rescue of these neurons, showed salt chemotaxis plasticity: mock-conditioned animals showed attraction behavior and NaCl-conditioned animals showed diminished attractive responses but did not show learned avoidance (Figure 5D). This result suggests that ASEL and ASER are sufficient to direct salt attraction but insufficient to direct salt avoidance after conditioning. Sensory neurons other than ASE are also required for the learned avoidance.
Next, we asked whether salt reception by ASER is sufficient to direct salt avoidance in daf-18 mutants. We tested salt chemotaxis of daf-18 mutants with disrupted chemosensory neurons except ASER. Expressing DYF-11 in ASER of dyf-11 mutant restored wild-type salt attraction under naive conditions (Figure 5E). However, under the dyf-11; daf-18 mutant background, expression of DYF-11 in ASER was insufficient to direct the salt avoidance (Figure 5E). This result is in contrast to daf-18 and daf-18; 2-ASER mutants where ASER-mediated salt avoidance behaviors were observed. These results suggest that ASER is not sufficient and chemosensory neurons other than ASER are required for the full expression of the PI 3-kinase-dependent avoidance behavior.
Enhanced Gq/DAG/PKC signaling impairs the learned salt avoidance:
As described above, the Gq/DAG/PKC signaling acts in ASER and promotes salt attraction. We further tested whether activity of the Gq/DAG/PKC signaling is crucial for the reorientation of chemotaxis in starvation-induced learning. Treatment with a DAG analog PMA during the conditioning caused attraction behavior even after NaCl conditioning (Figure 6A), an effect similar to overexpression of egl-30(pe914) in ASER (Tomioka et al. 2006). Enhanced activity of nPKC by TTX-4(A160E) expression in ASER disturbed the learned avoidance and caused strong salt attraction (Figure 6B). These results suggest that the normal regulation of the Gq/DAG/PKC signaling is required for switching of ASER function by the conditioning; the enhanced activity of the Gq/DAG/PKC signaling fixes ASER to the mode that promotes salt attraction. On the other hand, activation of TTX-4 in ASEL did not cause significant effects on the learned avoidance (Figure 6B), suggesting that the roles of TTX-4 are different depending on cell types. Notably, while loss of function mutants of ttx-4 showed defects in salt attraction, they showed salt avoidance after NaCl conditioning comparable to, or greater than, wild-type animals (Figure 6C). This suggests that TTX-4 is required for salt attraction but not for salt avoidance.
DISCUSSION
In this study, we reported that both ASEL and ASER generate starvation-induced salt chemotaxis plasticity. We showed that the PI 3-kinase signaling and the Gq/DAG/PKC signaling in ASER contribute to reversal of the orientation of salt chemotaxis. Attractive and aversive drives of ASER depend on GCY-22 function.
GCY-22 is essential for attractive and repulsive responses mediated by ASER:
GCY-22 is localized to the cilium and is required for calcium responses of ASER, suggesting that GCY-22 mediates sensory transduction of ASER (Ortiz et al. 2009). However, it has not been tested whether the activity of GCY-22 or cGMP level in ASER is regulated by changes in salt concentration. Another possibility that cannot be excluded at this point is that GCY-22 acts for modulation of ASER function. Because several guanylyl cyclases are expressed in interneurons or nonneuronal cells (Ortiz et al. 2006), guanylyl cyclases including GCY-22 may not be directly involved in odor or salt sensation. One prominent example is GCY-28, which regulates odor preference switching in the AWC neuron (Tsunozaki et al. 2008). GCY-28.d isoform is localized to the axon and may regulate synaptic transmission (Tsunozaki et al. 2008). The extracellular domain of GCY-22 is most homologous to mammalian guanylyl cyclase B (data not shown), which binds natriuretic peptides, raising a possibility that GCY-22 might be a receptor for hormonal peptides.
The fact that gcy-22 mutations suppress salt repulsion of daf-18 mutants can be explained by simply assuming that ASER promotes avoidance in daf-18 mutants and gcy-22 is required for ASER function. In addition, there might be functional interactions between GCY-22 and the PI 3-kinase signaling pathway, as well as the Gq pathway. For example, it is possible that the PI 3-kinase signaling and the Gq/DAG/PKC signaling may modulate the function of GCY-22 to help behavioral switching between attraction and avoidance. Conversely, it is also possible that GCY-22/cGMP signaling regulates the activities of the insulin/PI 3-kinase signaling and the Gq/DAG/PKC signaling.
