Abstract
PAS domains form a divergent protein superfamily with more than 20,000 members that perform a wide array of sensing and regulatory functions in all three domains of life. Only 9 residues are well-conserved in PAS domains, with an Asn residue at the start of α-helix 3 showing the strongest conservation. The molecular functions of these 9 conserved residues are unknown. We use static and time-resolved visible and FTIR spectroscopy to investigate receptor activation in the photosensor photoactive yellow protein (PYP), a PAS domain prototype. The N43A and N43S mutants allow an investigation of the role of side chain hydrogen bonding at this conserved position. The mutants exhibit a blue-shifted visible absorbance maximum and up-shifted chromophore pKa. Disruption of the hydrogen bonds in N43A PYP causes both a reduction in protein stability and a 3,400-fold increase in the lifetime of the signaling state of this photoreceptor. A significant part of this increase in lifetime can be attributed to the helical capping interaction of Asn43. This extends the known importance of helical capping for protein structure to regulating functional protein kinetics. A model for PYP activation has been proposed in which side chain hydrogen bonding of Asn43 is critical for relaying light-induced conformational changes. However, FTIR spectroscopy shows that both Asn43 mutants retain full allosteric transmission of structural changes. Analysis of 30 available high resolution structures of PAS domains reveals that the side chain hydrogen bonding of residue 43 but not residue identity is highly conserved, and suggests that its helical cap affects signaling kinetics in other PAS domains.
1. Introduction
Photoactive yellow protein (PYP) is a blue light receptor from the halophilic photosynthetic proteobacterium Halorhodospira halophila,1,2 and is a prototype PAS domain.3,4 PYP was the first PAS domain for which the three-dimensional structure was reported,5,6 and remains the PAS domain that is best understood in terms of biochemistry and biophysics at the protein level.7 The PAS domain is a ubiquitous protein module with a common three-dimensional fold involved in a wide range of regulatory and sensory functions in all domains of life.8 Amino acid sequence analysis indicates that over 20,000 proteins in the protein sequence database contain a PAS domain, with 43 PAS domains present in the human genome as listed in the SMART database.9 The term PAS is derived from the first letter of the name of the three founding members of the superfamily: the Drosophila period protein (Per), the aryl hydrocarbon receptor nuclear translocator protein (Arnt) and the Drosophila single-minded protein (Sim). PAS domains have been identified in a wide range of signaling proteins8,10,including transcription factors, circadian clock proteins, phytochrome, and other proteins involved in regulation, photosensing, and oxygen/redox sensing. Interestingly, all members of this diverse set of proteins appear to be involved in sensing or regulation. Medically important PAS domains include the human ERG potassium channel. Mutations in various domains of this protein, including its PAS domain, cause cardiac arrhytmias (long QT syndrome).11 The PAS domain protein hypoxia-inducible factor is involved in myocardial and cerebral ischemia12 and in tumor hypoxia.13
PYP consists of 125 residues divided into two regions (Fig. 1b): (i) a typical PAS domain fold14 with a central antiparallel 6-stranded β-sheet flanked by three α-helices (residue 28–125; referred to below as the PAS domain core of PYP); and (ii) two N-terminal α-helices (residues 1–27) not present in most other PAS domains (referred to below as the N-terminal region). The two N-terminal helices pack against the central β-sheet, forming a second, small hydrophobic core in PYP.5 Six of the nine residues that are fairly highly conserved in PAS domains are present in PYP. PYP has been studied extensively by site-directed mutagenesis (summarized in 15). A systematic study examining the effects of mutating each of the 13 Gly residues in PYP to Ala revealed that substitution of the three Gly residues in PYP that are conserved in the PAS domain superfamily (Gly31, Gly51, and Gly59) result in fairly small changes in its biochemical properties16,17.
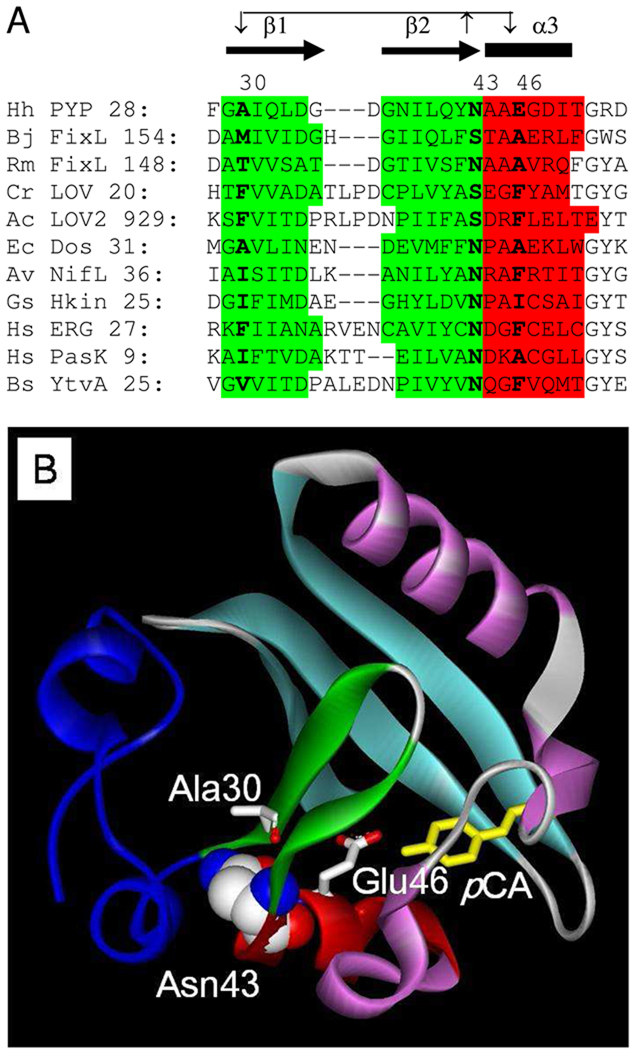
Figure 1.
Sequence conservation and and structural context of Asn43 in PYP. (A) Multiple sequence alignment of the N-terminal region of PAS domains with conserved side chain hydrogen bonding at residue 43. The PAS domain residue numbering used here is based on the PYP from Halorhodospira halophila. The number of the first depicted residue of each PAS domain is shown. The sequence alignment was adjusted based on the known three-dimensional structure of these PAS domains. Secondary structure is indicated: α-helices are shown in red and by the thick black bars; β-strands are indicated in green and by the black arrows. Residue 43 and its two key hydrogen bonding partners in β-strand 1 and α-helix 3 are highlighted in bold. The conserved side chain hydrogen bonds of residue 43 with the backbone amide groups of these two residues are indicated by arrows. Hh = Halorhodospira halophila; Bj = Bradyrhizobium japonicum; Rm = Rhizobium meliloti; Cr = Chlamydomonas reinhardtii; Ac =Adantium capillus-veneris; Ec = Escherichia coli; Av = Azotobacter vinelandii; Gs = Geobacter sulfurreducens; Hs = Homo sapiens; Bs = Bacillus subtilis. (B) Schematic representation of the crystal structure of PYP highlighting Asn43. The side chain of Asn43 is shown in CPK coloring. As in panel (A), α-helix 3 is shown in red, while β-strand 1 and 2 are indicated in green. The pCA chromophore is depicted in yellow, and the N-terminal region of PYP in dark blue.
PYP functions as the photoreceptor for negative phototaxis in H. halophila.18 It exhibits a light-triggered photocycle2 based on its p-coumaric acid (pCA) chromophore.19,20 In the initial pG dark state of PYP the pCA is in the trans conformation and its phenolic oxygen is deprotonated due to its pKa of 2.81,21, which is strongly down-shifted from the pKa of 8.8 for pCA in solution.22 The pCA forms functionally critical hydrogen bonds with active site residues Tyr42 and Glu46.23,24 Light initiates the PYP photocycle through the photoisomerization of the pCA25, followed by pCA protonation from Glu4630 and subsequent large protein conformational changes27–31 that result in the formation of the pB intermediate. This pB state is believed to be the signaling state of PYP, and spontaneously decays to the initial pG dark state of PYP in a few hundred milliseconds. Spectroscopic evidence indicates that upon pB formation the N-terminal region of PYP dissociates from its PAS domain core, and becomes largely unfolded.30,32,33 This is a striking example of allosteric transmission of a structural change across a protein, since the photoactive site of PYP and the N-terminal region are separated by the rigid central β-sheet in PYP. The changes in electrostatic interactions caused by proton transfer from Glu46 to the pCA have been identified as a key mechanism in triggering large conformational changes during the “protein quake” that results in pB formation.26,34 However, the mechanism for the allosteric transmission of conformational changes in PYP from the chromophore binding pocket to the N-terminal region has not yet been identified. Here we examine the role of Asn43 in this mechanism. The role of Asn43 in receptor activation in PYP is of interest for the more general question of allosteric signal transmission.35,36
The side chain of Asn43 in PYP forms four hydrogen bonds, all of which are buried: with the backbones of Leu23 in α-helix 2, Phe28 and Ala30 in β-strand 1, and Glu46 in α-helix 3. These side chain hydrogen bonds of Asn43 thus hold three secondary structure elements together. The side chain hydrogen bond between Asn43 and the backbone of Glu46 forms an α-helical cap37 (Fig. 1 and 2), which likely stabilizes α-helix 3. The term “helical cap” is occasionally used to describe a terminal helical domain in a protein. Here we use it to designate the specific hydrogen bonding pattern of a side chain at the end of an α-helix with a backbone amide in this helix. The side chain hydrogen bonds to Phe28 and Ala30 anchor β-strand 1 at the start of the PAS domain fold to the rest of PYP. Finally, the hydrogen bond to Leu23 contributes to the docking of the N-terminal region of PYP to the central β-sheet. Below we refer to the hydrogen bond between Asn43 and Ala30 as the structural bridging hydrogen bond and that between Asn43 and Glu46 as the helical capping hydrogen bond. Since the side chain hydrogen bonds of Asn43 are structurally implicated in linking the N-terminal region to the central β-sheet of PYP, light-induced changes in hydrogen bonding of this residue are a clear candidate for the mechanism of allosteric transmission of conformational changes at the photoactive site of PYP to its N-terminal region during pB formation. Evidence for such a model has been reported based on time-resolved X-ray crystallographic studies of PYP.38 Here we report experiments that provide an experimental test of this proposal.
