Abstract
Posttranslational modification is an important element in circadian clock function from cyanobacteria through plants and mammals. For example, a number of key clock components are phosphorylated and thereby marked for subsequent ubiquitination and degradation. Through forward genetic analysis we demonstrate that protein arginine methyltransferase 5 (PRMT5; At4g31120) is a critical determinant of circadian period in Arabidopsis. PRMT5 is coregulated with a set of 1,253 genes that shows alterations in phase of expression in response to entrainment to thermocycles versus photocycles in constant temperature. PRMT5 encodes a type II protein arginine methyltransferase that catalyzes the symmetric dimethylation of arginine residues (Rsme2). Rsme2 modification has been observed in many taxa, and targets include histones, components of the transcription complex, and components of the spliceosome. Neither arginine methylation nor PRMT5 has been implicated previously in circadian clock function, but the period lengthening associated with mutational disruption of prmt5 indicates that Rsme2 is a decoration important for the Arabidopsis clock and possibly for clocks in general.
Keywords: circadian rhythms, circadian clock, luciferase
The daily rotation of the earth on its axis creates a world characterized by dramatic environmental changes in light and temperature that occur at predictable intervals. Evidence from multiple taxa supports a fitness advantage associated with the possession of an internal timekeeping mechanism, the circadian clock, that allows an organism to anticipate these changes to coordinate biological processes temporally with diel cycles (1, 2). In plants, the fitness advantage of a clock that resonates with its environmental cycles has been attributed to the optimization of diurnal photosynthesis or nocturnal starch utilization (3, 4), but it also is clear that the circadian clock controls many biological processes (5, 6), and it seems likely that any fitness advantage associated with a functioning circadian clock accrues from multiple sources.
The plant circadian clock is composed of multiple, interlocked negative-feedback loops (5, 6) increasing in number and complexity as new components are identified and new interactions are established (7). Each loop involves transcriptional activation and repression. The central loop consists of two MYB transcription factors, Circadian clock-associated 1 (CCA1) and Late elongated hypocotyl (LHY), which repress expression of Timing of CAB expression 1 (TOC1), a pseudo-response regulator (PRR), through direct promoter binding. TOC1 is recruited to the CCA1 and LHY promoters and activates their expression, although the mechanistic details remain unclear. Three additional PRRs (PRR5, PRR7, and PRR9) in the morning loop repress CCA1 and LHY expression through direct promoter binding (8). Changes in expression of CCA1 and TOC1 are associated with changes in chromatin structure (9, 10). Posttranscriptional regulation is evident; for example, CCA1 mRNA stability is regulated by light (11). A number of clock proteins, including the PRR/TOC1 family, CCA1, and LHY, are posttranslationally phosphorylated (12, 13). The stability of many clock proteins is tightly regulated through ubiquitination and subsequent proteasomal degradation (14). Finally, subcellular localization of at least some clock components is tightly regulated; for example, the interaction of PRR5 with TOC1 promotes nuclear accumulation of TOC1, which is essential for its function (15).
The complexity of clock architecture and the multifaceted regulation of clock function argue that continued search for novel clock components and regulatory interactions is likely to remain productive. Many forward genetic screens for Arabidopsis clock mutants have relied on alteration in period of seedlings entrained to light-dark (LD) cycles and released into continuous light (LL), monitoring clock function via expression of a transcriptional fusion of firefly luciferase to the Chlorophyll a/b binding protein 2 gene promoter (pCAB2:LUC), which peaks at midday (16). Variations on this approach include first screening for a phenotype frequently associated with aberrant clock function, such as elongated hypocotyl, and secondarily screening for pCAB2:LUC expression (e.g., ref. 17). Although it is unlikely that such screens are yet saturated, use of another reporter construct with a different phase of peak expression might allow the identification of novel loci (18–21). Expression of some clock-controlled genes, including many involved in photosynthetic light harvesting and carbon fixation, is light induced, and rhythms in gene expression often dampen in the dark (22, 23). The rhythm in Catalase3 (CAT3) mRNA abundance also dampens in the dark, albeit to high constitutive expression (24). However, expression of a transcriptional fusion of LUC to the Catalase 3 promoter (pCAT3:LUC) maintains strong rhythms in continuous dark (DD), and the period does not lengthen compared with LL (25), in contrast to the rapid dampening and period lengthening of pCAB2:LUC in DD (26). Accordingly, we screened an ethylmethanesulfonate (EMS)-mutagenized population of seedlings expressing a pCAT3:LUC construct for altered period in DD. We identified a mutant defective in Protein arginine methyltransferase 5 (PRMT5). Loss of PRMT5 activity confers late flowering (27–29). We show that prmt5 mutants exhibit lengthened periods in several circadian rhythms, including cotyledon movement and the expression of multiple clock genes and clock-controlled output genes. Thus, protein methylation represents a posttranslational modification that modulates circadian clock function.
