Abstract
Porcine intestinal mucosa heparin was partially depolymerized by recombinant heparinase 1 (heparin lyase 1, originating from Flavobacterium heparinum and expressed in Escherichia coli) and then fractionated, leading to the isolation of 22 homogeneous oligosaccharides with sizes ranging from disaccharide to hexadecasaccharide. The purity of these oligosaccharides was determined by gel electrophoresis, strong anion exchange and reversed-phase ion-pairing high-performance liquid chromatography. The molecular mass of oligosaccharides was determined using electrospray ionization-mass spectrometry and their structures were elucidated using one- and two-dimensional nuclear magnetic resonance spectroscopy at 600 MHz. Five of the characterized oligosaccharides represent new compounds. The most prominent oligosaccharide comprises the common repeating unit of heparin, ΔUA2S-[-GlcNS6S-IdoA2S-]n-GlcNS6S, where ΔUA is 4-deoxy-α-l-threo-hex-4-eno-pyranosyluronic acid, GlcN is 2-deoxy-2-amino-d-glucopyranose, IdoA is l-idopyranosyluronic acid, S is sulfate and n = 0–7. A second prominent heparin oligosaccharide motif corresponds to ΔUA2S-[GlcNS6S-IdoA2S]n-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S (where n = 0–5 and GlcA is d-glucopyranosyluronic acid), a fragment of the antithrombin III binding site in heparin. The prominence of this second set of oligosaccharides and the absence of intact antithrombin III binding sites suggest that the -GlcNS3S6S-IdoA2S- linkage is particularly susceptible to heparinase 1.
Keywords: antithrombin III, enzyme specificity, heparin, heparin lyase, oligosaccharide structure
Introduction
Heparin (Figure 1) is a highly sulfated, linear polysaccharide with a wide range of important biological activities (Linhardt 2003). These activities are due to its ability to interact with a large number of proteins (Capila and Linhardt 2002), which in turn result in their activation, deactivation or stabilization. The interaction of heparin with antithrombin III takes place at specific oligosaccharide sequences present within the heparin polymer (Linhardt and Gunay 1999; Linhardt 2003). As a result of a major contamination crisis associated with the death of a number of patients, there has been an increased interest in recent years in determining the structure of heparin, due to its importance as an anticoagulant drug, and in the application of degradation enzymes, such as the heparin lyases, in the characterization of heparin (Guerrini et al. 2008, 2009). Due to its structural complexity, relatively high molecular weight (Mr average 12,000) and polydispersity (Mr 5000–40,000), direct determination of its structure is not currently possible (Ly et al. 2010). A common approach is, first, to break down it into smaller oligosaccharides using chemical or enzymatic methods (Shively and Conrad 1976; Linhardt 1992) and then to characterize the structure of these heparin-derived oligosaccharides (Pervin et al. 1995). In this work, we have used recombinant heparin lyases originating from Flavobacterium heparinum (Lohse and Linhardt 1992) to cleave specific glycosidic linkages in heparin with a β-eliminative mechanism yielding oligosaccharides with an unsaturated uronic acid at their nonreducing termini, without altering their fine structure. The specificity of each lyase was studied in detail (Desai et al. 1993a, b; Jandik et al. 1994; Shriver et al. 2000). As part of a continuing effort, in our laboratory, to understand the structure of heparin and the specificity of the heparin lyases, porcine intestinal mucosa heparin was partially depolymerized by heparinase 1, and large, heparin-derived oligosaccharides were recovered and characterized.
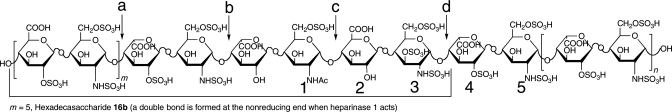
Fig. 1.
Heparin and its linkages susceptible to heparinase 1 cleavage. Linkage a and linkage b are susceptible to heparinase 1 cleavage, whereas linkage b and linkage c are resistant to heparinase 1. Residues labeled from 1 to 5 are the antithrombin pentasaccharide binding sites with a 3-O-sulfo group in residue 3. m + n = 16 for molecular weight of 12,000. When linkage d is cleaved by heparinase 1 and m = 0, 1, 2, 3, 4 or 5, the oligosaccharides released from heparin are corresponding to 6c, 8d, 10c, 12b, 14b and 16b, respectively.
Results
Isolation and purification of the oligosaccharides
Heparin was partially depolymerized (to approximately 30% reaction completion) using heparinase 1. The product was fractionated by Bio-gel P10 column chromatography to obtain oligosaccharides of uniform size from degree of polymerization (dp) 2 to dp20. After desalting and concentration, these uniform-sized oligosaccharides were further purified and separated by semi-preparative strong anion exchange (SAX)-high-performance liquid chromatography (HPLC) as described previously (Pervin et al. 1995). The most abundant peaks were recovered, and 22 oligosaccharides were prepared and their purities were confirmed to be >95% using analytical SAX-HPLC (Pervin et al. 1995), polyacrylamide gel electrophoresis (PAGE) analysis (Rice et al. 1987) and reversed-phase ion pairing (RPIP) HPLC-mass spectrometry (MS; Thanawiroon et al. 2004).
Oligosaccharide analysis using LC-MS
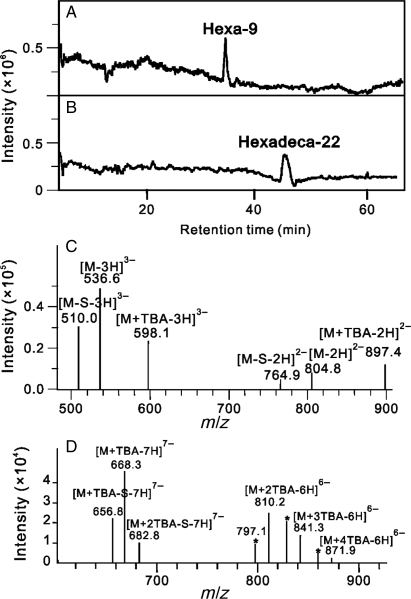
RPIP-HPLC-MS has been extensively used for the analysis of oligosaccharides derived from glycosaminoglycans (GAGs). In our laboratory, we routinely use an electrospray ionization (ESI) ion-trap mass spectrometer coupled with RPIP-HPLC for disaccharide compositional studies (Thanawiroon et al. 2004; Zhang et al. 2009; Volpi and Linhardt 2010). RPIP-HPLC-MS is also useful for the analysis of oligosaccharides as large as hexadecasaccharides (dp16) and provides easily interpretable mass spectra and molecular ion information (Figure 2 and Table I).
Fig. 2.
