Abstract
Designer cellulosomes are precision-engineered multienzyme complexes in which the molecular architecture and enzyme content are exquisitely controlled. This system was used to examine enzyme cooperation for improved synergy among Thermobifida fusca glycoside hydrolases. Two T. fusca cellulases, Cel48A exoglucanase and Cel5A endoglucanase, and two T. fusca xylanases, endoxylanases Xyn10B and Xyn11A, were selected as enzymatic components of a mixed cellulase/xylanase-containing designer cellulosome. The resultant mixed multienzyme complex was fabricated on a single scaffoldin subunit bearing all four enzymes. Conversion of T. fusca enzymes to the cellulosomal mode followed by their subsequent incorporation into a tetravalent cellulosome led to assemblies with enhanced activity (~2.4-fold) on wheat straw as a complex cellulosic substrate. The enhanced synergy was caused by the proximity of the enzymes on the complex compared to the free-enzyme systems. The hydrolytic properties of the tetravalent designer cellulosome were compared with the combined action of two separate divalent cellulase- and xylanase-containing cellulosomes. Significantly, the tetravalent designer cellulosome system exhibited an ~2-fold enhancement in enzymatic activity compared to the activity of the mixture of two distinct divalent scaffoldin-borne enzymes. These results provide additional evidence that close proximity between cellulases and xylanases is key to the observed concerted degradation of the complex cellulosic substrate in which the integrated enzymes complement each other by promoting access to the relevant polysaccharide components of the substrate. The data demonstrate that cooperation among xylanases and cellulases can be augmented by their integration into a single designer cellulosome.
IMPORTANCE
Global efforts towards alternative energy programs are highlighted by processes for converting plant-derived carbohydrates to biofuels. The major barrier in such processes is the inherent recalcitrance to enzymatic degradation of cellulose combined with related associated polysaccharides. The multienzyme cellulosome complexes, produced by anaerobic bacteria, are considered to be the most efficient systems for degradation of plant cell wall biomass. In the present work, we have employed a synthetic biology approach by producing artificial designer cellulosomes of predefined enzyme composition and architecture. The engineered tetravalent cellulosome complexes contain two different types of cellulases and two distinct xylanases. Using this approach, enhanced synergistic activity was observed on wheat straw, a natural recalcitrant substrate. The present work strives to gain insight into the combined action of cellulosomal enzyme components towards the development of advanced systems for improved degradation of cellulosic material.
INTRODUCTION
When enzymes attack polymeric substrates at different sites and create new sites for each other, they act synergistically, i.e., their combined activity is greater than the sum of the individual activities. Enhancing enzyme synergy to improve industrial processes is an important biotechnological challenge. Enhancing the synergy among different glycoside hydrolases (i.e., cellulases, xylanases, and other plant cell wall polysaccharide-degrading enzymes) could have a major impact on reducing environmental pollution and improving bioenergy production.
Various paradigms for plant cell wall-degrading enzymes have been described recently (1, 2), among which free-enzyme systems and multienzyme cellulosomes are prominent. Synergism has been demonstrated between cellulases from different microbial systems, between cellulosomal and noncellulosomal enzymes, between different types of enzymes from different families, and between enzymes that have different modes of action (i.e., exoglucanases and endoglucanases) (3–11).
Synergism among enzymes of the free-cellulase-producing, thermophilic, aerobic bacterium Thermobifida fusca has received much attention, owing to the potential industrial advantages of these enzymes, including high thermostability properties, broad pH range, and high activity. In this context, synergism among T. fusca xylan-degrading enzymes (12, 13), as well as their cooperation with cellulose-degrading enzymes (9, 14), has been demonstrated. Attempts to enhance T. fusca enzyme synergism also have been undertaken by integrating its enzymes into designer cellulosomes (3–5, 15). In the designer cellulosome format, enzymes are complexed together on a structural protein subunit (scaffoldin) via a tenacious and specific type of intermodular protein-protein interaction (between the cohesin modules of the scaffodin component and the dockerin-borne enzymes). In such designer cellulosome complexes, enzyme proximity, combined with substrate binding via a carbohydrate-binding module (CBM) contained in the scaffoldin subunit, can result in enhanced enzymatic activities (3, 8).
Previous research in this field has been limited to demonstrations of synergy between either xylanases or cellulases alone, while little is known about synergy between both types of enzymes within a single designer cellulosome. In a recent publication (16), we demonstrated the advantage of enzyme proximity in the degradation of a complex cellulosic substrate (wheat straw) by a mixed-enzyme designer cellulosome, comprising two xylanases and a single endoglucanase from T. fusca. Since that work focused on the contribution of a xylan/cellulose-binding module to overall activity, it was thus of interest to extend this line of research by examining the additional incorporation of an exoglucanase into a designer cellulosome. For this purpose, Cel48A (an exoglucanase from T. fusca) that has been shown previously to act synergistically with endoglucanase Cel5A (3) on crystalline cellulosic substrates was converted to the cellulosomal mode, and the two cellulases were combined with the two appropriate dockerin-containing xylanases used in the previous study (16). The present communication describes the capacity of the resultant designer cellulosome to degrade wheat straw.
RESULTS
Construction and expression of recombinant proteins.