Results
Forward Genetic Screen Based on pCAT3:LUC Expression.
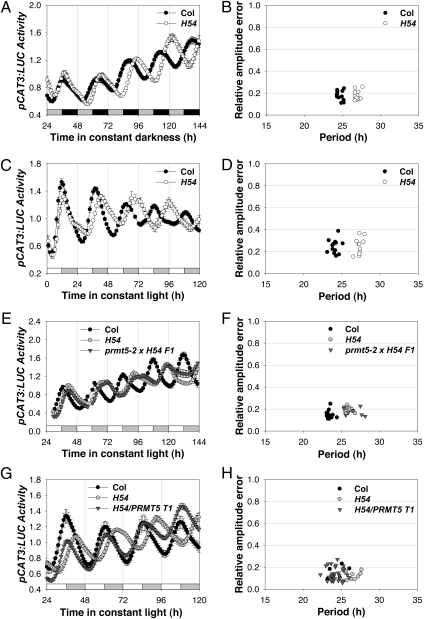
We screened an EMS-mutagenized M2 population of seedlings carrying pCAT3:LUC, which exhibits dusk-peaking expression (25), using seedlings entrained to LD cycles and released into DD, and we identified a long-period mutant, prmt5-54 (H54). In DD, Columbia 3 (Col-3) seedlings had a period (± SEM) of 24.7 ± 0.1 h (n = 11), whereas H54 seedlings had a period of 26.9 ± 0.1 h (n = 12; significant by two-tailed t test at P < 0.0001) (Fig. 1 A and B). The long period of pCAT3:LUC expression also was seen in seedlings entrained in LD cycles and released into LL. In experiments where wild-type Col-3 seedlings had a period of 24.8 ± 0.2 h (n = 15), an F3 line derived from a backcross of H54 to Col-3 had a period of 27.31 ± 0.1 h (n = 12; P < 0.0001) (Fig. 1 C and D).
Fig. 1.
prmt5 mutations lengthen the period of pCAT3:LUC in DD and LL. Seedlings of the indicated genotypes containing were entrained to photocycles (LD: 12/12 h) for 7 d before release into DD (A and B) or LL (C–H). Average traces of pCAT3:LUC expression (mean ± SEM; n = 11–13 in A–D; n = 8–36 in E–H) and period versus relative amplitude error plots show the long period of H54 (A–D), the allelism of H54 and prmt5-2 (E and F), and the rescue of the long period of H54 by PRMT5 (G and H). The gray and black bars in A and the white and gray bars in C, E, and G indicate subjective day and night, respectively.
The Gene Mutated in H54 Is Required for Multiple Circadian Rhythms.
To determine whether the gene mutated in H54 is required for multiple clock outputs, we examined the rhythm in cotyledon movement in seedlings. Consistent with the period lengthening seen in pCAT3:LUC expression, the period in cotyledon movement was lengthened. Col-3 seedlings had a period of 24.8 ± 0.2 h (n = 14), whereas H54 seedlings had a period of 26.9 ± 0.1 h (n = 12; P < 0.0001) (Fig. S1 A and B).
H54 Identifies a Role for PRMT5 in Circadian Clock Function.
