Abstract
The repair of chromosomal double-strand breaks (DSBs) can be accomplished through homologous recombination in most organisms. We report here that exogenous oligonucleotides can efficiently target for repair a single DSB induced in a chromosome of yeast. The efficiency of recombinational targeting leading to a desired DNA change can be as high as 20% of cells. The DSB was generated either by a regulatable I-SceI endonuclease just before transformation or appeared spontaneously at the site of a long inverted repeat composed of human Alu sequences. The approach used features of our previously described delitto perfetto system for selecting transformants with integrative recombinant oligonucleotides. The DSB repair mediated by pairs of complementary integrative recombinant oligonucleotides was efficient for targeting to homologous sequences that were close to or distant from the DSB and in the presence of a competing homologous chromosome in diploid cells. We also demonstrate that a DSB can strongly stimulate recombination with single-stranded DNA, without strand bias. These findings expand current models of DSB repair. In addition, we establish a high-throughput system for rapid genome-wide modification with oligonucleotides.
Unrepaired DNA double-strand breaks (DSBs) in chromosomal DNA, resulting from DNA damage or faulty DNA metabolism (1-3), can have a variety of negative consequences, including mutagenesis, loss of genetic information, chromosomal aberrations, cell death, and disease (3). To minimize these effects, cells use systems to repair DSBs by either homologous recombination or nonhomologous end joining (1, 2, 4-8).
Several approaches have been taken to address the consequences of a DSB and mechanisms of repair, including the introduction of a break at a unique site placed into the genome of the yeast Saccharomyces cerevisiae and other organisms (9-14). The systems typically use the yeast mating-type switching enzyme HO endonuclease (9) or the mitochondrial I-SceI endonuclease that cuts at a unique 18-bp sequence, creating four-base 3′ overhangs (10, 11).
DSB recombinational repair mechanisms have also been investigated by transfecting into cells linear or gapped plasmids where the ends, similar to ends of a chromosomal DSB, are highly recombinogenic with homologous sequences (15, 16) and can provide targeted integration and replacement of chromosomal DNAs (17, 18). Recombination of gapped plasmids with long linear molecules or short oligonucleotide is very efficient and has been used for rapid plasmid modification in yeast and in vivo cloning of large chromosomal fragments (16, 19-21). The typical level of chromosomal targeting by linear DNA (i.e., with DSB ends) in yeast is low, ≈10-6 per viable cell in a transformation mix when the homology at the tails is short (≈40-50 bp) and reaching 10-3 to 10-4 when the homology is long (22). Based on results in mammalian cells, targeting to chromosomes can be increased if a DSB is introduced into the chromosomal DNA before transfection (23-25).
Oligonucleotide targeting to homologous chromosomal sequences has also been reported in yeast with systems that rely on a selectable phenotype for the recombinants (26-28). Using a different procedure, referred to as delitto perfetto, we developed a system that provides direct selection for recombinants and requires the RAD52 recombination function (29), unlike other approaches (26-28, 30). The reported frequencies of genome targeting by oligonucleotides are between 10-3 and 10-6 per viable cell (31). In this study, we anticipated that a DSB might dramatically increase oligonucleotide targeting, which led to our development of a self-contained cassette-based system for generating a DSB that carries both a regulatable I-SceI enzyme and its cleavage site. The induction of a DSB in vivo before standard transformation procedures results in 5-20% of all cells in the population being targeted by small oligonucleotides. Using this system, we have addressed several issues relating to DSB recombinational repair, including the role of single-stranded (ss) DNA, targeting at a naturally occurring DSB generated by a large inverted repeat (IR), and the utility of oligonucleotides for creating a variety of genome modifications.
Materials and Methods
Strains and Plasmids. The yeast S. cerevisiae backgrounds used in this study were BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) (32) and E133a (isogenic to CG379) (MATa, ade5-1, lys2-12A, trp1-289, his7-2, leu2-3,112, ura3-52) (ref. 33 and references therein). The BY4742 wild-type and rad52 mutant strains containing the counterselectable KlURA3-kanMX4 reporter cassette counterselectable reporter marker cassette (CORE) in TRP5 (BY4742-TRP5-CORE) were described (29). The Alu-IR with 100% or 75% homology was inserted into the LYS2 gene in CG379 (34).
For experiments involving the TRP5 locus in CG379, the trp1-289 allele was converted to wild-type TRP1 by transformation with a TRP1 PCR product (CG379-TRP1). The diploid CG379-2n strain was derived by crossing haploid CG379-TRP1, containing the CORE-I-SceI cassette, with a hygromycin (Hygro)-resistance gene (hyg) (35) and KlURA3 in TRP5, with CG379 (MATα), where TRP5 was disrupted with LEU2 (details available on request). Genetic methods and standard media were as described (22, 29, 35, 36).
The plasmids pGSKU and pGSHU were constructed by cloning a 1.4-kb fragment, containing GAL1-I-SceI amplified with BglII tails from plasmid pKM4 (a gift from K. Lobachev, Georgia Institute of Technology, Atlanta) into the BglII site of plasmid pCORE-UK and pCORE-UH (31). As a result, GAL1-I-SceI was placed upstream of kanMX4 and KlURA3 or upstream of hyg and KlURA3, respectively.