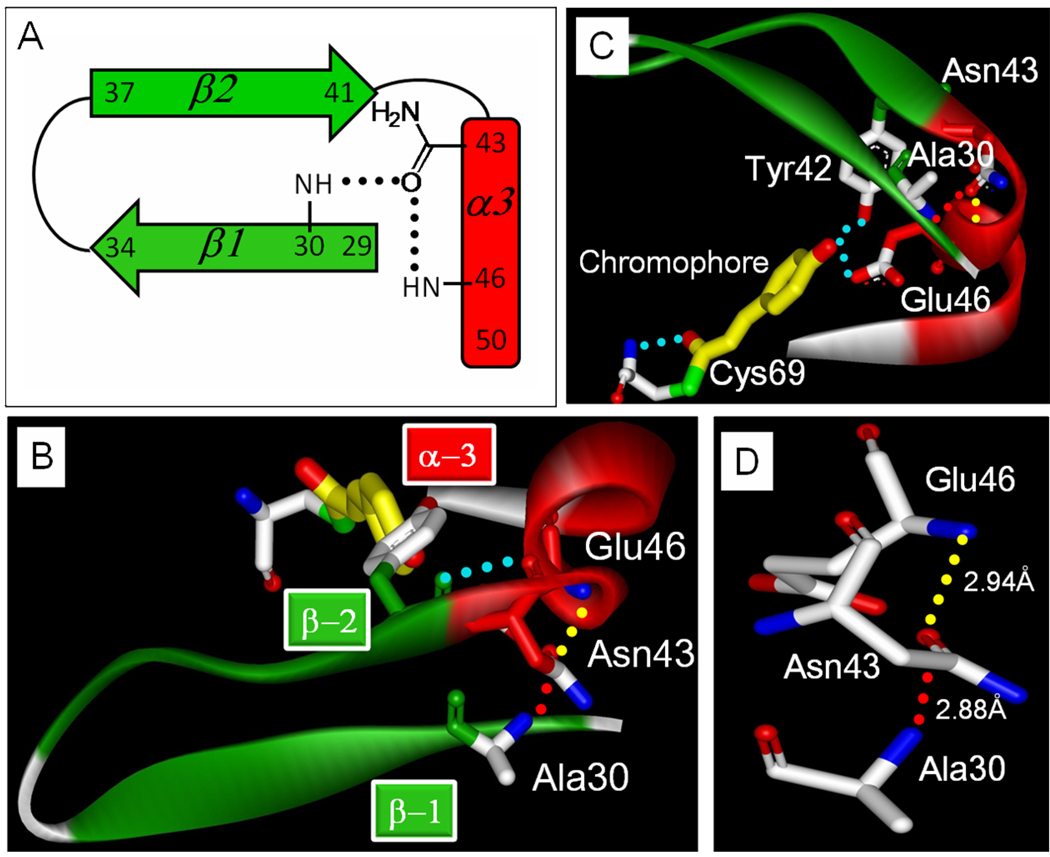
Figure 2.
Side chain hydrogen bonding of Asn43 in PYP. (A) Cartoon of the conserved hydrogen bonding pattern (residue numbering for PYP). (B and C) Residue 28 to 50 of PYP (1NWZ) shown in two orientations, together with the pCA. Selected hydrogen bonds are shown as dotted lines, α-helices are shown in red, and β-strands in green. The helical capping hydrogen bond is shown in yellow, the structural bridging hydrogen bond in red. (D) Zoom in to highlight the hydrogen binding interactions of Asn43.
A major recent development in the molecular life sciences is the determination of the amino acid sequences of large numbers of proteins due to improvements in DNA sequencing technology. Sensitive methods to identify weak amino acid sequence similarities in databases have revealed that many proteins contain short (~100 residues) conserved domains,9,39 and that shuffling of such domains is an important process in protein evolution.40 These conserved domains typically form large protein superfamilies that share a common three-dimensional fold, but are highly diverse with respect to both amino acid sequence and functional properties. Classical protein families have a common active site and therefore contain conserved active site residues. However, protein superfamilies often exhibit a high degree of variation in their active sites. This raises a novel question: what is the molecular role of the conserved amino acids in protein superfamilies? Are these residues required for stability and folding or are they functionally critical? It has been proposed that highly conserved residues form a folding nucleus, but this proposal is debated.41,42 We explore these issues for the most conserved residue in the PAS domain superfamily.
The PAS domain was initially identified through its weak sequence conservation.8,10 The subsequent determination of the three-dimensional structure of a number of proteins from this superfamily revealed that they share a common protein fold.3,14 While PAS domains are ~100 residues in length, only nine residues show fairly strong conservation. Three-quarters of PAS domains contain 4 to 7 of the 9 conserved residues, with Asn43 (residue numbering based on the PYP from Halorhodospira halophila) as the most conserved residue.8 PAS domains can bind a variety of cofactors, including heme and flavin, but some function in the absence of cofactors. Thus a conserved active site is not expected. The residues conserved in PAS domains could be required for the folding or stability of the PAS domain fold or be part of a conserved signaling mechanism. If these residues are functionally important, they may be involved in some aspect of a conserved allosteric switch for PAS domain signaling.
Since the residues conserved in PAS domains are not part of the active site, they have not been selected in structure-based site-directed mutagenesis studies. Therefore, the functional role that these 9 moderately conserved residues play in PAS domains is not known. This indicates that important gaps in understanding remain in current approaches to evaluating three-dimensional structures of proteins to identify residues that are critical for function. Here we report that Asn43, the most highly conserved residue in PAS domains (Fig. 1a), affects signaling kinetics in PYP.
The role of helical capping interactions, such as the side chain of Asn43, in determining protein stability had been extensively studied.43,44 The propensity for helical capping varies strongly for different side chains.37,45 Whereas Asn and Ser are often observed to form N-terminal α-helical capping interactions, this capacity is fully lacking in Ala. We therefore probe the role of side chain hydrogen bonding by residue 43 in PYP using the N43S and N43A mutants. The results show that the helical capping hydrogen bond strongly affects both the stability and signaling kinetics of PYP. This extends the importance of helical capping from protein structure and stability37,43–45 to regulating the kinetics of functional transitions, and provides a specific functional molecular role for the most highly conserved residue in PAS domains.
2. Experimental Section
Mutagenesis and protein purification
Mutagenesis was performed using Stratagene’s QuikChange site-directed mutagenesis kit with primers designed to introduce the N43A and N43S substitutions. A pQE-80A plasmid (QIAGEN) containing the pyp gene inserted between BamHI and HindIII sites was used as template. The PCR products were digested with DpnI and transformed into E. coli DH5α. DNA was purified using a QIAPrep Spin Miniprep kit (QIAGEN), used for DNA sequencing to confirm the mutation in the intact pyp gene, and transformed into E.coli BL21 (DE3) cells grown on LB agar with ampicillin (200 µg/mL). Overproduction of apoPYP was induced by the addition of 1 mM IPTG, and the protein was extracted from E. coli BL21 (DE3) using 8 M urea, and reconstituted with p-hydroxycinnamic anhydride (Sigma-Aldrich) following the procedure described in 23.
UV/vis absorbance and fluorescence spectroscopy
UV/vis absorption spectra were measured at room temperature using an HP-8453 (Hewlett-Packard) diode array spectrophotometer and a monochromator-based Cary300Bio (Varian) spectrophotometer. A 150 W halogen quartz light source (Cuda) with a broadband blue filter (band pass filter 59855, Oriel) was used to initiate the photocycle of PYP in 10 mM Tris-HCl pH 7.5. After 15 seconds of illumination the actinic light was shuttered, and the thermal recovery of the dark state was measured with a time resolution of up to 100 milliseconds. Photocycle kinetics at 445 nm were described as a monoexponential or biexponential decay as needed. A mixed buffer (glycine, succinate, MES and MOPS, 100 mM each) was used for pH titrations from pH 1.5 to 10.0. The acid titration was carried out in a darkened dark room using the minimal intensity of red light needed for handling of the samples. The pH dependence of the sample absorbance at its visible absorption maximum was described using the Henderson-Hasselbalch equation. Thermal unfolding curves were measured at pH 7.5 using an HP-8453 spectrophotometer equipped with a Peltier element. Denaturant titrations with guanidinium hydrochloride (Gdn-HCl) were performed at pH 7.5 in 10 mM Tris buffer at 25 °C in the same spectrophotometer and analyzed as described in 29.
Aromatic fluorescence emission measurements were performed at room temperature in a FluoroMax3 (Yobin Yvon) fluorimeter. Excitation was at 280 nm using a 3 nm slit width and PYP in 10 mM TrisHCl pH 7.5 with an OD of 0.2 at 441 nm. Titrations were performed using a buffered 6 M Gdn-HCl stock solution. For every measurement a fresh protein sample at a specific denaturant concentration was prepared.
FTIR spectroscopy
For FTIR spectroscopy PYP and its N43A and N43S mutants were used at 8 mM protein concentration in 50 mM phosphate buffer in D2O at pH* 6.6, obtained by washing and concentration with a Microcon (YM-10, 10,000 MWCO, Millipore) centrifuge filter. An FTIR sample consisted of 2.7 µl of PYP sandwiched between two CaF2 windows (15 mm diameter), separated by a 12 µm spacer. Sample temperature was controlled at 300 K. A Bruker IFS 66v spectrometer was utilized for static and time-resolved infrared measurements in the spectral range 4000-850 cm−1 using a liquid nitrogen-cooled Mercury Cadmium Telluride (MCT) detector. The spectrometer sample chamber was purged with dried nitrogen gas. Light-induced infrared absorption changes in PYP were measured using rapid-scan FTIR spectroscopy at 4.5 cm−1 spectral resolution with 200 kHz scanner velocity. The photocycle was triggered by 6 mJ laser pulses of 4 ns duration at 462 nm (Continuum Surelite-II pumped OPO laser). Laser repetition rate was set to 0.0033 Hz and 0.0166 Hz for N43A and N43S respectively, while the laser spot size on the FTIR sample was ~6 mm in diameter.
Time resolved FTIR measurements on N43A PYP were performed at room temperature and 4 cm−1 spectral resolution using a Nicolet 740 FTIR instrument with DTGS detector with dry air purging. A Cuda I-150 light source with a water filter and 420 nm cutoff filter was used to initiate the photocycle. Time resolved spectra were recorded on quasi-logarithmic time scale. Difference absorption spectra were calculated using single beam spectra of the sample immediately before illumination and single beam spectra at specific time points after shuttering the visible excitation. The resulting FTIR difference spectra were corrected for reproducible systematic baseline drift.
Analysis of three-dimensional structures
Swiss pdb viewer 3.7, WebLabViewerPro, and Pymol were used to examine three-dimensional structures of PAS domains, and for the automatic placement of hydrogen atoms in the structures. For four PAS domains (2O9C, 2OOL; 2VLG; 2Z6C) visual inspection of the automatic placement of the hydrogen atoms revealed an unfavorable hydrogen bonding geometry for the side chains of the residue corresponding to Asn43 in PYP. In these four cases, Swiss pdb viewer 3.7 was used for energy minimization, which converged to an alternative placement of the hydrogen atoms involved that resulted in a favorable hydrogen bonding geometry. Only hydrogen atoms moved in this optimization.