In this study, the suppression of the chemotaxis defect of daf-18 mutants by gcy-22 mutation was not fully rescued by the expression of gcy-22 under the control of the ASER-specific gcy-5 or gcy-22 promoters (Figure 3A). Several possibilities may explain the result. One is that gcy-22 may have several splicing isoforms. Another possibility is that GCY-22 also acts in cells other than ASER. Alternatively, expression level of GCY-22 was not sufficient for the rescue.
Activation of EGL-30/DAG/TTX-4 signaling in ASER promotes salt attraction:
Activation of EGL-30 or TTX-4 or treatment with a DAG analog PMA causes strong salt attraction even after starvation/NaCl conditioning and reverses salt avoidance in daf-18 mutants to attraction. EGL-30 was suggested to be coupled with several G-protein-coupled receptors, such as muscarinic acetylcholine receptors (mAChR) (Lackner et al. 1999), a DOP-1 dopamine receptor (Chase et al. 2004; Kindt et al. 2007), GAR-3 mAChR (Liu et al. 2007), and SER-3 octopamine receptor (Suo et al. 2006). Although activation of EGL-30 and addition of a DAG analog promote salt attraction, it is unknown whether EGL-30 is required for the production of DAG in ASER. GPA-12/G12/13α (Hiley et al. 2006), RHO-1 Rho (Mcmullan et al. 2006), and UNC-73 Rho GEF (Williams et al. 2007) also stimulate production of DAG. In motor neurons, EGL-30 activates phospholipase C-β EGL-8 phospholipase C-β, which results in production of DAG (Lackner et al. 1999). In ASER, it is unknown which phospholipase produces DAG. How environmental stimuli and signaling molecules control the DAG level in ASER is an important question, which is to be addressed in the future.
Although ttx-4 mutants display a defect in salt attraction, they show learned salt avoidance comparable to the wild-type animals. This observation suggests that TTX-4 is required for salt attraction but not for salt avoidance. In motor neurons of ttx-4 mutants, release of neuropeptides is impaired, whereas release of acetylcholine is intact (Sieburth et al. 2007). Therefore, release of neuropeptides regulated by TTX-4 in ASER may promote salt attraction. UNC-13 is also known as a presynaptic target of DAG and is essential for synaptic vesicle priming (Maruyama and Brenner 1991; Lackner et al. 1999; Richmond et al. 1999). In the ASER neuron, UNC-13 is likely to be involved in the release of classical neurotransmitters such as glutamate, and its roles in salt chemotaxis need to be determined.
Although activation of TTX-4 in ASER promotes salt attraction, activation of TTX-4 in ASEL did not promote attraction (Figure 6B). TTX-4 positively regulates function of the nociceptive neurons ASH and the olfactory neurons AWA, whereas TTX-4 negatively regulates function of the thermosensory neurons AFD (Okochi et al. 2005). These pieces of evidence imply that roles of the targets of TTX-4 are different among each sensory neuron. One possibility is that different sets of neuropeptides have distinct functions in each sensory neuron.
In mammals, diacylglycerol analog PMA augments release of synaptic vesicles and dense core vesicles in a Munc-13- and PKCs-dependent manner (Majewski and Iannazzo 1998; Brose et al. 2000). Although in vitro analyses revealed potential targets of PKCs (Leenders and Sheng 2005), little is known about targets of nPKC in vivo.
Neurons involved in salt chemotaxis plasticity:
Although ASEL, as well as ASER, contributes to salt chemotaxis plasticity, we have not found prominent roles of the insulin/PI 3-kinase signaling and the Gq/DAG/PKC signaling for regulation of ASEL. These results imply that signals of food/starvation and salt may be encoded by distinct molecules depending on cell types. Thus, it is important to investigate molecules that regulate the plasticity of each cell.