Integration of CORE-I-SceI Cassette. The CORE-I-SceI cassette contained, starting at the 5′ end, GAL1-I-SceI, kanMX4 (or hyg), and KlURA3 (Fig. 1). The cassette was amplified as a 4.6- or 4.8-kb DNA fragment from pGSKU or pGSHU, respectively, by using TaqDNA polymerase (Roche Diagnostics), with 32 cycles of 30 s at 94°C, 30 s at 56°C, and 4 min 30 s at 72°C. For integration of the cassette into chromosomal loci, chimeric primers were used, consisting of 50 nucleotides homologous to the genomic target and an additional 20 nucleotides shown below for the amplification of the CORE-I-SceI cassette and 18 nt for the induction of a DSB at the I-SceI site (underlined): 5′-... TAGGGATAACAGGGTAAT-TTGGATGGACGCAAAGAAGT-3′ for the GAL1-I-SceI side and 5′-... TTCGTACGCTGCAGGTCGAC-3′ for the KlURA3 side. To identify clones with the correct CORE-I-SceI cassette integration, colony PCR was performed as described (29). Primer sequences are available on request.
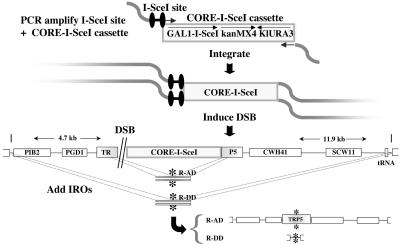
Fig. 1.
A self-contained inducible DSB system for the targeted integration of oligonucleotides into the yeast genome. Presented is a scheme for the PCR amplification and genomic integration of the CORE-I-SceI cassette. A segment of chromosome VII is shown with the CORE-I-SceI and a DSB induced at the I-SceI site within the TRP5 locus. Categories of IROs are defined by the position of the homologous tails relative to the position of the DSB. IROs that target to both sides of the break provide an opportunity for repair (R). They are classified as repair-adjacent distant (R-AD), repair-distant adjacent (R-DA), repair-distant distant (R-DD), or repair-adjacent adjacent (R-AA) (see Figs. 2, 3, and 5), according to whether the homology region is adjacent (A) (<15 bp from the I-SceI site) or distant (D) (up to 4.7 kb from one side and 11.9 kb from the other) to the DSB. Recombination with IROs during transformation leads to excision of the CORE-I-SceI cassette and the appearance of colonies with the desired mutation (asterisks) or deletion.
Induction of a DSB and Targeting Using Integrative Recombinant Oligonucleotides (IROs). One and one-half milliliters of an over-night culture grown in rich yeast extract/peptone/dextrose/adenine (2 mg/ml) (YPDA) medium were transferred into 50 ml of synthetic complete media containing 2% galactose and incubated with vigorous shaking at 30°C for 4 h to express GAL1-I-SceI and induce a DSB. In related experiments with CG379 background strains, the induction of GAL1-I-SceI results in the production of a DSB specifically at the target sequence, with up to 50% of growing cells experiencing a DSB within 2 h, reaching ≈90% by 6 h (K. Lobachev, personal communication). Transformation was done by a lithium acetate protocol (22), with modifications. Briefly, after galactose incubation, cells were washed with water and then with Solution 1 [lithium acetate 0.1 M/10 mM Tris/1 mM EDTA, pH 7.5 (TE), 1×] and resuspended in 250 μl of Solution 1. Fifty microliters of this cell suspension was mixed with IROs (20 μl of 50 μM solution; i.e., 1 nmol), and then 300 μl of Solution 2 (LiAc 0.1 M, TE 1× in polyethylene glycol 4000 50%) was added. Cells were incubated at 30°C for 30 min, heat-shocked at 42°C for 15 min, collected, and plated with the appropriate dilutions.
The IRO molecules (desalted custom primers) synthesized by Invitrogen were used in transformation experiments either singly or as pairs. IRO sequences are presented in Table 2, which is published as supporting information on the PNAS web site. Before addition of the IRO DNA to the cells, IROs were denatured at 100°C for 2 min and then placed immediately on ice to eliminate possible secondary structures. No IRO annealing or extension in vitro was performed in this study. Cells from each IRO transformation were appropriately diluted and spread directly to selective glucose Trp- (plating on galactose media did not increase the number of transformed cells; not shown) or Lys- plates (for some experiments) or YPDA plates. To select for Ura- cells, the YPDA plates were replica-plated to 5-fluoroorotic acid (5-FOA) media after the first day of incubation. After 3 days, colonies from 5-FOA were replica-plated to YPDA and YPDA containing geneticin (G418) or Hygro and Trp- (in experiments to identify Trp+ colonies) plates. In all transformation experiments, the 105-fold dilutions were plated directly to YPDA to determine viability of the cell population. Survival was ≈25% after transformation with or without IROs, in glucose or in galactose (not shown). To directly measure events resulting from a high frequency of targeting, the 10-5-fold diluted suspensions were plated to YPDA directly, and plates (containing 100-1,000 colonies) were replica-plated to 5-FOA and Trp-.