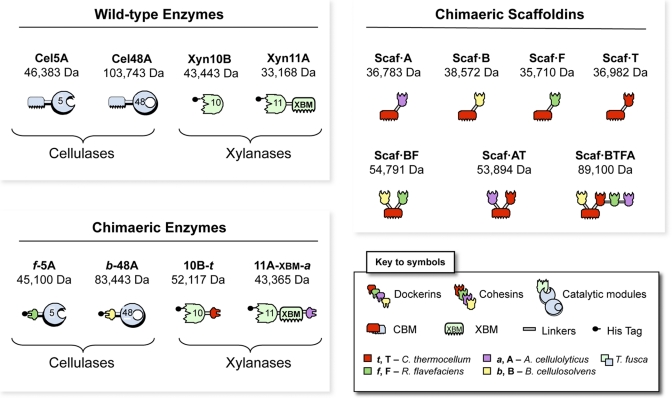
The recombinant proteins designed for use in this study are shown schematically in Fig. 1. Four different T. fusca enzymes were used, two xylanases, Xyn11A and Xyn10B, and two cellulases, family 5 endoglucanase Cel5A and family 48 exoglucanase Cel48A. The native T. fusca cellulases, Cel5A and Cel48A, are typical free (noncellulosomal) enzymes, each of which contains a family 2 cellulose-specific CBM. The native xylanase, Xyn11A, contains a CBM from the same family (named XBM [for xylan-binding CBM] for the purposes of this study), which shows binding specificity for both cellulose and xylan (17). Xyn10B lacks a CBM.
FIG 1 .
Schematic representations of the recombinant proteins used in this study. In shorthand notation for the engineered enzymes, the numbers 5, 10, 11, and 48 refer to the corresponding glycoside hydrolase (GH) family (GH5, GH10, GH11, and GH48) of the catalytic module; uppercase letters B, F, T, and A indicate the source of the cohesin module and lowercase letters b, f, t, and a indicate the source of the dockerin module from B. cellulosolvens, R. flavefaciens, C. thermocellum, and A. cellulolyticum, respectively. The source of the representative module (see key) is also indicated by color as follows: mint green or powder blue, T. fusca xylanase or cellulase, respectively; yellow, B. cellulosolvens; green, R. flavefaciens; red, C. thermocellum; lavender, A. cellulolyticus.
To convert these T. fusca enzymes into the cellulosomal mode, each was joined by recombinant DNA to a dockerin with a different specificity. These enzymes have been the topic of previous studies, and dockerins of different specificities were used to replace the N-terminal CBMs of the native cellulase (Cel5A and Cel48A), thereby generating f-5A and b-48A, or added at the C terminus of the xylanases, thus generating 11A-xbm-a and 10B-t.
The f-5A chimera is a recombinant cellulose-hydrolyzing enzyme consisting of two fused modules, a catalytic module of the family 5 endoglucanase Cel5A from T. fusca and a dockerin from the Ruminococcus flavefaciens ScaA scaffoldin (18). b-48 was designed to contain the catalytic module of T. fusca exoglucanase Cel48A ligated with a dockerin from the Bacteriodes cellulosolvens ScaA scaffoldin. In 11A-xbm-a, dockerin B from Acetovibrio cellulolyticus was appended at the C terminus of the original Xyn11A, thus retaining the original catalytic module and xylan-binding CBM (XBM). Indeed, the role of the intrinsic cellulose/xylan-binding module (XBM) of Xyn11A is essential for efficient substrate degradation in designer cellulosomes, presumably by correct orientation of the parent xylanase towards its preferred polysaccharide component of the complex wheat straw substrate (16). Consequently, this specific family 2 CBM was maintained in the chimeric enzyme. To integrate Xyn10B into an enzymatic complex, the dockerin from exoglucanase Cel48S of Clostridium thermocellum (19) was fused at its C terminus, resulting in 10B-t.
Scaf·BF carries two cohesins with different specificities, thereby allowing the possibility of binding two different dockerin-containing proteins selectively. The specific modules that comprise the construct are as follows: cohesin 3 from scaffoldin B of Bacteroides cellulosolvens (designated B for the purposes of this work), the family 3a CBM from C. thermocellum, which binds strongly to cellulose (20), and cohesin 1 from R. flavefaciens scaffoldin B (designated F) (18). Scaf·BF allows the specific incorporation of two of the above-described enzymes, i.e., cellulases b-48A and f-5A, and will direct the resultant divalent designer cellulosome complex to the substrate via the CBM.
Scaf·AT also has 2 different cohesins together with the same cellulose-binding family 3a CBM and was designed to incorporate the remaining enzymes (i.e., the xylanases) into an alternative divalent designer cellulosome complex. A. cellulolyticus cohesin 3 (designated A) (21) will interact specifically with an enzyme carrying the matching dockerin, i.e., 11A-xbm-a. At the C terminus, cohesin 3 from the CipA C. thermocellum scaffoldin (designated T) (22) will incorporate the dockerin-containing enzyme, 10B-t, into the complex.
In addition to the above-described chimeric scaffoldins, a more intricate tetravalent scaffoldin Scaf·BTFA was produced, which includes all four of the above-described cohesin types together with the cellulose-binding CBM. This 4-cohesin scaffoldin enables the integration of the two xylanases, 10B-t and 11A-xbm-a, and the two cellulases, f-5A and b-48A, into a single designer cellulosome complex.
All purified recombinant proteins showed a single major band on SDS-polyacrylamide gels (not shown), and in each case, their mobility was consistent with their molecular mass.
Affinity-based ELISA.
