Abstract
In Drosophila melanogaster, the gene Sex-lethal (Sxl) controls all aspects of female development. Since melanogaster males lacking Sxl appear wild type, Sxl would seem to be functionally female specific. Nevertheless, in insects as diverse as honeybees and houseflies, Sxl seems not to determine sex or to be functionally female specific. Here we describe three lines of work that address the questions of how, when, and even whether the ancestor of melanogaster Sxl ever shed its non-female-specific functions. First, to test the hypothesis that the birth of Sxl's closest paralog allowed Sxl to lose essential ancestral non-female-specific functions, we determined the CG3056 null phenotype. That phenotype failed to support this hypothesis. Second, to define when Sxl might have lost ancestral non-female-specific functions, we isolated and characterized Sxl mutations in D. virilis, a species distant from melanogaster and notable for the large amount of Sxl protein expression in males. We found no change in Sxl regulation or functioning in the 40+ MY since these two species diverged. Finally, we discovered conserved non-sex-specific Sxl mRNAs containing a previously unknown, potentially translation-initiating exon, and we identified a conserved open reading frame starting in Sxl male-specific exon 3. We conclude that Drosophila Sxl may appear functionally female specific not because it lost non-female-specific functions, but because those functions are nonessential in the laboratory. The potential evolutionary relevance of these nonessential functions is discussed.
THE X chromosome counting system used by Drosophila melanogaster (Erickson and Quintero 2007) is only one of a wide variety of primary sex-determining mechanisms known to operate among the Diptera (reviewed in Marin et al. 2000; Saccone et al. 2002; Shearman 2002; Pomiankowski et al. 2004; Sánchez 2008). The rapidity with which the genetic programs that determine population sex ratio can change and the extensive information available on how D. melanogaster determines sex are factors that recommend the fruit fly sex-signaling system for studies aimed at understanding the evolution of developmental pathways that determine cell fate.
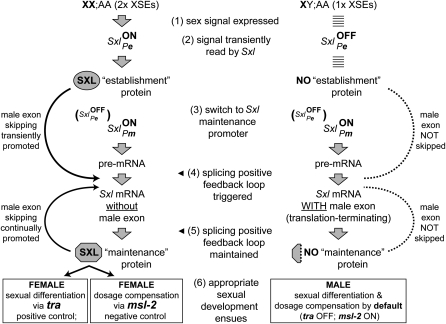
Here we examine aspects of the evolution of a central part of the sex-ratio determining mechanism of D. melanogaster: the feminizing developmental switch gene Sex-lethal (Sxl). Figure 1 outlines how sex is determined in this species by the products of a set of X-linked genes known as X chromosome signal elements (XSEs) that act in a dose-sensitive fashion on a single target, Sxl (most recently reviewed in Salz and Erickson 2010). The higher level of XSE proteins made in very young embryos with two X chromosomes (female) compared to those with one X (male) directly activates SxlPe, the sexual pathway establishment promoter, thereby generating a pulse of feminizing Sxl-f protein in diplo-X but not in haplo-X individuals. Soon thereafter, SxlPe shuts off and the sexual pathway maintenance promoter, SxlPm, turns on in both sexes and remains on thereafter. In contrast to transcripts from SxlPe, those from SxlPm are processed into mRNA that encodes more Sxl-f protein only if Sxl-f protein is already present to bind to SxlPm pre-mRNAs. This binding directs the splicing machinery to skip over the male-specific exon 3 that would otherwise introduce a translation stop signal into the N-terminal coding region of the Sxl-f ORF. In this way, the pulse of Sxl-f protein generated by transient expression of SxlPe in chromosomal females engages a positive feedback loop for SxlPm transcript splicing that epigenetically maintains the feminizing mode of Sxl expression thereafter. In chromosomal males, that feminizing mode of expression remains off by default, since males lack the initial pulse of Sxl-f protein required to engage the splicing positive feedback loop; hence, their corresponding SxlPm-derived mRNAs contain the translation-terminating exon 3. Since Sxl is an X-linked positively autoregulating gene, its dose is necessarily higher in diplo-X than in haplo-X animals, and that dose difference contributes to the fidelity of the sex-determination process. As a consequence, Sxl functions both as an XSE and as the target of the XSEs.
Figure 1.—
Molecular steps in the control of Drosophila sex determination by Sxl. Details of this process are described in the Introduction.
In D. melanogaster, Sxl-f proteins are known to direct all aspects of female development, including the vital process of X chromosome dosage compensation. They do so by binding to the RNA products of downstream gene targets, of which transformer (tra) and male-specific-lethal-2 are known. Because Sxl controls dosage compensation, strong loss-of-function Sxl alleles are recessive female-specific lethals, while strong gain-of-function alleles are dominant male-specific lethals. The fact that melanogaster males lacking Sxl are fully viable and fertile (Salz et al. 1987) is the strongest evidence that this key feminizing switch gene is functionally female specific in this species.
Sxl orthologs have been identified in insects as diverse as aphids, moths, beetles, and mosquitoes; however, outside of the family Drosophilidae, there seems to be no obvious somatic difference in Sxl expression level or products between the sexes and/or no DNA homologies to the key structural elements in Drosophila Sxl that are responsible for its female-specific functioning (referenced in Traut et al. 2006; and see Siera and Cline 2008 for Scaptodrosophila lebanonensis). In most insects, the transformer gene seems to occupy the position of Sxl as the master feminizing switch gene that responds to sex determination signals and epigenetically maintains the sexually determined state through a positive feedback loop on pre-mRNA splicing (Pane et al. 2002; Gempe et al. 2009; Hediger et al. 2010; Verhulst et al. 2010). In Drosophila, tra is a downstream target of Sxl and seems to have no direct positive autoregulatory character of its own. Thus on the basis of this evidence, the evolution of Sxl into a female-specific switch gene appears to have occurred over a relatively short evolutionary time (∼10 MY) as medfly and fruit fly ancestors went their separate ways and X chromosome number emerged as Drosophila's primary sex-determination signal. However, the extent to which Sxl functioning continued to change as various Drosophila species diverged was unknown, since Sxl mutant phenotypes had been determined only for D. melanogaster. In the present study, we subject Sxl in D. virilis to classical genetic analysis to determine whether Sxl regulation and functioning in these two distantly related Drosophila species are as similar as many have assumed purely on the basis of gene structure and expression. This is not a trivial point since, as we discuss, there is an important difference between melanogaster and most other Drosophila species including virilis that lies at the heart of the machinery for Sxl sex-specific expression.
The female-specific phenotype of loss-of-function mutations in D. melanogaster Sxl raises the question of whether Sxl's elevation to master sex-determination gene in the ancestors of this species was accompanied by the loss of vital ancestral, non-sex-specific functions. Here we report our exploration of this question from three complementary angles. Together these studies have led us to believe that this feminizing switch gene is not as functionally female specific as its null phenotype would suggest. While any functions that are not female specific would clearly have to be subtle, our experience with other equally subtle aspects of Sxl functioning that involve the regulatory relationship between Sxl and tra (Siera and Cline 2008) has shown that the least obvious aspects of a gene's activities may provide important clues to a gene's evolution.
First, we determined whether Sxl's closest paralog, CG3056, has a vital, non-sex-specific function that might reflect the ancestral, non-sex-specific role played by Sxl before the duplication generating this paralog occurred. On the basis of an estimate of when that event took place (Figure 2), it was hypothesized that Sxl was freed by this duplication to acquire a role in sex determination that was incompatible with its non-sex-specific functions (Traut et al. 2006). We tested this hypothesis, as well as an alternative hypothesis that this paralog is functionally redundant with Sxl with respect to ancestral functions, by phenotypically characterizing a null allele of CG3056. We then named this paralog sister-of-Sex-lethal (ssx).
Figure 2.—
The phylogenic relationship among Diptera of Sxl and its closest paralog ssx (CG3056). With a phorid fly (M.sca) Sxl as the outgroup among the “higher” Diptera (Brachycera), we aligned Sxl and CG3056 (sister-of-Sex-lethal) with respect to the two RRMs and 11 residues immediately downstream, as well as the 27 (in melanogaster) C-terminal residues corresponding to the exon 8 isoform of D.mel Sxl. See materials and methods for details. A previous estimate of when the paralog-generating Sxl duplication occurred (traut et al. 2006) could say only that it was sometime after the point indicated by the open arrow. Our analysis placed that duplication later (solid arrow, bootstrap value 99%), closer to the time at which Sxl became a master sex-determining switch gene. The evolutionary divergence scale bar is in substitutions per nucleotide. D.mel, Drosophila melanogaster; D.vir, D. virilis; M.dom, Musca domestica (house fly); Ch.Ruf, Chrysomya rufifacies (blow fly); C.cap, Ceratitis capitata (medfly); M.Sca, Megaselia scalaris (scuttle fly).
Second, we determined whether the phenotypic consequences of eliminating Sxl by mutation in a Drosophila species relatively distant from melanogaster are as female specific as they are for melanogaster. If not all species of Drosophila have yet lost or transferred their ancestral non-sex-specific Sxl functions, the Sxl null phenotype in males of such species could suggest what those ancestral functions might be. Moreover, the phenotypes of mutant Sxl alleles could show how well conserved the regulation and functioning of this gene's female-specific activities are among Drosophila species. We chose D. virilis for this genetic analysis not only because of its considerable evolutionary distance from melanogaster, but also because the species group to which it belongs was shown to produce remarkably large amounts of nearly full-length Sxl protein in males, and its male-specific Sxl exon contains a potential translation initiation site that may be responsible (Bopp et al. 1996). Thus, if any Drosophila species had not yet lost their ancestral, non-female-specific Sxl functions, we expected virilis would be among them. Moreover, D. virilis also had a useful variety of X-linked genetic markers and an X chromosome structure that would allow us to generate an attached-X chromosome for the genetic analysis planned (Alexander 1976).