Results
Experimental System. To position an inducible chromosomal DSB anywhere in the yeast genome, a cassette was developed containing a regulatable I-SceI endonuclease. The previously described CORE cassette containing the counterselectable KlURA3 and the reporter kanMX4 or hyg genes (29, 31) was modified to include the I-SceI gene under the control of the galactose-inducible GAL1 promoter (see Fig. 1). The CORE-I-SceI cassette, cloned into an Escherichia coli vector, could be amplified and integrated anywhere in the genome along with the 18-bp I-SceI cut site. We found that plasmids containing both the GAL1-I-SceI gene and its cleavage site could not be propagated in E. coli (not shown). To bypass this problem, we introduced the sequence of the I-SceI cut site with one of the primers used to amplify the integrating cassette. On changing the carbon source to galactose, I-SceI is expressed, and a DSB is generated at the I-SceI site (ref. 2; see Materials and Methods). This DSB system differs from those previously examined for chromosome targeting in yeast in that a chromosomal break is induced just before transformation. Therefore, it is possible to examine the ability of a DSB to be targeted for recombinational repair by exogenously added oligonucleotides, IROs, which have homology to regions in the vicinity of the break. Recombination was identified by loss of both CORE markers (Fig. 1) (29, 31). In the case of high efficiency of targeting, frequencies could be determined directly by replica-plating colonies arising on YPDA plates to 5-FOA and Trp- media.
Although the observations have proven to be general for different genomic loci, most of the experiments were based on an integrated CORE cassette at the TRP5 gene (Fig. 1). IROs with homology to the TRP5 ORF not only eliminated the cassette but also restored the Trp+ phenotype and introduced a silent mutation generating a new BamHI site. Therefore, the IRO-mediated targeting could be distinguished from reversion via deletion by nonhomologous end joining.
A Single DSB Is Efficiently Targeted for Recombination Repair by IROs. The impact of a single-chromosome DSB on targeting by IROs was examined in haploid yeast that contained the CORE-I-SceI cassette and the I-SceI recognition site at the TRP5 locus (Fig. 1; Table 1). The cells were incubated in galactose to express I-SceI and induce a DSB and then transformed with a pair of fully complementary 95-nt IROs. There were two sets of controls: cells containing the original CORE cassette (29), where there was no opportunity to induce a DSB, and cells containing the CORE-I-SceI cassette and the I-SceI recognition site grown only in glucose.
Table 1. Effect of DSBs on the frequency of IRO targeting.
| Transformation frequency × 107*
|
|||
|---|---|---|---|
| Insert (background) | Selection† | No DSB‡ | DSB |
| TRP5::CORE (BY4742) | ΔCORE | 241 ± 30 | NA§ |
| Trp+ | 227 ± 17 | NA | |
| TRP5::CORE-I-SceI (BY4742) | ΔCORE | 482 ± 69 | 2,143,000 ± 61,000 |
| Trp+ | 386 ± 53 | 1,784,000 ± 75,000 | |
| TRP5::CORE-I-SceI (CG379) | ΔCORE | 658 ± 175 | 681,000 ± 174,000 |
| Trp+ | 507 ± 126 | 561,000 ± 147,000 | |
| BAR1::CORE-I-SceI (CG379) | ΔCORE | 321 ± 24 | 995,000 ± 232,000 |
| TRP5::CORE-I-SceI (BY4742 Δrad52) | Trp+ | 0 ± 0 | 304 ± 136 |
| TRP5::CORE-I-SceI (CG379—2n) | ΔCORE | 13,000 ± 2,700 | 1,110,000 ± 331,000 |
| Trp+ | 64 ± 16 | 44,200 ± 5,400 | |
| LYS2::Alu-IR (CG379) | Lys+ | 305 ± 67 | 9,100 ± 1,900 |
Mean numbers and standard deviations were calculated from three to six determinations. One nanomole of fully complementary 95-mer IROs was used. CORE-loss events per 107 viable cells obtained for the no-DNA controls (not shown) were 0 without the induction of a DSB and <100 when a DSB was induced. For the diploid cells (CG379—2n), the CORE-loss events were mostly due to recombination with the homologous chromosome
Selection conditions are described in Materials and Methods. Only ≈0.01% of the CORE-I-SceI transformants were both 5-FOAR and resistant to G418/Hygro after DSB induction, whereas ≈50% of the colonies had this phenotype when there was no DSB induction (not shown; ref. 29)
The no-DSB controls are explained in Results
NA, not applicable
Remarkably, up to 20% of all cells (By4742) in the transformation mix could undergo targeted recombination by the IROs (Table 1). This level of recombination is 2-5 orders of magnitude higher than any other type of targeted recombination reported for yeast, and it was >4,000-fold greater than IRO targeting when there was no induction of a DSB. Among clones that lost the cassette the number that were Trp- was low, with or without the induction of a DSB (6-23%). Thus, additional TRP5 inactivating mutations were not increased by the DSB and the majority of targeting events occurred via a homology-driven process. All of the 64 Trp- clones that were sequenced showed the correct change (BamHI site); however, there was at least one additional point mutation within the region of the IRO sequence (data not shown).