LC-MS analysis of hexasaccharide 6c and hexadecasaccharide 16b. Total-ion chromatograms of 6c (A) and 16b (B) show high purities of the two oligosaccharides. Molecular ions marked with charges ranging from −3 to −2 for 6c (C) and −7 to −6 for 16b (D) are the major signals observed from negative-ion ESI-MS spectra scanned from 150 to 1500 Da, respectively. A set of ions at m/z 797.1, 828.0 and 859.0, marked by stars, are molecular ions lost one SO3 moiety each from the protonated complex of 16b with two, three or four TBA molecules, respectively.
Table I.
Major molecular ions of oligosaccharides observed in the negative-ion ESI mass spectra
|
m/z |
||||
|---|---|---|---|---|
| No. | Structure | Molecular ion | Calculated | Observed |
| 2a | ΔUA2S-GlcNS | [M-H]− | 496.0 | 495.7a |
| 2b | ΔUA2S-GlcNS6S | [M-H]− | 576.0 | 575.6a |
| 4a | ΔUA2S-GlcNS6S-IdoA2S-GlcNS | [M-2H]2− | 536.0 | 535.8a |
| 4b | ΔUA2S-GlcNS6S-GlcA-GlcNS6S | [M-2H]2− | 536.0 | 535.8a |
| 4c | ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S | [M-2H]2− | 576.0 | 575.7a |
| 5 | ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA | [M-2H]2− | 605.0 | 604.8a |
| 6a | ΔUA2S-GlcNS6S-IdoA2S-GlcNAc-GlcA-GlcNS6S | [M-2H]2− | 765.5 | 765.0 |
| [M-3H]3− | 510.0 | 509.9a | ||
| 6b | ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-GlcA-GlcNS6S | [M-2H]2− | 824.5 | 824.2 |
| [M-3H]3− | 549.3 | 549.3a | ||
| 6c | ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S | [M-2H]2− | 805.5 | 804.8 |
| [M-3H]3− | 536.7 | 536.6a | ||
| 8s | ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS | [M-3H]3− | 741.7 | 741.6 |
| [M-4H]4− | 556.0 | 556.0a | ||
| 8b | ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-GlcA-GlcNS6S | [M-3H]3− | 741.7 | 742.1b |
| [M-4H]4− | 556.0 | 556.4a | ||
| 8c | ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S | [M-4H]4− | 576.0 | 576.1a |
| [M-3H]3− | 768.3 | 768.1b | ||
| 8d | ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S | [M-4H]4− | 546.5 | 546.7a |
| [M-3H]3− | 729.0 | 728.9b | ||
| 10a | ΔUA2S-GlcNS6S-[-IdoA2S-GlcNS6S-]3-GlcA-GlcNS6S | [M-5H]5− | 561.0 | 559.9a |
| [M-4H]4− | 700.3 | 700.2b | ||
| 10b | ΔUA2S-[-GlcNS6S-IdoA2S-]4-GlcNS6S | [M-5H]5− | 576.0 | 576.4a |
| [M-S-5H]5− | 560.0 | 560.1 | ||
| 10c | ΔUA2S-[-GlcNS6S-IdoA2S-]2-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S | [M-5H]5− | 552.4 | 553.0a |
| [M-S-5H]5− | 536.4 | 537.0 | ||
| 12a | ΔUA2S-[-GlcNS6S-IdoA2S-]5-GlcNS6S | [M-6H]6− | 576.0 | 575.9a |
| [M-5H]5− | 691.4 | 691.1b | ||
| 12b | ΔUA2S-[-GlcNS6S-IdoA2S-]3-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S | [M-6H]6− | 556.3 | 556.3 |
| [M-5H]5− | 667.8 | 667.6b | ||
| [M-S-6H]6− | 543.0 | 543.1a | ||
| 14a | ΔUA2S-[-GlcNS6S-IdoA2S-]6-GlcNS6S | [M-7H]7− | 576.0 | 575.7 |
| [M-2S-7H]7− | 553.1 | 553.1a | ||
| 14b | ΔUA2S-[-GlcNS6S-IdoA2S-]4-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S | [M-7H]7− | 559.1 | 559.0 |
| [M-6H]6− | 652.5 | 652.6 | ||
| [M-2S-7H]7− | 536.2 | 536.3a | ||
| 16a | ΔUA2S-[-GlcNS6S-IdoA2S-]7-GlcNS6S | [M-8H]8− | 576.0 | 576.4 |
| [M-7H]7− | 658.4 | 658.6a,b | ||
| 16b | ΔUA2S-[-GlcNS6S-IdoA2S-]5-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S | [M-7H]7− | 641.5 | 641.9a,b |
| [M-6H]6− | 748.7 | 748.5b | ||
aBase peak.
bMass deduced from the protonated complex of oligosaccharides with TBA.
-S, SO3 moiety (80 U) lost from molecular ion.
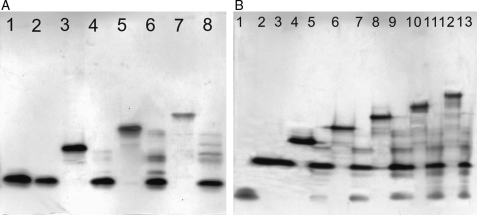
PAGE analysis of oligosaccharides and their enzymatic digested products
PAGE analysis, on a gel with a polyacrylamide concentration of 22%, clearly demonstrates a high level of sample purity and confirms the size of each oligosaccharide (Figure 3). An assessment of the primary structure of oligosaccharides was also made by PAGE analysis after treatment with heparinase 1. Tetrasaccharide ΔUA2S-GlcNS6S-GlcA-GlcNS6S (4b) and hexasaccharide ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S (6c) were resistant to heparinase 1, which is consistent with previous reports (Desai et al. 1993b; Yamada et al. 1994; Shriver et al. 2000). The structural specificity of heparinase 1 was further examined by exhaustively treating these oligosaccharides with heparinase 1 under similar protocol. The starting materials and their digested products were subjected to 22% PAGE gels and developed by Alcian blue and later by silver staining. The resulting gels clearly showed that the oligosaccharides could be generally divided into three groups of structures: disaccharide ΔUA2S-GlcNS6S (2b), tetrasaccharide ΔUA2S-GlcNS6S-GlcA-GlcNS6S (4b) and hexasaccharide ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S (6c).
Fig. 3.
PAGE analysis of oligosaccharides and their products digested by heparinase 1. (A) Lanes 1–8: 4b, heparinase 1-treated 4b, 6b, digested 6b, 8b, digested 8b, 10b and digested 10b, respectively. These oligosaccharides share the common sequence of 4b at the reducing end. (B) Lanes 1–13: di-2, 6b, heparinase 1-treated 6b, 8d, digested 8d, 10c, digested 10c, 12b, digested 12b, 14b, digested 14b, 16b and digested 16b, respectively. These oligosaccharides have a partially intact antithrombin III binding site -GlcNAc6S-GlcA-GlcNS3S6S at the reducing end. Oligosaccharides 4b and 6c were resistant to heparinase 1, and the other large oligosaccharides, which have either of them at the reducing end, show corresponding bands on PAGE.