Third, through comparative genomics followed by RACE and RT–PCR, we discovered possible sources of the non-female-specific Sxl protein isoforms. Although melanogaster Sxl is generally thought of as an “on–off” developmental switch that operates by a mechanism that aborts translation of all Sxl proteins in males, it has long been known that even melanogaster males generate Sxl isoforms only slightly shorter than full-length Sxl-f proteins, albeit at levels estimated to be 20–40 times lower than those of Sxl-f isoforms (Bopp et al. 1991). Here we describe the first Sxl mRNAs known to be non-sex-specific and show them to include a highly conserved Sxl exon, not previously noted, that is located in the genome just upstream of the male-specific exon 3. Like exon 3, this new exon Z is alternatively spliced. The mRNA species that include exon Z are likely to encode at least some of the nearly full-length Sxl proteins found in Drosophila males. The fact that exon Z corresponds to an alternatively spliced, non-sex-specific exon in flies that seem not to use Sxl as a sex switch suggests that mRNAs including this exon may encode ancestral non-sex-specific Sxl activities that have not been lost by Drosophila. We suggest that exon Z may have contributed to the evolution of Sxl female-specific functioning by duplicating to form the neighboring exon 3. We believe that our study illustrates the principle that aspects of gene function that have no obvious phenotypic consequences when disrupted in the lab may nevertheless provide valuable clues to how functions that are obvious have evolved.
MATERIALS AND METHODS
Drosophila culture and genetics:
Flies were raised in uncrowded conditions on a standard cornmeal, yeast, sucrose, and molasses medium at 25° unless otherwise stated. Markers, balancers, and transgenes not mentioned in this section are described at http://flybase.org/ for melanogaster and https://stockcenter.ucsd.edu/info/welcome.php for virilis.
Phylogenetic analysis of Sxl and ssx:
The following sequences were used: ssx_D. mel (NM_130552.2), ssx_D. vir (XM_002058083 with exon 1 and exon 5 splice sites modified according to the genome data NW_002014442.1), Sxl_D. mel (NM_080052), Sxl_D. vir (XM_002056740), Sxl_C. cap (AF026145.1), Sxl_M. dom (AF025690.1), Sxl_Ch. Ruf (S79722.1), and Sxl_M. sca (AJ245662.1). Sequences were aligned using ClustalW in MegAlign. The alignable region includes RNA recognition motif (RRM)1 (79 aa for Sxl_D. mel, etc.), the linker between RRM1 and -2 (GGESIKD in Sxl_D. mel), RRM2 (80 aa for Sxl_D. mel, etc., or 81 aa for Sxl_M. sca), 11 amino acids after RRM2 (EHGKAKAAHFM in Sxl_D. mel), and a conserved C-terminal domain of 27 amino acids and a stop codon (MMHRGRSIKSQQRFQNSHPYFDAKKFI* in Sxl_D. mel). Phylogenetic analysis was performed in MEGA3.1. Neighbor-joining trees were constructed using cDNA sequences with the Kimura two-parameter distances on the first and second codon positions.
P-element excision alleles of ssx:
The starting chromosome was y1 P{w+mC y+mDint2 = EPgy2}CG3056EY14203 w67c23 and the transposase source was P{Δ2-3}99B. Excisions, recognized by loss of the transposon markers, were induced in the male germ line and recovered in balanced daughters, from which lines were generated. DNA from each line was screened in initial pools of 10 by long-template genomic PCR using the following primers flanking ssx: forward 1, 5′-AATATCGGAAGGGGGTTTGTTA-3′ (3.9 kb upstream of ssx); forward 2, 5′-CTGCCCGATCATTAGTGCTTGTCC-3′ (1.8 kb upstream of ssx); and reverse, 5′-GAGGGGAGCGGTGGTAAGGTCGTT-3′ (1.1 kb downstream of ssx).
Precise excision events generated 11,011- and 7100-bp fragments with F1/R and F2/R primer pairs, respectively. Deletions were signaled by the appearance of smaller bands, separated on a 0.6% agarose gel. From 57 independent excision lines, deletions within the amplified area were found for 4 lines. DNA sequencing defined the deletion breakpoints and also identified a precise excision line to serve as a wild-type control.
Fecundity determinations for ssx mutant animals:
For females, single virgins were mated 12–24 hr after eclosion to four aged Canton-S males. Progeny were collected for a total of 10 days with two transfers of parents to fresh media. For males, total progeny were counted from the matings of single newly eclosed males to three successive pairs of aged Canton-S virgins. Males were removed from the first females after 4 days, and those females were allowed to lay for two more 3-day collections. Males were removed from their second mates after 3 days, and those females laid for two more 3-day collections. Males were removed from the third batch after 3 days and those females laid for two more 3-day collections.
Precise duplication of Sxl+ via transgene DNA gap repair:
Following the technique of Takeuchi et al. (2007), we generated P-element–mediated insertions of a transgene that carried two fragments of genomic DNA from the flanks of Sxl, joined by an I-SceI nuclease target site. Neither of the flanking fragments contained a functional gene or any part of the Sxl transcription unit. The fragments provided the homology necessary so that a cut induced at the I-SceI site would stimulate in vivo repair using the endogenous Sxl region as a template, thereby duplicating Sxl but no other gene. The centromere distal 3468-bp genomic fragment (X: 6,966,160–6,969,628) included the 3′ 17% of Sxl's nearest distal neighbor, CG4607, while the centromere proximal 3,389-bp fragment (X: 6,992,119–6,995,507) included the 3′ 79% of Sxl's nearest proximal neighbor, CG4615. The distal fragment ended 69 bp from Sxl's most distal 3′ end, while the proximal fragment ended 32 bp from Sxl's 5′ end. Thus fully templated gap repair would require that all of the missing 22,489 bp between these regions of homology be filled in, all but 101 bp of which belonged to Sxl. Functionally wild-type Sxl duplications were obtained from two homozygous viable and fertile chromosome III template target transgene insertion lines as described below.
For the successful conversion attempt, we subjected each y w/w cm Sxlf7,M1 ct v; P{ryt7.2=hsp70FLP}11 P{v+t1.8-hsp70-ISce1}2B Sco/+; P{I-SceI target, w+mC}2 or 9/+ developing female to three 45-min 38° heat shocks [72–96 hr after egg laying (AEL), 96–120 hr AEL, and 104–138 hr AEL] to induce the I-SceI nuclease. These females were then mated, one or two per vial, to five y w cm Sxlf1ct sn/Y males and their progeny were collected for 8 days. From an estimated 336 fertile Sxl+/Sxlf7,M1 mothers heterozygous for transgene insertion line 2, we screened an estimated 7800 Sxlf7,M1/Sxlf1 transgene-carrying zygotes for their ability to develop into fertile females. For line 9, the corresponding numbers were 392 mothers and 14,100 zygotes. Only gap repair using the Sxl+ template in the Sxl+/Sxlf7,M1 mothers would yield fertile Sxlf7,M1/Sxlf1 daughters; hence, if one assumes that the homology search for DNA gap repair is not biased against Sxlf7,M1 by the 9.5-kb roo transgene it carries (Bernstein et al. 1995), the number of zygotes screened for Sxl+ duplications is effectively only 50% of the numbers given. Four independent Sxl+ transgenes were recovered, two from each line. One of the four conversion events generated a cluster of rescued progeny, indicating a germ-line clone.
Generation of the 2XSE D. virilis transgenes:
The extent of the minimal genomic fragment providing full sc XSE function for melanogaster was known (Erickson and Cline 1991). From virilis, we cloned and sequenced a 20-kb genomic region containing the sc transcription unit and identified a 9.9-kb region within it that corresponded to the 5.1-kb melanogaster minimal XSE region. For sisA, a 6.4-kb genomic fragment from melanogaster that also carried the neighboring upstream gene was known to provide full XSE function (Erickson and Cline 1993), but the size of the minimal fully rescuing sisA fragment was not determined. Since the sisA transcription unit is <1 kb, with <100 bp upstream of the protein-coding region and only a little >100 bp downstream, we gambled that full sisA function would be provided by a 4.3-kb fragment from melanogaster that included 1.7 kb upstream and 2.0 kb downstream of the sisA transcription unit and a 4.3-kb virilis fragment with 3.0 kb upstream and 0.7 kb downstream. The sc and sisA genes in both transformation constructs were oriented tail to tail. As expected, the transformation frequency in D. virilis (G0 flies with w+mC progeny) for the 9.5-kb transformation vector alone (eight flies = 4%) was higher than that for the 18.6-kb mel2XSE construct (five flies = 1%). The 23.5-kb vir2XSE construct generated transformants even less frequently (one fly = 0.2%).
The Minos transformation vector was pMiw1 (Loukeris et al. 1995a) modified by cleanly replacing the w-promoter–driven mini-white marker that had been inserted at an EcoRI site by a 4.5-kb EcoRI fragment from pW8 (Klemenz et al. 1987) that contained the mini-white gene driven by the hsp70 promoter instead. The Mi{mel/sisAsisB,w+mC} transgene carried a 5.1-kb EcoRI-XbaI sc fragment from pJEP200 (Erickson and Cline 1991) and a 4.2-kb ScaI sisA fragment from pJE301 (Erickson and Cline 1993) inserted at a NotI site just upstream of the w+marker. The Mi{vir/sisAsisB,w+mC} transgene carried a 9.9-kb BamHI-XmnI sc fragment (recovered from genomic virilis DNA libraries) and a 4.2-kb EcoRV-EcoRI sisA fragment from pSF1 (Erickson and Cline 1998) inserted at the same NotI site. Standard techniques were used for germ-line transformation (Spradling 1986), although we found it essential for virilis that the parents generating transformation host w− embryos be kept highly outbred and be allowed to lay in complete darkness. Laying by parents aged at least 1 week after eclosion was at 25°, while DNA injection was at 18°. The Minos transposase source was pHSS6hsMi2 (Loukeris et al. 1995a). Vector and transposase DNA were mixed together in injection buffer (5 mm KCL and 0.1 mm Na Phosphate, pH 6.8) at a final concentration of 800 and 200 ng/μl, respectively.