The high transformation frequency was observed for both the TRP5 and BAR1 loci, independent of the strain background (Table 1). Also, >1,000-fold increases over the no-DSB control were obtained with IROs designed to create deletions upstream of the MSH2 or PMS1 promoter and to produce point mutations in the RAD27 ORF (data not shown). Similarly, there was more than a 1,000-fold stimulation when a much larger PCR fragment (≈1 kb long) was used to replace the CORE-I-SceI cassette cloned upstream of the RAD27 gene (not shown). The strong dependence on the RAD52 gene (Table 1) supports the idea that the targeting process involves recombinational repair of the DSB. Targeting without a DSB was also RAD52-dependent in our system (29).
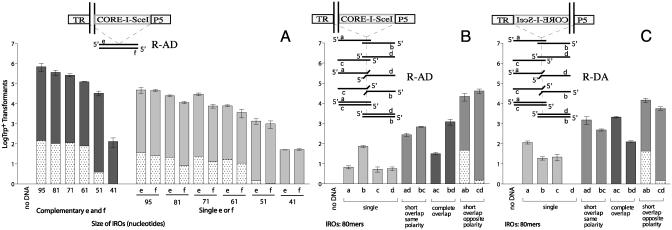
A high level (≈0.1%) of DSB-stimulated IRO targeting was observed with as little as 0.001 nmol of double-stranded molecules (Fig. 5A, which is published as supporting information on the PNAS web site), reaching a plateau at ≈1 nmol of IROs (≈30 μg). The cutoff size for efficient stimulation by complementary IROs was between 41 and 51 nucleotides; no transformants were detected with 31-mers (Fig. 2A Left). Because there was only a 10-fold difference in frequency between the 61- and 95-mer R-AD complementary oligonucleotides, further increases in size are expected to have relatively little impact.
Fig. 2.
Factors affecting IRO targeting efficiency to a DSB (structure and homology of IROs relative to the position of the DSB). Presented are numbers of Trp+ transformants per 107 viable cells resulting from targeting by the various IROs to an I-SceI DSB induced in the TRP5 gene. The vertical bars correspond to the average values from three to six determinations; the range identifies the standard deviations. Dark gray bars represent values from pairs of fully complementary IROs, medium gray bars represent values from pairs of overlapping IROs, and light gray bars represent values for ss IROs. When no bars are present, values were ≤1 transformant/107 viable cells. “No-DSB” control values (corresponding to cells incubated in glucose media only) are indicated by stippled bars. The types of IROs analyzed are indicated below the bars. The amount of total IRO DNA used in each transformation is 1 nmol (≈10-30 μg) unless otherwise indicated. The IROs examined in A are referred to as e and f: fully complementary with equal lengths of homology to both sides of the DSB (i.e., symmetrical for homology) and from 31 to 95 nucleotides in length. The oligonucleotides examined in B and C are various combinations of 80-mers referred to as a, b, c, and d, where the homology to each side of the DSB is asymmetric (i.e., 69 nucleotides to one side and 10 nucleotides to the other). The position of the CORE-I-SceI cassette and the DSB in TRP5, together with the kinds of IROs used, are shown in the small diagrams. In A and B, the DSB is between the 5′ region of TRP5 and the CORE-I-SceI cassette; in C, the DSB is between the CORE-I-SceI cassette and the 3′ region of TRP5. (A) Effect of size of fully complementary and ss e and f IROs on transformation efficiency in the R-AD conformation. (B and C) Effect of positions of homology relative to the DSB, using the asymmetric a, b, c, and d IROs in the R-AD and R-DA conformation, respectively.
Based on the results with haploid strains, we tested the ability of IROs to target a single DSB in diploids where the oligonucleotides would be competing with the unbroken homologous chromosome for repair of the break. The diploid strain had a Trp- phenotype, which could not revert spontaneously but only through recombination with the IROs (see Materials and Methods). However, the loss of the cassette (5-FOAR and HygroS colonies) could occur by recombination with either the targeting oligonucleotides or the homologous chromosome. Although the CORE-loss events due to IRO targeting (Trp+ colonies) were only 4% of the total, as shown in Table 1, targeting was stimulated up to 700-fold by the induction of the DSB.
We also examined the ability of oligonucleotides to target a different type of site-specific DSB that is induced in association with a noncanonical structure. A closely spaced IR composed of 100% identical human Alu-IR DNA can result in ≈2% of chromosomes experiencing a hairpin-capped DSB at the site of the IR (37). A strain containing the Alu-IR sequence integrated in the LYS2 gene was transformed with fully complementary 95-mers that could delete the Alu-IR sequence and restore a functional LYS2 gene. The no-DSB control for the Alu-IR DSB system was determined from cells containing an inverted pair of Alus with only 75% identity, which does not lead to a DSB (34). The presence of the 100% identical IR resulted in a 30-fold higher frequency of IRO targeting (Table 1).