Low-resolution mapping by bulked segregant analysis (30) places H54 on the lower arm of chromosome 4, in a 42-cM region between ciw7 and nga1107 (Fig. S2A). In organisms with smaller genomes, resequencing has proven a fast and economical alternative to map-based cloning for mutation identification (31–33). This approach is particularly attractive when the phenotypic characterization necessary for conventional positional cloning is made difficult by high individual variation among genotypically identical individuals, requiring confirmation in F3 lines, as is the case for circadian phenotypes. In Arabidopsis, high-throughput short-read sequencing has identified spontaneous and chemically induced mutations in the reference (34, 35) and nonreference genomes (36). Accordingly, we first resequenced the Col-3 parent used for the mutagenesis to an average base coverage of 25-fold (122 million reads of 35 bp were mapped) and identified ∼6,000 heterozygous and homozygous SNPs relative to the Col-0 sequence (Fig. S2 B and C and Table S1). We then resequenced a pool of F2 progeny from a backcross of H54 to Col-3 that were long period and, hence, homozygous for the H54 mutation. Thirty-two homozygous SNPs were clustered in the region of chromosome 4 to which H54 mapped; 15 of these SNPs were intergenic, and 17 mapped to annotated genes, although one was in an intron. Two intragenic and one intergenic SNPs also were seen in Col-3 and therefore were unlikely to be responsible for the H54 mutant long-period phenotype. Eight of the intragenic SNPs were predicted to alter the coding sequence of eight distinct genes (Table S1). One of these, a G642A transition that is consistent with EMS mutagenesis, is predicted to confer the nonsense mutation W214* and to result in a truncated form of PRMT5 (At4g31120).
Two aspects of the H54 phenotype are consistent with published reports of prmt5 mutants: H54 has dark green, curled leaves and flowers late (Fig. S3A) (27–29). However, clock defects had not yet been associated with loss of PRMT5 function. We therefore obtained additional prmt5 alleles and tested for defective clock function. Consistent with our observations with H54, prmt5-2 showed long period in cotyledon movement: Col-3 seedlings had a period of 23.4 ± 0.2 h (n = 10), whereas prmt5-2 seedlings had a period of 26.4 ± 0.6 h (n = 5; P < 0.01) (Fig. S1 C and D).
To confirm that the H54 mutant phenotype results from the W214* nonsense mutation in PRMT5, we crossed H54 to prmt5-2 and observed that the F1 progeny all showed a long period in pCAT3:LUC expression. In LL, Col-3 seedlings had a period of 23.5 ± 0.1 h (n = 12), whereas H54 seedlings had a period of 25.9 ± 0.1 h (n = 12; P < 0.0001), and F1 seedlings also exhibited a long period of 26.5 ± 0.4 h (n = 8; P < 0.0001) (Fig. 1 E and F). In addition, the F1 progeny of a cross of H54 to prmt5-2 were all late flowering (Fig. S3A), indicating allelism. Finally, we introduced a wild-type genomic copy of PRMT5 driven from its endogenous promoter into H54 and observed full rescue of the wild-type period in pCAT3:LUC expression. In LL, Col-3 seedlings had a period of 24.3 ± 0.3 h (n = 9), and T1 transgenic lines also exhibited a wild-type period of 24.2 ± 0.1 h (n = 36; P = 0.7), whereas H54 seedlings had a period of 26.3 ± 0.1 h (n = 11; P < 0.001) (Fig. 1 G and H). We tested the expression of PRMT5 in the H54 background by quantitative real-time PCR (qPCR); expression was significantly decreased (Fig. S4A). Accordingly, we conclude that H54 is a loss-of-function allele of PRMT5 that we name “prmt5-54.”
prmt5 Mutants Are Impaired in Light Inhibition of Hypocotyl Elongation.
Disruption of circadian clock function has many manifestations. Many clock mutants show defects in light inhibition of hypocotyl elongation, and we observed that prmt5-54 and prmt5-2 show long hypocotyls relative to their respective wild-type parents in blue but not in red light (Fig. S3 B and C).
PRMT5 Is Broadly Expressed, and PRMT5 Protein Is Found in both the Nucleus and Cytoplasm.
