Abstract
Myristoylation, the covalent linkage of a saturated, C14 fatty acyl chain to the N-terminal glycine in a protein, plays a vital role in reversible membrane binding and signaling by the modified proteins. Currently, little is known about the effects of myristoylation on protein folding and stability, or about the energetics and molecular mechanisms of switching involving states with sequestered versus accessible myristoyl group. Our analysis of these effects in hisactophilin, a histidine-rich protein that binds cell membranes and actin in a pH-dependent manner, shows that myristoylation significantly increases hisactophilin stability, while also markedly increasing global protein folding and unfolding rates. The switching between sequestered and accessible states is pH dependent, with an apparent pKswitch of 6.95, and an apparent free energy change of 2.0 kcal·mol-1. The myristoyl switch is linked to the reversible uptake of ∼1.5 protons, likely by histidine residues. This pH dependence of switching appears to be the physical basis of the sensitive, pH-dependent regulation of membrane binding observed in vivo. We conclude that an increase in protein stability upon modification and burial of the attached group is likely to occur in numerous proteins modified with fatty acyl or other hydrophobic groups, and that the biophysical effects of such modification are likely to play an important role in their functional switches. In addition, the increased global dynamics caused by myristoylation of hisactophilin reveals a general mechanism whereby hydrophobic moieties can make nonnative interactions or relieve strain in transition states, thereby increasing the rates of interconversion between different states.
Keywords: thermodynamic cycle, switch dynamics, switch energetics
Myristoylation is a common cotranslational modification found in ∼0.5–0.8% of eukaryotic proteins (1). This modification involves the covalent linkage of a saturated C14 fatty acyl chain to the N-terminal glycine residue in a protein (1). Myristoylated proteins play vital roles in many biological processes and commonly undergo reversible switches. The “flipping” of myristoyl switches typically involves interconversion between a myristoyl-sequestered state, myrseq, where the myristoyl group is located in a hydrophobic binding pocket within the protein, and a myristoyl-accessible state, myracc, where the myristoyl group is available for binding to membranes or other proteins. Switching may be associated with relatively large or subtle structural and/or dynamic changes in the myristoylated protein (2, 3). It can also be regulated by binding of various ligands (e.g., H+, Ca2+, GTP, or regulatory protein) (3–5). Some examples of proteins that undergo myristoyl switching include: Ca2+-dependent recoverin, which mediates photoresponses in the retina (3); Ca2+-dependent guanylate cyclase activating protein (GCAP), which regulates the function of guanylate cyclase (2, 6); oligomerization-dependent HIV-1 Gag, which orchestrates HIV-1 viral proliferation (5); GTP-dependent ADP ribosylation factor proteins, which are involved in membrane trafficking (7); and pH-dependent hisactophilin, involved in controlling cytoskeletal changes during cellular movement and osmotic stress (8, 9). Thus, ligand-regulated myristoyl switching is a versatile mechanism for controlling a wide range of biologically important processes. Myristoyl switches have been characterized extensively at the functional level, but remain poorly understood at the energetic and molecular levels (1). Furthermore, there is a paucity of data on the effects of myristoylation on protein folding and stability.
In this study, we use hisactophilin as a model to analyze the effects of myristoylation. Hisactophilin is a pH-dependent, myristoylated, histidine-rich actin- and membrane-binding protein from the model organism, Dictyostelium discoideum. This small (13.5 kDa) protein facilitates cell shape changes and movements in response to chemotactic signals and osmotic stress, which result in cellular pH changes. In vivo, hisactophilin reversibly switches between a cytoplasmic form at pH 7.5 to a membrane-bound form at pH 6.5, which also anchors actin filaments to the inner leaflet of the cellular membrane. In vitro, hisactophilin undergoes a reversible myristoyl switch driven by pH. Hisactophilin contains an unusually large proportion of histidines (31 of 118 residues), with average apparent pKa values of ∼6.8 (10). Reversible proton binding by histidines has been implicated in regulating the equilibrium between the cytosolic and membrane-bound forms (8). In-depth biophysical analyses have been conducted for the nonmyristoylated form of the protein (11, 12); the myristoylated form is amenable to similar analyses, as described in this report. We describe the quantitative analyses of thermodynamic stability and kinetics of folding–unfolding for a myristoylated protein, combined with NMR analyses of switching. Our results reveal dramatic effects of myristoylation on folding, which are mediated by nonnative interactions, and provide unique insights into the energetics and mechanism of pH-dependent myristoyl switching in hisactophilin. The results and methodology have important implications for understanding the interplay between ligand binding and interactions of hydrophobic moieties in many other switching systems.
Results
Dependence on pH of Increased Stability upon Myristoylation and the Energetics of the Myristoyl Switch.