Surprisingly, the hsp∷w+mC eye color marker, which reliably exhibits dose effects in melanogaster, failed to do so in virilis for autosomal insertions, even though eye colors of the transgenes at different sites were different and all lighter than wild type. Lightening eye color further with sepia or peach mutations did not reveal any cryptic dose effect. In light of this, it was even more puzzling that X-linked hsp∷w+mC insertions displayed both a dose effect within a sex and dosage compensation between the sexes: in a w mutant background, the eye color of one-copy males was the same as that of two-copy females and darker than that of one-copy females.
Generation and maintenance of the D. virilis attached-X chromosome:
Since mutant alleles of the X-linked virilis white (w) gene were available, we could easily recognize a new compound X chromosome by the altered pattern of w allele inheritance that it would cause. Consequently, we exposed w/w virgin females to gamma rays (1.7 to 3.3 kR) 7–9 days after eclosion and then mated them en mass to w+ males. Their rare matroclinous exceptional w/w daughters were crossed to w+/Y males, and their progeny were examined for the telltale reversed pattern of X-linked inheritance. Of 39 exceptional females recovered among ∼18,000 w+ daughters, 1 showed the expected attached-X pattern. The one compound-X chromosome recovered is somewhat unstable, but we designed a stock that appears to self-select against its breakdown: Mi{mel/sisAsisB,w+mC}X1 y virSxlf1 cv w/Y & C(1),w/Y; Mi{mel/sisAsisB,w+mC}A1. Although this attached-X chromosome proved to be useful, the particular X-linked genetic background that the rearrangement kept intact prevented us from using this new chromosome in the first step of our screen for suppressors of XSE-induced male lethality. The maternal genetic background effect of this chromosome allowed 14% of X1/Y; A1/+ sons to survive relative to their X1/Y; +/+ brothers (n = 479) at 18°, in contrast to 0.13% survival for the same males generated by the scheme illustrated in Figure 3.
Figure 3.—
A positive genetic selection scheme for mutations in D. virilis Sxl as suppressors of XSE-duplication–induced male lethality. Each Minos (Mi) transgene carried a copy of D. melanogaster sc and sisA in tandem,. The w+ marker on the X-linked transgene X1 generated orange eyes, while that of the autosomal transgene A1 generated red eyes. F0 males were exposed to 2700 rad. Of the 39,000 F1 females, 50% were mated in groups of 10, 20% in groups of 5, and 30% in groups of 2. The attached-X chromosome C(1)w was generated for the purpose of this scheme.
Molecular mapping of virSxl mutant lesions:
Each line was subjected to a panel of PCR primer pairs that would potentially amplify all the DNA between SxlPm and the end of the long form of exon 8. DNA corresponding to each of the exons was then sequenced. The location of indels in virSxlf2 and virSxlf3 was further localized with additional PCR primer pairs, and in the case of virSxlf2 the sequence of the genomic fragment spanning the deletion was determined to be …TCAAATGAGTGTTCT|||CACAAATATCCCAGATGAAAA.
Western blots of virSxl male protein:
Ten testes or heads were dissected, resuspended in 100 μl of 1× protein loading dye, and physically disrupted. The crude extracts loaded correspond to 0.25 testes or 0.5 heads. The nitrocellulose was probed with either polyclonal rabbit anti-Sxl antibody against the two RRMs (used for the antibody in Blanchette et al. 2009; construct in Lee et al. 1997) at a 1:5000 dilution or DM1A anti-α-tubulin (Sigma, St. Louis) at a 1:10,000 dilution. For detection of the Sxl antibody, an HRP-conjugated protein A secondary (GE Lifesciences) was used at 1:5000. For detection of tubulin, HRP-conjugated anti-mouse secondary (Bio-Rad, Hercules, CA) was used at 1:5000.
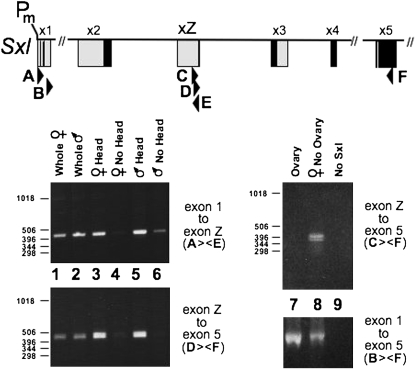
Identification and characterization of exon Z mRNAs:
We initially used 5′- and 3′-RACE on poly(A)+ RNA from adult females to discover the exon 1/exon Z/exon 4 structure of exon Z mRNAs. We then sequenced cloned RT–PCR products to discover that the corresponding mRNAs were present in males in a variety of species besides melanogaster. For Figure 7, the initial RT–PCR reactions were on total RNA primed from exon 8 (5′-GAAATGGCCTCCTGGGCCTCCTCACG-3′). The PCR primers used subsequently on these RT products are listed below, and their products were separated on a 2% agarose gel: A, 5′-ACTGCTTTGTTGTTGCCGAAGAAG-3′; B, 5′-CCCATGCAATCCGTGTAGCTACAG-3′; C, 5′-TCTCATCGTGCGGATTGTGCAAC-3′; D, 5′-CCTAAACAGTCTCACAATGTACCG-3′; E, 5′-CGGTACATTGTGAGACTGTTTAGG-3′; and F, 5′-CGGTCATGTCCTGGGGCAAG-3′.
Figure 7.—
Analysis of D. melanogaster mRNAs showing that exon Z is not sex specific and is disproportionately expressed in heads. Primers for this RT–PCR analysis are positioned on the Sxl schematic and described in materials and methods. The pair used for each gel is indicated. One mRNA preparation was used for the two gels on the left and another for the two gels on the right. Estimated sizes of the bands are consistent with expectations for Exon Z mRNAs being generated in both sexes by a 1-Z-4-5 splicing pattern that skips the male-specific exon 3. Predicted sizes of the various RT–PCR products are presented in materials and methods. The CF primer pair indicated that exon Z mRNA is not present in D. melanogaster ovaries. The same was found to be true for the scuttle fly (Sievert et al. 2000).
The expected ranges in size of the expected PCR products are as follows:
1-Z: 422–560 bp (AE), depending on which exon 1 5′ ss is used. The data in Figure 7 exclude the alternative of 871–1057 bp expected for inclusion of exon 2. The data support preferential use of the exon 1 5′ splice site (ss) at +53 (cDNA MS3 in Samuels et al. 1991) in exon Z mRNA, rather than that at +191 discovered in our 5′-RACE studies; however, data for the BF primer pair in Figure 7 argue that the previously unreported +191 site is used.
Z-4-5: 335–359 bp (DF) or 357–381 bp (CF), with the differences reflecting use of the alternative 3′ splice sites in exon 5. The data exclude alternatives of 507–549 bp and 529–571 bp respectively expected for inclusion of the male-specific exon 3.
1-(2-±3)/(Z)-5: 713–863 bp (BF) with the differences reflecting the mutually exclusive splicing to exon 2 vs. Z, as well as the alternative splice acceptor sites in exons 2 and 5. Note that primer B runs from +163 to +186, well downstream of the standard exon 1 splice donor site at +53, but just upstream of the +191 donor site we describe in this article. The fact that the BF pair generate RT–PCR products is additional evidence that the +191 site is used.
RESULTS
The closest paralog of Sxl, sister-of-Sex-lethal, is a nonessential gene that interacts only very weakly with Sxl:
Traut et al. (2006) estimated that the closest paralog to Sxl, CG3056, was generated by a duplication event that occurred some time after the separation of the two suborders of Diptera, the Brachycera (the “higher” Diptera, which includes Drosophila) and Nematocera (which includes mosquitoes and midges). They suggested that this event may have allowed Sxl to lose its ancestral functions because those functions could be maintained by CG3056 from that point on. Using additional regions of sequence conservation that were not included in the previous analysis (see materials and methods), we pinned down the timing of this duplication event further. We found that it likely occurred well after the separation of the Acalypterate and Calypterate subsections of the Schizophora and hence remarkably close to the time at which Sxl appears to have acquired its sex-determining function (Figure 2).
If the Traut et al. hypothesis were true, the phenotype of null mutations in CG3056 could reveal ancestral Sxl functions. Another possibility is that the two genes remained partially redundant because neither one completely lost the non-female-specific ancestral Sxl functions. In that case, both genes would have to be knocked out to expose those ancestral functions. To explore these possibilities, we generated and characterized a null allele of CG3056. We subsequently named the gene sister-of-Sex-lethal (ssx).
We generated deletions of ssx by imprecise excision of a P-element transposon in the gene's first intron (supporting information, Figure S1). All deletions extended into or beyond the upstream (centromere distal) neighbor, CG14770, a small gene with no recognized sequence motifs and no known function. The deletion most likely to be a ssx null was Df(1)ssx55. It eliminated DNA on both sides of the P-element insertion (3268 bp total). The deletion extended upstream to just beyond CG14770 and downstream into ssx exon 6, eliminating half of the ssx open reading frame and destroying the gene's two RRMs, anticipated to be the functional heart of the gene, by analogy to Sxl. We also recovered a precise excision allele, ssx+Rev26, to serve as a wild-type control in our studies.