3. Results and Discussion
Differential effects of the N43A and N43S mutations on the stability of PYP
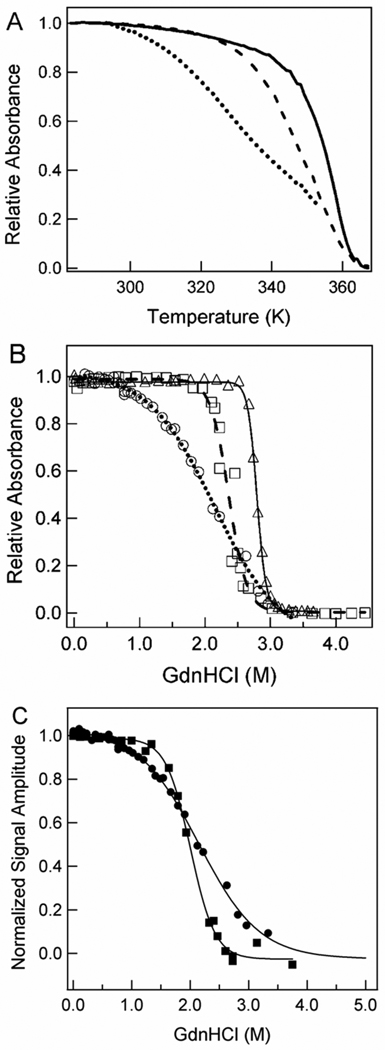
The effects of the N43A and N43S mutations on various functional properties of the initial pG state of PYP were determined, as summarized in Table 1. We first studied the effect of the N43A and N43S mutations on the stability of PYP (Table 1; Fig. 3; supplemental Fig. 1 and 2). In these experiments UV/vis absorbance spectroscopy was used to monitor thermal melting and equilibrium denaturant titrations. All three samples exhibited a thermally induced transition with an isosbestic point near 385 nm in which the native absorbance band near 445 nm is converted into an unfolded species with an absorbance maximum near 335 nm (supplemental Fig. 1). For wtPYP and the N43S mutant the entire thermal unfolding transition could be monitored. Thermal unfolding of the N43A mutant caused aggregation at temperatures above 80 °C. The midpoint temperature for thermal melting Tm of wtPYP (80.5°C) was only slightly reduced in the N43S mutant (76.3°C), but significantly decreased by the N43A mutation (57.4°C).
Table 1.
Effect of the N43S and N43A mutations and deletion of the N-terminal 23 residues on the properties of PYP.
| Property | wt | N43S | N43A | Δ23 PYPa |
|---|---|---|---|---|
| Tm (°C) | 80.5 ± 0.4 | 76.3 ± 0.2 | 57.4 ± 0.3 | nd |
| ΔGU (kJ/mol) | 44.9 ± 0.2 | 41.0 ± 1.8 | (11.2 ± 1.1)d | nd |
| [Gdm]½ | 2.8 ± 0.05 | 2.4 ± 0.05 | (2.2 ± 0.05)d | nd |
| m (M−1) | 6.6 ± 0.5 | 7.1 ± 0.3 | (4.5 ± 0.9)d | nd |
| λmax of pG (nm) | 446 | 441 | 441 | 442 |
| pKa of pCA in pG | 3.0 | 3.4 | 4.6 | 2.7 |
| pB – pG amide I | 100% | 133% | 138% | 60% |
| pG Glu46 C=O (cm−1) | 1737b/1726c | 1725c | 1725c | 1736b |
| pB lifetime (s) | 0.5 | 17 | 1,640 | 590 |
Figure 3.
Effect of the N43A and N43S mutations on the stability of PYP. Thermal unfolding (A) and denaturant titration (B) of wtPYP (solid line), N43S PYP (dashed line), and N43A PYP (dotted line) probed using visible absorbance at 446 nm for wtPYP and 441 nm for the mutants. The data for the thermal unfolding of N43A PYP are truncated at the point where aggregation started to occur. The lines in (B) are fits of the denaturant titration data as single transitions. (C) Comparison of the denaturant titration of N43A PYP as detected by absorbance at 441 nm (circles) and aromatic fluorescence emission at 350 nm (squares).
In the denaturant titrations wtPYP and the N43S mutant exhibited apparent two-state behavior33 with an isosbestic point near 380 nm (supplemental Fig. 2). The free energy for unfolding ΔGU of wt PYP (44.9 ± 0.2 kJ/mol) derived from Gdn-HCl titrations with UV/vis absorbance detection (Fig. 3b) was decreased only slightly in N43S PYP (41.0 ± 1.8 kJ/mol). The ~4 kJ/mol reduction in ΔGU caused by the N43S mutation is smaller than the destabilization of ~8 kJ/mol that is typically observed for mutants that disrupt a buried hydrogen bond.46–51 This indicates that hydrogen bonding remains largely intact in N43S PYP.
In the case of the N43A mutant two distinct isosbestic points were observed: at 385 nm up to 0.8 M Gdn-HCl and at 375 nm at higher denaturant concentrations (supplemental Fig. 3). The spectral changes during the first part and last part of the denaturant titration were distinctly different (Supplemental Fig. 3B). As for wtPYP and the N43S mutant, the denaturant dependence of the absorbance at 445 nm could be fitted as a single transition. However, this yielded a significantly reduced denaturant m value (Table 1). Based on this observation and on the non-isosbestic nature of the transition, we conclude that unfolding of N43A PYP by Gdn-HCl involves one or more partially unfolded intermediates. To test this proposal, we used fluorescence emission from aromatic amino acids to probe the unfolding of N43A PYP. These data reveal a transition that deviates from the one observed using 441 nm absorbance as a probe (Fig. 3C). This confirms that the denaturant-induced unfolding of N43A PYP deviates from two-state folding. While these data do not allow a complete description of the denaturant-induced unfolding of N43A PYP, they reveal that the N43A mutation strongly impairs the stability of PYP and disrupts the cooperativity of the folding process in PYP.
These results indicate that the side chain hydrogen bonds at position 43 contribute significantly to the stability of PYP, and that the stabilizing side chain hydrogen bonding interactions of Asn43 can also be performed by a serine at this position. This is in line with the shared propensity of Asn and Ser to form a helical capping hydrogen bond to residue i + 3.37,43 In addition, in various PAS domains containing a Ser at this position the helical capping hydrogen bond is maintained (see below). Part of the reduction in stability of N43A PYP can be attributed to the loss of the helical capping hydrogen bond of Asn43.43,44
Tuning of the properties of the pG dark state of PYP by Asn43
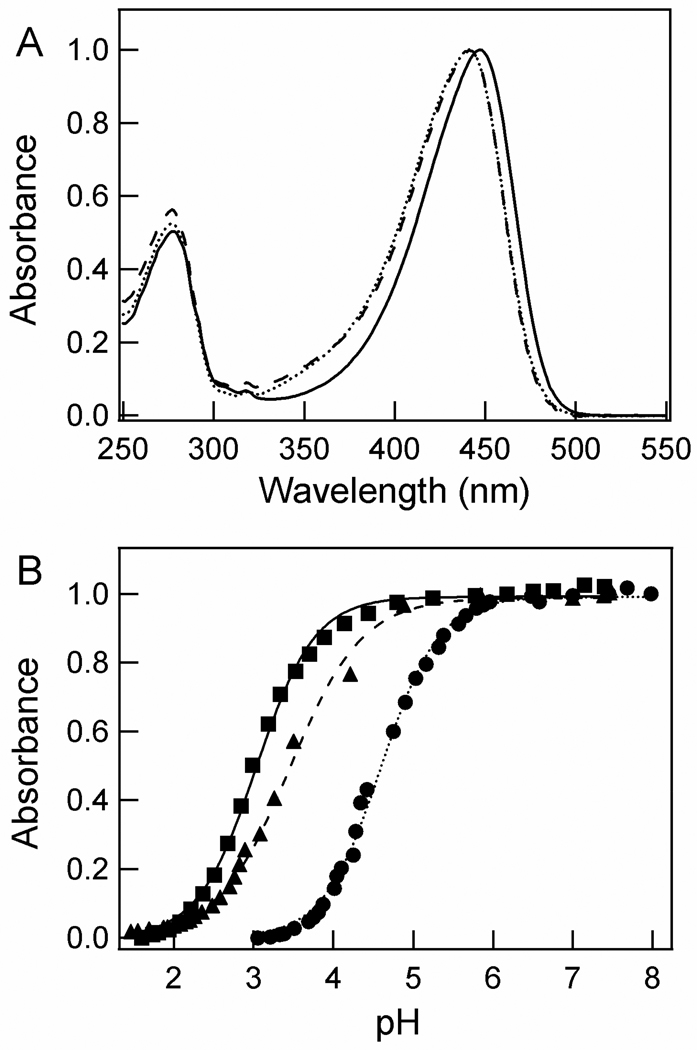
The absorbance spectrum (λmax) of N43S and N43A PYP is blue-shifted by 5 nm, from 446 nm in wtPYP to 441 nm in the mutants (Fig. 4a). The overall shape of the absorbance spectra is essentially unchanged in the mutants. This shows that the side chain of Asn43 is involved in tuning the λmax of PYP. Since the same blue-shift in the λmax of the pG state is observed in N43A and N43S PYP, this spectral tuning is caused by an effect for which the hydrogen bonding interactions offered by the Ser side chain are not sufficient.
Figure 4.
Effect of the N43A and N43S mutations on the absorbance maximum and pKa of the pCA in PYP. Absorbance spectra at pH 7.1 (A) and pH titrations fit with the Henderson-Hasselbalch equation (B) are depicted, with data for wtPYP (solid lines and squares), N43S PYP (dashed lines and triangles), and N43A PYP (dotted lines and circles). The titration curves were obtained at 446 nm for wtPYP and 441 nm for the mutants. The pKa and n values for these three proteins are 3.0/1.2, 3.4/1.0, and 4.6/1.2, respectively. The amplitudes of the data were normalized at their visible absorbance maximum.
The protonation of the pCA chromophore at low pH in the mutants was monitored by UV/vis absorbance spectroscopy as the loss of the native absorbance band near 445 nm and the concomitant population of a new species with a λmax near 345 nm (supplemental Fig. 4). This transition is described by the Henderson-Hasselbalch equation, revealing that the N43A mutation causes an up-shift in the pKa of the pCA by 1.6 units, from 3.0 in wtPYP to 4.6 in the mutant (Fig. 4b). In contrast, the pKa of the N43S mutant is up-shifted by only 0.4 pH units to 3.4. Thus, the removal of side chain hydrogen bonding at position 43 in the N43A mutant significantly increases the pKa of the pCA in the pG state of PYP.