A DSB in one chromosome did not improve targeting of oligonucleotides homologous to a site located on another chromosome. In a strain containing both the Alu-IR in LYS2 and the CORE-I-SceI plus I-SceI target site in TRP5, targeting to TRP5 was not increased on glucose or galactose media. Similarly, targeting to LYS2 was not increased when cells were incubated in galactose (data not shown).
A DSB Is Efficiently Repaired by Recombination with ss IROs. We previously reported that in the absence of a site-specific DSB, ss oligonucleotides could target homologous sequences in yeast chromosomes albeit at a very low efficiency (29). The introduction of a DSB greatly increased the targeting by ss oligonucleotides (Fig. 2 A). Single 95-base oligomers e and f (from the R-AD category) were only 10-fold less effective than a mix of complementary IROs. Importantly, no significant strand bias was revealed between e and f, regardless of the size. With the shorter 41-base IROs, where targeting was very inefficient, there was little difference in efficiencies between ss and fully complementary IROs. Similar results were obtained in the CG379 background with IROs targeted to the TRP5 or to the BAR1 locus (not shown). Moving the position of the I-SceI site to the opposite side of the cassette in TRP5 did not change transformation frequencies for IRO e and f (R-DA) single species (data not shown).
Impact of Homology and Distance on Targeting of IROs to a DSB. In the above experiments, homologous sequences of 20 bases to either side of the DSB were sufficient to stimulate targeting and repair. To better understand the requirements for IRO repair of a DSB, several 80-base oligonucleotides (a, b, c, and d) were investigated in different combinations or individually for their ability to target the I-SceI-induced DSB at two different CORE-I-SceI orientations in TRP5 in the BY4742 background, as described in Fig. 2 B and C. All of the transformations involving pairs of oligonucleotides were added to the mix without prehybridization. Each oligonucleotide had 69 bases of homology to the chromosome at one side of the break (indicated by the dashed vertical line) and 10 bases of homology to the other. The homologies were to chromosomal sequences that were either immediately adjacent to the break or ≈4.7 kb (the region included in the I-SceI cassette) distant from the break. In some cases, the pairs of IROs were fully complementary (ac and bd pairs), so that one double-stranded end had only 10 bases of homology to one side of the DSB, and the other double-stranded end had 69 bases. Two of the IRO pairs had 20-base complementary overlaps at their 3′ (ab) or 5′ (cd) ends, so that if the pairs hybridized in the cell, there would be 69 bases of homology to either side of the DSB. Two pairs of IROs (ad and bc) had 20-base overlaps with the same polarity and, thus, they could not hybridize directly.
Transformation with the partially complementary IRO pairs that had 69 bases of homology on each side of the break resulted in ≈0.1% of all cells being targeted for both orientations of the CORE-I-SceI. The fully complementary pairs that had 69 bases of homology on one side of the break but only 10 bases of homology to the other side (ac and bd) were also capable of repair, although the efficiency was reduced at least 10-fold. Targeting appeared to be better if the shorter 10-base homology with the chromosome was adjacent to the break (compare bd vs. ac in Figs. 2 B and C and 5B). The ss a, b, c, and d IROs were also able to target the DSB, but at a much lower level. However, pairs of IROs of the same polarity (ad and bc) were more efficient at targeting than either ss for the R-AD and the R-DA configurations of IROs. Thus, the presence of an overlap (even if not complementary) within an IRO pair enhances the DSB repair process. In the absence of a break, targeted transformation was detected only for the ab and cd strand combinations (Fig. 2 B and C and Fig. 5B). The results were similar with comparable single IROs or combinations of a, b, c, and d R-AD and R-DA oligonucleotides targeted to a DSB at the TRP5, BAR1, and MSH2 loci in the CG379 strain (data not shown).
Targeting of IROs Can Occur at Distant Sites. Because IROs could be targeted when one of the homologous tails was 4.7 kb from the break, we examined the ability of a DSB to stimulate repair when both homologous tails were distant from the break. In the strain containing the CORE-I-SceI cassette and the I-SceIsitein TRP5, transformation frequencies of the R-AD ef 81-mer oligonucleotides (containing 40-nt symmetrical homologous tails) were compared with those obtained with R-DD (83-mers designed to delete 16.6 kb around TRP5) containing 40-nt symmetrical homologous tails (Fig. 1 and Table 2). Both the complementary and the ss IROs were examined (Fig. 3).
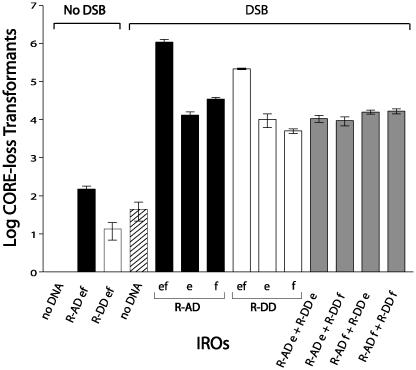
Fig. 3.