The specificities of the cohesins for the chimeric dockerin-bearing enzymes were examined semiquantitatively by a sensitive enzyme-linked affinity assay in microtiter plates (23). All the cohesins in each scaffoldin bound their respective dockerin in a specific manner and failed to bind (or bound very poorly) other nonmatching dockerin-bearing molecules (data not shown). The scaffoldin-borne cohesins bound their matching dockerins as efficiently as the individual monovalent scaffoldins did, indicating that the binding capabilities of the scaffoldins were reliable and selective. All specific cohesin-dockerin interactions, for each scaffoldin, were of similar intensity as judged by the affinity enzyme-linked immunosorbent assay (ELISA) procedure, thus indicating that similar amounts of protein were bound in each well, suggesting a molar equivalent of the 1:1 scaffoldin (cohesin)-to-dockerin ratio.
Analysis of complex formation.
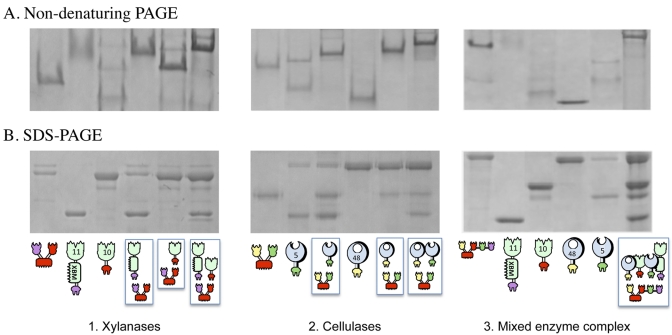
For each chimeric designer cellulosome, the formation of complexes was tested by nondenaturing PAGE. Denaturing SDS-PAGE was used as a control for verification of sample content. Stoichiometric mixtures of the enzymes and the scaffoldin resulted in a single major band with altered mobility (the band became bigger and its position shifted), thus indicating that complete or near-complete complexation was achieved in all cases (Fig. 2).
FIG 2 .
Electrophoretic mobility of components and assembled complexes on nondenaturing and denaturing gels. For the two xylanase gels, equimolar concentrations of the chimeric enzymes (11A-xbm-a and 10B-t) and their matching divalent scaffoldin (Scaf·AT) were combined. For the two cellulase gels, equimolar concentrations of the chimeric enzymes (f-5A and b-48A) and their matching divalent scaffoldin (Scaf·BF) were combined. For the two mixed-enzyme complex gels, equimolar concentrations of the chimeric enzymes (f-5A, b-48A, 11A-xbm-a, and 10B-t) and their matching tetravalent scaffoldin (Scaf·BTAF) were combined. The single fusion proteins and the mixtures were subjected to nondenaturing PAGE (A) and denaturing SDS-PAGE (B). Analysis of the matching components by nondenaturing PAGE indicates their near-complete interaction as a single major band formed. The schematic representations below the gels are explained in the symbol key in Fig. 1 and in the legend to Fig. 1.
As in previous designer cellulosome studies, some of the chimeric scaffoldin and enzyme preparations were characterized by residual contaminating bands, viewed either by denaturing PAGE and/or by nondenaturing PAGE. These bands were generally minor, and for the purposes of these studies, the quality of the different designer cellulosome preparations and their suitability for use in activity experiments were judged by the banding pattern of the final product—i.e., the formation of a major band on a nondenaturing polyacrylamide gel.
Enzymatic activity on wheat straw.
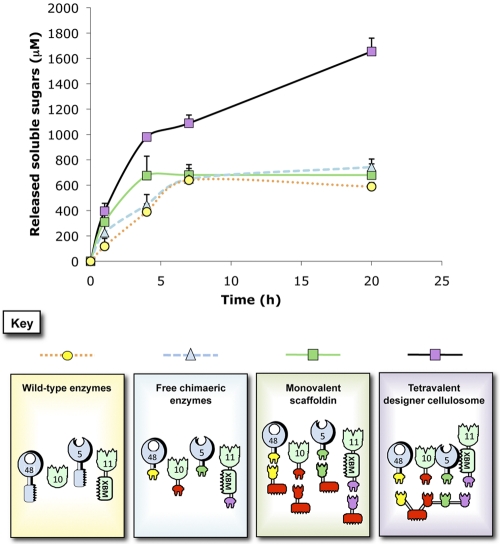
As observed in Fig. 3, the various mixtures of free enzymes were found to be less active on wheat straw than the designer cellulosome complex, which exhibited an ~2.4-fold enhancement compared to the other enzyme mixtures after 20 h of degradation. No significant difference was observed in activity on the wheat straw substrate of the wild-type enzymes, the free chimeric enzymes containing dockerins, and the monovalent scaffoldin-bearing enzymes, after 7 h of degradation. At earlier time points, the monovalent scaffoldin-bearing enzymes appeared to be more efficient than the other controls, presumably owing to the well-established targeting effect (4–6).
FIG 3 .
Kinetics studies of wheat straw hydrolysis by the free-enzyme systems versus the tetravalent cellulosome. The kinetics curves for scaffoldin-bound enzyme complex (designer cellulosomes), free-chimeric-enzyme system (lacking CBMs), chimeric enzymes attached to monovalent scaffoldins (containing CBM), and wild-type enzymes (containing native CBMs) are shown. Each reaction was performed three times. Error bars represent standard deviations.