Structural characterization of oligosaccharides by 1D proton and 2D NMR experiments
The 600 MHz 1D proton and 2D NMR spectra of all the oligosaccharides were acquired for their structural elucidation. Oligosaccharides 4c, 8c, 10b, 12a, 14a and 16a yielded similar 1D and 2D NMR spectra, demonstrating that they were structural analogs, differing only in the number of central trisulfated GlcNS6S-IdoA2S disaccharide units. The major spectral difference between these oligosaccharides was in the integration of the anomeric protons of internal GlcNS6S and IdoA2S residues compared with the ΔUA2S nonreducing-end residue and the GlcNS6S reducing-end residue. The NMR data of 4c, 8c, 10b, 12a and 14a (Table II) were consistent with structures previously reported by our laboratory (Pervin et al. 1995). The 600 MHz 1D spectrum of 16a showed the presence of 16 anomeric protons that resonate as doublets at δ 5.46 (A-1, J 1,2 = 2.2 Hz), 5.36 (P-1, J1,2 = 2.4 Hz), 5.30 (7H, broad singlet) and 5.28 ppm (7H, broad singlet). The other signals of cross-peaks were confirmed by 1H–1H homonuclear COrrelation SpectroscopY (COSY) spectrum and Heteronuclear Multiple-Quantum Coherence (HMQC) spectrum.
Table II.
Proton chemical shift assignments of heparin-derived oligosaccharidesa
| α-l-IdoA/β-d-GlcA |
α-d-GlcN |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 6 | 6′ | ||
| 2a | |||||||||||||
| A | 5.45 | 4.47 | 4.22 | 5.91 | B | 5.34 | 3.16 | 3.66 | 3.70 | 3.88 | 3.77 | 3.77 | |
| 2b | |||||||||||||
| A | 5.44 | 4.50 | 4.28 | 5.89 | B | 5.38 | 3.21 | 3.69 | 3.77 | 4.08 | 4.30 | 4.15 | |
| 4a | |||||||||||||
| A | 5.41 | 4.53 | 4.21 | 5.89 | B | 5.28 | 3.20 | 3.54 | 3.75 | 3.93 | 4.26 | 4.15 | |
| C | 5.11 | 4.20 | 4.13 | 3.98 | 4.70 | D | 5.35 | 3.15 | 3.59 | 3.61 | 3.84 | 4.26 | 4.15 |
| 4b | |||||||||||||
| A | 5.45 | 4.58 | 4.27 | 5.94 | B | 5.51 | 3.27 | 3.59 | 3.78 | 3.94 | 4.30 | 4.15 | |
| C | 4.54 | 3.33 | 3.79 | 3.37 | 3.76 | D | 5.41 | 3.21 | 3.67 | 3.66 | 4.11 | 4.31 | 4.29 |
| 4c | |||||||||||||
| A | 5.45 | 4.57 | 4.25 | 5.93 | B | 5.39 | 3.25 | 3.58 | 3.79 | 4.00 | 4.30 | 4.20 | |
| C | 5.15 | 4.27 | 4.15 | 4.06 | 4.69 | D | 5.39 | 3.21 | 3.65 | 3.69 | 4.08 | 4.32 | 4.25 |
| 5 | |||||||||||||
| A | 5.44 | 4.51 | 4.27 | 5.99 | B | 5.24 | 3.20 | 3.65 | 3.78 | 3.92 | 4.23 | 4.12 | |
| C | 5.03 | 3.74 | 4.07 | 4.00 | 4.95 | D | 5.28 | 3.84 | 3.73 | 3.75 | 3.83 | 4.27 | 4.10 |
| E(α) | 5.14 | 3.50 | 3.79 | 4.11 | 3.78 | E(β) | 4.54 | 3.21 | 3.58 | 3.66 | 3.67 | ||
| 6a | |||||||||||||
| A | 5.47 | 4.53 | 4.25 | 6.06 | B | 5.24 | 3.22 | 3.63 | 3.77 | 3.88 | 4.25 | 4.12 | |
| C | 5.20 | 4.25 | 4.13 | 4.01 | 4.70 | D | 5.29 | 3.84 | 3.70 | 3.63 | 3.90 | 3.76 | 3.76 |
| E | 4.53 | 3.27 | 3.66 | 3.74 | 3.70 | F | 5.38 | 3.18 | 3.62 | 3.65 | 4.07 | 4.29 | 4.24 |
| 6b | |||||||||||||
| A | 5.44 | 4.51 | 4.22 | 6.01 | B | 5.27 | 3.20 | 3.58 | 3.75 | 3.88 | 4.23 | 4.12 | |
| C | 5.18 | 4.24 | 4.22 | 3.99 | 4.94 | D | 5.48 | 3.16 | 3.61 | 3.66 | 3.81 | 4.23 | 4.12 |
| E | 4.52 | 3.29 | 3.75 | 3.87 | 3.73 | F | 5.36 | 3.17 | 3.58 | 3.61 | 4.05 | 4.24 | 4.22 |
| 6c | |||||||||||||
| A | 5.41 | 4.51 | 4.22 | 6.01 | B | 5.27 | 3.20 | 3.58 | 3.75 | 3.88 | 4.23 | 4.12 | |
| C | 4.98 | 3.73 | 4.06 | 3.97 | 4.73 | D | 5.29 | 3.83 | 3.70 | 3.68 | 3.93 | 4.27 | 4.12 |
| E | 4.55 | 3.29 | 3.61 | 3.70 | 3.69 | F | 5.37 | 3.36 | 4.43 | 3.94 | 4.15 | 4.35 | 4.22 |
| 8a | |||||||||||||
| A | 5.45 | 4.52 | 4.25 | 5.94 | B | 5.29 | 3.21 | 3.61 | 3.77 | 3.87 | 4.29 | 4.17 | |
| CE | 5.18 | 4.25 | 4.18 | 4.00 | 4.90 | DF | 5.32 | 3.22 | 3.61 | 3.76 | 3.89 | 4.27 | 4.15 |
| G | 5.18 | 4.27 | 4.16 | 4.11 | 4.91 | H | 5.37 | 3.23 | 3.63 | 3.62 | 3.85 | 3.78 | 3.77 |
| 8b | |||||||||||||
| A | 5.42 | 5.49 | 4.21 | 5.95 | BD | 5.26 | 3.15 | 3.58 | 3.73 | 3.87 | 4.22 | 4.11 | |
| CE | 5.20 | 4.24 | 4.16 | 3.99 | 4.90 | F | 5.47 | 3.16 | 3.59 | 3.66 | 3.80 | 4.22 | 4.11 |
| G | 4.50 | 3.26 | 3.74 | 3.80 | 3.70 | H | 5.33 | 3.18 | 3.62 | 3.60 | 4.01 | 4.22 | 4.23 |