We used the PRMT5 promoter to drive β-glucuronidase (GUS) expression and observed widespread expression throughout the shoot and root (Fig. S4 B–G). To address the subcellular localization of PRMT5, we fused GFP in frame to the carboxyl terminus of PRMT5 (driven by the cassava vein mosaic virus promoter) and observed PRMT5-GFP accumulation in both nucleus and cytoplasm of transfected protoplasts (Fig. S4 H–J).
PRMT5 Expression Is Regulated by the Circadian Clock and Responds to both Light and Temperature Cues.
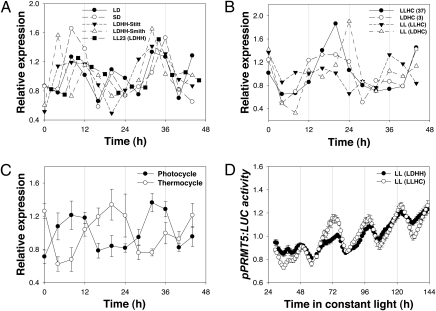
To establish whether PRMT5 mRNA abundance is circadian and/or diurnally regulated, we used the publically available DIURNAL (http://diurnal.cgrb.oregonstate.edu/) database of Arabidopsis microarrays sampling a range of time courses (37–40). PRMT5 mRNA abundance exhibits daily oscillations during growth in photocycles (LD), thermocycles (hot/cold, HC), and during growth in constant conditions following entrainment to either LD or HC cycles, indicating control by the circadian clock (Fig. 2 A–C). In plants entrained to photocycles, PRMT5 is phased to dusk (∼8–12 h after dawn or subjective dawn) during driven conditions of long (LD: l6/8 h), intermediate (LD: 12/12 h), or short (LD: 8/16 h) days as well as during free-run conditions of constant light and temperature following entrainment to photocycles (Fig. 2 A and C). Consistent with this observation, qPCR analysis using seedlings entrained to photocycles (LD: 12/12 h) before release into LL shows a dusk-specific peak of PRMT5 mRNA accumulation (Fig. S4A). In contrast, the expression of PRMT5 is phased to late evening/early dawn (∼20 h after dawn or subjective dawn) during driven conditions of thermocycles or during free-run conditions following entrainment to thermocycles (Fig. 2 B and C). This altered phase in response to thermocycles is seen both with and without concomitant exposure to photocycles (Fig. 2B) and indicates that PRMT5 is sensitive to both light and temperature but is preferentially responsive to temperature entrainment.
Fig. 2.
PRMT5 transcript abundance and transcription is clock regulated and sensitive to both light and temperature cues, but PRMT5 phase is preferentially responsive to temperature entrainment. (A–C) PRMT5 mRNA accumulates maximally in the late afternoon during photocycles and constant temperature or following entrainment to photocycles alone (A and C) but accumulates maximally in the subjective night during thermocycles or following entrainment to thermocycles or to both thermo- and photocycles (B and C). Data are from DIURNAL (http://diurnal.cgrb.oregonstate.edu/) and for each experiment are normalized to the average value to facilitate comparison of the temporal patterns despite differences in absolute signal strengths. LD, 16 h light/8 h dark; SD, 8 h light/6 h dark; LDHH-Stitt, 12 h light/12 h dark; LDHH-Smith, 12 h light/12 h dark; LL23 (LDHH), LL following entrainment to 12 h light/12 h dark; LLHC, constant light, 12 h at 22 °C/12 h at 12 °C; LDHC, 12 h in light at 22 °C/12 h in dark at 12 °C; LL (LLHC), constant conditions following entrainment to LLHC; LL (LDHC), constant conditions following entrainment to LDHC. (C) The data of A and B are averaged and plotted as mean ± SEM. (D) pPRMT5:LUC expression (mean ± SEM; n = 25) in seedlings entrained to photocycles [LL (LDHH)] or to thermocycles [LL (LLHC)] for 7 d before release into constant light and temperature.