How do neural circuits including ASE neurons direct the learned avoidance? There are at least two possible mechanisms. One model is that unidentified sensory neurons (such as ASH neurons) direct the learned avoidance and these avoidance neurons are modulated by ASE neurons. In accordance with this model, Hukema et al. (2006) showed that ASI, ADF, and ASH neurons are required for salt avoidance and proposed that ASEs sensitize ASH, which directs salt avoidance. ASH, as well as ASE, is activated by changes in salt concentration (Suzuki et al. 2008; Thiele et al. 2009), making ASH neurons good candidates for salt receptors in the learned avoidance. However, it is still unknown whether ASH directs salt avoidance. Another model is that ASE neurons act as salt receptors and other supporting neurons modulate ASEs to direct the learned avoidance. In this model, supporting neurons modulate signaling such as the PI 3-kinase signaling or the Gq/DAG/PKC signaling in ASEs. At present, there is no direct evidence that discriminates these two possibilities. Our results point out the importance of GCY-22 in ASER for salt avoidance behavior, which supports the second model.
Since the PI 3-kinase signaling may mediate a starvation signal, food-sensing neurons may be also important for regulating the salt preference directed by ASER, for example, through secretion of an insulin-like ligand. Apart from possible roles for interaction between sensory neurons, it is also possible that signals from several sensory neurons are integrated at interneurons to establish the learned avoidance.
The salt-avoidance behavior in daf-18 mutants required functions of neurons other than ASER (Figure 5E). Although expression of DAF-18 in ASER is sufficient to restore wild-type salt attraction (Tomioka et al. 2006), it is possible that DAF-18 also acts in cells other than ASER. Alternatively, the supporting neurons might be required to induce the salt avoidance caused by loss of DAF-18 in ASER.
The preference switch as a common property of sensory neurons:
In mouse, fly, and C. elegans, characteristics of each sensory neuron define each innate behavior (Yarmolinsky et al. 2009). C. elegans shows dramatic changes of preference for AWC-sensed odors and ASE-sensed salts; when worms are conditioned with odor (salts) and starvation, they show avoidance behavior to the same odorants (salts) (Hukema et al. 2006; Tomioka et al. 2006; Tsunozaki et al. 2008). ASER and AWC have some common features (summarized in Figure 6D). Both neurons use cGMP in sensory transduction (Coburn and Bargmann 1996; Komatsu et al. 1996; Birnby et al. 2000; L'etoile and Bargmann 2000; Ortiz et al. 2009). Calcium concentration is elevated upon decreases of odor or salt concentration (Chalasani et al. 2007; Suzuki et al. 2008). Furthermore, AWC and ASER have common synaptic targets (White et al. 1986). Therefore, ASER and AWC may share common molecular mechanisms that switch preferences for the chemicals.
In ASER, the insulin/PI 3-kinase signaling and the Gq/DAG/PKC signaling regulate salt preference. Similarly, the insulin/PI 3-kinase signaling and the DAG/PKC signaling regulate odor preference in AWC neurons (Matsuki et al. 2006; Tsunozaki et al. 2008; Lin et al. 2010). It is interesting that gcy-28 and ttx-4 mutants show repulsive behavior to odors in an AWC-dependent manner (Tsunozaki et al. 2008). From analysis of these mutants, Tsunozaki et al. (2008) proposed that AWC neurons can switch between attractive and repulsive signaling modes. However, it was not tested whether reception of odors by AWC directs the learned odor avoidance. It is important to determine gustatory or olfactory neurons that act as receptors in the learned behavior. Nevertheless, ASER and AWC contribute to both attraction and avoidance according to past experience and have potential to act as receptors for bidirectional chemotaxis.
Acknowledgments
We specially thank O. Hobert and C. O. Ortiz for sharing with us the lsy-6(ot71), OH7621, OH8585, and OH8593 strains; G. Ruvkun for the daf-18(mg198) strain; S. Mitani for the gcy-22(tm2364) strain; M. Doi for the egl-30 cDNA; members of the Iino Lab for discussion and comments on the manuscript; and the C. elegans Knockout Consortium for the akt-1(ok525) strain. All other nematode strains used in this study were provided by the Caenorhabditis Genetics Center (CGC). This work was supported by the Grant-in-Aid for Scientific Research on Priority Area “Molecular Brain Sciences,” Innovative Area “Systems Molecular Ethology,” and Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.119768/DC1
References
- Bargmann, C., 2006. Chemosensation in C. elegans (October 25, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.123.1, http://www.wormbook.org.