| 8c | |||||||||||||
| A | 5.46 | 4.55 | 4.28 | 6.05 | B | 5.31 | 3.21 | 3.65 | 3.70 | 3.86 | 4.28 | 4.16 | |
| CE | 5.22 | 4.26 | 4.22 | 4.01 | 4.96 | DF | 5.30 | 5.32 | 3.66 | 3.78 | 3.87 | 4.28 | 4.18 |
| G | 5.23 | 4.29 | 4.23 | 4.10 | 4.94 | H | 5.37 | 3.24 | 3.67 | 3.69 | 4.08 | 4.30 | 4.25 |
| 8d | |||||||||||||
| A | 5.33 | 4.41 | 4.15 | 5.88 | B | 5.21 | 3.10 | 3.49 | 3.67 | 3.82 | 4.16 | 4.02 | |
| C | 5.10 | 4.14 | 4.09 | 3.92 | 4.80 | D | 5.11 | 3.03 | 3.53 | 3.60 | 3.77 | 4.12 | 4.02 |
| E | 4.95 | 3.98 | 3.67 | 3.86 | 4.81 | F | 5.15 | 3.73 | 3.60 | 3.63 | 3.72 | 4.18 | 4.03 |
| G | 4.41 | 3.19 | 3.50 | 3.63 | 3.69 | H | 5.26 | 3.23 | 4.27 | 3.63 | 4.01 | 4.26 | 4.12 |
| 10a | |||||||||||||
| A | 5.48 | 4.55 | 4.27 | 6.04 | BDF | 5.31 | 3.20 | 3.59 | 3.77 | 3.90 | 4.28 | 4.15 | |
| CEG | 5.21 | 4.26 | 4.20 | 4.05 | 5.00 | H | 5.52 | 3.21 | 3.60 | 3.69 | 3.92 | 4.29 | 4.16 |
| I | 4.56 | 3.32 | 3.79 | 3.90 | 3.78 | J | 5.38 | 3.22 | 3.68 | 3.64 | 3.84 | 4.25 | 4.27 |
| 10b | |||||||||||||
| A | 5.34 | 4.42 | 4.15 | 5.93 | B | 5.20 | 3.07 | 3.48 | 3.57 | 3.76 | 4.20 | 4.06 | |
| CEG | 5.10 | 4.16 | 4.09 | 3.93 | 4.85 | DFH | 5.22 | 3.09 | 3.49 | 3.65 | 3.80 | 4.19 | 4.04 |
| I | 5.12 | 4.17 | 4.10 | 3.94 | 4.86 | J | 5.23 | 3.05 | 3.53 | 3.55 | 3.95 | 4.13 | 4.15 |
| 10c | |||||||||||||
| A | 5.34 | 4.41 | 4.14 | 5.86 | BD | 5.20 | 3.12 | 3.50 | 3.64 | 3.80 | 4.13 | 4.01 | |
| CE | 5.10 | 4.15 | 3.10 | 3.91 | 4.79 | F | 5.19 | 3.05 | 3.49 | 3.57 | 3.82 | 4.15 | 4.02 |
| G | 4.93 | 3.66 | 3.97 | 3.86 | 4.82 | H | 5.15 | 3.74 | 3.60 | 3.58 | 3.72 | 4.16 | 4.00 |
| I | 4.42 | 3.20 | 3.51 | 3.62 | 3.66 | J | 5.25 | 3.22 | 4.28 | 3.78 | 4.00 | 4.26 | 4.20 |
| 12a | |||||||||||||
| A | 5.42 | 4.55 | 4.23 | 5.90 | B | 5.37 | 3.19 | 3.59 | 3.74 | 3.93 | 4.26 | 4.16 | |
| CEGI | 5.13 | 4.27 | 4.12 | 4.00 | 4.75 | DFHJ | 5.34 | 3.20 | 3.59 | 3.67 | 3.94 | 4.33 | 4.15 |
| K | 5.14 | 4.22 | 4.09 | 3.95 | 4.67 | L | 5.37 | 3.20 | 3.60 | 3.68 | 3.95 | 4.30 | 4.20 |
| 12b | |||||||||||||
| A | 5.48 | 4.53 | 4.26 | 6.04 | BDF | 5.29 | 3.20 | 3.60 | 3.78 | 3.88 | 4.26 | 4.13 | |
| CEG | 5.25 | 4.27 | 4.21 | 4.03 | 4.95 | H | 5.23 | 3.21 | 3.61 | 3.77 | 3.86 | 4.27 | 4.14 |
| I | 5.07 | 3.80 | 4.11 | 3.98 | 4.96 | J | 5.25 | 3.85 | 3.71 | 3.73 | 3.92 | 4.28 | 4.15 |
| K | 4.55 | 3.30 | 3.63 | 3.71 | 3.85 | L | 5.36 | 3.34 | 4.39 | 3.88 | 4.14 | 4.37 | 4.20 |
| 14a | |||||||||||||
| A | 5.43 | 4.53 | 4.23 | 5.90 | B | 5.34 | 3.18 | 3.57 | 3.71 | 3.93 | 4.33 | 4.15 | |
| CEGIK | 5.15 | 4.26 | 4.12 | 4.03 | 4.76 | DFHJL | 5.33 | 3.19 | 3.56 | 3.70 | 3.94 | 4.32 | 4.14 |
| M | 5.14 | 4.24 | 4.13 | 4.01 | 4.67 | N | 5.36 | 3.19 | 3.59 | 3.71 | 4.04 | 4.31 | 4.21 |
| 14b | |||||||||||||
| A | 5.42 | 4.54 | 4.21 | 5.90 | BDFH | 5.33 | 3.18 | 3.56 | 3.69 | 3.95 | 4.34 | 4.17 | |
| CEGI | 5.14 | 4.26 | 4.11 | 4.02 | 4.73 | J | 5.25 | 3.19 | 3.57 | 3.70 | 3.94 | 4.33 | 4.16 |
| K | 4.94 | 3.70 | 4.03 | 4.00 | 4.71 | L | 5.30 | 3.84 | 3.68 | 3.67 | 3.89 | 4.32 | 4.15 |
| M | 4.54 | 3.28 | 3.59 | 3.74 | 3.70 | N | 5.37 | 3.36 | 4.42 | 3.91 | 4.13 | 4.33 | 4.25 |
| 16a | |||||||||||||
| A | 5.46 | 4.51 | 4.25 | 5.91 | B | 5.28 | 3.20 | 3.65 | 3.72 | 4.00 | 4.30 | 4.16 | |
| CEGIKM | 5.30 | 4.29 | 4.19 | 4.01 | 4.82 | DFHJLN | 5.27 | 3.21 | 3.66 | 3.73 | 4.05 | 4.31 | 4.17 |
| O | 5.31 | 4.30 | 4.20 | 4.05 | 4.83 | P | 5.36 | 3.22 | 3.67 | 3.78 | 3.93 | 4.25 | 4.20 |
| 16b | |||||||||||||
| A | 5.46 | 4.52 | 5.26 | 5.92 | BDFHJ | 5.29 | 3.21 | 3.66 | 3.75 | 4.00 | 4.33 | 4.15 | |
| CEGIK | 5.29 | 4.28 | 4.18 | 4.01 | 4.84 | L | 5.20 | 3.22 | 3.71 | 3.76 | 3.99 | 4.34 | 4.16 |
| M | 5.11 | 3.80 | 4.08 | 3.95 | 4.85 | N | 5.29 | 3.84 | 3.75 | 3.70 | 3.98 | 4.34 | 4.16 |
| O | 4.54 | 3.29 | 3.63 | 3.78 | 3.77 | P | 5.37 | 3.37 | 4.41 | 3.93 | 4.14 | 4.27 | 4.20 |
aChemical shifts observed from 1D proton and 2D NMR spectra. Protons in the N-acetyl group of glucosamine residues resonate at 2.03 ppm are not presented.