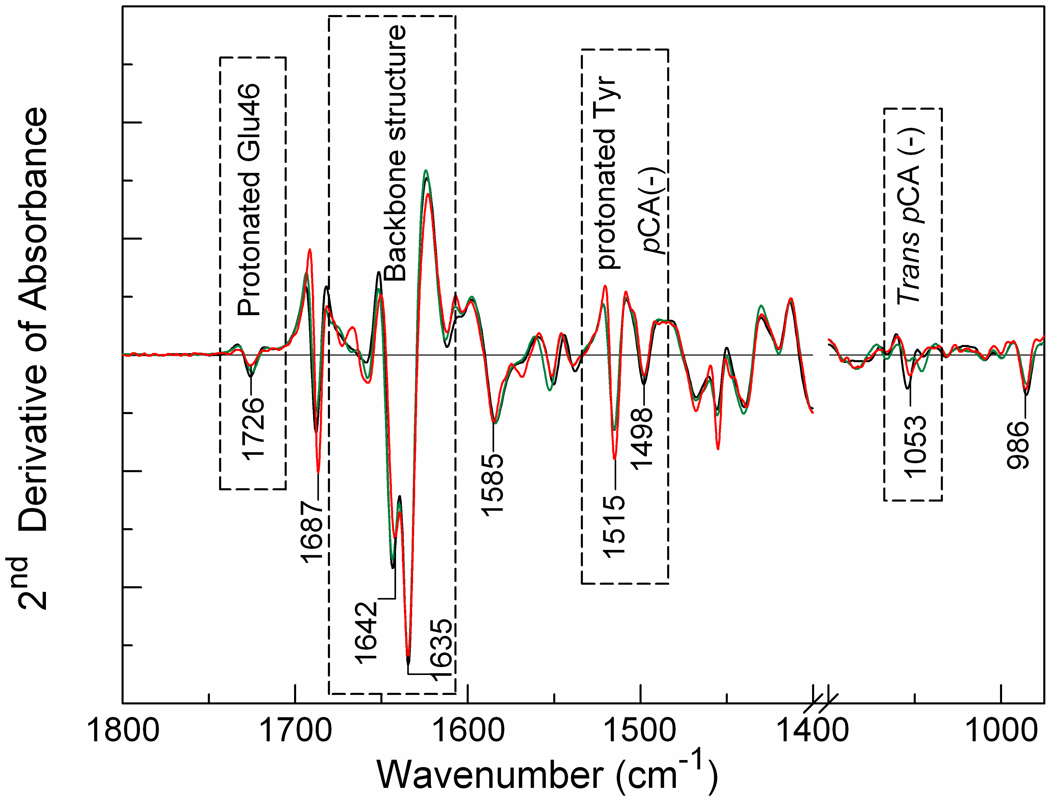
The effect of the N43S and N43A mutations on the structure of the pG state was studied by second derivative FTIR absorbance spectroscopy. The FTIR spectrum of a protein in the 1800-950 cm−1 region contains contributions from the protein backbone and from all polar and charged side chains. The peak positions of these overlapping bands are better resolved in the second derivative of the FTIR spectrum.52 Thus, the second derivative of the FTIR absorbance of a protein provides a sensitive “fingerprint” for the structure of a protein.52,53 This approach has been extensively applied to the amide I region, revealing that it provides a sensitive probe for changes in secondary structure.54,55 The N43A mutation could result in a release and partial unfolding of the two N-terminal helices of PYP. This would be sensitively detected in the amide I region of the second derivative FTIR spectrum.
The second derivative of the infrared absorbance spectrum of the pG dark state of wtPYP is very similar to that for the N43S and N43A mutants of PYP (Fig. 5). Specifically, the strong amide I signals at 1644 and 1635 cm−1 are essentially unchanged. This is strong evidence that the N43A mutation does not significantly perturb the secondary structure of PYP, indicating that the two N-terminal helices are still packed against PYP in the pG state of these two mutants. Most signals from side chains, for example at 1726 and 1689 cm−1, are also largely unaltered. These results indicate that the side chain hydrogen bonds provided by Asn43 are not required for the docking of the N-terminal region of PYP in the pG dark state. Apparently, the N43A mutant reduces the stability of the pG state (see Fig. 3) while not significantly altering the structural properties of the folded state.
Figure 5.
Probing structural deformations of the pG dark state of wtPYP (black) by the N43A (red) and N43S (green) mutations using second derivative FTIR spectroscopy. The FTIR absorbance spectrum of the three protein samples was measured at 2 cm−1 resolution, and was used to calculate the second derivative to facilitate comparison of spectral features in the three spectra. Selected peak positions are indicated.
Careful inspection of the second derivative spectra indicates that the amplitude of the peak at 1726 cm−1, which originates from the C=O stretching mode of the side chain of Glu4626 , is reduced in the N43A mutant. Since the amplitude of signals in the second derivative spectrum is highly sensitive to the width of the peak in the infrared absorbance spectrum, this observation indicates a slight increase in the width of the Glu46 C=O stretching mode. This suggests a slight increase in the structural heterogeneity of the Glu46 side chain, without a change in its average properties.
Mechanism of active site tuning in the pG state of PYP by Asn43
Free pCA thioester compounds in water exhibit a pKa near 8.8, and a λmaxnear 400 nm in the ionized state.22 These values are significantly shifted compared to the pKa of 2.81,21 and λmax of 446 nm13 in native PYP. The mechanisms involved in tuning the pK56,57 and λmax22,57–60 in PYP have attracted significant attention, and are of more general relevance.61,62 , Both of these properties are affected by the N43A and N43S substitutions. The X-ray structure of the pG state PYP5,6 provides a framework to evaluate the structural basis for these effects. The side chain of Asn43 is 7.5 Å removed from the pCA, indicating that the effects of the N43A and N43S mutations on the properties of the pCA in the pG state are caused by an indirect mechanism.
Since Asn43 forms hydrogen bonds with the backbones of Leu23 in α-helix 2, Phe28 and Ala30 in β-strand 1, and Glu46 in α-helix 3, the observed effects caused by the N43A and N43S substitutions may be caused by three possible effects: (i) deformation of α-helix 3 due to a change in helical capping, (ii) a change in the docking of the N-terminal region, or (iii) a structural change in the PAS domain core of PYP by the disruption of the structural bridge between α-helix 3 and β-strand 1. We consider these three possibilities for altering the λmax and pKa of the pCA in the pG dark state of the mutants. In the first option the loss of the helical capping interaction of α-helix 3 would result in an altered structure/position of this helix and thus of the critical active site residues Glu46 and Tyr42 that it contains. This would likely alter the λmax and pKa of the pCA, since substitutions at these two residues significantly alter these properties.23,24,63,64 A reduction in the hydrogen bonding strength between the pCA and residue 46 in the E46Q mutant causes a red-shift in the λmax of PYP.23,24 The absorbance spectra of the N43A and N43S mutants are essentially identical, and both are blue-shifted by 5 nm compared to wtPYP. This implies that it is not a change in the side chain hydrogen bonding interactions of residue 43 that blue-shifts the absorbance spectra in the mutants. In addition, FTIR spectroscopic results presented below show that the position of the negative signal at 1726 cm−1 in the pB – pG difference spectrum caused by Glu46 deprotonation26 is unaltered in both mutants (see Fig. 7), demonstrating that the hydrogen bond between Glu46 and the pCA is not changed in the pG state of the two mutants. This argues against a change in the location of α-helix 3 or a structural disruption of the PAS domain core of PYP in the N43A and N43S mutants.
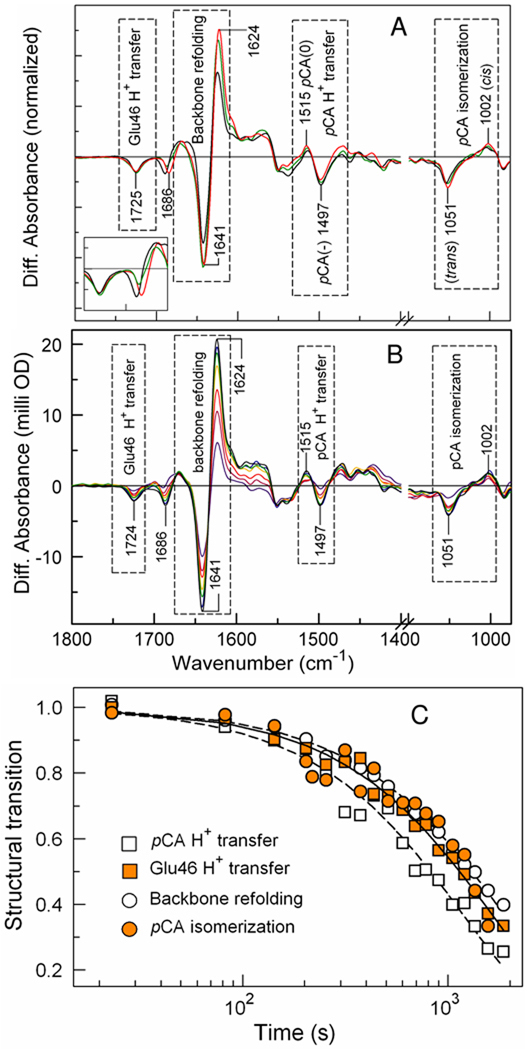
Figure 7.
Structural changes upon pB formation in wtPYP and its N43A and N43S mutants detected by FTIR difference spectroscopy. (A) The pB – pG infrared difference spectra at 4 cm−1 resolution for wt (blue), N43A (red), and N43S (green) PYP (pH* = 6.6) were derived from flash-induced time-resolved rapid scan FTIR spectra. The inset shows a magnification of the peaks near 1726 and 1689 cm−1. (B) Time-resolved FTIR difference spectroscopy during PB decay in N43A PYP. The plotted spectra range from 8 to 1,862 seconds. The region 1100–1140 cm−1 is not shown because it does not contain relevant vibrational marker signals. (C) Amplitude-normalized kinetics of structural events during pB decay in N43A PYP. The data from (B) were used to extract the kinetics of Glu46 protonation (1724 cm−1), pCA deprotonation (1515 and 1497 cm−1), pCA reisomerization (1051 cm−1), and backbone conformational changes (1641 and 1624 cm−1). The transitions were fitted as monoexponential decays (lines), yielding lifetimes of 1,640, 1,190, 1,630, and 1,880 s (see main text).
We also considered the possibility that mutations at position 43 cause a change in the strength of the hydrogen bond between the pCA and Tyr42. Spectroscopic analysis of active site mutants of PYP has indicated that mutations that weaken the pCA-Tyr42 hydrogen bond also affect the pCA-Glu46 hydrogen bond.65,66 Thus, the observation that the position of the C=O stretching mode of Glu46 at 1726 cm−1 is unaltered in the Asn43 mutants makes it unlikely that the Tyr42-pCA H-bond is significantly changed in these mutants.
Mutants lacking the N-terminal 23–27 residues of PYP exhibit a blue-shifted λmax (442 nm)30,32,33, similar to that of the N43A and N43S mutants. This suggests that the blue-shift in the two mutants reported here is caused by an altered interaction between the PAS domain core of PYP and its N-terminal region. The second derivative FTIR spectra indicate that the N-terminal helices of PYP remain docked on the rest of PYP in the N43A and N43S mutants. This implies that an altered docking of the N-terminal region of PYP is involved in the blue-shifted λmax of N43A and N43S PYP.