Effect on IRO targeting of position of homology relative to a DSB. Presented are numbers of 5-FOAR/HygroS transformants per 107 viable cells resulting from targeting to a DSB induced in the TRP5 gene by oligonucleotides that are fully complementary and symmetrical for homology (equal size of target sequences; see Fig. 1 and Table 2). Shown are the e and f R-AD oligonucleotides (81-mers that restore TRP5) and the R-DD oligonucleotides (83-mers that delete the CORE-I-SceI cassette plus 11.9 kb around the TRP5 locus). Vertical bars along with the standard deviations correspond to the average values from three to six determinations. Black, gray, and white bars are values for R-AD, R-AD + R-DD, and R-DD IROs, respectively. The striped bar represents the no-DNA control in galactose. The no-DNA control value (for conditions of no-DSB induction) is ≤1 transformant/107 viable cells. The types of IROs analyzed are indicated below the bars, and the amount of total IRO DNA used in each transformation is 1 nmol.
The efficiency of the R-DD pair with homologies to chromosome sequences 4.7 kb upstream and 11.9 kb downstream of the DSB, respectively, was reduced only 5-fold in comparison with the R-AD pair, even if the IROs generated a large 16.6-kb deletion. The ss IROs were also efficiently targeted to the sites distant from the break, although less than the corresponding complementary pair. No pairwise combinations of R-AD with R-DD ss increased the targeting frequencies over the level of the individual ss oligonucleotides. This result indicates that the increased targeting with the pairs of complementary R-AD or R-DD IROs is not simply due to targeting each of the two broken strands of the DSB.
Discussion
Oligonucleotide Targeting and Implications for DSB Repair. We have demonstrated that a single DSB that is induced or arises spontaneously in the chromosome can be found and repaired by exogenous oligonucleotides with high efficiency. DSB induction at an I-SceI site caused an ≈5,000-fold increase in targeting over that found by simply adding IROs, resulting in recombinational changes in 5-20% of the cells. If all of the cells have a break available at the time of IRO uptake, this corresponds to a minimum estimate of DSBs that are repairable by IROs. However, if DSB induction is not complete (see Materials and Methods), the efficiency will be higher.
The Alu-IR sequence caused a 30-fold stimulation of targeting, and the frequency of transformation with oligonucleotides was ≈0.1%. The Alu-IR site can lead to a spontaneous hairpin-capped break in ≈2% of asynchronous cells (37). Thus, there was also a high level (at least 5%) of targeting toward this form of spontaneous DSB. Targeting of oligonucleotides was stimulated (≈700-fold) even in the presence of a homologous chromosome, i.e., in diploid cells. These data demonstrate that the small oligonucleotides can efficiently compete with a large homologous chromosome for repair of the DSB.
The high frequencies of oligonucleotide targeting to a DSB could result from the large number (1 nmol) of IROs used and/or by the lack of chromatin structure of the IROs. Also, the small molecular weight may significantly reduce physical barriers to movement into and through the nucleus.
Based on results obtained with various IRO constructs, we conclude that the targeting and recovery of transformants are the result of homology-driven recombinational repair of a DSB. Although it is well established in yeast that a chromosomal DSB can be efficiently repaired by recombination with a homologous chromosome or sister chromatid, we show that exogenous short DNAs, such as pairs of overlapping oligonucleotides or even ss oligonucleotides, can also efficiently repair the DSB by homologous recombination. This conclusion follows from a comparison between transformants obtained with two different types of selection: selection for CORE loss and selection for the restoration of TRP5 function (Table 1). The oligonucleotides were designed in such a way that homologous recombination would restore function of TRP5. Even in the case of selection for CORE loss, the vast majority (≈80%) of transformants restored TRP5. Direct sequencing of the CORE-loss events established that all of them occur via homologous recombination, and the small fraction of Trp- events is due to mutations. Appearance of additional mutations could be explained by the occurrence of random errors in the preparation of commercial oligonucleotides or during the course of recombination, because all point mutations found were confined to the region of the oligonucleotide sequence.
Targeting depended strongly on RAD52 function (Table 1), further supporting the role for homology-driven recombination. Rad52 also appears to slow down degradation of 5′ ends (5′-resection) and prevent degradation of 3′ ends, which normally stay intact long after formation of a DSB (38). This dual function may explain the severe reduction of oligonucleotide transformation in the rad52 mutant observed in our experiments. However, even in the absence of RAD52, induction of a DSB could still stimulate IRO targeting at least 300-fold by complementary 95-mers. These less frequent oligonucleotide-targeting events might still involve the development of recombinational intermediates. Similarly, a residual RAD52-independent pathway of recombination was described in diploids after HO-endonuclease cutting of one of two homologous chromosomes (14).
The efficient targeting to positions up to 11.9 kb from the DSB (Fig. 3) indicates that regions distant from the DSB can be activated for recombinational interactions, in agreement with reports showing that sequences up to 8.6 kb from the DSB could successfully participate in a search for homology (39, 40).