Similarly, oligosaccharides 4b, 6b, 8b and 10a yielded similar 1D and 2D NMR spectra, demonstrating that they were structural analogs, differing only in the numbers of central GlcNS6S-IdoA2S disaccharide units. Cross-peaks, in the 1H–1H COSY spectra corresponding to the H-3/H-4 of the 3-O-sulfo glucosamine residues and in the HMQC spectra corresponding to the H-6/C-6 of the glucosamine residues without 6-O-sulfo groups, as well as the signals of N-acetyl groups in the GlcNAc residues, were absent in the NMR analysis of these oligosaccharides. These NMR data together with molecular data from mass spectral analysis were important for the structural characterization of these oligosaccharides. The sequences of these four oligosaccharides (Table I) begin with an identical disaccharide unit, GlcA-GlcNS6S, at their reducing end, as deduced by 1H–1H COSY, HMQC, Rotating-frame Overhauser Effect SpectroscopY (ROESY), Nuclear Overhauser Effect SpectroscopY (NOESY) and TOtal Correlation SpectroscopY (TOCSY) spectra. The NMR data for these oligosaccharides are also consistent with those reported previously (Pervin et al. 1995).
Oligosaccharides 6c, 8d, 10c, 12b, 14b and 16b showed similar 1D NMR spectra and HMQC spectra, demonstrating that they were structural analogs differing only in the number of central GlcNS6S-IdoA2S disaccharide units. The major spectral difference between these oligosaccharides was in the integration of the anomeric protons of internal GlcNS6S and IdoA2S residues when compared with the ΔUA2S nonreducing-end residue and the glucosamine reducing-end residue. An increase in the molecular weight of oligosaccharides resulted in a decrease in the resolution of various internal sugar residues. The 1H–1H COSY spectrum of hexasaccharide 6c clearly resolves the coupling connectivity within all the saccharide residues. Although the chemical shift differences for the H-3 and H-4 resonances of IdoA are very small, the spin systems for internal IdoA residues in 6c are clearly visible. The 2D 1H–1H COSY spectra of 8d, 10c, 12b, 14b and 16b, however, showed a superimposition of the various signals of the internal IdoA and GlcN residues. This is because the chemical shift differences of H-2, H-3 and H-4 of these internal IdoA residues are less than their coupling constants; hence, they appeared as overlapping multiplets. Similarly, the H-6, H-6′, H-3 and H-4 of the internal GlcN residues also appear as overlapping multiplets.
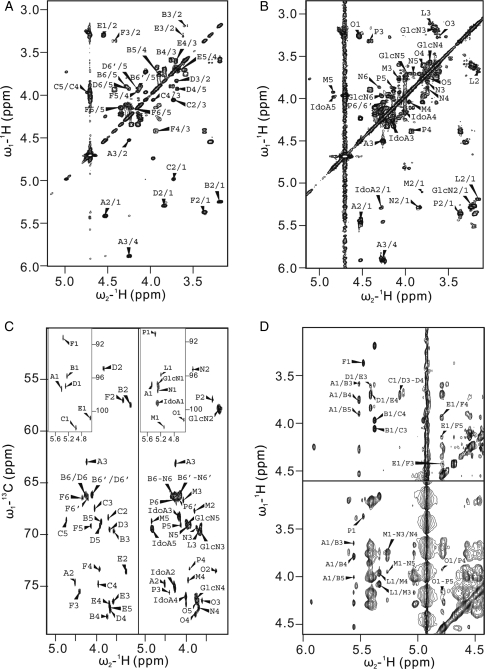
Hexasaccharide 6c had been reported previously (Linhardt et al. 1992). The 1D spectrum of 6c shows six anomeric protons in the downfield region (Table II) and the homonuclear COSY spectrum provides complete coupling information for all the protons within the same spin system. The protons of GlcN (residue B), IdoA (residue C), GlcN (residue D), GlcA (residue E) and GlcN (residue F) were unambiguously assigned from the 1H–1H COSY spectrum (Figure 4A) and the HMQC spectrum (Figure 4B). The sequence of 6c was further determined by 2D ROESY (Figure 4C) and 1H–1H TOCSY (data not shown). 1H–1H TOCSY showed intrasugar residue couplings and ROESY showed both intra- and intersugar residue couplings. The 2D ROESY spectrum showed that the H-1 of the glucosamine (residue F), at δ 5.37 ppm, coupled with none of the inter-residue protons and that the carbon chemical shift of C-1 in residue F, at δ 91.3 ppm, in HMQC spectrum is relatively upfield-shifted compared with the internal anomeric protons of GlcN, further confirming the position of residue F. H-1 of GlcA (residue E) coupled with H-4 in residue F, at δ 3.93 ppm, in ROESY determined the position of the GlcA residue. Similarly, the anomeric proton in residue D, at δ 5.29 ppm, and the anomeric proton in residue C, at δ 4.98 ppm, coupled with H-4 of residue E and H-4 of residue D, respectively. All other cross-peaks (Figure 4C) were consistent with the structure of 6c (Figure 5).
Fig. 4.