- Bargmann, C., and H. Horvitz, 1991. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7 729–742. [DOI] [PubMed] [Google Scholar]
- Bargmann, C., E. Hartwieg and H. Horvitz, 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74 515–527. [DOI] [PubMed] [Google Scholar]
- Birnby, D., E. Link, J. Vowels, H. Tian, P. Colacurcio et al., 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose, N., C. Rosenmund and J. Rettig, 2000. Regulation of transmitter release by Unc-13 and its homologues. Curr. Opin. Neurobiol. 10 303–311. [DOI] [PubMed] [Google Scholar]
- Brundage, L., L. Avery, A. Katz, U. Kim, J. Mendel et al., 1996. Mutations in a C. elegans Gqalpha gene disrupt movement, egg laying, and viability. Neuron 16 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani, S., N. Chronis, M. Tsunozaki, J. Gray, D. Ramot et al., 2007. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 450 63–70. [DOI] [PubMed] [Google Scholar]
- Chase, D., J. Pepper and M. Koelle, 2004. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat. Neurosci. 7 1096–1103. [DOI] [PubMed] [Google Scholar]
- Coburn, C., and C. Bargmann, 1996. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron 17 695–706. [DOI] [PubMed] [Google Scholar]
- Dekker, L., P. McIntyre and P. Parker, 1993. Mutagenesis of the regulatory domain of rat protein kinase C-eta: a molecular basis for restricted histone kinase activity. J. Biol. Chem. 268 19498–19504. [PubMed] [Google Scholar]
- Gil, E., E. Malone Link, L. Liu, C. Johnson and J. Lees, 1999. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc. Natl. Acad. Sci. USA 96 2925–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiley, E., R. McMullan and S. Nurrish, 2006. The Galpha12-RGS RhoGEF-RhoA signalling pathway regulates neurotransmitter release in C. elegans. EMBO J. 25 5884–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukema, R., S. Rademakers, M. Dekkers, J. Burghoorn and G. Jansen, 2006. Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. EMBO J. 25 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, G., D. Weinkove and R. Plasterk, 2002. The G-protein gamma subunit gpc-1 of the nematode C.elegans is involved in taste adaptation. EMBO J. 21 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R., and O. Hobert, 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 845–849. [DOI] [PubMed] [Google Scholar]
- Kimura, K., H. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 942–946. [DOI] [PubMed] [Google Scholar]
- Kindt, K., K. Quast, A. Giles, S. De, D. Hendrey et al., 2007. Dopamine mediates context-dependent modulation of sensory plasticity in C. elegans. Neuron 55 662–676. [DOI] [PubMed] [Google Scholar]
- Komatsu, H., I. Mori, J. Rhee, N. Akaike and Y. Ohshima, 1996. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17 707–718. [DOI] [PubMed] [Google Scholar]
- Kunitomo, H., and Y. Iino, 2008. Caenorhabditis elegans DYF-11, an orthologue of mammalian Traf3ip1/MIP-T3, is required for sensory cilia formation. Genes Cells 13 13–25. [DOI] [PubMed] [Google Scholar]
- Lackner, M., S. Nurrish and J. Kaplan, 1999. Facilitation of synaptic transmission by EGL-30 Gqalpha and EGL-8 PLCbeta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24 335–346. [DOI] [PubMed] [Google Scholar]
- Leenders, A., and Z. Sheng, 2005. Modulation of neurotransmitter release by the second messenger-activated protein kinases: implications for presynaptic plasticity. Pharmacol. Ther. 105 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Etoile, N., and C. Bargmann, 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25 575–586. [DOI] [PubMed] [Google Scholar]
- Lin, C., M. Tomioka, S. Pereira, L. Sellings, Y. Iino et al., 2010. Insulin signaling plays a dual role in Caenorhabditis elegans memory acquisition and memory retrieval. J. Neurosci. 30 8001–8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., B. LeBoeuf and L. Garcia, 2007. G alpha(q)-coupled muscarinic acetylcholine receptors enhance nicotinic acetylcholine receptor signaling in Caenorhabditis elegans mating behavior. J. Neurosci. 27 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski, H., and L. Iannazzo, 1998. Protein kinase C: a physiological mediator of enhanced transmitter output. Prog. Neurobiol. 55 463–475. [DOI] [PubMed] [Google Scholar]