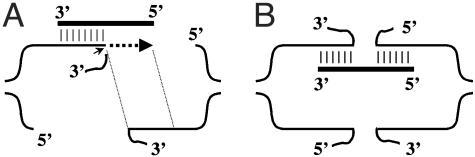
Based on current recombination models, the ends of a DSB are subject to 5′ resection, creating extended ss 3′ tails (2, 38). Resection could impair or even prevent recombination with ss DNA, especially if the ssDNA can pair only with one recessed end. In our experiments, the highest efficiency of DSB repair was obtained with the pair of fully complementary oligonucleotides (>50 bases) and with symmetrical homologous tails. However, the efficiency of repair by symmetrical ss oligonucleotides was also high, and there was no significant strand bias. These results raise a question about the structure of the broken ends at the time of interaction with an oligonucleotide. If a ss oligonucleotide interacts with a DSB after 5′ resection, it will be able to anneal with only one of the 3′ ends. However, after extension of this 3′ end by using the oligonucleotide molecule as a template, the opposite 3′ end of the DSB could be involved in the interaction (Fig. 4A). This “template” model contrasts with a “bridge” model (Fig. 4B), where the ss IRO is able to pair with homologous regions on both sides of the DSB. In both models, the bridge and template intermediates can be matured into final repair products after exonucleolytic trimming of nonhomologous chromosomal tails, DNA synthesis, and ligation.
Fig. 4.
Models of IRO targeted interactions and repair of a DSB. Homologous pairing interactions between the ss oligonucleotide (thick line) and the DSB ends are presented as short thin parallel vertical lines. (A) Template intermediate model. The IRO pairs with the homologous region on the 3′ strand after resection of the 5′ strand. The small arrow identifies the position at which nonhomologous sequence is cut. DNA synthesis (dotted line) can occur to copy the rest of the oligonucleotide. Homologous sequences between the 3′ ends of the DSB appear between the thin dotted lines. (B) Bridge intermediate model. As a result of unwinding of the DSB ends, annealing of the oligonucleotide can occur with the homologous regions on either side of the DSB creating a “bridge” between the two ends.
In light of the models described in Fig. 4 and those generally invoked in DSB repair, the efficiency of all of the IROs can be explained in terms of the ability to generate long complementary regions. Pairs of overlapping oligonucleotides of either opposite or the same polarity might work better than single IROs in both models, because they could extend the sequence available for homologous interaction between the IROs and the chromosomal ends (bridge model) or between the two 3′-chromosomal ends (template model). Regions of overlap that provide for complementarity between two oligonucleotides may also prevent strand degradation by ss exonucleases. IROs with two different lengths of homology to target sequences worked better when the short homologous tail was adjacent to the DSB. In both models, this might represent an advantage if there were oligonucleotide degradation. Alternatively, this could be explained by short homologous regions (i.e., 10 bases) being able to interact much more easily with homologous sequences close to the DSB than regions several kilobases distant from it.
Because of the high efficiency of targeting, it will be possible to use this system to examine new aspects of DSB repair, the impact of various mutants, and possibly characterize physical intermediates.
High-Throughput Technology for Genome Modification. The IRO targeting that we developed takes advantage of our previously described delitto perfetto approach (29), which provides for genomic modifications with oligonucleotides without the retention of any heterologous sequence. The present system with the CORE-I-SceI cassette containing a target DSB site utilizes the same straightforward procedures to generate site-directed mutations, replacements, or deletions and involves transformation by easily designed oligonucleotides.
Given the extremely high transformation frequencies, the possibilities for directed genomic changes are widely expanded in haploid and diploid yeast, making break-mediated delitto perfetto a high-throughput and versatile system. It can provide large-scale mutational scanning analyses as well as isolation of mutants without selection. Because even large deletions (16.6 kb) were obtained with high efficiency by using oligonucleotides, we are exploring the development of large genome rearrangements, such as development of circular chromosomes, chromosome fusions, and translocations (unpublished results). Moreover, this approach could be extended to the modification of genomes other than yeast where homologous recombination is also proficient.
Supplementary Material
Acknowledgments
We thank Kirill Lobachev for plasmid pKM4 and, together with Kerry Bloom and Gregory Stuart, for helpful suggestions and critical reading of the manuscript. We also thank all members of the Resnick laboratory for discussions and comments during the preparation of this work.
Abbreviations: DSB, double-strand break; IROs, integrative recombinant oligonucleotides; IR, inverted repeat; CORE, counterselectable reporter marker cassette; ss, single stranded; R-AD, repair-adjacent distant; R-DA, repair-distant adjacent; R-DD, repair-distant distant; R-AA, repair-adjacent adjacent; YPDA, yeast extract/peptone/dextrose/adenine medium; 5-FOA, 5-fluoroorotic acid; Hygro, hygromycin.