Two-dimensional NMR analysis of a hexasaccharide and a hexadecasaccharide. 1H–1H COSY spectra of 6c (A) and 16b (B). GlcN and IdoA stand for the overlapped spin systems of glucosamine and iduronic acid residues, respectively. (C) HMQC spectra of 6c (left panel) and 16b (right panel). Insets show the anomeric proton regions. (C) ROESY spectra of 6c (upper panel) and NOESY spectra of 16b (lower panel). All these spectra show the similarities between these two structures.
Fig. 5.
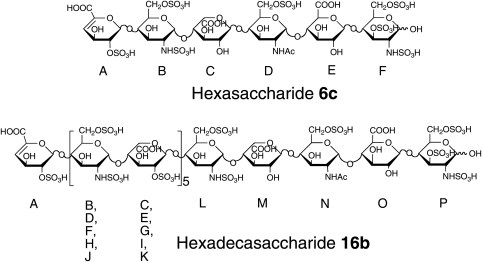
The chemical structures of hexasaccharide 6c and hexadecasaccharide 16b. In 6c, the residues are labeled A–F, starting with the ΔUA2S residue at the nonreducing end, and has a 3-O-sulfate in F residue. In contrast to 6c, 16b has an octasaccharide [GlcNS6S-IdoA2S]4 in the middle of its sequence. The M, N, O and P residues at the reducing end of 16b correspond to the C, D, E and F residues in 6c.
The same analysis can be successfully performed on larger oligosaccharides. The chemical shifts of H-3 in the GlcNS3S6S residues shift ∼0.8 ppm downfield compared with those in the GlcNS6S residues, resulting in characteristic cross-peaks of H-3 and H-2 in the 1H–1H COSY spectra of 8d, 10c, 12b and 14b. In the HMQC spectra, the cross-peaks of anomeric protons and carbons in the GlcNS3S6S residues of these four oligosaccharides resonated at relative upfield in the 13C dimension. The 5–11 ppm upfield shifts allow for their assignments as the anomeric signals at the reducing-end residues. The presence of the anomeric protons of the GlcA residues and the N-acetyl methyl groups in the GlcNAc6S residues was confirmed by the characteristic signals in their 1H–1H COSY spectra and HMQC spectra, respectively (Table II). The sequence of -IdoA-GlcNAc6S-GlcA-GlcNS3S6S at the reducing end of each of these oligosaccharides was confirmed by ROESY and NOESY experiments assisted by 1H–1H TOCSY (Yang et al. 2000).
Hexadecasaccharide (16b) also has a partially intact antithrombin III binding site at its reducing end. Its molecular size and molecular weight were confirmed, in part, by oligosaccharide analysis using LC-MS and PAGE analysis before and after complete enzymatic digestion. NMR experiments were performed to confirm its primary structure and sequence. From the 1D NMR spectrum, signals corresponding to the anomeric protons of 16b were observed at δ 5.48 (A-1), 5.37 (P-1), 5.20 (L-1), 5.12 (M-1) and 4.55 ppm (O-1). Spin systems belonging to the GlcNS6S and IdoA2S residues, the signals of which overlapped completely, showed a large broad peak at δ ∼ 5.3 ppm with integration of 11H. From 1H–1H COSY and HMQC, the overlapped anomeric protons could easily be assigned. The N1 resonated at δ 5.29 ppm, whereas the other 10 anomeric protons of glucosamine and iduronic acid appeared as two individual cross-peaks in the 1H–1H COSY and HMQC (Figure 4A and B). Compared with the 1H–1H COSY and HMQC spectra of 6c, the 2D spectra of 16b were very similar except for some additional cross-peaks arising from the additional internal GlcN2S6S and IdoA2S residues. From HMQC, the cross-peak of the anomeric proton at the reducing end resonated at δ 13C 91 ppm. The anomeric proton of the reducing-end residue could be confirmed by the absence of an interring cross-peak in the NOESY spectrum (Figure 4C). From the NOESY spectrum of 16b, residues of L, M, N, O and P could be assigned as a contiguous saccharide sequence in this order by their anomeric protons coupled with the H-4 protons in the adjacent residues.
Discussion
This study clearly demonstrates that heparinase 1 cleaves heparin at both the GlcNS6S (1 → 4) IdoA2S and GlcNS3S6S (1 → 4) IdoA2S linkages. The GlcNS3S6S (1 → 4) IdoA2S linkage, found primarily within the antithrombin III binding site, is more susceptible to heparinase 1 than the major heparin disaccharide repeating unit GlcNS6S (1 → 4) IdoA2S. Thus, although heparinase 1 can be utilized in the preparation of oligosaccharides with uniform GlcNS6S (1 → 4) IdoA2S repeating structures and oligosaccharides having partially intact antithrombin III binding sites, containing a trisaccharide GlcNAc6S-GlcA-GlcNS3S6S sequence at their reducing end, it is not very useful in the preparation of intact antithrombin III binding sites. Our study using viscosometric and PAGE analysis reported that heparinase 1 acts on heparin in a random endolytic fashion (Linhardt et al. 1982; Cohen and Linhardt 1990; Jandik et al. 1994), whereas other studies have suggested that heparinase 1 was an exolytic lyase because it left intact hexasaccharide 6c (Ernst et al. 1998; Shriver et al. 2000). In the current study, we find that heparinase 1 preferentially forms larger oligosaccharides (dp ≥ 8) having reducing-end sequences of -GlcNAc6S-GlcA-GlcNS3S6S, identical to that present at the reducing-end of hexasaccharide 6c, and these larger oligosaccharides (dp ≥ 8) are still sensitive to heparinase 1 and can be degraded to 6c on prolonged enzymatic treatment. In light of these observations, we suggest that heparinase 1 is an endolytic enzyme that preferentially cleaves GlcNS3S6S (1 → 4) IdoA2S linkages. The presence of linkages, within heparin, having very different heparinase 1 sensitivity results in product distributions that can be easily mistaken for an exolytic action pattern. The preference of heparinase 1 for GlcNS3S6S (1 → 4) IdoA2S linkages over GlcNS6S (1 → 4) IdoA2S linkages fortuitously affords a similar distribution of oligosaccharides in each size grouping that can be used to guide for the collection of fractions having similar structures but different molecular-size oligosaccharides. Among the most convincing data we find for the predominant endolytic activity of heparinase 1 is the kinetic analysis of oligosaccharide products by gel permeation chromatography (Linhardt et al. 1982; and Supplementary data).