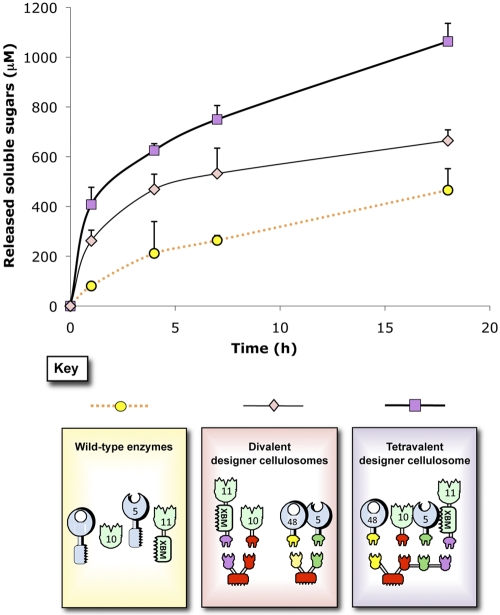
To evaluate potential proximity effects between cellulases and xylanases, two separate divalent designer cellulosomes were produced. For this purpose, the chimeric xylanases 10B-t and 11A-xbm-a were incorporated into an appropriate divalent scaffoldin, and the chimeric cellulases f-5A and b-48A were integrated into a second scaffoldin. We were able to estimate the differences in substrate degradation between the mixture of the two divalent scaffoldins (containing cellulase or xylanase) and a third scaffoldin bearing all four enzymes (Fig. 4). After 17 h of degradation, the tetravalent cellulosome exhibited a significant increase in activity compared to the mixture of the two distinct scaffoldins (~2-fold enhancement), while the mixture of the two designer cellulosomes also showed an enhanced rate of degradation compared to the wild-type enzymes (~1.5-fold enhancement). Interestingly, the data showed that the production of soluble sugars by the tetravalent designer cellulosomes continued to increase significantly as a function of time.
FIG 4 .
Kinetics studies of wheat straw hydrolysis by the different complexes versus the free wild-type enzyme system. Degradation by a single tetravalent designer cellulosome complex bearing all four enzymes, degradation by two divalent designer cellulosomes complexes, one bearing the cellulases and the other bearing the xylanases, and degradation by the free wild-type enzymes are shown. Each reaction was performed three times. Error bars represent standard deviations.
Sugar analysis.
Sugar concentrations and identification were determined using known concentrations of standards, and the relative amounts of products following enzymatic action of the different preparations on the wheat straw substrate were calculated via integration of the identified peaks. Combinations of free and scaffoldin-borne enzymes were applied to samples of hatched wheat straw, and the degradation products were analyzed after 17 h of incubation. Different quantities of arabinose, cellobiose, xylose, xylobiose, and xylotriose were found in each sample (Table 1). Note that the differences between the total sugars for each sample identified by high-performance liquid chromatography (HPLC) are not as large as those seen in Fig. 3 and 4, as HPLC analysis did not detect all the reducing sugars measured by the dinitrosalicylic acid (DNS) reagent.
TABLE 1 .
Soluble sugar production following digestion of hatched wheat straw (17-h incubation period) by various enzyme combinationsa
| Enzyme combination | Amt of sugar producedb |
||||
|---|---|---|---|---|---|
| Arabinose | Xylose | Xylobiose | Cellobiose | Xylotriose | |
| Wild-type free enzymes | 6.8 ± 0.1 (45.7) | 6.8 ± 0.3 (45.7) | 19 ± 0.6 (68.6) | 46.7 ± 1.4 (137.1) | 4.5 ± 0.4 (10.9) |
| Divalent cellulase- and xylanase-containing designer cellulosomes | 6.5 ± 0.5 (42.8) | 5.3 ± 0.3 (34.2) | 12.7 ± 1.5 (45.7) | 58.7 ± 1.9 (168.6) | 11.3 ± 0.7 (27.1) |
| Tetravalent cellulase/xylanase-containing designer cellulosome | 6.8 ± 0.2 (45.7) | 7.8 ± 0.1 (51.4) | 31.6 ± 0.7 (111.4) | 61.5 ± 1 (180) | 12.7 ± 0.6 (30.6) |
Data were obtained by HPLC analysis. The absence of glucose was confirmed by using a glucose assay kit. Values for xylose were corroborated using a xylose assay kit. An unidentified peak, present only after enzymatic treatments, eluted at ~3.9 min (between the xylose and xylobiose peaks) and was possibly a modified monosaccharide.
Values are shown in milligrams per gram of substrate (mean ± standard deviation). The values in parentheses are micromoles per gram of substrate.
Incorporation of the dockerin-containing enzyme derivatives into two chimeric scaffoldins enhanced the levels of cellobiose and xylotriose at the apparent expense of xylobiose and arabinose relative to the degradation products released by the wild-type enzymes, while the levels of all sugars were higher in samples treated with the tetravalent cellulose/xylanase-containing designer cellulosome, in accordance with the levels of reducing sugars shown in Fig. 4.
Yield calculations.
In an earlier report (8), the washed, untreated, commercially obtained hatched wheat straw substrate was found to contain 3.3 mmol of acid-extracted reducing sugar/g (dry matter), which is made up of approximately 2.3 mmol of glucose, 0.8 mmol of xylose, and 0.1 mmol of arabinose (all three measurements per gram [dry matter]). In the present work, the reaction yields after 4.5 h of enzyme treatment (initial rates of the reaction) comprised 8.1% for the tetravalent cellulose/xylanase designer cellulosome system versus 4.3% for the mixture of wild-type enzymes; after 20 h of reaction, the yields increased to 13.8% versus 6.1%, respectively.