To determine whether the circadian clock regulates transcription of PRMT5, we placed LUC under the control of the PRMT5 promoter (pPRMT5:LUC) and observed robust circadian oscillations in LUC activity (Fig. 2D). LUC activity is minimal before dusk and then increases throughout the subjective night to peak late in the subjective night, before declining before subjective dawn. This expression pattern is seen in seedlings in constant conditions following entrainment to either photocycles or thermocycles and is consistent with the peak in mRNA abundance seen following entrainment to thermocycles (Fig. 2 B and C) but contrasts with the peak in mRNA abundance seen at dusk following entrainment to photocycles (Fig. 2 A and C). Although our data are inadequate to explain this apparent discrepancy, we speculate that changes in mRNA stability may contribute to the differences between transcription rates as measured with pPRMT5:LUC and transcript accumulation. The stability of a number of transcripts has been shown to be under circadian control (41), and the stability of others, including CCA1 and LHY, is light regulated (11). The stabilization of CAT3 mRNA in extended dark obscures an underlying rhythm in CAT3 transcription (24, 42). Perhaps elevated temperature during the subjective night destabilizes PRMT5 mRNA, preventing the continued accumulation of PRMT5 mRNA at night in the absence of thermocycles. It also is possible that our pPRMT5:LUC construct lacks some sequence(s), presumably downstream of the transcription start site, necessary to recapitulate fully the transcriptional regulation of the endogenous PRMT5 gene.
Using a large set of circadian and diel expression experiments (http://diurnal.cgrb.oregonstate.edu/), we established a list of 1,253 genes (Pearson correlation coefficient = 0.9) that share the same expression profile as PRMT5 under cycling conditions (Table S2). These coexpressed genes are enriched for genes associated with ribosomal structural molecules, cytosol, cell organization and biogenesis, mitochondria, intracellular components, nucleus, cytoplasmic components, and DNA/RNA binding (Fig. S5). Using the PHASER tool (http://phaser.cgrb.oregonstate.edu/), we asked at what time of day expression of the top 500 genes peaks. Like PRMT5, this set of coregulated genes peaks before dawn after entrainment to thermocycles or to concurrent thermocycles and photocycles, but in the absence of entrainment to thermocycles (entrainment to photocycles alone), these genes exhibit altered phase and peak between midday and dusk (Fig. S6). These results support the notion that PRMT5 is part of an environmentally controlled transcription module.
Loss of PRMT5 Function Affects both the Period and Strength of Expression of Multiple Clock Genes.
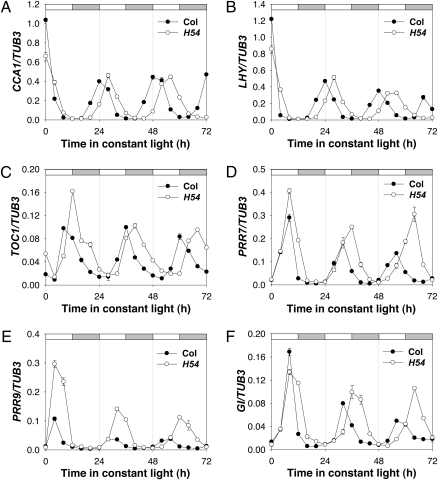
To determine if the effect of loss of PRMT5 function on period length results from the misexpression of specific clock genes, we measured the mRNA abundance of a number of clock and clock-regulated transcripts by qPCR and with promoter:LUC fusions. In each case the period was lengthened relative to wild type (Fig. 3 and Fig. S1 E–L). The amplitude and waveform of expression was relatively unaffected for several clock genes, including CCA1, LHY, and TOC1. However, the amplitude of mRNA abundance rhythms was increased dramatically for two PRR genes (PRR7 and PRR9) and for an evening-expressed clock gene, GIGANTEA (GI) (Fig. 3), suggesting that these genes may be targets of PRMT5 activity.
Fig. 3.
prmt5 mutation affects the period of all and the amplitude of expression of a subset of clock genes. Col and prmt5-54 seedlings were entrained to photocycles (LD: 12/12 h) for 10 d before release into LL. Transcript levels (mean ± SEM from two independent experiments) of (A) CCA1, (B) LHY, (C) TOC1, (D) PRR7, (E) PRR9, and (F) GI were estimated by qPCR and normalized to tubulin (TUB3) expression. White and gray bars indicate subjective day and night, respectively.