We found that loss of ssx+ and its upstream neighbor had no adverse effect on viability or fecundity in either sex, even in combination with mutations in Sxl (Table 1). The 89% number for ssx− female viability in cross A was not significantly lower than the 97% number for ssx+Rev/ssx− females in the control cross B (χ2 P = 0.15). The corresponding comparison of male viability likewise showed no significant decrement (92% vs. 96%, χ2 P = 0.48). Female fecundity was no lower for the mutant females than for their nonmutant sisters (Mann–Whitney P = 0.31). Although the mutant males were somewhat less fertile than their control sibs (76%), the fact that they were not significantly less fertile than the ssx+Rev male controls (90%, Mann—Whitney P = 0.25) allowed us to attribute that small difference to extraneous factors on the parental chromosome.
TABLE 1.
Phenotypic analysis of a sister-of-Sex-lethal null mutant allele
| Crossa | Genotype | % relative viability (no.) | % relative fecundityb (no. vs. control) | Control sibs for viability and fecundity determination |
|---|---|---|---|---|
| A | ssx−/ssx−♀ | 89 (940) | 102 (7 vs. 10) | +/ssx−♀ |
| A | ssx−/Y ♂ | 92 (913) | 76 (12 vs. 10) | +/Y ♂ |
| B | ssx+Rev/ssx−♀ | 97 (1075) | ND | +/ssx−♀ |
| B | ssx+Rev/Y ♂ | 96 (1031) | 90 (12 vs. 12) | +/Y ♂c |
| C | ssx−Sxl−/ssx− Sxl+♀ | 75 (376) | 96 (9 vs. 12) | ssx−Sxl−/ssx− +; Dp(Sxl+)/+♀ |
| C | ssx−Sxl−/ssx− Sxl+; Dp(Sxl+)/+♀ | 102 (504) | ND | Balancer/ssx−♀ |
| C | ssx− Sxl−/Y ♂ | 90 (462) | 139 (15 vs. 14) | ssx−Sxl−/Y; Dp(Sxl+)/+ ♂ |
| D | ssx− Sxl−/ssx+Rev +♀ | 85 (377) | 76 (11 vs. 11) | ssx−Sxl−/ssx+Rev +; Dp(Sxl+)/+♀ |
Full genotypes of crosses: (A) y ssx−55 w/+ ♀♀ × ♂♂ y ssx−55 w/Y; (B) y ssx+Rev26 w/+ ♀♀ × ♂♂ y ssx−55 w/Y; (C) Binsinscy/y ssx−55 w cm Sxlf1 ct; P[Sxl+w+mC]2A/+♀♀ × ♂♂ y ssx−55 w/Y; (D) same mothers as C × ♂♂ y ssx−+Rev26 w/Y.
Ratio of the median progeny recovered per parent to that for the control sibs.
In this one case, the controls for fecundity determination were cousins from cross A rather than sibs.
Loss of ssx+ was not deleterious even in combination with loss of Sxl+. While the viability of ssx−/ssx− mutant females that were also heterozygous for Sxl− was somewhat lower than that of their Sxl+/Sxl+ sisters (75%, χ2 P < 0.001), the fact that they were not significantly less viable than the Sxl−/Sxl+ females in cross D that were ssx−/ssx+Rev (85%, χ2 P = 0.36) showed that the difference in viability between the sisters in cross C simply reflected a commonly encountered weak dominant female-lethal effect of Sxl−, notably an effect shown here not to be significantly enhanced by the loss of ssx+. As expected, ssx− females from cross C that were not also heterozygous for Sxl−were as viable as their ssx+/ssx− sisters (102%). Heterozygosity for Sxl− also did not reduce the fecundity of ssx− females (96% vs. the control sisters). For ssx− males, neither viability (cross C, 90%, χ2 P =0.08) nor fecundity (139%) was significantly reduced by the loss of Sxl+. Even when the five ssx− to ssx+Rev comparisons were considered together, there was no statistically significant bias against ssx− (P = 0.19 by the “sign test”).
To have the best Sxl+ internal controls for these experiments, we took advantage of Sxl+ duplications that we generated by the DNA-gap–induced gene conversion method of Takeuchi et al. (2007) (see materials and methods). These duplications fully rescue Sxl− female viability and fertility but have the important advantages over previously available duplications of not affecting the dose or functioning of any other gene except white (the transgene marker) and of allowing randomization of genetic background on the duplication-bearing chromosome. With such duplications, one can increase Sxl+ dose almost without limit, a feature likely to be important in future studies of Sxl germ-line functioning (Hager and Cline 1997).
Although loss of ssx+ did not enhance the semidominant female-specific lethal effect of Sxl−, a weak relationship between ssx functioning and the female-specific functioning of Sxl was revealed by an effect of the ssx null on SxlM12 males. M12 is a gain-of-function Sxl mutation so weak that it does not reduce male viability on its own, but does cause etching of distal tergites due to abdomen-specific upsets in dosage compensation that probably occur late in development (Cline et al. 1999). This etching caused by low-level female splicing of Sxl pre-mRNA in males is extremely sensitive to anything that increases or decreases Sxl-positive autoregulation. Comparing the phenotype of SxlM12/Y sons of ssx− SxlM12/ssx+ SxlM12 mothers, we found that while 92% of the ssx+ sons (n = 82) had one or more etched abdominal tergites 5 or 6, only 16% of their ssx− brothers (n = 67) displayed this abnormality. This suppression of SxlM12 suggests that loss of ssx+ reduces Sxl-positive autoregulation and hence that ssx function is at least weakly related to that of Sxl. How direct this relationship may be is unclear. Of course, it is formally possible that the effect is due to the simultaneous deletion of ssx's upstream neighbor.
Function and regulation of virilis Sxl and melanogaster Sxl are similar despite their evolutionary distance and difference in male Sxl protein expression:
To further explore the evolution of Sxl female-specific functioning, we determined whether Sxl is as female specific in D. virilis as it is in melanogaster. In this effort, we used the most definitive test possible: isolation and characterization of a virilis null Sxl allele. We also used mutations in virilis Sxl (virSxl) to determine whether virilis females require Sxl for both dosage compensation and sex determination, as do melanogaster females. Since it seemed likely that regulation of virSxl expression by X chromosome dose would be similar to that known for melSxl (Bopp et al. 1996; Erickson and Cline 1998; and see Jinks et al. 2003), we gambled that a strategy we used for isolating loss-of-function Sxl mutations in melanogaster would work in virilis: selecting for suppressors of the effects of inappropriate expression of feminizing Sxl-F protein in males by duplications of XSEs (Sefton et al. 2000; Wrischnik et al. 2003). If virSxl were also required for essential non-sex-specific functions, our strategy might yield only partial loss-of-function mutant Sxl alleles. If virSxl were required for sex determination but not dosage compensation, we would expect to see that increasing XSE dose feminized males but did not kill them. For this genetic selection, we needed to generate virilis transgenes with which we could increase XSE gene dose, and we anticipated that a virilis attached-X chromosome would be tremendously useful for maintaining mutant lines.
Generating a virilis attached-X chromosome:
We were successful in generating a compound-X chromosome (see materials and methods) that we anticipated would facilitate both the generation and the maintenance of virSxl mutants and prove useful to others contemplating virilis genetic analysis. Since this new chromosome necessarily lacks the design features that contribute to the stability of melanogaster attached-X chromosomes, it does spontaneously break down, but not at a rate that seriously detracts from its utility in experimental crosses. Moreover, we subsequently exploited our virilis transgenes and Sxl mutant alleles to design a stock that self-selects against such breakdown events, thereby facilitating maintenance of this genetic tool (see materials and methods).
The male-lethal effect of increased XSE dose in D. virilis reveals evolutionary conservation of Sxl regulation:
To increase the dose of XSEs in D. virilis, we exploited the Minos transposon system. Minos has already been used to transform the germ lines of D. melanogaster and the medfly (Loukeris et al. 1995a,b). We designed transformation constructs that carried the XSEs scute (sc) and sisterlessA (sisA) together in tandem. One version of the “2XSE” construct had the XSE pair from virilis (vir2XSE) while the other had the melanogaster pair (mel2XSE).
The one vir2XSE transgene recovered caused some virilis male-specific lethality in a single copy, but only one of the five mel2XSE transgenes did. In neither case was the effect sufficient to serve reliably for the planned genetic selection. Although vir2XSE/+ males were only 8% as viable as their +/+ brothers overall (n = 1097, 25°), there was considerable variation among single-female crosses. Although this variation was presumably due to genetic background differences, attempts to reduce those differences by inbreeding were unsuccessful. One copy of the most potent mel2XSE transgene reduced virilis male viability only to 60% overall (n = 453 sibs, 25°), and again there was considerable variation among single-female crosses.
Fortunately for the purposes of the planned suppressor screen, the 2XSE transgenes synergized: the viability of males carrying a copy of two different transgenes was far below that of males carrying only either one. Even with two copies present, however, the magnitude of the male-specific viability effect depended strongly on temperature and genetic background. Curiously, the temperature dependence of the vir2XSE transgene was opposite to that of the mel2XSE transgenes, with the former causing less male lethality at 18° and the latter less at 25°. Consequently, for the Sxl mutagenesis effort described in Figure 3, we settled on a male-lethal combination of two mel2XSE transgenes at 18°: an X-linked transgene (X1) whose w+ marker generated orange eyes and an autosomal transgene (A1) that generated red eyes, with red being epistatic to orange.
In a cross of mel2XSE-X1 females to mel2XSE-A1/+ males, the viability of mel2XSE-X1/Y; mel2XSE-A1/+ sons (red) relative to their mel2XSE-X1/Y; /+ control brothers (orange) was only 0.13% overall at 18° (n = 1685 control males). Moreover, all escaper males were missing part or all of their terminalia. We refer to this as the “Mickey Mouse” phenotype and have encountered it previously among escaper melanogaster males who survived despite carrying extra copies of XSEs (Cline 1988; Erickson and Cline 1993). For melanogaster we concluded that these malformations were due to upsets in dosage compensation rather than perturbations of sexual differentiation, since the phenotype was not suppressed by eliminating Sxl's downstream feminizing target, transformer (data not shown).