DISCUSSION
The tetravalent cellulose/xylanase-containing designer cellulosome exhibited enhanced degradation of a natural recalcitrant substrate compared to the combined action of the corresponding free wild-type enzymes. Enzyme proximity was shown to be mainly responsible for the improvement of substrate degradation, and little or no contribution could be attributed to the substrate-targeting effect, which would be expected, as the free enzymes contained CBMs.
Earlier reports (3–8, 16) have demonstrated that both proximity and CBM targeting effects can occur simultaneously in designer cellulosomes. Indeed, both effects play a role in enhancing enzyme synergy by designer cellulosomes, and the effects were demonstrated to be cumulative. Nevertheless, one study (10) reported the dual action of fungal and bacterial enzymes on designer cellulosomes, whereby either a proximity effect or a targeting effect, but not both, was observed. The authors suggested that the origin of the enzymes from different microbial systems may have been responsible for apparent antagonism between the proximity and CBM targeting effects and that the benefit of combined effects may occur in designer cellulosomes composed only of bacterial enzymes. Moreover, in a recent article (24), the designer cellulosome approach was used to examine the interplay of prominent cellulosomal and noncellulosomal cellulases from C. thermocellum on crystalline cellulose. In that case, the targeting effect was found to be the major factor responsible for the synergism among the enzyme combinations, whereas the proximity effect appeared to play a negligible role.
From these studies, we can conclude that any given designer cellulosome complex may exhibit either of these effects, either singly or in combination, depending on the characteristics (specific enzymes, composition and organization of scaffoldin, linker regions, etc.) of the individual system. The phenomena that cause the synergistic effect seem to depend on the characteristics of the specific enzyme combination used to fabricate the designer cellulosome, and the properties of the component parts should be examined carefully in every study. Another important consideration is the type of substrate employed. Indeed, the type of substrate influences enzyme synergy and thus represents a fundamental parameter of the experimental system. In the present work, wheat straw was chosen as the substrate. This natural complex cellulosic substrate was subjected only to physical pretreatment, and the experimental conditions are thus closer to those in nature, as opposed to synthetic cellulosic substrates such as Avicel. Wheat straw was selected in lieu of other possible complex natural substrates (e.g., switchgrass or poplar) to allow comparison with the results of previous studies on designer cellulosomes (8, 16).
We have recently demonstrated the advantageous property of enzyme proximity in wheat straw degradation by a designer cellulosome, comprising T. fusca xylanases Xyn10B and Xyn11A and endoglucanase Cel5A (16). This complex lacked an exoglucanase—a key type of enzyme which acts in concert with appropriate endoglucanases for synergistic degradation of the cellulosic component of the straw substrate. In the current work, the chimeric scaffoldin, which contains four cohesin modules, now enabled the inclusion of the Cel48A exoglucanase to provide a more complete tetravalent cellulase/xylanase cellulosome. The close proximity between cellulases and xylanases is key to concerted degradation of the substrate, whereby the activities of the different enzymes facilitate the activities of their counterparts by promoting access to appropriate portions of the complex insoluble substrate, since the release of sugar products was higher for the tetravalent designer cellulosome than the combined effects of the two distinct divalent designer cellulosomes. These results demonstrate that cooperation among xylanases and cellulases can be promoted by their integration into a single designer cellulosome complex. The fact that the mixture of the two distinct divalent designer cellulosomes exhibited enhanced activity relative to the wild-type enzymes is an indication that the proximity between two enzymes of the same type (i.e., cellulases or xylanases) is also advantageous for efficient straw degradation.
Because they contain matching chimeric cohesin and dockerin pairs, designer cellulosomes provide an exquisite experimental tool for precision incorporation of selected enzymes into a distinct multienzyme assembly. They differ from other types of artificial complexes (minicellulosomes) in which the scaffoldin-borne cohesins are of uniform specificity, thereby generating heterogeneous populations of complexes (25, 26). The artificial tetravalent cellulase/xylanase-containing designer cellulosome described in this study includes a defined number of the known enzymes produced by T. fusca. By characterizing its degradation properties on the wheat straw substrate, we now have a basis with which to examine the effects of additional enzymes—either cellulosomal or noncellulosomal in origin, from T. fusca or from other microbial sources. New enzymatic activities can thus be evaluated in the future to introduce desirable synergistic contributions and to increase the rate of hydrolysis of complex polysaccharides. The long-term objective of such studies is to approach and hopefully surpass the rates of plant cell wall degradation exhibited by natural cellulosomes.
The cellulose in wheat straw is not highly crystalline (27), but enzyme attack on the cellulose is blocked physically by hemicellulose, consisting mainly of substituted xylan, and by lignin (28). In the present work, enhanced degradation is observed for both the cellulose and hemicellulose components, which appear to be solubilized in roughly equal amounts. This is in contrast to the effect of mild acid and hydrothermal pretreatments, which lead to selective hydrolysis of hemicellulose (28). The enzymatic content of designer cellulosomes can thus be geared to account for the structural composition of the substrate and to overcome some of the barriers (including the chemical bonding of hemicellulose to lignin) that impede access of the cellulases to their more recalcitrant cellulosic substrate. Additional benefit may be achieved by combining mixed-enzyme designer cellulosomes with pretreatment techniques, e.g., aqueous ammonia, that serve to selectively delignify plant biomass while retaining the cellulose and hemicellulose components (29).