While the absorbance spectra of the N43A and N43S mutants are essentially identical, their pCA pKa is significantly different: 3.4 for the N43S mutant and 4.6 for the N43A mutant. This result indicates that the side chain hydrogen bonds of residue 43 reduce the pKa of the pCA by up to ~1.6 pH units. This could be due either to the capping function of residue 43 or to its role in holding the N-terminal region of PYP in place by its hydrogen bonds to Leu23, Phe28, and Ala30. Since the pKa of the pCA in mutants lacking the N-terminal region is essentially unperturbed (below 3.0)32, we propose that a change in the helical capping interaction is a key factor in the altered pKa of the N43A mutant. Protonation of the pCA at low pH causes significant structural changes that have been described as acid denaturation.21 The capping interaction likely stabilizes α-helix 3, and the reduced stability of this helix in the N43A mutant can explain the reduced acid stability of the protein (Fig. 4b).
The side-chain hydrogen bonds of residue 43 strongly affect the lifetime of the pB state
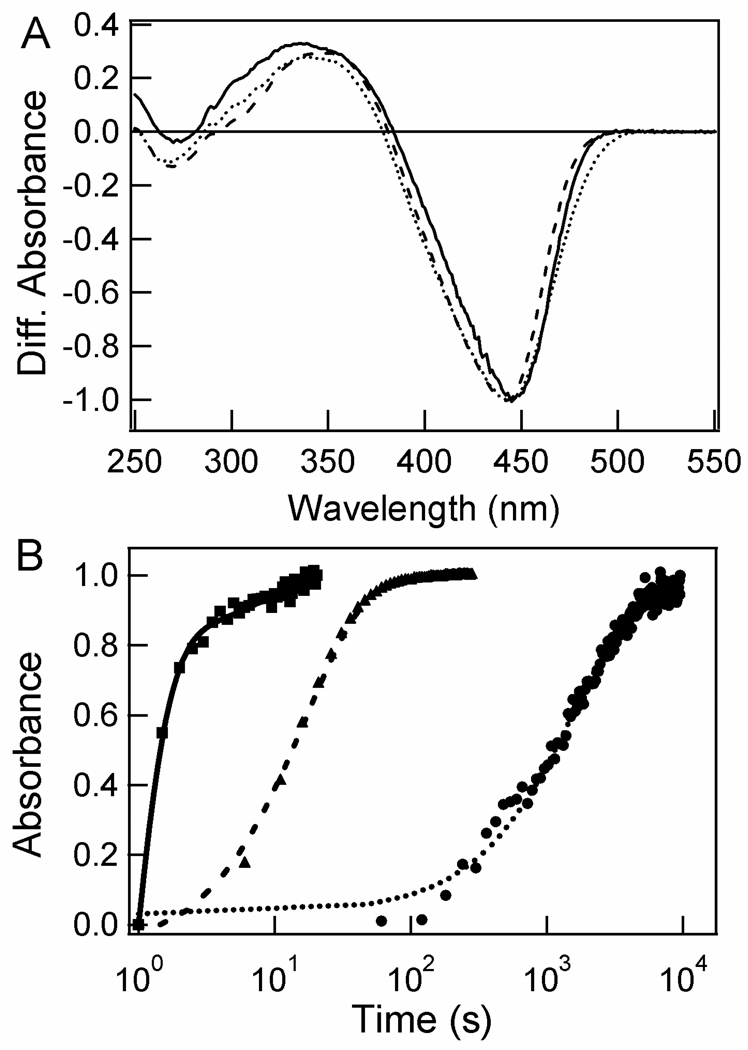
The kinetics of the last photocycle step in the N43A and N43S mutants was determined. The light – dark UV/Vis absorbance difference spectra of the two mutants show the formation of a pB state with a characteristic λmax near 350 nm, very similar to that in wtPYP (Fig. 6a). For both mutants the decay of the pB state is significantly slowed down (Fig. 6b). However, the degree of deceleration greatly depends on the substitution at position 43. In wtPYP the decay of the pB state is biphasic, with the major fast component (90%) decaying with a time constant of 0.48 ± 0.04 seconds. In the N43S mutant the lifetime of the pB state is extended to 17 ± 0.2 seconds, while in N43A PYP it is 1,640 ± 19 seconds. These results reveal that the side chain hydrogen bonds of residue 43 have a strong effect on the lifetime of the pB state.
Figure 6.
Substitutions at Asn43 slow down the kinetics of the last PYP photocycle step. The pB-pG UV/vis absorbance difference spectra (A) and kinetics of the pB to pG transition (B) for wtPYP (solid lines and squares), N43S PYP (dashed lines and triangles), and N43A PYP (dotted lines and circles) at pH 7.1 are shown. The difference spectra were normalized for the extent of bleaching of the pG state. The kinetics of the wtPYP photocycle were fitted as a biexponential decay, those of N43A and N43S PYP as a monoexponential decay. The amplitudes of the kinetic traces were normalized; the optical densities of the samples at their visible absorbance maxima near 445 nm were in the 0.35 – 0.45 range.
Substitutions at four positions have been reported to greatly slow down the kinetics of pB decay: Met10067,68, Glu4669, Phe9670, and Gly2916. For the G29A, E46D, M100K, and F96A mutants the lifetime of pB is 210, 385, 600, and 1,075 seconds, respectively. Thus, the N43A mutant of PYP exhibits the most dramatic increase in pB lifetime reported to date.
Structural changes during pB formation in the N43A and N43S mutants
To examine proton transfer events and structural changes during the formation of the pB intermediate in the N43A and N43S mutants, we performed time-resolved rapid-scan FTIR difference spectroscopy (Fig. 7). The negative signal at 1726 cm−1 caused by the deprotonation of Glu46 and the signals at 1497 and 1515 cm−1 caused by the protonation of the chromophore26 are unchanged in the two mutants (Fig. 7), indicating that the proton transfer process from Glu46 to the pCA proceeds in the mutants as in wtPYP. The pB – pG FTIR difference spectra of wtPYP and the two Asn43 mutants can be convincingly scaled using these signals. This comparison reveals that the amplitude of the amide I difference signals at 1643 and 1623 cm−1 are increased by ~35% in the two mutants. This result shows that photoexcitation still triggers a large protein quake in the two Asn43 mutants, and that the amplitude of the protein quake is somewhat larger than in wtPYP. The increase in the amplitude of the amide I signals indicates that the Asn43 side chain somewhat constrains the structural changes that occur upon pB formation in a manner that cannot be achieved by Ala or Ser.
Close inspection of the pB – pG FTIR difference spectra indicates that the negative feature at 1689 cm−1 and the positive band at 1669 cm−1 are somewhat altered by the N43S and N43A mutations. Since these vibrational modes are still present in the mutants, they do not originate from the C=O stretching mode of the side chain of Asn43. These signals have not yet been assigned, precluding a further structural interpretation at this point.
Since the side chain of Asn43 forms multiple hydrogen bonds that bring together multiple regions of PYP, it is possible that pB decay in the N43A mutant is greatly slowed down because of a lack of cooperativity for the multiple structural changes that occur during the last part of the PYP photocycle. This possibility is supported by the loss of cooperativity in the unfolding of N43A PYP (Fig. 3C). To test this possibility we performed time-resolved FTIR measurements during the decay of the pB state in N43A PYP (Fig. 7B). In these measurements PYP in D2O buffer was used to allow accurate measurements in the amide I region. We probed pB decay using infrared signals for four key events in the PYP photocycle: reprotonation of Glu46 using the signal at 1725 cm−1; protein conformational changes at 1641 and 1624 cm−1, pCA protonation at 1515 and 1497 cm−1, and pCA re-isomerization at 1051 cm−1 (Fig. 7C). Each of these processes followed a monoexponential decay, with an average time constant of 1,590 s. This is essentially identical to the rate observed using changes in visible absorbance at 441 nm. The rates for the four processes varied within a factor 1.25 (Fig. 7C). Since the very slow photocycle of N43A PYP reduces the quality of the FTIR spectra due to contamination of the sample with H2O and water vapor signals, we conclude that the N43A mutations leaves the cooperativity of the pB decay process essentially intact.
Role of Asn43 in PYP receptor activation and deactivation
We examined the role of Asn43 in determining two aspects of the pB signaling state: its lifetime and its structural properties. Elimination of side chain hydrogen bonding in the N43A mutant results in a dramatic increase in the lifetime of the pB intermediate. We first consider altered docking of the N-terminal region as a factor in the affecting the lifetime of the pB state in N43A PYP. Two lines of evidence indicate that docking of the N-terminal region accelerates pB decay. First, substitutions at Phe6 in the minor hydrophobic core of PYP that weaken docking of the N-terminal region prolong the lifetime of the pB state by ~25-fold71 Second, mutants lacking 6, 15, 23, 25, or 27 N-terminal residues exhibit a reduced rate for pB decay ranging from 18 to 590 seconds.30,32,72 These results show that the N-terminal region is not required for pB decay but does accelerate it. Above we concluded that the blue-shift in λmax in the Asn43 mutants is caused by an altered docking of the N-terminal region of PYP, which could affect pB lifetime However, the effect of the N43A mutation on the lifetime of the pB state is at least a factor ~3 stronger than that of mutants lacking the entire N-terminal region. This indicates that factors other than docking of the N-terminal region are involved in causing the prolonged pB lifetime in N43A PYP.
Both the structural bridging hydrogen bond of Asn43 to Ala30 and its helical capping hydrogen bond to Glu46 may contribute to the increased pB lifetime in N43A PYP. The large conformational changes that occur upon pB formation27–30 have been studied in detailed by NMR spectroscopy.31 This revealed that formation of the pB state involves unfolding of α-helix 3 and disruption of the helical capping hydrogen bond of Asn43, while the hydrogen bond from Asn43 to Ala 30 remains intact. We therefore propose that refolding of α-helix 3, which contains key active site residue Glu46, is facilitated by the capping interaction of Asn43 during the reformation of this helix upon pB decay. Thus, the formation of the helical capping hydrogen bond of Asn43 during pG recovery is a key interaction regulating the lifetime of the pB state. In this analysis both the refolding/re-docking of the N-terminal region and the re-establishment of the helical cap in α-helix 3 are key events in determining the lifetime of the pB signaling state. This provides a specific molecular functional role for a PAS-conserved residue.
The N43A mutation strongly affects both ΔGU and pB lifetime. While no general trend has been reported that destabilizing mutations slow down functional turnover, the possibility exists that pB lifetime is directly related to the ΔGU of PYP. Published data on the M100A and R52A mutant argue against this proposal. The ΔGU of M100A PYP is reduced by 3.8 kJ/mol, while its pB lifetime is ~550 seconds.67 The R52A mutant exhibits a very similar reduction in ΔGU, but its pB lifetime is 0.8 seconds.24 We conclude that the N43A mutation has a specific effect on the kinetics of the PYP photocycle.