- Maruyama, I., and S. Brenner, 1991. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 88 5729–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki, M., H. Kunitomo and Y. Iino, 2006. Goalpha regulates olfactory adaptation by antagonizing Gqalpha-DAG signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan, R., E. Hiley, P. Morrison and S. Nurrish, 2006. Rho is a presynaptic activator of neurotransmitter release at pre-existing synapses in C. elegans. Genes Dev. 20 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C., J. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova, V., C. Borland, L. Manjarrez, M. Stern and H. Sun, 1999. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc. Natl. Acad. Sci. USA 96 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J., H. Tissenbaum and G. Ruvkun, 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382 536–539. [DOI] [PubMed] [Google Scholar]
- Ogg, S., and G. Ruvkun, 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2 887–893. [DOI] [PubMed] [Google Scholar]
- Okochi, Y., K. Kimura, A. Ohta and I. Mori, 2005. Diverse regulation of sensory signaling by C. elegans nPKC-epsilon/eta TTX-4. EMBO J. 24 2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, C., J. Etchberger, S. Posy, C. Frøkjaer-Jensen, S. Lockery et al., 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, C., S. Faumont, J. Takayama, H. Ahmed, A. Goldsmith et al., 2009. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr. Biol. 19 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis, S., and G. Ruvkun, 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura, J., S. Faumont, M. Gaston, B. Pearson and S. Lockery, 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410 694–698. [DOI] [PubMed] [Google Scholar]
- Richmond, J., W. Davis and E. Jorgensen, 1999. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault, J., P. Kuwabara, O. Sinilnikova, L. Duret, D. Thierry-Mieg et al., 1999. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr. Biol. 9 329–332. [DOI] [PubMed] [Google Scholar]
- Saeki, S., M. Yamamoto and Y. Iino, 2001. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J. Exp. Biol. 204 1757–1764. [DOI] [PubMed] [Google Scholar]
- Schade, M., N. Reynolds, C. Dollins and K. Miller, 2005. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G α(s) pathway and define a third major branch of the synaptic signaling network. Genetics 169 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth, D., J. Madison and J. Kaplan, 2007. PKC-1 regulates secretion of neuropeptides. Nat. Neurosci. 10 49–57. [DOI] [PubMed] [Google Scholar]
- Solari, F., A. Bourbon-Piffaut, I. Masse, B. Payrastre, A. Chan et al., 2005. The human tumour suppressor PTEN regulates longevity and dauer formation in Caenorhabditis elegans. Oncogene 24 20–27. [DOI] [PubMed] [Google Scholar]
- Suo, S., Y. Kimura and H. Van Tol, 2006. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J. Neurosci. 26 10082–10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., T. Thiele, S. Faumont, M. Ezcurra, S. Lockery et al., 2008. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 454 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele, T., S. Faumont and S. Lockery, 2009. The neural network for chemotaxis to tastants in Caenorhabditis elegans is specialized for temporal differentiation. J. Neurosci. 29 11904–11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka, M., T. Adachi, H. Suzuki, H. Kunitomo, W. Schafer et al., 2006. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51 613–625. [DOI] [PubMed] [Google Scholar]
- Troemel, E., B. Kimmel and C. Bargmann, 1997. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91 161–169. [DOI] [PubMed] [Google Scholar]
- Tsunozaki, M., S. Chalasani and C. Bargmann, 2008. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 59 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, O., H. Nakano, M. Koga and Y. Ohshima, 2003. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130 1215–1224. [DOI] [PubMed] [Google Scholar]
- White, J., E. Southgate, J. Thomson and S. Brenner, 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314 1–341. [DOI] [PubMed] [Google Scholar]
- Wicks, S., R. Yeh, W. Gish, R. Waterston and R. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28 160–164. [DOI] [PubMed] [Google Scholar]
- Williams, S., S. Lutz, N. Charlie, C. Vettel, M. Ailion et al., 2007. Trio's Rho-specific GEF domain is the missing Galpha q effector in C. elegans. Genes Dev. 21 2731–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky, D., C. Zuker and N. Ryba, 2009. Common sense about taste: from mammals to insects. Cell 139 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]