One of the major strengths of the designer cellulosome system is also an inherent limitation. The fact that this system affords controlled incorporation of the desired enzymes implies that the resultant complexes are homogeneous in content. While this is a necessity for the purposes of basic science, this is clearly at variance with natural cellulosome systems, which are heterogeneous. Moreover, it has been shown that stoichiometric amounts of endoglucanases and exoglucanases in a given system are suboptimal for efficient degradation of crystalline cellulosic substrates, where large excesses of exoglucanases are required (9, 11, 30–33). This is true for both cellulosomal and noncellulosomal systems. To achieve efficient conversion of cellulosic biomass, future studies using designer cellulosomes need to address these issues.
In this work, we have investigated the advantages of cellulosomes versus free-enzyme systems in the degradation of plant cell wall polysaccharides in their natural setting. Our study provides additional evidence that chimeric designer cellulosome technology can include enzymes that are optimized for degradation of recalcitrant natural complex cellulosic substrates but do not form complexes in vivo. Designer cellulosomes thus hold promise as powerful biotechnological tools for improvement of enzymatic systems, and our capacity to control the specific incorporation of enzymatic and nonenzymatic components into defined complexes has considerable potential for a broad variety of applications, notably for conversion of cellulosic biomass to soluble saccharides en route to biofuels.
MATERIALS AND METHODS
Cloning.
Wild-type enzymes, chimeras, and recombinant scaffoldins were cloned as described previously (16, 17, 34–36). All enzyme constructs were designed to contain a His tag for the subsequent purification. PCRs were performed using ABgene Reddymix x2 (Advanced Biotechnologies Ltd., United Kingdom), and DNA samples were purified using a HiYield gel/PCR fragment extraction kit (Real Biotech Corporation [RBC], Taiwan).
Scaf·BTFA was assembled from cohesin modules and CBM, which were cloned from the appropriate genomic DNAs. Cohesin B (cohesin 3 from Bacteroides cellulosolvens scaffoldin B) was amplified using 5′ CCATGGCGGGGAAAAGTTCACCAG 3′ (five random bases were added before the restriction site in each primer) and 5′ GGTACCTTAGTTACAGTAATGCTTCC 3′ primers (NcoI and KpnI sites in boldface type). CBM-T (CBM3a and cohesin 3 from the cellulosomal scaffoldin subunit Clostridium thermocellum YS) was amplified using 5′ GGTACCGACAAACACACCGACAAACACA 3′ and 5′ GGATCCCTATATCTCCAACATTTACTCCAC 3′ primers (KpnI and BamHI sites in boldface type). Cohesin F (cohesin 1 from Ruminococcus flavefaciens strain 17 scaffoldin B) was amplified using 5′ GGATCCCGCCGGTGGTTTATCCGCTGTG 3′ and 5′ GCTAGCTTAATGGTGATGGTGATGGTGAACAATGATAGCGCCATCAGT 3′ primers (BamHI and NheI sites in boldface type), and cohesin a (cohesin 3 from Acetovibrio cellulolyticus scaffoldin C) was cloned using 5′ GCTAGCATTTACAGGTTGACATTGGAAGT 3′ and 5′ CTCGAGGATGCAATTACCTCAATTTTTCC 3′ primers (NheI and XhoI sites in boldface type). The different modules were assembled in the linearized pET28a plasmid to form pScaf·BTFA.
Protein expression and purification.
Wild-type enzymes and chimeras were prepared as described previously (3, 5, 16, 17, 35, 36). Recombinant scaffoldins were purified on phosphoric acid swollen cellulose (PASC) by the previously described method (34). The purity of the recombinant proteins was tested by SDS-PAGE on 12% acrylamide gels. The concentration of the purified protein was estimated by absorbance (280 nm) based on the known amino acid composition of the desired protein using the Protparam tool (http://www.expasy.org/tools/protparam.html). The proteins were then stored in 50% (vol/vol) glycerol at −20°C.
Affinity-based ELISA.
The matching fusion protein procedure of Barak et al. (23) and Caspi et al. (5) was followed to determine cohesin-dockerin specificity.
Nondenaturating PAGE.
To check the extent of interaction between the cohesin-bearing scaffoldin and dockerin-bearing enzymes, a differential mobility assay on nondenaturing gels was used. Protein samples (4 to 8 µg each) were added in a predetermined equimolar manner to Tris-buffered saline (TBS) (pH 7.4) supplemented with 10 mM CaCl2 and 0.05% Tween 20 to a total volume of 30 µl. The tubes were incubated for 2 h at 37°C. Sample buffer (7.5 µl in the absence of SDS) was added to 15 µl of the reaction mixture, and the samples were loaded onto nondenaturating gels (4.3% stacking gels/9% separating gels). A parallel SDS-PAGE gel (10%) was performed on the remaining 15-µl sample.
Enzymatic activity.