References
- 1.Resnick, M. A. (1978) J. Theor. Biol. 71, 339-346. [DOI] [PubMed] [Google Scholar]
- 2.Paques, F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeiffer, P., Goedecke, W. & Obe, G. (2000) Mutagenesis 15, 289-302. [DOI] [PubMed] [Google Scholar]
- 4.Jackson, S. P. (2002) Carcinogenesis 23, 687-696. [DOI] [PubMed] [Google Scholar]
- 5.Pastink, A., Eeken, J. C. & Lohman, P. H. (2001) Mutat. Res. 480-481, 37-50. [DOI] [PubMed] [Google Scholar]
- 6.Prudden, J., Evans, J. S., Hussey, S. P., Deans, B., O'Neill, P., Thacker, J. & Humphrey, T. (2003) EMBO J. 22, 1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung, P., Trujillo, K. M. & Van Komen, S. (2000) Mutat. Res. 451, 257-275. [DOI] [PubMed] [Google Scholar]
- 8.Lewis, L. K. & Resnick, M. A. (2000) Mutat. Res. 451, 71-89. [DOI] [PubMed] [Google Scholar]
- 9.Haber, J. E. (2002) Methods Enzymol. 350, 141-164. [DOI] [PubMed] [Google Scholar]
- 10.Plessis, A., Perrin, A., Haber, J. E. & Dujon, B. (1992) Genetics 130, 451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belfort, M. & Roberts, R. J. (1997) Nucleic Acids Res. 25, 3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong, W. J. & Golic, K. G. (2003) Proc. Natl. Acad. Sci. USA 100, 2556-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jasin, M. (1996) Trends Genet. 12, 224-228. [DOI] [PubMed] [Google Scholar]
- 14.Malkova, A., Ivanov, E. L. & Haber, J. E. (1996) Proc. Natl. Acad. Sci. USA 93, 7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, X. B. & Wilson, J. H. (1987) Proc. Natl. Acad. Sci. USA 84, 4959-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr-Weaver, T. L. & Szostak, J. W. (1983) Proc. Natl. Acad. Sci. USA 80, 4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothstein, R. J. (1983) Methods Enzymol. 101, 202-211. [DOI] [PubMed] [Google Scholar]
- 18.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duno, M., Bendixen, C., Krejci, L. & Thomsen, B. (1999) Nucleic Acids Res. 27, e1. [PMC free article] [PubMed] [Google Scholar]
- 20.Larionov, V., Kouprina, N., Graves, J., Chen, X. N., Korenberg, J. R. & Resnick, M. A. (1996) Proc. Natl. Acad. Sci. USA 93, 491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, H., Kunes, S., Schatz, P. J. & Botstein, D. (1987) Gene 58, 201-216. [DOI] [PubMed] [Google Scholar]
- 22.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- 23.Donoho, G., Jasin, M. & Berg, P. (1998) Mol. Cell. Biol. 18, 4070-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, D. G., Petek, L. M. & Russell, D. W. (2003) Mol. Cell. Biol. 23, 3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porteus, M. H., Cathomen, T., Weitzman, M. D. & Baltimore, D. (2003) Mol. Cell. Biol. 23, 3558-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barre, F. X., Mir, L. M., Lecluse, Y. & Harel-Bellan, A. (1998) BioTechniques 25, 294-296. [DOI] [PubMed] [Google Scholar]
- 27.Barre, F. X., Ait-Si-Ali, S., Giovannangeli, C., Luis, R., Robin, P., Pritchard, L. L., Helene, C. & Harel-Bellan, A. (2000) Proc. Natl. Acad. Sci. USA 97, 3084-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moerschell, R. P., Tsunasawa, S. & Sherman, F. (1988) Proc. Natl. Acad. Sci. USA 85, 524-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storici, F., Lewis, L. K. & Resnick, M. A. (2001) Nat. Biotechnol. 19, 773-776. [DOI] [PubMed] [Google Scholar]
- 30.Liu, L., Cheng, S., van Brabant, A. J. & Kmiec, E. B. (2002) Nucleic Acids Res. 30, 2742-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storici, F. & Resnick, M. A. (2003) in Genetic Engineering Principles and Methods, ed. Setlow J. K. (Kluwer/Plenum, Upton, NY), Vol. 25, pp. 189-207. [PubMed] [Google Scholar]
- 32.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P. & Boeke, J. D. (1998) Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- 33.Storici, F., Henneke, G., Ferrari, E., Gordenin, D. A., Hubscher, U. & Resnick, M. A. (2002) EMBO J. 21, 5930-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobachev, K. S., Stenger, J. E., Kozyreva, O. G., Jurka, J., Gordenin, D. A. & Resnick, M. A. (2000) EMBO J. 19, 3822-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein, A. L. & McCusker, J. H. (1999) Yeast 15, 1541-1553. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, F., Fink, G. R. & Hicks, J. B. (1986) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 37.Lobachev, K. S., Gordenin, D. A. & Resnick, M. A. (2002) Cell 108, 183-193. [DOI] [PubMed] [Google Scholar]
- 38.Aylon, Y., Liefshitz, B., Bitan-Banin, G. & Kupiec, M. (2003) Mol. Cell. Biol. 23, 1403-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inbar, O. & Kupiec, M. (1999) Mol. Cell. Biol. 19, 4134-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray, A., Machin, N. & Stahl, F. W. (1989) Proc. Natl. Acad. Sci. USA 86, 6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.