Receptor deactivation kinetics are of direct importance for in vivo signal transduction. The lifetime of signaling states in microbial sensory rhodopsins has been show to alter the sensitivity of the in vivo response73, and appears to match the typical time scale of the biological response triggered by the receptor.74 Two-component regulatory systems also show a correspondence between the lifetime of an activated signaling component and the time scale of the biological response involved.75,76 Thus, the effects of the N43A and N43S mutations on pB lifetime would be expected to be biologically relevant.
Regarding the role of Asn43 in determining the structure of the pB state, the finding that the N43A mutant exhibits light-induced structural changes that are somewhat larger than in wtPYP (Fig. 7) has important implications for the mechanism of receptor activation in PYP. X-ray crystallographic studies of PYP have suggested a role for the side chain hydrogen bonds of Asn43 in the allosteric transmission of conformational changes at the photoactive site of PYP to its N-terminal region during pB formation.38 However, large light-induced conformational changes are observed in the N43A mutant. If the N43A mutation would cause the release of the N-terminal region in the pG state, the amplitude of the light-induced amide I signals would be expected to be smaller, contrary to the rapid-scan FTIR results. In addition, the negative signals in the pB – pG FTIR difference spectrum, which originate from the pG state and thus provide a probe for the structure of the pG state, are largely unaffected by the two substitutions. This confirms the conclusion from the analysis of the second derivative FTIR absorbance spectrum of the pG state of the N43A mutant that the N-terminal region remains bound to the rest of PYP before photoexcitation. The data reported here show that even in the absence of side chain hydrogen bonding at residue 43 (the N43A mutant), large light-induced structural changes occur upon pB formation. This argues against a major role for photocycle-associated changes in side chain hydrogen bonds of residue 43 in triggering the release of the N-terminal region of PYP during pB formation.
The FTIR difference spectra of the N43A and N43S mutants of PYP are highly similar, while their pB lifetime is very different. This indicates that the structure of the pB intermediate in these two mutants is very similar, and that it is the transition state for pB decay that is differentially affected by the two mutations. Apparently it is not the altered pB structure of the two mutants detected in the FTIR difference spectrum but the lack of side chain hydrogen bonds to promote recovery of the pG state that greatly reduces the rate of pB decay in N43A PYP. In summary, the data reported here indicate that side chain hydrogen bonding at position 43 is a key factor in regulating the lifetime of the pB state, but not in triggering allosteric conformational changes upon pB formation.
A functionally important helical capping hydrogen bond
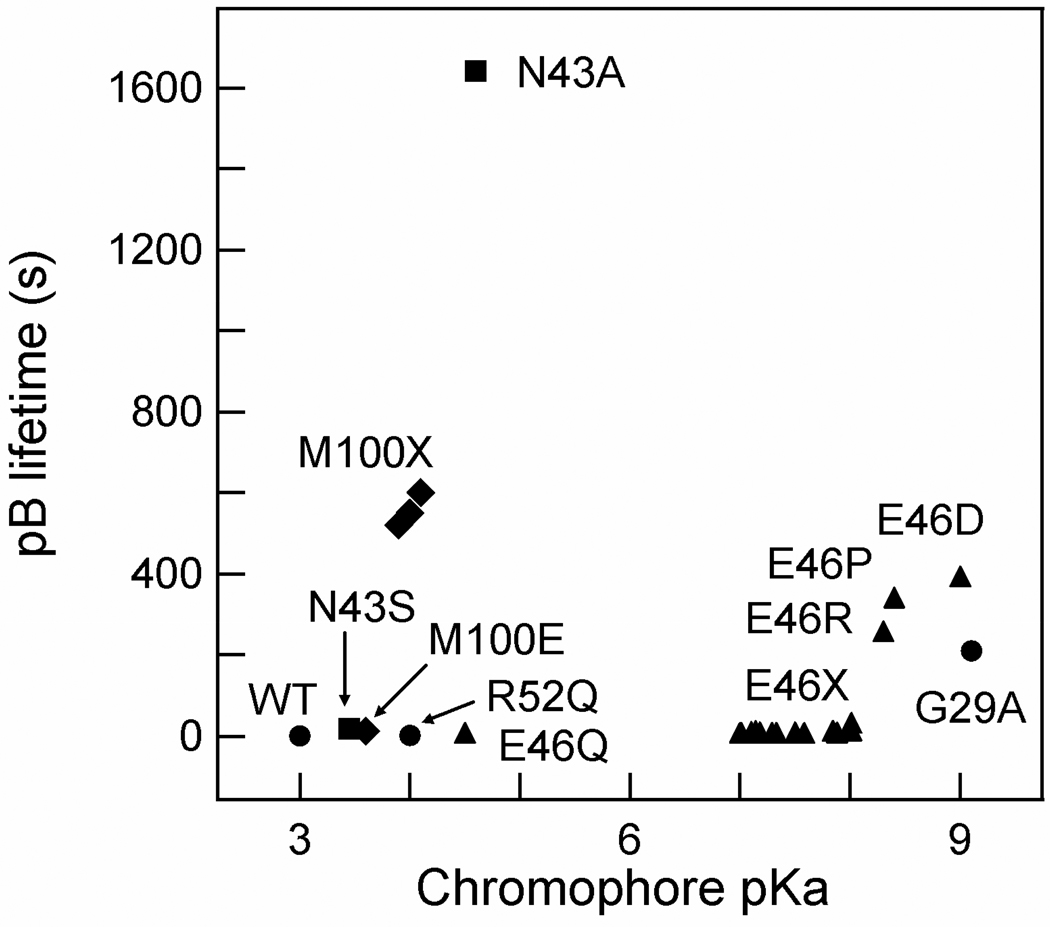
The data reported here allow a structural dissection of the role of Asn43 in determining key functional properties of PYP. The results indicate that the blue-shift in the λmax of PYP in the N43A and N43S mutations is not caused by the loss of the helical capping hydrogen bond of Asn43, and that allosteric transmission of structural changes upon pB formation can proceed in the absence of this helical capping hydrogen bond. Our results provide evidence for the importance of the helical capping hydrogen bond of Asn43 in determining two functional properties of PYP. The analysis presented above indicates that the upshift in the pKa of the pCA in the pG state of N43A PYP is caused by the disruption of the helical capping hydrogen bond of Asn43. In addition, a significant fraction of the dramatic increase in the lifetime of the pB state caused by the N43A mutation can be attributed to the removal of this helical capping hydrogen bond. This interpretation is strongly supported by the observation that the pCA pKa and pB lifetime are largely unchanged in the N43S mutant but greatly altered in the N43A mutant (Fig. 8).
Figure 8.
Comparison of functional properties of PYP mutants. The pKa of the pCA in the pG state and the lifetime of the pB state for all mutants for which both of these properties have been reported is depicted. Substitution mutants at position Asn43 are indicated as squares, those at Glu46 as triangles, and at Met100 as diamonds; the remaining proteins are depicted by circles. The cluster of mutants indicated as M100X consists of the A, L and K substitution mutants of Met100; cluster E46X contains the A, C, G, H, I, K, L, N, S, T, V, W and Y substitutions of Glu46.
A number of studies have shown the importance of N-terminal α-helical capping interactions on protein stability.43,44 Asn43 in PYP provides an example of an N-terminal helical capping hydrogen bond with functional importance. Since helical capping interactions are common in proteins, the importance of this interaction in regulating functional properties as reported here for PYP is likely to also be applicable to other proteins.
PYP has been studied extensively by site-directed mutagenesis, resulting in the identification of a number of mutations that alter its functional properties15,16. A summary of the effects of these mutations on the pKa of the pCA in the pG state of PYP and on the lifetime of the pB state is provided in Figure 8. This analysis reveals that the N43A mutation causes a unique combination of strong changes in these two properties of PYP.
Side chain hydrogen bonding of residue 43 is highly conserved in PAS domains
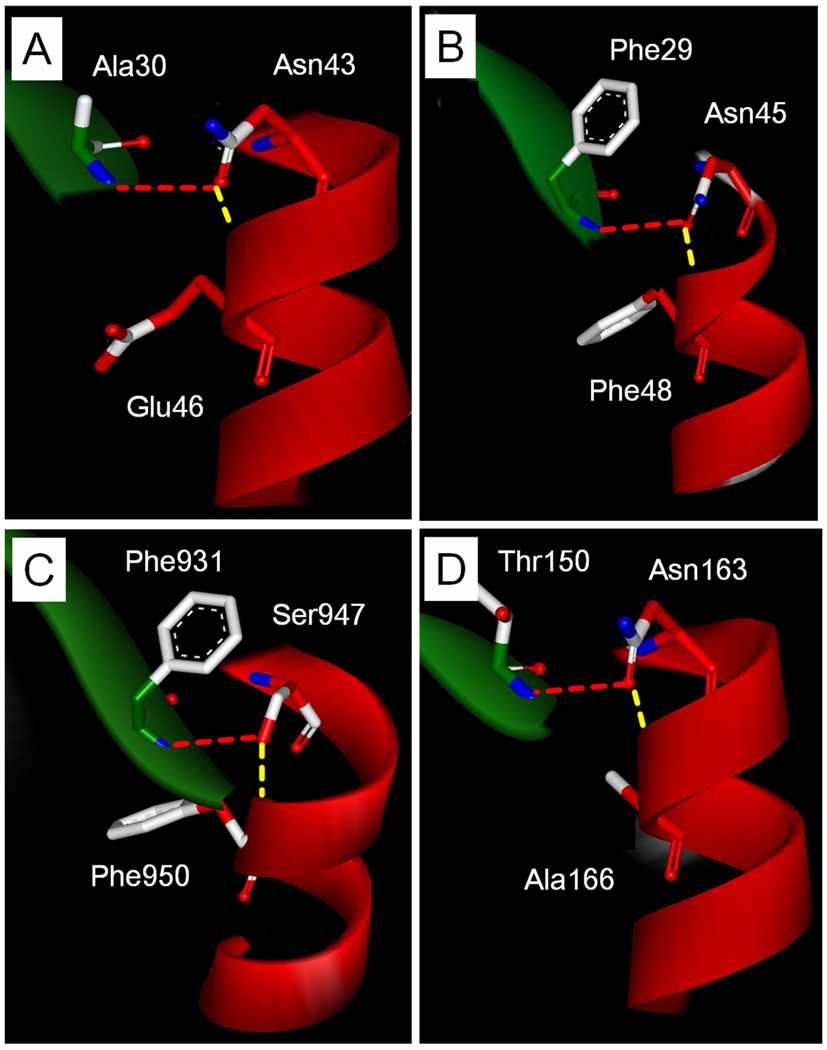
To examine a possible general role of side chain hydrogen bonding of residue 43 in the PAS domain superfamily, we used the DALI server77 to identify proteins that are structurally related to PYP. We examined the three-dimensional structure of each of the 28 different PAS domains identified in this search. Six of these structures were determined by the Protein Structure Initiative.78 This analysis revealed that in 66% of the PAS domain structures the hydrogen bonding pattern of residue 43 (and the equivalent position in other PAS domains) is identical to that observed in PYP (Table 2 and supplemental Table 1). In an additional 28% of the structures both hydrogen bonds were observed, but in an n+1, n+2, or n+4 capping configuration. The average dihedral angle Θ at the hydrogen atom for the structural bridging hydrogen bonds in this set of PAS domain structures is at an energetically favorable value of 156 ± 14. In the case of the n+3 helical capping hydrogen bond the values of Θ are 146 ± 16, slightly lower than ideal but still clearly in the allowed region.79 The three-dimensional structures of the region involved in the interactions of residue 43 with α-helix 3 and β-strand 3 are highly similar in most PAS domain structures (Fig. 9). Thus, the pattern of side chain hydrogen bonding interactions of residue 43, forming a helical cap for α-helix 3 and a structural bridge to β-strand 1, is highly conserved in the PAS domain superfamily. These conserved helical capping and structural bridging hydrogen bonds are indicated in Figs. 1, 2, and 9.