Hatched wheat straw (0.2 to 0.8 mm) provided by Valagro (Poitiers, France) was treated as described previously (8, 37): 5 g of prehatched wheat straw was placed in 500 ml of double-distilled water and subjected to 1-h intermittent mechanical treatment (10-min blend, 5-min pause) with a blender (PowerBlend, Braun, Kronburg, Germany). The crude substrate was then incubated in distilled water with gentle stirring for 3 h at room temperature, vacuum filtered on a 2.7-µm glass filter, resuspended in water, and incubated for 16 h with gentle stirring at 4°C. The suspension was filtered and washed three times with water, and a sample was dried at 100°C overnight to estimate the dry weight. A typical assay mixture consisted of 100 µl of buffer (50 mM citrate buffer [pH 6.0], 12 mM CaCl2, 2 mM EDTA), the concentration of the straw was 3.5 g/liter, and the concentration of each enzyme added was 0.2 µM. The reaction mixtures were incubated at 50°C, and the reactions were terminated at predetermined time points by transferring the tubes to an ice-water bath. After a centrifugation step (5 min at 14,000 rpm), 100-µl portions of the supernatant fluids were added to 150 µl of dinitrosalicylic acid (DNS) reagent, the tubes were boiled for 10 min, and absorbance was measured at 540 nm (38). The release of soluble sugars (reducing sugar) was determined using glucose as a standard. Dockerin-containing enzymes were subjected to 2 h of incubation (37°C in the absence of substrate) in the presence of equimolar concentrations of scaffoldin before the assay for binding interaction was performed. All assays were performed in triplicate.
Sugar identification and analysis.
Analysis of sugar content was performed using a high-performance anion-exchange chromatography (HPAEC) system equipped with a PA1 column (Dionex, Sunnyvale, CA). Reaction mixtures were loaded onto the column and eluted with NaOH (200 mM). Sugar concentrations were determined by integration of the chromatographic peaks based on the peaks of arabinose, xylose, xylobiose, xylotriose, and cellobiose standards. Low levels of arabinose and xylose were observed in blanks (double-distilled water); these values were deducted in all the samples.
Xylose concentrations were confirmed by a d-xylose assay kit purchased from Megazyme (Wicklow, Ireland); glucose (absence thereof) was determined using a glucose assay kit (product code GAGO20; Sigma-Aldrich); both kits were used following the manufacturer’s instructions.
ACKNOWLEDGMENTS
We are grateful for the critique and assistance of Ely Morag (Designer Energy, Rehovot, Israel), Dan Goldman (Technion, Haifa, Israel), and Maria Cernis throughout the stages of preparation of the manuscript.
S.M. greatly appreciates a scholarship received from the Ministry of Immigrant Absorption, Jerusalem, Israel. This research was supported by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, by the Brazilian friends of the Weizmann Institute of Science Alternative Energy Research Initiative (research grants from Charles Rothschild, Mario Fleck, and Roberto and Renata Ruhman), by the Technion-Niedersachsen Research Cooperation Program, and by the Israel Science Foundation (grants 966/09 and 159/07). Y.S. holds the Erwin and Rosl Pollak Chair in Biotechnology at the Technion-Israel Institute of Technology, and E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry at The Weizmann Institute of Science.
Footnotes
Citation Moraïs, S., Y. Barak, J. Caspi, Y. Hadar, R. Lamed, et al. 2010. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. mBio 1(5):e00285-10. doi:10.1128/mBio.00285-10.
REFERENCES
- 1. Himmel M. E., Xu Q., Luo Y., Ding S.-Y., Lamed R., Bayer E. A. 2010. Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels 1:323–341 [Google Scholar]
- 2. Wei H., Xu Q., Taylor L. E., Baker J. O., Tucker M. P., Ding S. Y. 2009. Natural paradigms of plant cell wall degradation. Curr. Opin. Biotechnol. 20:330–338 [DOI] [PubMed] [Google Scholar]
- 3. Caspi J., Barak Y., Haimovitz R., Irwin D., Lamed R., Wilson D. B., Bayer E. A. 2009. Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl. Environ. Microbiol. 75:7335–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caspi J., Irwin D., Lamed R., Fierobe H.-P., Wilson D. B., Bayer E. A. 2008. Conversion of noncellulosomal Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J. Biotechnol. 135:351–357 [DOI] [PubMed] [Google Scholar]
- 5. Caspi J., Irwin D., Lamed R., Shoham Y., Fierobe H.-P., Wilson D. B., Bayer E. A. 2006. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocat. Biotransform. 24:3–12 [Google Scholar]
- 6. Fierobe H.-P., Bayer E. A., Tardif C., Czjzek M., Mechaly A., Belaich A., Lamed R., Shoham Y., Belaich J.-P. 2002. Degradation of cellulose substrates by cellulosome chimeras: substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621–49630 [DOI] [PubMed] [Google Scholar]
- 7. Fierobe H.-P., Mechaly A., Tardif C., Belaich A., Lamed R., Shoham Y., Belaich J.-P., Bayer E. A. 2001. Design and production of active cellulosome chimeras: selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257–21261 [DOI] [PubMed] [Google Scholar]
- 8. Fierobe H.-P., Mingardon F., Mechaly A., Belaich A., Rincon M. T., Lamed R., Tardif C., Belaich J.-P., Bayer E. A. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined tri-functional scaffoldin. J. Biol. Chem. 280:16325–16334 [DOI] [PubMed] [Google Scholar]
- 9. Irwin D., Walker L., Spezio M., Wilson D. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002–1013 [DOI] [PubMed] [Google Scholar]