Discussion
Our results demonstrate a crucial role for PRMT5 in the Arabidopsis circadian clock. PRMT5 encodes a type II protein arginine methyltransferase that catalyzes the symmetric dimethylation of arginine residues (Rsme2). A number of posttranslational modifications have been established as important elements in the plant circadian clock. For example, many clock components are phosphorylated, as has been seen in other taxa from cyanobacteria through humans (43, 44). A second posttranslational modification, ubiquitination, also is encountered often and typically targets a protein for proteasomal degradation (14). Phosphorylation often serves as a prelude to ubiquitination and degradation. However, phosphorylation also can alter protein–protein and protein–DNA interactions as well as protein activity without concomitant effects on protein stability. Posttranslational decoration with N-acetylglucosamine also plays a significant role in Arabidopsis clock function, because mutational loss of or overexpression of the O-linked β-N-acetylglucosamine transferase, Spindly (SPY), lengthens or shortens, respectively, the circadian period (45). The period lengthening associated with mutational disruption of prmt5 indicates that Rsme2 represents a decoration important for the functioning of the plant clock and possibly for clocks in general. In general terms, the methylation of arginine residues in proteins can be expected to alter binding interactions, because methylation alters both the shape and hydrogen-bonding potential of the modified arginine (46).
At this time we do not know the key targets for PRMT5-mediated methylation that are of functional significance to the circadian clock, although several can be proposed. PRMT5 methylates components of the transcription complex, including the transcription elongation factor SPT5, altering its interaction with RNA polymerase II and potentially affecting global transcription rates (47). PRMT5 also interacts with Switch/Sucrose nonfermentable (SWI/SNF) chromatin remodeling complexes to act as a transcriptional coactivator (46). PRMT5 is a transcriptional corepressor that methylates R8 of histone H3 and R3 of histone H4, modifications that repress gene expression (46, 47). In this role, PRMT5 can be targeted to specific genes as well as exerting more ubiquitous effects. In Arabidopsis, PRMT5 has been shown to methylate R3 of histone H4 (H4R3sme2) at the Flowering locus C (FLC) promoter, which decreases expression of FLC, a flowering repressor, to promote flowering (27–29). Loss of PRMT5 function elevates FLC expression and therefore would lengthen period, which lengthens in response to increased FLC expression (39, 48, 49). However, the period lengthening seen in prmt5 mutants is greater than might be expected based on increased FLC expression, suggesting additional consequences of loss of PRMT5 function. Thus, if the period lengthening associated with loss of PRMT5 function is mediated entirely through histone modification and concomitant changes in gene expression, there are likely to be expression changes for genes in addition to FLC.
In addition to methylating histones, PRMT5 methylates Sm proteins, constituents of small nuclear ribonucleoprotein (snRNP) components of the spliceosome, and inhibition of methylation disrupts premRNA splicing (47). It has been shown recently that histone modifications, including methylation of lysine residues in H3, regulate alternative splicing of a number of human genes (50). This observation may be of relevance to the period effects of prmt5 mutations in Arabidopsis. CCA1 premRNA undergoes alternative splicing which alters the balance between productive and unproductive mRNAs that cannot be translated to give full-length protein (51). There are alternatively spliced isoforms of other clock gene mRNAs, including at least two alternatively spliced isoforms of PRR9 mRNA. Thus, the period-lengthening effects of the loss of PRMT5 function could stem, at least in part, from either global or targeted changes in splicing.
Among the genes with altered expression in prmt5 mutants, the morning loop components PRR9 and PRR7 and the evening loop component GI show increased amplitude as well as long period and are potential direct or indirect targets of PRMT5 activity. In contrast, central loop components CCA1, LHY, and TOC1 are relatively unaffected in terms of amplitude and exhibit only long period. Together with the sensitivity of PRMT5 phase to entrainment by photocycles versus thermocycles, this finding suggests that PRMT5 may be an important conduit of environmental input, via the morning and evening loops, to modulate clock entrainment and performance. An exciting possibility is that PRMT5 exerts this effect through environmentally induced epigenetic genome decorations.