In this study, five new oligosaccharides were isolated and structurally characterized. Although the structures of these new oligosaccharides were not novel, they offer additional reagents for future activity studies and as potential defined substrates for biosynthetic and catabolic enzymes. The α-anomeric form of the reducing terminal N-sulfoglucosamine is expected to be more stable than the β-anomeric form based on the α-anomeric effect. In this study, we observe only the α-anomeric form (>95%) in both 1D and 2D NMR spectra. The characterization of these larger structures demonstrates that both sophisticated NMR spectroscopy and LC-MS can be applied to very large, highly charged structures. The largest that has been characterized so far are 16a and 16b hexadecasaccharides with a molecular mass of 5240–5320 having eight carboxylate and 23–24 sulfate groups. These oligosaccharides are approximately of the same size as the typical chains found in many low-molecular weight heparins (Linhardt and Gunay 1999). One of these low-molecular weight heparins, tinzaparin, is prepared by partial β-eliminative cleavage of porcine intestinal heparin using heparinase 1. Thus, this study describes components likely contained in this low-molecular weight heparin and explains their presence based on the specificity of heparinase 1.
Materials and methods
Preparation of oligosaccharides derived from heparin by gel permeation chromatography and semi-preparative SAX-HPLC
Heparin sodium salt was obtained from porcine intestinal mucosa (Celsus Laboratories, Cincinnati, OH). Recombinant heparinase 1 (EC 4.2.2.7) from F. heparinum (also known as Pedobacter heparinus) and expressed in Escherichia coli was a gift from Dr. Jian Liu. Heparin (6 g) was digested by 10 U of heparinase 1 at 30°C and the reaction was quenched in a 100°C water bath for 10 min when it reached 30% of the maximum ultraviolet absorbance, as predetermined in a small-scale reaction. The reaction mixture was then concentrated by a vacuum rotary evaporation and filtered through a 0.22 µm Millipore membrane prior to loading onto a P-10 (BioRad, Hercules, CA) column (1.5 m × 5 cm) that was pre-equilibrated and eluted with 0.2 M NaCl solution. The uniform-sized oligosaccharide fractions collected from the P-10 column were pooled prior to desalting on a P-2 column. The lyophilized uniform-sized oligosaccharides with the degree of polymerization from dp2 to dp20 were purified on a semi-preparative SAX-HPLC (Shimadzu, Kyoto, Japan) equipped with a ∼5 µm Spherisorb column of dimension 2.0 × 25 cm2 from Waters (Milford, MA). A series of gradient elution programs used 2 M NaCl and water (pH 3.5) as combined elution solutions were used to fractionate the various uniform-sized oligosaccharide mixtures. The capacity of the column varied and ranged from 5 mg for the dp2 mixture to 1 mg for the dp20 mixture. Fractions were combined from repeated separations based on their similar chromatographic profiles as detected at 232 nm. Repurification was carried out for some fractions that were determined impure by an analytical SAX-HPLC on a 5 µm Spherisorb 0.46 × 25 cm2 analytical column.
Analysis of oligosaccharides by RPIP-LC-MS
Oligosaccharides were analyzed by an RPIP-LC-MS (Agilent 1100 LC/MSD; Agilent Technologies, Inc., Wilmington, DE) under conditions described previously (Zhang et al. 2008). Generally, ∼5 µg of each sample was loaded through a 5 µm Agilent Zorbax SB-C18 (0.5 × 250 mm2) column. Eluent A was water/acetonitrile (85:15, v/v) and eluent B was water/acetonitrile (35:65, v/v). Both eluents contained 12 mM tributylamine (TBA) and 38 mM ammonium acetate with pH adjusted to 6.5 by acetic acid. A gradient of 0% B for 15 min and 0–100% B over 85 min was used at a flow rate of 10 µL/min. The column effluent entered the source of the ESI-MS for continuous detection by MS. In addition, another 5 µL/min of acetonitrile was added just after column and before MS to make the solvent and TBA easy to spray and easy to evaporate in the ion source. The electrospray interface was set in negative ionization mode with a skimmer potential of −40.0 V, a capillary exit of −40.0 V and a source temperature of 325°C to obtain maximum abundance of the ions in a full scan spectrum (150–1500 Da, 10 full scans/s). Nitrogen was used as a drying (5 L/min) and nebulizing gas (20 psi). Total ion chromatograms and mass spectra were processed using Data Analysis 2.0 (Bruker Software, Billerica, MA).
Oligosaccharide digestion and PAGE analysis
PAGE analysis was carried out to determine the purity of oligosaccharides and the structural motif in heparin oligosaccharides digested by heparinase 1. Each oligosaccharide (∼50 µg) was individually incubated with 50 mU heparinase 1 at 30°C overnight. Polyacrylamide gel was prepared in 22% of total acrylamide for resolving gel (0.75 mm × 6.8 cm × 8.6 cm) and 5% of total acrylamide for stacking gel. The gel was loaded with 5 µg of each sample and subjected to electrophoresis for 80 min at 200 V. The gel was visualized first with 0.5% (w/v) Alcian blue in 2% (v/v) acetic acid aqueous solution, followed by silver nitrate staining.
1D. and 2D NMR experiments (600 MHz)
Each oligosaccharide (approximately 2.0–4.0 mg) for NMR analysis was repeatedly exchanged in deuterium oxide through lyophilization. 1D and 2D NMR spectra were measured at 298 K on a Bruker Avance 600 MHz (14.1 T) NMR spectrometer installed with the software Bruker Topspin 2.1. The water peak served as a reference (HO2H, 4.76 ppm) was confirmed by acetone (2.22 ppm). For the 1H–1H COSY, 1H–1H TOCSY, ROESY and NOESY spectra, 512 experiments resulting in 4096 data points for a spectral width of 10 ppm were measured. Inverse- or proton-detected HMQC experiments used 10- and 78-ppm spectral widths in the 1H dimension and 13C dimension, respectively. A mixing time of 300 ms was used in NOESY experiments with 1.5 s relaxation delay. The 2D NMR data sets were processed by Topspin and cross-peak assignments were carried out using an NMR assignment software Sparky (Goddard and Kneller 2001).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the US National Institutes of Health Grants HL101721, HL096972 and GM38060 and Scott A. McCallum at NMR Core Facility, Center for Biotechnology and Interdisciplinary Studies, RPI, for the assistance with NMR experiments. Z. Xiao is a recipient of a Chinese scholarship (File No. 2008633078) from the State Scholarship Fund to pursue his study in the United States as a joint PhD student.
Conflict of interest statement
None declared.
Abbreviations
COSY, COrrelation SpectroscopY; dp, degree of polymerization; ΔUA, 4-deoxy-α-l-threo-hex-4-eno-pyranosyluronic acid; ESI, electrospray ionization; GAG, glycosaminoglycan; GlcA, d-glucopyranosyluronic acid; GlcN, 2-deoxy-2-amino-d-glucopyranose; GlcNAc, 2-deoxy-2-acetylamino-d-glucopyranose; HMQC, Heteronuclear Multiple-Quantum Coherence experiment; HPLC, high-performance liquid chromatography; IdoA, l-idopyranosyluronic acid; MS, mass spectrometry; NOESY, Nuclear Overhauser Effect SpectroscopY; PAGE, polyacrylamide gel electrophoresis; ROESY, Rotating-frame Overhauser Effect SpectroscopY; RPIP, reversed-phase ion pairing; S, sulfate; SAX, strong anion exchange; TBA, tributylamine; TOCSY, TOtal Correlation SpectroscopY.