- 10. Mingardon F., Chanal A., López-Contreras A. M., Dray C., Bayer E. A., Fierobe H.-P. 2007. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73:3822–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker L. P., Belair C. D., Wilson D. B., Irwin D. C. 1993. Engineering cellulase mixtures by varying the mole fraction of Thermomonospora fusca E5 and E3, Trichoderma reesei CBHI, and Caldocellum saccharolyticum beta-glucosidase. Biotechnol. Bioeng. 42:1019–1028 [DOI] [PubMed] [Google Scholar]
- 12. Bachmann S. L., McCarthy A. J. 1991. Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl. Environ. Microbiol. 57:2121–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tuncer M., Ball A. S. 2003. Co-operative actions and degradation analysis of purified xylan-degrading enzymes from Thermomonospora fusca BD25 on oat-spelt xylan. J. Appl. Microbiol. 94:1030–1035 [DOI] [PubMed] [Google Scholar]
- 14. Wilson D. B. 2004. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4:72–82 [DOI] [PubMed] [Google Scholar]
- 15. Caspi J., Barak Y., Haimovitz R., Gilary H., Irwin D., Lamed R., Wilson D. B., Bayer E. A. 2010. Thermobifida fusca exoglucanase Cel6B is incompatible with the cellulosomal mode in contrast to endoglucanase Cel6A. Syst. Synth. Biol. 4:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moraïs S., Barak Y., Caspi J., Hadar Y., Lamed R., Shoham Y., Wilson D. B., Bayer E. A. 2010. Contribution of a xylan-binding module to the degradation of a complex cellulosic substrate by designer cellulosomes. Appl. Environ. Microbiol. 76:3787–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irwin D., Jung E. D., Wilson D. B. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding S.-Y., Rincon M. T., Lamed R., Martin J. C., McCrae S. I., Aurilia V., Shoham Y., Bayer E. A., Flint H. J. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang W. K., Kruus K., Wu J. H. D. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morag E., Lapidot A., Govorko D., Lamed R., Wilchek M., Bayer E. A., Shoham Y. 1995. Expression, purification and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61:1980–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu Q., Gao W., Ding S.-Y., Kenig R., Shoham Y., Bayer E. A., Lamed R. 2003. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell-surface anchoring protein. J. Bacteriol. 185:4548–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yaron S., Morag E., Bayer E. A., Lamed R., Shoham Y. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121–124 [DOI] [PubMed] [Google Scholar]
- 23. Barak Y., Handelsman T., Nakar D., Mechaly A., Lamed R., Shoham Y., Bayer E. A. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491–501 [DOI] [PubMed] [Google Scholar]
- 24. Vazana Y., Moraïs S., Barak Y., Lamed R., Bayer E. A. 2010. Interplay between Clostridium thermocellum family 48 and family 9 cellulases in cellulosomal versus noncellulosomal states. Appl. Environ. Microbiol. 76:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arai T., Matsuoka S., Cho H. Y., Yukawa H., Inui M., Wong S. L., Doi R. H. 2007. Synthesis of Clostridium cellulovorans minicellulosomes by intercellular complementation. Proc. Natl. Acad. Sci. U. S. A. 104:1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamaru Y., Miyake H., Kuroda K., Ueda M., Doi R. H. 2010. Comparative genomics of the mesophilic cellulosome-producing Clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing. Environ. Technol. 31:889–903 [DOI] [PubMed] [Google Scholar]
- 27. Liu R., Yu H., Huang Y. 2005. Structure and morphology of cellulose in wheat straw. Cellulose 12:25–34 [Google Scholar]
- 28. Kristensen J. B., Thygesen L. G., Felby C., Jørgensen H., Elder T. 2008. Cell-wall structural changes in wheat straw pretreated for bioethanol production. Biotechnol. Biofuels 1:5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta R., Lee Y. Y. 2010. Investigation of biomass degradation mechanism in pretreatment of switchgrass by aqueous ammonia and sodium hydroxide. Bioresour. Technol. 101:8185–8191 [DOI] [PubMed] [Google Scholar]
- 30. Boisset C., Petrequin C., Chanzy H., Henrissat B., Schulein M. 2001. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 72:339–345 [DOI] [PubMed] [Google Scholar]
- 31. Chen S., Wilson D. B. 2007. Proteomic and transcriptomic analysis of extracellular proteins and mRNA levels in Thermobifida fusca grown on cellobiose and glucose. J. Bacteriol. 189:6260–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moraïs S., Heyman A., Barak Y., Caspi J., Wilson D. B., Lamed R., Shoseyov O., Bayer E. A. 2010. Enhanced cellulose degradation by nano-complexed enzymes: synergism between a scaffold-linked exoglucanase and a free endoglucanase. J. Biotechnol. 147:205–211 [DOI] [PubMed] [Google Scholar]
- 33. Vuong T. V., Wilson D. B. 2009. Processivity, synergism, and substrate specificity of Thermobifida fusca Cel6B. Appl. Environ. Microbiol. 75:6655–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haimovitz R., Barak Y., Morag E., Voronov-Goldman M., Lamed R., Bayer E. A. 2008. Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8:968–979 [DOI] [PubMed] [Google Scholar]
- 35. Irwin D. C., Zhang S., Wilson D. B. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988–4997 [DOI] [PubMed] [Google Scholar]
- 36. Kim J. H., Irwin D., Wilson D. B. 2004. Purification and characterization of Thermobifida fusca xylanase 10B. Can. J. Microbiol. 50:835–843 [DOI] [PubMed] [Google Scholar]
- 37. Tabka M. G., Herpoël-Gimbert I., Monod F., Asther M., Sigoillot J. C. 2006. Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enzyme Microb. Technol. 39:897–902 [Google Scholar]
- 38. Miller G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 31:426–428 [Google Scholar]