The spectrum of mutant virSxl alleles recovered as suppressors of XSE-induced male lethality reveals evolutionary conservation of Sxl function:
The scheme shown in Figure 3 yielded four independent suppressed lines from ∼40,000 F1 irradiated females. Suppression in three of the four lines was complete, and those three suppressors roughly mapped to crossveinless (cv), a marker near virSxl at cytological position 4D1–3 (Bopp et al. 1996). The mel2XSE-X1 transgene itself mapped near yellow (y: >22 cM distal to cv). The fourth suppressor rescued only 25% of the X1/Y; A1/+ males. It proved to be autosomal and was discarded due to difficulty keeping the line.
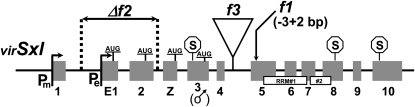
All three X-linked suppressors proved to be mutations in virSxl (Figure 4). The change in the first allele, virSxlf1, is expected to destroy all Sxl functioning. It substitutes 2 bases for 3 in a region of exon 5 present in all Sxl mRNAs. The change from …gaa ttT ACa ttt cca… to …gaa ttA Aat ttc ca… shifts the open reading frame and introduces a premature stop codon before the region coding for the highly conserved RNA-binding heart of Sxl that is present in all Sxl isoforms (RRM1 and RRM2 in Figure 4). Consistent with expectations for a null allele, the Western blot of virSxlf1 male extracts illustrated in Figure 5 shows that this mutation eliminates virSxl protein. The second allele, virSxlf2, carries a 6.8-kb deletion whose left breakpoint is 1440 bp downstream of the SxlPm start site and therefore leaves SxlPm and its associated first exon intact. Its right breakpoint is 115 bp downstream of exon 2. Hence this allele is missing SxlPe and its associated exon E1, as well as exon 2 (see materials and methods for sequence across the breakpoints). Although virSxlf2 eliminates the only established translation start sites for Sxl in females, it leaves intact both the hypothesized translation (re)start in the male-specific exon 3 (see discussion) and the putative start site in a previously unknown exon (Z) described below. This allele seems to modestly increase the amount of virSxl proteins that males make without affecting their molecular weight (Figure 5). The third allele, virSxlf3, carries a 2.5-kb insertion of uncharacterized DNA between exons 4 and 5.
Figure 4.—
Lesions in three new D. virilis Sxl mutant alleles. The exon/intron structure of Sxl is shown. Exon 3 is male specific. Established (exons E1 and 2) and proposed (exons Z and 3) translation start sites are labeled “AUG.” Translation termination sites are labeled S. RRM1 and -2 refer to the two RNA recognition motifs. The frameshifting lesion in vSxlf1 is predicted to destroy all Sxl functions, and the genetic behavior of this allele is consistent with it being a null.
Figure 5.—
Western blot of Sxl proteins from wild-type and Sxl mutant D. virilis male heads and testes. No Sxl proteins were found in extracts of adult males hemizygous for the predicted null allele, vSxlf1 (vF1). Sxl proteins from males hemizygous for the intragenic deletion allele vSxlf2/Y (vF2) matched those from wild-type males (vWT) in mobility, but seemed somewhat more abundant. Note the difference between testes and heads with respect to the male Sxl proteins generated. The doublet for Sxl is likely due to the use of alternative exon 5 3′-splice sites (see Figure 6) (Bopp et al. 1991).
Wild-type virilis males make a Sxl protein in testes that appears to be different from those made in male heads and considerably less abundant (Figure 6). The fact that this testes protein is also eliminated by virSxlf1 shows that it is a bona fide product of virSxl. In melanogaster, testes were reported to lack Sxl protein (Hager and Cline 1997), but it may simply have been missed due to the much lower abundance of male Sxl proteins in that species.
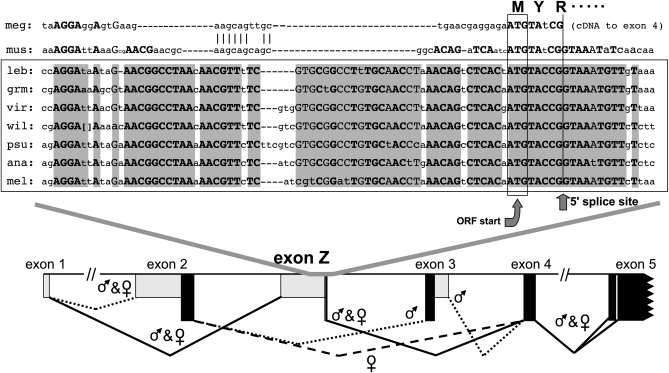
Figure 6.—
DNA sequence conservation near the 5′-splice site of the newly discovered Sxl exon Z. Uppercase letters in boldface type indicate positions completely conserved in a diverse collection of six Drosophila and one Scaptodrosophila species [namely, Scaptodrosophila lebanonensis (leb) and the Drosophila species grimshawi (grm), virilis (vir), wilistoni (wil), pseudoobscura (psu), ananassae (ana), and melanogaster (mel)] (Drosophila 12 Genomes Consortium 2007; Siera and Cline 2008). Uppercase letters in regular type signify positions at which all but one of the seven sequences are identical. For D. wilistoni, the sequence CTCTCTGTAAAGAG was present between the brackets. The boxed ATG is the only potential translation start site for each species that would encode nearly full-length Sxl proteins from mRNAs containing exon Z. A corresponding region was apparent in a cDNA from the scuttle fly, Megaselia scalaris (meg) (Sievert et al. 2000) and in genomic DNA from the housefly, Musca domestica (mus) (our sequence). The splice site shown for Megaselia exon Z is based on the authors' revised genomic sequence (AF110846.1) near exon 4 (only cDNA sequence was available near exon Z). The schematic in the bottom section is for D. melanogaster and shows the sex in which the splice sites for each of the indicated Sxl exons are active. Solid bars indicate protein-coding regions. Note that exon Z mRNAs are made by skipping the male-specific exon 3 in both sexes (see Figure 7).
We removed the 2XSE transgene from the Sxl mutant X chromosomes so that we could determine the phenotype of these Sxl alleles in an otherwise normal genetic background. The viability results in Table 2, cross A, show that the putative null allele, virSxlf1, behaves just like a melanogaster null allele: recessive lethal for females, but fully viable in males. Moreover, the mutant males were fully fertile (data not shown). Indeed there was no significant difference (χ2 P = 0.37) in male viability among all three of the virSxl mutant alleles and the virSxl+ parental y cv w chromosomes (Table 2, bottom four rows). virSxlf2 was also recessive, female-specific lethal (cross B), but virSxlf3 females (cross C) were fully viable (and fertile). Nevertheless, virSxlf3 was clearly deficient for Sxl function: no more than 1 in 1000 f3/f1 females survived (cross D). The one apparent escaper female in this cross was likely a matroclinous exception (i.e., f3/f3/Y).
TABLE 2.
Viability of vSxl mutant animals
| virSxl genotype (y cv w except § = w) | % relative viability | Control sibs for viability determination (y cv w except § = w) |
||
|---|---|---|---|---|
| Crossa | vSxl genotype | No. | ||
| A | +/f 1 § ♀ | 101 | +/Y § ♂ | 174 |
| A | f 1/f 1♀ | 0 | f 1/Y ♂ | 179 |
| B | f 2/f 2♀ | 0 | f 2/Y ♂ | 80 |
| C | f 3/f 3♀ | 100 | f 3/Y ♂ | 125 |
| D | f 3/f 1♀ | 0.1 | f 3/Y ♂ | 1275 |
| E | f 3/f 2♀ | 9 | f 3/Y ♂ | 2076 |
| E | + f 3/Mi{2XSE}X1 f 2♀ | 28 | f 3/Y ♂ | 2076 |
| F | +/Y ♂ | 79 | +/Y § ♂ | 73 |
| A | f 1/Y ♂ | 103 | +/Y ♂ | 174 |
| B | f 2/Y ♂ | 88 | +/Y ♂ | 91 |
| C | f 3/Y ♂ | 80 | +/Y ♂ | 157 |
Full genotypes of crosses: (A) y virSxlf1 cv w/+ + + w ♀♀ × ♂♂ y virSxlf1 cv w/Y; (B) same as A but virSxlf2; (C) same as A but virSxlf3; (D) y virSxlf3 cv w ♀♀ × ♂♂ y virSxlf1 cv w/Y; (E) Mi{mel/sisAsisB, w+mC}X1 y virSxlf3 cv w/y virSxlf3 cv w ♀♀ × ♂♂ y virSxlf1 cv w/Y; (F) y cv w/+ + w ♀♀ × ♂♂ y cv w/Y.
The viability of f3/f2 females (cross E) was nearly two orders of magnitude higher than that of f3/f1 females, though still not wild type. This difference established that virSxlf2 is not a null allele, despite lacking both previously known translation start sites for female mRNAs. Complementation between virSxlf3 and virSxlf2 was stimulated more than threefold by a 2XSE transgene (cross E). This XSE dose effect is likely acting on 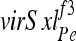 , since virSxlf2 has no SxlPe. The effect suggests that virSxlf3 is specifically defective in its sexual pathway “establishment” function–the process by which X chromosome dose leads to the engagement of the Sxl positive feedback loop. This conclusion is also supported by the molecular similarity between virSxlf3 and the melanogaster mutant Sxlfb, considered in light of the similarity between Sxlfb and the canonical “early defective” allele Sxlf9 in their ability to complement Sxlf7,M1(compare Granadino et al. 1991 to Siera and Cline 2008).