Materials and Methods
Bioluminescence and Cotyledon Movement Assays.
Rhythm assays were performed as described (52), except that LUC measurement in the dark was recorded with a Hamamatsu digital CCD camera (C4742-98 ERG; Hamamatsu Photonics) using MetaMorph software. Seedlings were entrained in photocycles (LD: 12/12 h) for 6 (cotyledon movement) or 7 (LUC) d. Rhythms were analyzed by fast-Fourier transform nonlinear least-squares (53).
Generation of Constructs and Transgenic Plants.
For complementation of H54, ∼5.6 kb of the PRMT5 gene including 1 kb of promoter was amplified by PCR from genomic DNA using gene-specific primers (Table S3), cloned into pCR8/GW/TOPO (Invitrogen), and recombined into pEarleyGate 301 (54). For pPRMT5:GUS or pPRMT5:LUC lines, ∼1.1 kb of the PRMT5 promoter was amplified from genomic DNA using gene-specific primers (Table S3), cloned into pCR8/GW/TOPO (Invitrogen), and recombined into pMDC 163 (55) or subcloned into pZPΩLUC+ (56). The resulting binary vectors were introduced into Agrobacterium tumefaciens strain AGL1 by electroporation and transformed into Col (52).
Expression Analysis by Quantitative RT-PCR.
Seedlings were entrained for 10 d in photocycles (LD: 12/12 h) and transferred to LL. Samples were collected every 4 h for the following 3 d. RNA was extracted using Qiagen RNeasy Plant Mini Kit (Qiagen). First-strand cDNA synthesis used 2 μg of total RNA with the SuperScript III first-strand synthesis system (Invitrogen). The cDNA was diluted 10× with water, and 1 μL was used for PCR amplification using a SYBR Premix Ex Taq II (Takara) with gene-specific primers (Table S3). mRNA abundances were calculated using the comparative CT method, with TUB3 (At5g62700) as the normalization control.
Plant Growth and DNA Preparation for Sequencing.
H54 and its parent (Columbia, Col-3) were grown under intermediate-length days (LD: 12/12 h) and constant temperature (22 °C) for 7–12 d on 1/2 strength Murashige and Skoog plates without sucrose. Whole plants were harvested, and nuclear DNA was extracted using a modified protocol (57). Briefly, groups of 20–50 seedlings were frozen in liquid nitrogen, homogenized in a Retsch mixer (MM301), and filtered through cheesecloth, and nuclei isolated using a sucrose gradient were used for DNA extraction. SOLiD fragment libraries were prepared according to manufacturer's protocol (Life Technologies).
SOLiD Sequence by Ligation Sequencing.
Sequencing template beads were generated by emulsion PCR, affixed to a microscope slide, and sequenced by oligonucleotide ligation and detection according to the SOLiD manufacturer's instructions (Life Technologies). We generated 35-bp reads for Col-3 and 50-bp reads for H54. Primary images were processed, color bases were called, and colorspace quality values were generated on the SOLiD onboard cluster. Reads were rejected if any colorbase could not be called in a read; usually this problem resulted from poor quality toward the end of the read or beads that were out of focus during a specific ligation cycle.
SOLiD Colorspace Alignment, SNP and INDEL Detection.
Colorspace SOLiD reads (.csfasta) were mapped to the TAIR7 annotation of the Arabidopsis genome (arabidopsis.org). Raw colorspace reads that included both polyclonal reads and low quality were mapped (in colorspace) to the TAIR7 Arabidopsis genome using the corona lite pipeline (http://solidsoftwaretools.com/) allowing three mismatches (mm, -e 3) for 35-bp reads and 5 mm (-e 5) for 50-bp reads, counting adjacent errors as one error (-a 1), and aligning a maximum of 10 hits across the genome (-z 10). Both uniquely and randomly (more than one time in the genome) matching reads were used to call SNPs, thus leading to an inflated number of heterozygous SNP calls. In colorspace every base is queried twice by two separate ligation reactions (two base pair encoding) and provides two independent supports for SNP calling. Thus in colorspace mapping, an SNP results in two adjacent mismatches with the reference genome; two colorspace changes are required for each SNP. To maximize the number of reads that map to the genome, adjacent errors are considered as one error. This approach allows the detection of up to one SNP and one error, two SNPs or two errors per read. To handle reads that map to multiple places in the genome (random), usually the result of repetitive sequences, we limited the number of times that these reads can map to 10.