Table 2.
Conservation of hydrogen bonding interactions of Asn43 in PYP and other PAS domains.
| Helical cap helix 3 |
Link to β-strand 1 |
||||
|---|---|---|---|---|---|
| Proteina (PBD) | Residueb | lengthc | lengthc | ||
| Hh PYP (1NWZ) | Asn43 | Glu46 | 2.9 | Ala30 | 2.9 |
| Rm FixL (1D06) | Asn163 | Ala166 | 2.8 | Thr150 | 2.8 |
| Av NifL (2GJ3) | Asn51 | Phe54 | 2.9 | Ile38 | 3.0 |
| Ec DOS – O2 (1VB6) | Asn46 | Ala49 | 2.9 | Ala33 | 2.8 |
| Ec DOS + O2 (1S67) | Asn46 | H2O | 2.6 | - | |
| Hs ERG (1BYW) | Asn45 | Phe48 | 2.9 | Phe29 | 3.2 |
| Hs PasK (lLL8) | Asn25 | Ala28 | 3.2 | Ile11 | 3.4 |
| Bs YtvA (2PR5) | Asn43 | Phe46 | 2.9 | Val27 | 2.9 |
| Bj FixL (1DP6) | Ser169 | Ala172 | 3.4 | Met156 | 3.2 |
| Ac LOV2 (1G28) | Ser947 | Phe950 | 3.3 | Phe931 | 3.0 |
| Cr LOV1 (1N9L) | Ser38 | Phe41 | 3.2 | Phe22 | 3.1 |
Hh = Halorhodospira halophila; Rm = Rhizobium meliloti; Av = Azotobacter vinelandii; Ec = Escherichia coli; Hs = Homo sapiens; Bs = Bacillus subtilis; Bj = Bradyrhizobium japonicum; Ac =Adantium capillus-veneris; Cr = Chlamydomonas reinhardtii.
Hydrogen bond from the side chain of Asn43 and corresponding residues to the backbone NH group of the indicated residue.
Hydrogen bonding length is indicated in Å.
Figure 9.
Conservation of the side chain hydrogen bonding interactions of residue 43 in the PAS domain superfamily. Depicted are the relevant regions of the PYP from H. halophila (A), human ERG (B), LOV2 from Adantium capillus-veneris (C), and FixL from B. japonicum (D). α-Helix 3 is shown in red and β-strand 1 in green. The residues in the PAS domains corresponding to Asn43 in PYP are indicated with their side chain hydrogen bonds (dashed lines). The helical capping hydrogen bond is shown in yellow, the structural bridging hydrogen bond in red.
The 28 PAS domains with conserved side chain hydrogen bonding for residue 43 (PYP numbering) have very little sequence similarity. The residue corresponding to Asn43 in PYP is conserved as Asn in 13 of these proteins, and is substituted by Ser, Asp, and Thr in 11, 2, and 1 PAS domain, respectively (Table 2 and supplemental Table 1). These substitutions retain side chain hydrogen bonding capability at position 43. Within the PYP family, Asn43 is fully conserved15, except for the PYP from Thermochromatium tepidum,80 where it is substituted with Ser. Multiple sequence alignments6 of the PAS domain superfamily show that Ser is the most common substitution for Asp at position 43, providing support for an important role of side chain hydrogen bonding by residue 43 in the PAS domain superfamily. This analysis indicates that the side chain hydrogen bonding interactions of residue 43 are strongly conserved in PAS domains despite significant variation in the side chain found at this position. Thus, not residue identity at position 43 but the pattern of side chain hydrogen bonding interactions is conserved in this protein superfamily.
The high level of evolutionary conservation of the side chain hydrogen bonds of residue 43 in PAS domains suggests the possibility that it plays a conserved role in the function of this diverse protein superfamily as a whole. This is confirmed by analysis of hydrogen bonding in the heme-containing PAS domain from the oxygen sensor DOS from E. coli.81 The side chain hydrogen bonds of Asn46 that form the helical cap and the structural bridge to β-strand 1 in this PAS domain (Table 2) are intact in the absence of oxygen. However, O2 binding causes conformational changes that disrupt both of these hydrogen bonds. This supports the notion that the functional importance of the side chain hydrogen bonds of Asn43 in PYP is conserved in other members of the PAS domain superfamily. This analysis indicates the possibility that the side chain hydrogen bonds of residue 43 modulate the kinetics of signaling in many members of the PAS domain superfamily.
4. Conclusions
Disruption of the side chain hydrogen bonds of Asn43 in N43A PYP significantly reduces the stability of PYP and causes an up-shift in the pKa of the pCA (from 3.0 to 4.6), likely because of the removal of the helical capping hydrogen bond to the backbone of Glu46. In both the N43A and N43S mutants the λmax is blue-shifted to 441 nm, which is attributed to a slightly altered docking of the N-terminal region.
The N43A mutation causes a dramatic increase in the lifetime of the pB state, largely attributed to the loss of side chain hydrogen bonding at this position. Analysis of the available data indicates that the helical capping hydrogen bond of residue 43 plays an important role in regulating the kinetics of receptor resetting in PYP. Thus, α-helical capping is not only important for protein structure and stability, but can also be a key factor for regulating functional kinetics. These results provide a clear molecular role for the most conserved residue in the PAS domain superfamily.
It has been proposed that the side chain hydrogen bonds of Asn43 are a key part of the mechanism of allosteric transmission of structural changes from the active site of PYP to its N-terminal region. However, time-resolved FTIR difference spectroscopy shows that the N43A mutant still exhibits large light-induced conformational change. Thus, side chain hydrogen bonding at this position is not an essential part of the allosteric trigger but affect the lifetime of the pB state.
Analysis of the structure of PYP together with a set of 28 PAS additional domains shows that the two side chain hydrogen bonds of the residue corresponding to Asn43 in PYP are highly conserved despite significant variation in the residue at this position: the structural bridging hydrogen bond to the start of β-strand 1 and the helical capping hydrogen bond. Analysis of published crystal structures of the oxygen sensor DOS from E. coli shows that O2 binding disrupts these side chain hydrogen bonds, suggesting that they play a functional role in a number of different PAS domains.
Supplementary Material
Acknowledgements
WDH gratefully acknowledges support from NIH grant GM063805 and OCAST grant HR07-135S, and from startup funds provided by Oklahoma State University. AX was supported by OCAST grant HR02-137R and the Oklahoma State Regents for Higher Education. We acknowledge help of Zhouyang Kang in data collection for the N43A time-resolved FTIR measurements. WDH acknowledges helpful discussions with Peter T. S. van der Gulik.
Footnotes
Supporting Information Available. UV/vis absorbance spectra during pH titrations, Gdn-HCl titrations, and thermal melts of the N43A and N43S mutants; table with hydrogen bonding interactions of Asn43 (and corresponding residues) in 29 PAS domains with known structure.