, since virSxlf2 has no SxlPe. The effect suggests that virSxlf3 is specifically defective in its sexual pathway “establishment” function–the process by which X chromosome dose leads to the engagement of the Sxl positive feedback loop. This conclusion is also supported by the molecular similarity between virSxlf3 and the melanogaster mutant Sxlfb, considered in light of the similarity between Sxlfb and the canonical “early defective” allele Sxlf9 in their ability to complement Sxlf7,M1(compare Granadino et al. 1991 to Siera and Cline 2008).
The fact that loss of virSxl function specifically kills females, while increasing XSE dose specifically kills males and causes the Mickey Mouse phenotype mentioned earlier in those that do survive, shows that Sxl controls the vital, sex-specific process of X chromosome dosage compensation in virilis, as it does in melanogaster. We can conclude that Sxl also controls sex determination from the fact that 2% of virSxlf3/virSxlf2 escaper females displayed mosaic intersexual terminalia–clasper teeth and lateral plates. Molecular analysis of the effect of mutations in virSxl on the sex-specific splicing of pre-mRNA from transformer (tra) was consistent with these morphological effects on sexual phenotype. In melanogaster, tra is a direct target of Sxl that controls sexual differentiation (McKeown et al. 1987). To analyze tra regulation in the mutant situation, we performed RT–PCR on mRNA extracted from unsexed populations of 0- to 24-hr AEL embryos grown at 25°. The control population was from wild-type parents, while the virSxl mutant embryos were from homozygous virSxlf3 females crossed to virSxlf1/Y males. Recall that almost all virSxlf3/virSxlf1 females (the only genotype of female produced in this cross) die before the adult stage. While the controls showed the expected mixture of functional (female-specific) and nonfunctional (non-sex-specific) tra mRNA, the Sxl mutant embryos produced the nonfunctional form almost exclusively (data not shown).
Non-sex-specific Sxl mRNA containing a previously unrecognized, highly conserved exon Z is a likely source of male Sxl protein:
When Sievert et al. (2000) described Sxl cDNAs from the scuttlefly, Megaselia scalaris, they thought they had identified a non-sex-specific, alternatively spliced Sxl exon that corresponded to the male-specific alternatively spliced exon 3 of Drosophila. Although there was no DNA sequence homology between these two exons, we noted a subtle but telling DNA sequence similarity between that scuttlefly exon and a region of DNA sequence conservation in Drosophila Sxl that ended just 1.4 kb upstream of the male-specific exon 3 (Figure 6). The scuttlefly is a higher Dipteran (Brachycera), but only distantly related to Drosophila (cf. Figure 2). We sequenced the corresponding genomic region of the housefly, Musca domestica, a Dipteran more closely related to Drosophila than is the scuttlefly, and found that the degree of homology of the Musca region is intermediate between that for Drosophila and the scuttlefly, as expected from Musca's phylogenetic position. Data below show that this conserved region is the 3′ end of an ancestral exon that encodes an alternative Sxl N terminus in a variety of flies, including Drosophila. If Drosophila used the translation start site and splicing pattern that Megaselia seems to use (the box and vertical line, respectively, in Figure 6), the Sxl isoforms produced would be approximately the same size as those seen in male Drosophila. Moreover, their N termini would all have tyrosine as their second residue, just like the N termini encoded by the predicted exon 2 homologous translation start sites for all Diptera.
Using 5′-RACE and RT–PCR of adult male Sxl mRNA from a variety of Drosophila species including melanogaster, pseudoobscura, virilis, americana, and robusta (data not shown), we confirmed that the highly conserved region shown in Figure 6 does indeed correspond to the 3′ end of an exon that is spliced to exon 4 even in males. We named it exon Z. In melanogaster, this exon covers 371 bp. Its 5′ end is considerably less conserved than its 3′ end, but in all species examined, that 5′ end was spliced directly to exon 1 (data not shown). Although we do not provide a direct demonstration that the perfectly conserved open reading frame that begins at the position indicated on Figure 6 does indeed correspond to a translation start site, we can infer as much from the fact that for none of the species in Figure 6, nor for any of the other Drosophila species whose genomes have been sequenced, is there another potential translation start site upstream of this position in exon Z whose open reading frame would extend correctly into exon 4.
Our small-scale 5′-RACE study of exon Z mRNA indicated use of a 5′-splice site for melanogaster in exon 1 at +191. Although this site is downstream of any described by Samuels et al. (1991), it does appear in the Celniker group modENCODE RNAseq splice-junction data (see Celniker et al. 2009). For all the other species we examined, the exon Z splice was to a standard exon 1 donor site corresponding to +53 in melanogaster. The +53 site was the splice donor for the only exon-Z–containing melanogaster cDNA sequence in FlyBase, CG18350-RI. Curiously the exon 4 splice acceptor in that cDNA was a noncannonical site (CG rather than AG), which would add eight residues to the protein encoded. While our 5′-RACE sequence established that the standard exon 4 acceptor site is used, the Celniker group modENCODE data mentioned above show that this unusual splice acceptor is not likely to be an artifact and is not uniquely associated with the Z-4 splice.
Analysis of exon Z in melanogaster adults by RT–PCR is shown in Figure 7. The first point to note is that exon Z mRNAs are not sex specific: the size of the RT–PCR products shows that exon Z splicing to exon 4 skips the male-specific exon 3 in both sexes (see materials and methods for the size predictions that lead to this conclusion). Second, exon Z mRNAs appear to be far more abundant in heads than in the rest of the body. Bopp et al. (1991) showed that non-sex-specific melanogaster Sxl protein isoforms are at highest levels in adult heads. Finally, exon Z mRNAs do not appear to be present in ovaries.
If most Sxl protein made in virilis male heads is due to mRNAs derived primarily from exon 1-Z-4 splicing rather than the exon 1-2-3-4 splicing proposed by Bopp et al. (1996), the observation mentioned above that loss of exon 2 in virSxlf2 leads to an increase in the level of male Sxl proteins without any obvious change in their molecular weight (Figure 5) could be explained easily. One would expect the level of wild-type exon Z mRNAs to increase if exon Z no longer had to compete with exon 2 for exon 1 5′-splice sites. If instead most Sxl protein in wild-type male heads were due to exon 3-containing mRNAs, to account for the increase with no change in protein mobility, one would have to attribute the increase to a novel exon 1–3 splicing pattern in virSxlf2. Although translation initiation in exon Z is expected to generate proteins that are only eight residues shorter (1.1 kDa lighter) than those initiating in exon 3, any large change in the ratio of exon Z to exon 3 mRNAs would probably have blurred the doublet on this blot.
DISCUSSION
If Sxl in D. melanogaster were truly female specific in its functioning, it would be a singular exception among all the X chromosome signal element genes and among all the sex signal transduction genes such as daughterless with which those XSEs work. All these other genes seem to have acquired their roles in Drosophila sex determination without losing their ancestral functions unrelated to sex determination. Before the work we reported here, the only hint that Drosophila Sxl might not be as much of an exception as its null phenotype in melanogaster would suggest was the observation that the males of many Drosophila species make nearly full-length Sxl protein, sometimes in amounts approaching the level of full-length protein in females (Bopp et al. 1991, 1996). While this slightly shortened protein was believed to be of no functional significance, its very existence was paradoxical in light of the fact that all male Sxl mRNAs were believed to contain a translation-terminating exon that should make them sensitive to nonsense-mediated decay if translation initiated in males at the same codon as it did in females.
In the present study, we addressed three general questions regarding both this apparent evolutionary distinction between Sxl and its regulators and the related mystery of the nearly full-length Sxl protein that so many Drosophila males make:
Did melanogaster Sxl, in the course of becoming the master sex-determining gene for this species, transfer full, or even partial responsibility for vital non-female-specific functions to its closest paralog, the gene we named sister-of-Sxl (ssx)? Having generated and characterized a ssx null allele, we found no indication that it did.
Is Sxl in Drosophila species only distantly related to melanogaster as functionally female specific as it appears to be in melanogaster itself, and are those female-specific functions as extensive? Having generated and characterized various loss-of-function alleles of Sxl in D. virilis, we conclude that it is as female specific and the female-specific functions are as extensive.
Might Sxl in Drosophila have retained ancestral, non-sex-specific functions that have been unrecognized because they are not essential for viability or fertility under standard laboratory conditions? Having discovered the alternatively spliced exon Z and the non-sex-specific Sxl mRNAs that contain it, and having discovered that exon Z mRNAs correspond to mRNAs made in a fly even less closely related to Drosophila than the housefly, we conclude that it likely has. The fact that exon Z is adjacent to the male-specific exon 3—an exon unique to Drosophila and its close relatives—and the fact that both of these alternatively spliced exons appear to have a highly conserved translation initiation site, together have implications for how Sxl might have acquired its sex-determination role (see below).
The ssx null phenotype fails to support the idea that this Sxl paralog has responsibility for Drosophila Sxl's non-sex-specific ancestral functions: our DNA-sequence–based estimate of when ssx might have arisen by duplication made even more attractive the suggestion by Traut et al. (2006) that this duplication event allowed Sxl to delegate important non-female-specific ancestral functions to ssx that otherwise would have impeded Sxl's ability to acquire its current central role in sex determination. Nevertheless, our genetic analysis of ssx, our discovery of non-sex-specific transcripts from Sxl, and our recognition that ssx is more highly diverged than Sxl from their shared orthologs, make us suspect that the similar timing of these two events may have been coincidental. If not simply coincidental, perhaps the similar timing reflected two unrelated responses of the genome to whatever crisis led the fruit fly ancestor to change its sex-determination mechanism so radically.