Supplementary Material
References
- Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. Angew Chem. 114: 426–450] doi:10.1002/1521-3773(20020201)41:3<390::AID-ANIE390>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Cohen DM, Linhardt RJ. Randomness in the heparin polymer: computer simulations of alternative action patterns of heparin lyase. Biopolymers. 1990;30:733–741. doi: 10.1002/bip.360300708. doi:10.1002/bip.360300708. [DOI] [PubMed] [Google Scholar]
- Desai UR, Wang HM, Linhardt RJ. Specificity studies on the heparin lyases from Flavobacterium heparinum. Biochemistry. 1993a;32:8140–8145. doi: 10.1021/bi00083a012. doi:10.1021/bi00083a012. [DOI] [PubMed] [Google Scholar]
- Desai UR, Wang HM, Linhardt RJ. Substrate specificity of the heparin lyases from Flavobacterium heparinum. Arch Biochem Biophys. 1993b;306:461–468. doi: 10.1006/abbi.1993.1538. doi:10.1006/abbi.1993.1538. [DOI] [PubMed] [Google Scholar]
- Ernst S, Rhomberg AJ, Biemann K, Sasisekharan R. Direct evidence for a predominantly exolytic processive mechanism for depolymerization of heparin-like glycosaminoglycans by heparinase 1. Proc Natl Acad Sci USA. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. doi:10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY. 3rd ed. San Francisco: CA: University of California; 2001. [Google Scholar]
- Guerrini M, Beccati D, Shriver Z, Naggi AM, Bisio A, Capila I, Lansing J, Guglieri S, Fraser B, Al-Hakim A, et al. Oversulfated chondroitin sulfate is a major contaminant in heparin associated with adverse clinical events. Nat Biotechnol. 2008;26:669–775. doi: 10.1038/nbt1407. doi:10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini M, Zhang Z, Shriver Z, Masuko S, Langer R, Casu B, Linhardt RJ, Torri G, Sasisekharan R. Orthogonal analytical approaches to detect potential contaminants in heparin. Proc Natl Acad Sci USA. 2009;106:16956–16961. doi: 10.1073/pnas.0906861106. doi:10.1073/pnas.0906861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandik KA, Gu K, Linhardt RJ. Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology. 1994;4:289–296. doi: 10.1093/glycob/4.3.289. doi:10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ. Chemical and enzymatic methods for the depolymerization and modification of heparin. In: Ogura H, Hasegawa A, Suami T, editors. Carbohydrates—synthetic methods and applications in medicinal chemistry. Tokyo/Weinheim: Kodansha/VCH Publishers; 1992. pp. 387–403. Chap. 20. [Google Scholar]
- Linhardt RJ. Perspective: 2003 Claude S. Hudson Award Address in carbohydrate chemistry. Heparin: Structure and Activity. J Med Chem. 2003;46:2551–2554. doi: 10.1021/jm030176m. doi:10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Fitzgerald GL, Cooney CL, Langer R. Mode of action of heparin lyase on heparin. Biochim Biophys Acta. 1982;702:197–203. doi: 10.1016/0167-4838(82)90503-9. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Gunay NS. Production and chemical processing of low molecular weight heparins. Semin Thromb Hemost. 1999;25:5–16. [PubMed] [Google Scholar]
- Linhardt RJ, Wang HM, Loganathan, Bae JH. Search for the heparin antithrombin III-binding site precursor. J Biol Chem. 1992;267:2380–2387. [PubMed] [Google Scholar]
- Lohse DL, Linhardt RJ. Purification and characterization of heparin lyases from Flavobacterium heparinum. J Biol Chem. 1992;267:24347–24355. [PubMed] [Google Scholar]
- Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: recent progress and future challenges. Omics. 2010;14:389–399. doi: 10.1089/omi.2009.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervin A, Gallo C, Jandik KA, Han X, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. doi:10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- Rice KG, Rottink MK, Linhardt RJ. Fractionation of heparin-derived oligosaccharides by gradient polyacrylamide-gel electrophoresis. Biochem J. 1987;244:515–522. doi: 10.1042/bj2440515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JE, Conrad HE. Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry. 1976;15:3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- Shriver Z, Sundaram M, Venkataraman G, Fareed J, Linhardt RJ, Biemann K, Sasisekharan R. Cleavage of the antithrombin III binding site in heparin by heparinases and its implication in the generation of low molecular weight heparin. Proc Natl Acad Sci USA. 2000;97:10365–10370. doi: 10.1073/pnas.97.19.10365. doi:10.1073/pnas.97.19.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawiroon C, Rice KG, Toida T, Linhardt RJ. LC/MS sequencing of highly sulfated heparin-derived oligosaccharides. J Biol Chem. 2004;279:2608–2615. doi: 10.1074/jbc.M304772200. doi:10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- Volpi N, Linhardt RJ. High performance liquid chromatography-mass spectrometry for mapping and sequencing glycosaminoglycan-derived oligosaccharides. Nat Protoc. 2010;5:993–1004. doi: 10.1038/nprot.2010.48. doi:10.1038/nprot.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Sakamoto K, Tsuda H, Yoshida K, Sugahara K, Khoo KH, Morris HR, Dell A. Structural studies on the tri- and tetrasaccharides isolated from porcine intestinal heparin and characterization of heparinase/heparitinases using them as substrates. Glycobiology. 1994;4:69–78. doi: 10.1093/glycob/4.1.69. doi:10.1093/glycob/4.1.69. [DOI] [PubMed] [Google Scholar]
- Yang HO, Gunay NS, Toida T, Kuberan B, Yu G, Kim YS, Linhardt RJ. Preparation and structural determination of dermatan sulfate-derived oligosaccharides. Glycobiology. 2000;10:1033–1039. doi: 10.1093/glycob/10.10.1033. doi:10.1093/glycob/10.10.1033. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xie J, Liu J, Linhardt RJ. Tandem MS can distinguish hyaluronic acid from N-acetylheparosan. J Am Soc Mass Spectrom. 2008;19:82–90. doi: 10.1016/j.jasms.2007.10.012. doi:10.1016/j.jasms.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xie J, Liu H, Liu J, Linhardt RJ. Quantification of heparan sulfate and heparin disaccharides using ion-pairing, reverse-phase, micro-flow, high performance liquid chromatography coupled with electrospray ionization trap mass spectrometry. Anal Biochem. 2009;81:4349–4355. doi: 10.1021/ac9001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.