To identify the causative SNP in H54, we used several filtering criteria. First we removed all SNPs that were identified in both Col-3 and H54 as compared with the TAIR7 reference. Second, we only used homozygous SNPs, which were mapped back to annotated genes in the TAIR7 assembly to determine if they changed amino acids.
Additional materials and methods are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Grants IOS-0960803 from the National Science Foundation (to C.R.M.) and DE-FG02-08ER64630 from the Department of Energy (to T.P.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011987107/-/DCSupplemental.
References
- 1.Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12:970–981. doi: 10.1111/j.1461-0248.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 2.Resco V, Hartwell J, Hall A. Ecological implications of plants ability to tell the time. Ecol Lett. 2009;12:583–592. doi: 10.1111/j.1461-0248.2009.01295.x. [DOI] [PubMed] [Google Scholar]
- 3.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 7.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a novel component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perales M, Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yakir E, Hilman D, Hassidim M, Green RM. CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol. 2007;145:925–932. doi: 10.1104/pp.107.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugano S, Andronis C, Green RM, Wang Z-Y, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara S, et al. Post-translational regulation of the circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 14.Somers DE, Fujiwara S. Thinking outside the F-box: Novel ligands for novel receptors. Trends Plant Sci. 2009;14:206–213. doi: 10.1016/j.tplants.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millar AJ, Carré IA, Strayer CA, Chua N-H, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 17.Hazen SP, et al. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onai K, et al. Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J. 2004;40:1–11. doi: 10.1111/j.1365-313X.2004.02191.x. [DOI] [PubMed] [Google Scholar]
- 19.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007;143:473–486. doi: 10.1104/pp.106.088757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Tryon EL, Harmer SL. XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell. 2008;20:1244–1259. doi: 10.1105/tpc.107.056655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin PF, Kay SA. Circadian photoperception. Annu Rev Physiol. 2001;63:677–694. doi: 10.1146/annurev.physiol.63.1.677. [DOI] [PubMed] [Google Scholar]
- 23.Salomé PA, McClung CR. What makes Arabidopsis tick: Light and temperature entrainment of the circadian clock. Plant Cell Environ. 2005;28:21–38. [Google Scholar]
- 24.Zhong HH, Resnick AS, Straume M, Robertson McClung C. Effects of synergistic signaling by phytochrome A and cryptochrome1 on circadian clock-regulated catalase expression. Plant Cell. 1997;9:947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis thaliana. Plant Physiol. 2002;130:627–638. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei Y, et al. Mutations in the type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007;144:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz RJ, Sung S, Amasino RM. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:411–416. doi: 10.1073/pnas.0710423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukowitz W, Gillmor CS, Scheible W-R. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 32.Sarin S, Prabhu S, O'Meara MM, Pe'er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods. 2008;5:865–867. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DR, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneeberger K, et al. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 35.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laitinen RAE, Schneeberger K, Jelly NS, Ossowski S, Weigel D. Identification of a spontaneous frame shift mutation in a nonreference Arabidopsis accession using whole genome sequencing. Plant Physiol. 2010;153:652–654. doi: 10.1104/pp.110.156448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith SM, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bläsing OE, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards KD, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michael TP, Salomé PA, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA. 2003;100:6878–6883. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 44.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tseng T-S, Salomé PA, McClung CR, Olszewski NE. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell. 2004;16:1550–1563. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: Cellular functions and methods of analysis. Biochim Biophys Acta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 48.Swarup K, et al. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 1999;20:67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- 49.Salathia N, et al. FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol. 2006 doi: 10.1186/1471-2229-6-10. 10.1186/1471-2229-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 54.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 55.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.