By the hypothesis of Traut et al. the null phenotype of ssx would reflect disruption of the ancestral, presumably non-female-specific Sxl functions. Alternatively, if neither ssx nor Sxl had shed all ancestral functionality following the duplication event, the two genes might still be redundant today with respect to those ancestral activities. If redundant, both genes might have to be knocked out simultaneously to generate an obvious phenotype reflecting loss of ancestral functions. However, our results supported neither of these alternatives. Not only did we find that the ssx null allele had no adverse effect on the viability or fertility of either sex, but also we found that males missing both ssx and Sxl were similarly unaffected. Hence if ssx is currently responsible for ancestral non-female-specific Sxl functions, whether alone or in tandem with Sxl, those functions must be nonessential under standard laboratory conditions.
Although ssx seems to have no obvious functions that are not related to Sxl's current role as a sex-determination gene, we did observe that loss of ssx negatively affected Sxl positive autoregulation, albeit only in a highly sensitized genetic situation. This effect on Sxl female-specific functioning was sufficiently weak to suggest that it may simply be a vestige of the much stronger ancestral similarity that once existed between these two genes' RNA-binding protein products.
Exon 3, exon Z, and the virSxl knockout—evidence for a subtle Sxl function in Drosophila males and its implications for Sxl functions in other insects:
In a very different approach to searching for indications of the ancestral role of Sxl, we turned our attention to D. virilis. The work on virSxl had the added benefit of telling us how similar Sxl's sex-specific regulation and functions were in a Drosophila species only distantly related to melanogaster. Bopp et al. (1996) observed that the much higher level of slightly truncated Sxl proteins made by virilis vs. melanogaster males correlated with the presence in virilis of an ORF in the male-specific exon 3 that was in frame with the rest of the gene and began with an AUG only 29 bp upstream of that exon's 3′ end. They suggested that leaky scanning for translation initiation that skipped the normal start site in exon 2 and started instead at this AUG could account for the unusually large amount of male Sxl protein in virilis.
Subsequent sequencing of the corresponding region of Sxl in S. lebanonensis (Siera and Cline 2008) and in a diverse collection of 10 additional Drosophila species (Drosophila 12 Genomes Consortium 2007) revealed that what was true for virilis was true for all these other sequenced species except for the 4 closest relatives of melanogaster (Figure S2). Those exceptional 4 had all lost the same single base pair that disrupted this ORF. Of the remaining 9 species whose ORF was intact, only wilistoni had undergone a change in the number of base pairs in that part of exon 3, and that change maintained the reading frame. For none of these “intact” species did the ORF extend upstream to another AUG. Moreover, the DNA sequence of that region upstream of the putative translation start site was markedly more variable than the region downstream. These facts made it hard to escape the conclusion that the ORF beginning in exon 3 is functionally significant. They also suggested that D. virilis would be a far more representative Drosophila species than melanogaster with which to pursue the possibility of non-female-specific, potentially ancestral functions for Sxl.
From the fact that Sxl proteins encoded by exon 3 would necessarily be male specific, it does not follow that their functions would necessarily be male specific. If Sxl in most Drosophila species were still responsible for a non-sex-specific ancestral function, the exon-3–initiated proteins might provide males with that function, while the full-length Sxl proteins serve that purpose for females. On the other hand, this male protein's role might be to antagonize any Sxl-F protein that was generated inappropriately in males, thereby increasing the fidelity of the gene's sex-specific control. In connection with the question of whether male Sxl proteins have functions, it is worth noting that the second residue encoded by all nine intact male-exon ORFs is phenylalanine, the most conservative substitute for tyrosine, which is the second residue in all Dipteran Sxl proteins whose translation is initiated in the homologs of exons 2 and Z.
Because D. melanogaster and its close relatives lack the translation start site in exon 3 that all the more distantly related species share, some other translation start site must be responsible for generating their male Sxl proteins. Bopp et al. (1991) suggested that both sexes of melanogaster might occasionally initiate translation in exon 4 instead of in exon 2. This possibility is supported by the discovery in the scuttle fly, M. scalaris, of several mRNA species that could initiate translation only in their exon 4 homolog (Sievert et al. 2000). Interestingly, there is an ATG in exon 4 that begins a sequence of six codons that specify the same amino acids in all known Dipteran Sxl genes (Traut et al. 2006).
Our discovery of a previously unrecognized, alternatively spliced Sxl exon that we call Z suggests a different and perhaps more straightforward explanation for the source of the Sxl protein seen in both sexes of melanogaster. This exon contains a potential translation initiation site that is present not just in all Drosophila, but also in higher Diptera (Brachycera) that are not likely to be using Sxl as a sex switch. Consequently, those Sxl isoforms whose translation initiates in this exon are likely to be providing the ancestral, non-sex-specific functions. By this hypothesis, those ancestral functions would have been nonessential even before Sxl became a sex switch. The lack of an obvious mutant phenotype for the ssx null allele is also consistent with the idea that Sxl's ancestral functions were nonessential.
Sievert et al. (2000) understandably assumed that the alternatively spliced Megaselia exon that we now know is exon Z corresponded instead to the Drosophila male-specific exon 3, despite the fact that the exons shared no significant DNA homology. As we showed here, the evidence that the Megaselia exon really does correspond to exon Z goes beyond DNA sequence homology to include the fact that the mRNAs carrying this exon are unlike other Sxl mRNAs in not being expressed in the ovaries of either species.
Sxl protein isoforms generated by translation initiation in either exon Z or exon 3 would disrupt male development unless they lacked the full-length proteins' ability to positively regulate tra and negatively regulate male-specific-lethal-2 (msl-2). Yanowitz et al. (1999) studied the functionality in vivo of a Sxl protein whose N terminus was trimmed, but their trimming eliminated not just all the residues we can now predict to be missing in all Drosophila males, but also 14 of the 15 residues encoded by exon 4, the majority of which are conserved among all Dipteran Sxls that have been sequenced. While their protein did lack the ability to regulate tra, its ability to negatively regulate the dosage compensation switch gene msl-2 seemed surprisingly intact. On the other hand, it is not straightforward to relate the Yanowitz et al. studies to expectations for expression of the wild-type truncated proteins in males, since the two situations differ with respect to the timing, level, and complexity of the protein isoforms expressed. Yanowitz et al. drove the expression of only a single Sxl isoform from an hsp83-driven transgene whose putative translation start site had been optimized. Since Sxl proteins in wild-type males would have been selected by evolution to provide ancestral functions while specifically not influencing msl-2, the possibility remains that their unique wild-type N termini might have an active role in preventing such interference.
In those insect species where there seems to be no difference in Sxl protein expression between males and females, one can fairly infer that Sxl must not be functioning as a feminizing switch gene, but it does not necessarily follow that it has a function in both sexes. For a gene that functions in only one sex, evolution need not have bothered to limit expression to that sex so long as expression in the opposite sex is not deleterious. For this reason, the idea that Sxl is functioning in both sexes in all these Dipterans is supported at least as much by our finding a conserved ORF that begins in the Drosophila male-specific exon as it is by our discovery that exon Z mRNAs are not female specific. The fact that the male-specific exon 3 ORF is conserved in most Drosophila species shows that evolution has gone to some length to keep Sxl functioning in both sexes even as Sxl acquired new functions that are demonstrably female specific. Of course this argument does not hold if the possibility mentioned earlier is true; namely, that the role of male-specific Sxl protein is to antagonize any Sxl-F protein that males might inadvertently make. The latter alternative could be tested by observing whether overexpression of bona fide wild-type male-specific Sxl isoforms generated a dominant negative phenotype.
Only a thorough molecular analysis of the variety of Sxl N termini actually present as a function of species, sex, developmental stage, and tissue type will reveal the extent to which the myriad alternative potential translation initiation sites for this gene are actually used. But regardless of how complex that pattern proves to be, our analysis of the virSxl null allele showed that such proteins are not essential for males under standard laboratory conditions. Moreover, our characterization of partial loss-of-function mutant virSxl alleles showed that Sxl has the same general female-specific functions in virilis as it does in melanogaster: control of sexual phenotype and X chromosome dosage compensation. The method by which all these mutant Sxl alleles were generated—suppression of the male-lethal effects of XSE duplications—shows that the regulation of Sxl by X chromosome dose is also very similar between these two species. Since virilis is phylogenetically distant from melanogaster within the Drosophila genus, these similarities argue that the current system of Drosophila sex determination evolved relatively soon after the last common ancestor of the medfly and the fruit fly diverged, but then remained remarkably stable throughout the subsequent evolution of the family Drosophiladae.
Relevance of nonessential aspects of gene function to understanding the evolutionary takeover from tra by Sxl of the role of master feminizing gene:
The location of exon 3 just downstream of exon Z raises the possibility that the first step by Sxl toward the evolution of sex-specific alternative splicing, and presumably thereby toward its ultimate role in sex determination, might have been a tandem duplication of exon Z. That duplication might have allowed the downstream copy of exon Z to have evolved in a direction where the N terminus it encoded became subtly beneficial to males, perhaps at the same time as the ancestral exon 2 on the opposite side of exon Z evolved toward encoding an N terminus specifically useful to females. The upstream copy of exon Z would have remained responsible to this day for functions needed equally by both sexes, functions that are not obvious because they are nonessential under standard laboratory conditions, at least in the absence of other genetic changes.
None of the subtle complexities introduced by the potential translation initiation sites in the non-sex-specific exon Z and the male-specific exon 3 detract from the simple textbook view that Sxl functions in Drosophila sex determination as a straightforward developmental switch that is on in females and off in males. That on/off switch character of Sxl was inferred from the striking phenotypes of various individual mutations in the gene. But while strong mutant phenotypes are a good place to start the study of a developmental pathway, we believe that developing an understanding of how such pathways might have evolved is likely to require recognizing and understanding subtle aspects of gene functioning that may not be readily apparent from individual mutant phenotypes.