References
- 1.Meyer TE. Biochim. Biophys. Acta. 1985;806:175–183. doi: 10.1016/0005-2728(85)90094-5. [DOI] [PubMed] [Google Scholar]
- 2.Meyer TE, Yakali E, Cusanovich MA, Tollin G. Biochemistry. 1987;26:418–423. doi: 10.1021/bi00376a012. [DOI] [PubMed] [Google Scholar]
- 3.Pellequer J-L, Wagner-Smith KA, Kay SA, Getzoff ED. Proc. Natl. Acad. Sci. USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusanovich MA, Meyer TE. Biochemistry. 2003;42:4759–4770. doi: 10.1021/bi020690e. [DOI] [PubMed] [Google Scholar]
- 5.Borgstahl GEO, Williams DR, Getzoff ED. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 6.Getzoff ED, Gutwin KN, Genick UK. Nat. Struct. Biol. 2003;10:663–668. doi: 10.1038/nsb958. [DOI] [PubMed] [Google Scholar]
- 7.Hellingwerf KJ, Hendriks J, Gensch T. J. Phys. Chem. A. 2003;107:1082–1094. [Google Scholar]
- 8.Taylor BL, Zhulin IB. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz J, Milpetz F, Bork P, Ponting CP. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Aravind L. Curr. Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Zou AR, Splawski I, Keating MT, Sanguinetti MC. J. Biol. Chem. 1999;274:10113–10118. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. J. Clin. Invest. 2000;106:809–812. doi: 10.1172/JCI11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Proc. Natl. Acad. Sci. USA. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hefti MH, Françoijs KJ, de Vries SC, Dixon R, Vervoort J. Eur. J. Biochem. 2004;271:1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x. [DOI] [PubMed] [Google Scholar]
- 15.Kumauchi M, Hara M, Stalcup P, Xie A, Hoff WD. Photochem. Photobiol. 2008;84:956–969. doi: 10.1111/j.1751-1097.2008.00335.x. [DOI] [PubMed] [Google Scholar]
- 16.Imamoto Y, Tatsumi S, Harigai M, Yamazaki Y, Kamikubo H, Mataoka M. Biophys. J. 2008;94:3620–3628. doi: 10.1529/biophysj.107.123414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Aalten DMF, Haker A, Hendriks J, Hellingwerf KJ, Joshua-Tor L, Crielaard W. J. Biol. Chem. 2002;277:6463–6468. doi: 10.1074/jbc.M109313200. [DOI] [PubMed] [Google Scholar]
- 18.Sprenger WW, Hoff WD, Armitage JP, Hellingwerf KJ. J. Bacteriol. 1993;175:3096–3104. doi: 10.1128/jb.175.10.3096-3104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff WD, Düx P, Hård K, Devreese B, Nugteren-Roodzant IM, Crielaard W, Boelens R, van Beeumen J, Hellingwerf KJ. Biochemistry. 1994;33:13959–13962. doi: 10.1021/bi00251a001. [DOI] [PubMed] [Google Scholar]
- 20.Baca M, Borgstahl GEO, Boissinot M, Burke PM, Williams DR, Slater KA, Getzoff ED. Biochemistry. 1994;33:14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 21.Hoff WD, van Stokkum IHM, Gural J, Hellingwerf KJ. Biochim. Biophys. Acta. 1997;1322:151–162. [Google Scholar]
- 22.Kroon A, Hoff WD, Fennema H, Koomen G-J, Verhoeven JW, Crielaard W, Hellingwerf KJ. J. Biol. Chem. 1996;271:31949–31956. doi: 10.1074/jbc.271.50.31949. [DOI] [PubMed] [Google Scholar]
- 23.Mihara K, Hisatomo O, Imamoto Y, Kataoka M, Tokunaga F. J. Biochem. 1997;121:876–880. doi: 10.1093/oxfordjournals.jbchem.a021668. [DOI] [PubMed] [Google Scholar]
- 24.Genick UK, Devanathan S, Meyer TE, Canestrelli IL, Williams E, Cusanovich MA, Tollin G, Getzoff ED. Biochemistry. 1997;36:8–14. doi: 10.1021/bi9622884. [DOI] [PubMed] [Google Scholar]
- 25.Kort R, Vonk H, Xu X, Hoff WD, Crielaard W, Hellingwerf KJ. FEBS Lett. 1996;382:73–78. doi: 10.1016/0014-5793(96)00149-4. [DOI] [PubMed] [Google Scholar]
- 26.Xie A, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. Biochemistry. 2001;40:1510–1517. doi: 10.1021/bi002449a. [DOI] [PubMed] [Google Scholar]
- 27.van Brederode ME, Hoff WD, van Stokkum IHM, Groot ML, Hellingwerf KJ. Biophys. J. 1996;71:365–380. doi: 10.1016/S0006-3495(96)79234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoff WD, Xie A, van Stokkum IHM, Tang X-J, Gural J, Kroon AR, Hellingwerf KJ. Biochemistry. 1999;38:1009–1017. doi: 10.1021/bi980504y. [DOI] [PubMed] [Google Scholar]
- 29.Lee B-C, Pandit A, Croonquist PA, Hoff WD. Proc. Natl. Acad. Sci. USA. 2001;98:9062–9067. doi: 10.1073/pnas.111153598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imamoto Y, Kamikubo H, Harigai M, Shimizu N, Kataoka M. Biochemistry. 2002;41:13595–13601. doi: 10.1021/bi0264768. [DOI] [PubMed] [Google Scholar]
- 31.Bernard C, Houben K, Derix NM, Marks D, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NAJ. Structure. 2005;13:953–962. doi: 10.1016/j.str.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 32.van der Horst MA, van Stokkum IHM, Crielaard W, Hellingwerf KJ. FEBS. Lett. 2001;497:26–30. doi: 10.1016/s0014-5793(01)02427-9. [DOI] [PubMed] [Google Scholar]
- 33.Harigai M, Imamoto Y, Kamikubo H, Yamazaki Y, Kataoka M. Biochemistry. 2003;42:13893–13900. doi: 10.1021/bi034814e. [DOI] [PubMed] [Google Scholar]
- 34.Derix NM, Wechselberger RW, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NAJ. Biochemistry. 2003;42:14501–14506. doi: 10.1021/bi034877x. [DOI] [PubMed] [Google Scholar]
- 35.Goodey NM, Benkovic SJ. Nat. Chem. Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 36.Tsai CJ, Del Sol A, Nussinov R. Mol. Biosystems. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aurora R, Rose GD. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal S, Anderson S, Srajer V, Schmidt M, Pahl R, Moffat K. Structure. 2005;13:55–63. doi: 10.1016/j.str.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orengo CA, Thornton JM. Annu. Rev. Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 41.Mirny L, Shakhnovich E. J. Mol. Biol. 2001;308:123–129. doi: 10.1006/jmbi.2001.4602. [DOI] [PubMed] [Google Scholar]
- 42.Larson SM, Ruczinski I, Davidson AR, Baker D, Plaxco KW. J. Mol. Biol. 2002;316:225–233. doi: 10.1006/jmbi.2001.5344. [DOI] [PubMed] [Google Scholar]
- 43.Doig AJ, Baldwin RL. Protein Sci. 1995;4:1325–1336. doi: 10.1002/pro.5560040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serrano L, Sancho J, Hirshberg M, Fersht AR. J. Mol. Biol. 1992;277:544–559. doi: 10.1016/0022-2836(92)90906-z. [DOI] [PubMed] [Google Scholar]
- 45.Doig AJ, MacArthur MW, Stapley BJ, Thornton JM. Protein Sci. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fersht AR, Shi J-P, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 47.Shirley BA, Stanssens P, Hahn U, Pace CN. Biochemistry. 1992;31:725–732. doi: 10.1021/bi00118a013. [DOI] [PubMed] [Google Scholar]
- 48.Byrne MP, Manuel RL, Lowe LG, Stites WE. Biochemistry. 1995;34:13949–13960. doi: 10.1021/bi00042a029. [DOI] [PubMed] [Google Scholar]
- 49.Takano K, Yamagata Y, Kubota M, Funahashi J, Fujii S, Yutani K. Biochemistry. 1999;38:6623–9662. doi: 10.1021/bi9901228. [DOI] [PubMed] [Google Scholar]
- 50.Pace CN, Horn G, Hebert EJ, Bechert J, Shaw K, Urbanikova L, Scholtz JM, Sevcik J. J. Mol. Biol. 2001;312:393–404. doi: 10.1006/jmbi.2001.4956. [DOI] [PubMed] [Google Scholar]
- 51.Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 52.Dong A, Huang P, Caughey WS. Biochemistry. 1992;31:182–189. doi: 10.1021/bi00116a027. [DOI] [PubMed] [Google Scholar]
- 53.He WZ, Newell WR, Haris PI, Chapman D, Barber J. Biochemistry. 1991;30:4552–4559. doi: 10.1021/bi00232a027. [DOI] [PubMed] [Google Scholar]
- 54.Dong A, Huang P, Caughey WS. Biochemistry. 1990;29:3303–3308. doi: 10.1021/bi00465a022. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Yan YB. Anal. Biochem. 2005;340:89–98. doi: 10.1016/j.ab.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 56.Yoda M, Inoue Y, Sakurai M. J. Phys. Chem. B. 2003;107:14569–14575. [Google Scholar]
- 57.Philip AF, Eisenman KT, Papadantonakis GA, Hoff WD. Biochemistry. 2008;47:13800–13810. doi: 10.1021/bi801730y. [DOI] [PubMed] [Google Scholar]
- 58.Yoda M, Houjou H, Inoue Y, Sakurai M. J. Phys. Chem. 2001;105:9887–9895. [Google Scholar]
- 59.Yamato T, Ishikura T, Kakitani T, Kawaguchi K, Watanabe H. Photochem. Photobiol. 2007;83:323–327. doi: 10.1562/2006-06-16-RA-930. [DOI] [PubMed] [Google Scholar]
- 60.Philip AF, Nome RA, Papadantonakis GA, Scherer NF, Hoff WD. Proc. Natl. Acad. Sci. USA. 2010;107:5821–5826. doi: 10.1073/pnas.0903092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. Trends Biochem. Sci. 1999;24:300–305. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- 62.Harris TK, Turner GJ. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 63.Imamoto Y, Koshimizu H, Mihara K, Hisatomi O, Mizukami T, Tsujimoto K, Kataoka M, Tokunaga F. Biochemistry. 2001;40:4679–4685. doi: 10.1021/bi002291u. [DOI] [PubMed] [Google Scholar]
- 64.Meyer TE, Devanathan S, Woo T, Getzoff ED, Tollin G, Cusanovich MA. Biochemistry. 2003;42:3319–3325. doi: 10.1021/bi020702w. [DOI] [PubMed] [Google Scholar]
- 65.Sigala PA, Tsuchida MA, Herschlag D. Proc. Natl. Acad. Sci. USA. 2009;106:9232–9237. doi: 10.1073/pnas.0900168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joshi CP, Otto H, Hoersch D, Meyer TE, Cusanovich MA, Heyn MP. Biochemistry. 2009;48:9980–9993. doi: 10.1021/bi9012897. [DOI] [PubMed] [Google Scholar]
- 67.Devanathan S, Genick UK, Canestrelli IL, Meyer TE, Cusanovich MA, Getzoff ED, Tollin G. Biochemistry. 1998;37:11563–11568. doi: 10.1021/bi9803776. [DOI] [PubMed] [Google Scholar]
- 68.Kumauchi M, Hamada N, Sasaki J, Tokunaga F. J. Biochem. 2002;132:205–210. doi: 10.1093/oxfordjournals.jbchem.a003211. [DOI] [PubMed] [Google Scholar]
- 69.Devanathan S, Brudler R, Hessling B, Woo TT, Gerwert K, Getzoff ED, Cusanovich MA, Tollin G. Biochemistry. 1999;38:13766–13772. doi: 10.1021/bi991634p. [DOI] [PubMed] [Google Scholar]
- 70.Morishita T, Harigai M, Yamazaki Y, Kamikubo H, Kataoka M, Imamoto Y. Photochem. Photobiol. 2007;83:280–285. doi: 10.1562/2006-06-15-RA-929. [DOI] [PubMed] [Google Scholar]
- 71.Harigai M, Kataoka M, Imamoto Y. J. Am. Chem. Soc. 2006;128:10646–10647. doi: 10.1021/ja062125v. [DOI] [PubMed] [Google Scholar]
- 72.Harigai M, Yasuda S, Imamoto Y, Yoshihara F, Tokunaga F, Kataoka M. J. Biochem. 2001;130:51–56. doi: 10.1093/oxfordjournals.jbchem.a002961. [DOI] [PubMed] [Google Scholar]
- 73.Yan B, Takahashi T, Johnson R, Spudich JL. Biochemistry. 1991;30:10686–10692. doi: 10.1021/bi00108a012. [DOI] [PubMed] [Google Scholar]
- 74.Jung KH, Trivedi VD, Spudich JL. Mol. Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- 75.Janiak-Spens F, Sparling JM, Gurfinkel M, West AH. J. Bacteriol. 1999;181:411–417. doi: 10.1128/jb.181.2.411-417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas SA, Brewster JA, Bourret RB. Mol. Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grabowski M, Joachimiak A, Otwinowski Z, Minor W. Curr. Opin Struct. Biol. 2007;17:347–353. doi: 10.1016/j.sbi.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kortemme T, Morozov AV, Baker D. J. Mol. Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 80.Kyndt JA, Meyer TE, Cusanovich MA. Photochem. Photobiol. Sci. 2004;3:519–530. doi: 10.1039/b315731h. [DOI] [PubMed] [Google Scholar]
- 81.Kurokawa H, Lee DS, Watanabe M, Sagami I, Mikami B, Raman CS, Shimizu T. J. Biol. Chem. 2004;279:20186–20193. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.