That principle has already been established for Sxl in a study of the regulatory relationship between Sxl and its downstream target transformer (tra) in Drosophila (Siera and Cline 2008), the gene whose regulatory role Sxl seems to have usurped, at least in part. As mentioned in the Introduction, in most insects tra rather than Sxl appears to be the master feminizing switch gene that responds to sex determination signals and then epigenetically maintains the sexually determined state through a positive autoregulatory feedback loop on pre-mRNA splicing. Moreover, expression of tra in the mother's germ line seems to be a key element in how progeny respond to their sex signals. By contrast in melanogaster, nothing in the standard phenotypes of loss-of-function or gain-of-function tra and Sxl alleles suggested that tra was anything more than a passive downstream target of Sxl, at least in somatic cells. Nor did those phenotypes suggest that tra expression in the germ cells of Drosophila females even took place, much less was relevant to the development of those females' offspring. Only in a highly contrived and complex sensitized genotype was it revealed that both maternally and zygotically expressed tra contribute to Sxl positive autoregulation in D. melanogaster. Hence even in melanogaster, tra is at least indirectly autoregulatory. Moreover, it is expressed in melanogaster germ cells and that expression is relevant to sex signaling in the next generation. Once this unanticipated tra–Sxl regulatory relationship was observed phenotypically, it became evident that the molecular signal of the relationship had been hiding in plain sight for decades: two highly conserved Tra binding sites in Sxl just upstream of exon 3.
It seems hard to believe that this echo in Drosophila of the developmental role tra has in other insects is not a reflection of the evolutionary path both tra and Sxl took as the Drosophila ancestor changed its primary sex-determination signal. Indeed the discovery led to a hypothesis for how tra might have passed off its direct autoregulatory character to Sxl. Similarly, the more nuanced view we develop here of the extent to which Sxl has evolved as a female-specific gene in Drosophila suggests that the functioning of Sxl in the fruit fly has far more in common with its functioning in other insects than anyone would have imagined. Those commonalities seem a good starting place from which to explore the evolution of this particular developmental pathway.
Acknowledgments
We thank Maren Bell for assisting with some of the cloning and sequencing work, Jessica Chow for assisting with the ssx deletion screen, and Barbara J. Meyer for helpful comments on the manuscript. This work was supported by National Institutes of Health grant GM23468 (to T.W.C.), American Cancer Society postdoctoral fellowship PF0717901DCC (to M.M.H.), and a Wellcome Trust and Human Frontier Science Program postdoctoral fellowship 038110Z93Z (to L.S.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121202/DC1.
DNA sequence data from this article for the M. domestica Sxl genomic region between exons 2 and 5 have been deposited with the EMBL/GenBank Data Libraries under accession no. HM776132.
References
- Alexander, M. L., 1976. The Genetics and Biology of Drosophila virilis. Academic Press, London.
- Bernstein, M., R. A. Lersch, L. Subrahmanyan and T. W. Cline, 1995. Transposon insertions causing constitutive Sex-lethal activity in Drosophila melanogaster affect Sxl sex-specific transcript splicing. Genetics 139 631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., R. E. Green, S. MacArthur, A. N. Brooks, S. E. Brenner et al., 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA binding specificities of the Drosophila hnRNP A/B family members. Mol. Cell 33 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp, D., L. R. Bell, T. W. Cline and P. Schedl, 1991. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 5 403–415. [DOI] [PubMed] [Google Scholar]
- Bopp, D., G. Calhoun, J. I. Horabin, M. Samuels and P. Schedl, 1996. Sex-specific control of Sex-lethal is a conserved mechanism for sex determination in the genus Drosophila. Development 122 971–982. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., L. A. Dillon, M. B. Gerstein, K. C. Gunsalus, S. Henikoff et al., 2009. Unlocking the secrets of the genome. Nature 459 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, T. W., 1988. Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics 119 829–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, T. W., D. Z. Rudner, D. A. Barbash, M. Bell and R. Vutien, 1999. Functioning of the Drosophila integral U1/U2 protein Snf independent of U1 and U2 small nuclear ribonucleoprotein particles is revealed by snf+ gene dose effects. Proc. Natl. Acad. Sci. USA 96 14451–14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 203–218. [DOI] [PubMed] [Google Scholar]
- Erickson, J. W., and T. W. Cline, 1991. Molecular nature of the Drosophila sex determination signal and its link to neurogenesis. Science 251 1071–1074. [DOI] [PubMed] [Google Scholar]
- Erickson, J. W., and T. W. Cline, 1993. A bZIP protein, SISTERLESS-A, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 7 1688–1702. [DOI] [PubMed] [Google Scholar]
- Erickson, J. W., and T. W. Cline, 1998. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development 121 3259–3268. [DOI] [PubMed] [Google Scholar]
- Erickson, J. W., and J. J. Quintero, 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5 2821–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe, T., M. Hasselmann, M. Schiøtt, G. Hause, M. Otte et al., 2009. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 7 e1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino, B., M. Torres, D. Bachiller, E. Torroja, J. Barbero et al., 1991. Genetic and molecular analysis of new female-specific lethal mutations at the gene Sxl of Drosophila melanogaster. Genetics 129 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager, J. H., and T. W. Cline, 1997. Induction of female Sex-lethal RNA splicing in male germ cells: implications for Drosophila germline sex determination. Development 124 5033–5048. [DOI] [PubMed] [Google Scholar]
- Hediger, M., C. Henggeler, N. Meier, R. Perez, G. Saccone et al., 2010. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 184 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks, T. M., G. Calhoun and P. Schedl, 2003. Functional conservation of the Sex-lethal sex determining promoter, Sxl-Pe, in Drosophila virilis. Dev. Genes Evol. 213 155–165. [DOI] [PubMed] [Google Scholar]
- Klemenz, R., U. Weber and W. J. Gehring, 1987. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 15 3947–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A. L., B. F. Volkman, S. A. Robertson, D. Z. Rudner, D. A. Barbash et al., 1997. Chemical shift mapping of the RNA-binding interface of the multiple-RBD protein, Sex-lethal. Biochemistry 36 14306–14317. [DOI] [PubMed] [Google Scholar]
- Loukeris, T. G., B. Arca, I. Livadaras, G. Dialektaki and C. Savakis, 1995. a Introduction of the transposable element Minos into the germ line of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 92 9485–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukeris, T. G., I. Livadaras, B. Arca, S. Zabalou and C. Savakis, 1995. b Gene transfer into the medfly, Ceratitis capitata, with a Drosophila hydei transposable element. Science 270 2002–2005. [DOI] [PubMed] [Google Scholar]
- Marin, I., M. L. Siegal and B. S. Baker, 2000. The evolution of dosage compensation mechanisms. BioEssays 22 1106–1114. [DOI] [PubMed] [Google Scholar]
- McKeown, M., J. M. Belote and B. S. Baker, 1987. A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell 48 489–499. [DOI] [PubMed] [Google Scholar]
- Pane, A., M. Salvemini, P. DelliBovi, C. Polito and G. Saccone, 2002. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129 3715–3725. [DOI] [PubMed] [Google Scholar]
- Pomiankowski, A., R. Nöthiger and A. Wilkins, 2004. The evolution of the Drosophila sex-determination pathway. Genetics 166 1761–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone, G., A. Pane and L. C. Polito, 2002. Sex determination in flies, fruitflies, and butterflies. Genetica 116 15–23. [DOI] [PubMed] [Google Scholar]
- Salz, H. K., and J. W. Erickson, 2010. Sex determination in Drosophila. The view from the top. Fly 4 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz, H. K., T. W. Cline and P. Schedl, 1987. Functional changes associated with structural alterations induced by mobilization of a P element inserted in the Sex-lethal gene of Drosophila. Genetics 117 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, M. E., P. Schedl and T. W. Cline, 1991. The complex set of late transcripts from the Drosophila sex determination gene Sex-lethal encodes multiple related polypeptides. Mol. Cell. Biol. 11 3584–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton, L., J. R. Timmer, Y. Zhang, F. Beranger and T. W. Cline, 2000. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature 405 970–973. [DOI] [PubMed] [Google Scholar]
- Shearman, D. C. A., 2002. The evolution of sex determination sytems in dipteran insects other than Drosophila. Genetica 116 25–43. [DOI] [PubMed] [Google Scholar]
- Siera, S. G., and T. W. Cline, 2008. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene Sex-lethal by its target transformer. Genetics 180 1963–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert, V., S. Kuhn, A. Paululat and W. Traut, 2000. Sequence conservation and expression of the Sex-lethal homologue in the fly Megaselia scalaris. Genome 43 382–390. [DOI] [PubMed] [Google Scholar]
- Sánchez, L., 2008. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52 837–856. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., 1986. P-element-mediated transformation, pp. 175–197 in Drosophila, A Practical Approach., edited by D. B. Roberts. IRL Press, Oxford.
- Takeuchi, H., O. Georgiev, M. Fetchko, M. Kappeler, W. Schaffner et al., 2007. In vivo construction of transgenes in Drosophila. Genetics 175 2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut, W., T. Niimi, K. Ikeo and K. Sahara, 2006. Phylogeny of the sex-determining gene Sex-lethal in insects. Genome 49 254–262. [DOI] [PubMed] [Google Scholar]
- Verhulst, E. C., L. W. Beukeboom and L. van de Zande, 2010. Maternal control of haplodiploid sex determination in the Wasp Nasonia. Science 328 620–623. [DOI] [PubMed] [Google Scholar]
- Wrischnik, L. A., J. R. Timmer, L. A. Megna and T. W. Cline, 2003. Recruitment of the proneural gene scute to the Drosophila sex-determination pathway. Genetics 165 2007–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanowitz, J. L., G. Deshpande, G. Calhoun and P. D. Schedl, 1999. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of Sex-lethal. Mol. Cell. Biol. 19 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]