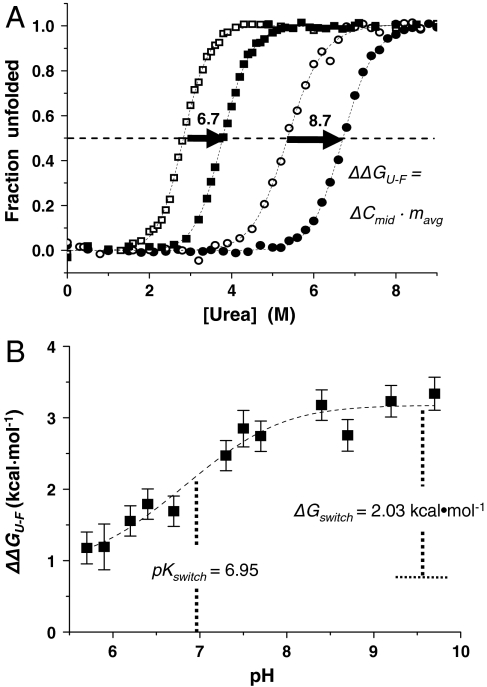
The equilibrium stabilities of myristoylated and nonmyristoylated hisactophilin were measured by circular dichroism (CD) and fluorescence-monitored urea denaturation curves at pH 5.7–9.7. The data can be well fit by a reversible two-state folding transition between folded (F) and unfolded (U) states of the protein (see SI Materials and Methods) to determine the Gibbs free energy of unfolding, ΔGU-F( = GU - GF), and the denaturant dependence of ΔGU-F or m value (Fig. 1A, and Tables S1 and S2). There is good agreement between fitted values determined by CD and fluorescence, and between equilibrium and kinetic (see below) measurements, which supports the applicability of the two-state folding model (13). The lack of observable three-state behavior in the equilibrium curves indicates that there is rapid interchange between the two folded states, myrseq and myracc, which was confirmed by NMR lineshape analysis (see below).
Fig. 1.
Effects of myristoylation on protein stability. (A) Fluorescence-monitored equilibrium denaturation curves showing fraction of unfolded protein as a function of urea concentration for nonmyristoylated (open symbols) and myristoylated (filled symbols) hisactophilin at pH 6.7 (squares) and pH 8.7 (circles). Dashed lines represent fits of the data using the binomial extrapolation method two-state model (11). The horizontal dotted line is at the transition midpoint, Cmid, where half of the protein is unfolded. (B) ΔΔGU-F as a function of pH. The magnitude of the change in ΔΔGU-F at limiting pH values corresponds to the ΔGswitch of 2.03 ± 0.17 kcal·mol-1 (Figs. S1 and S2). The pKswitch of 6.95 ± 0.15 is the pH at the midpoint of the switch. The dashed line represents the fit of the data to Eq. 1.
Myristoylated hisactophilin is more stable than the nonmyristoylated protein at all pH values (Fig. 1). Notably, the increase in stability upon myristoylation, ΔΔGU-F( = ΔGU-F,myr - ΔGU-F,nonmyr), varies significantly with pH in the physiological range, from 3.15 to 1.13 kcal·mol-1, corresponding to predominantly myrseq at high pH and myracc at low pH, respectively. These changes in energetics can be interpreted using thermodynamic cycles (Fig. S1 and SI Results) (8). The pH dependence of ΔΔGU-F fits an apparent pKswitch of 6.95 ± 0.15 (Eq. 1 and Fig. 1B), and has an apparent switch energy for flipping from the sequestered to the accessible state, ΔGswitch( = ΔΔGU-F,low pH - ΔΔGU-F,high pH), of 2.03 ± 0.17 kcal·mol-1 (Eq. 1 and Fig. 1B). The pKswitch is in the typical range of pKa values for histidine residues in proteins, further supporting involvement of these residues in controlling the switch (10), which is also supported by NMR data (see below).
The pH dependence of ΔΔGU-F can also be fit in terms of the number of ionizable groups involved in controlling the switch and the associated pKa values in the myrseq and myracc states (14) (Fig. 1B and Fig. S1). This fitting reveals that the apparent pKa of ionizable groups increases from pKa,seq of ∼6–6.5 to pKa,acc ∼ 7.6–7.0 when the myristoyl group switches from myrseq to myracc, respectively. Thus, decreasing pH favors increased population of myracc because this state more readily binds protons owing to its higher pKa value (Figs. S1 and S2). There is a net uptake of ∼1.5 protons associated with the switch from myrseq to myracc and the quality of the fits is slightly better for smaller numbers of ionizable groups (Fig. S2 and SI Results).
Myristoylation Stabilizes the Transition State of Folding and Increases Global Dynamics.
The kinetics of protein folding and unfolding for myristoylated and nonmyristoylated hisactophilin were also measured at various pH values (Fig. 2 and Table S1), and these data are also well fit by the two-state model. Surprisingly, despite its higher stability, myristoylated hisactophilin unfolds ∼10 times faster than the nonmyristoylated protein, at all pH values. However, the folding rates are also increased within error, to a larger extent than the unfolding rates, resulting in increased stability. The effects of myristoylation on protein energetics can be understood using free energy diagrams and Φ-value analysis (13) (Fig. 3). Φ is defined as ΔΔG‡-U/ΔΔGF-U, where ΔΔG‡-U and ΔΔGF-U represent the free energy change upon myristoylation (analogous to mutation) of the transition state and folded state, respectively, relative to the unfolded state. Classical Φ-values range from 0 to 1, corresponding to the energetic effects of the myristoyl group in the transition state being the same as in the folded state or unfolded state, respectively. Strikingly, the Φ-value for myristoylation is larger than one. This unusual, nonclassical Φ-value suggests that the myristoyl group stabilizes the transition state more than the folded and unfolded states (Fig. 3) (15). Such nonclassical Φ-values have often been interpreted as evidence for the formation of nonnative interactions in the transition state, which can decrease kinetic energy barriers and so contribute to increased folding and unfolding rates. Additional factors to consider when interpreting energy changes are possible structural reorganization in the protein or solvent. Structural changes in folded hisactophilin appear to be small based on no substantial changes in CD spectrum (11) or protein NOEs upon myristoylation (see below). Further details regarding interpretation of energetic changes are considered in the Discussion.
Fig. 2.
Chevron plots of the natural logarithm of the observed rate constants, kobs, as a function of urea concentration for nonmyristoylated (open symbols) and myristoylated (closed symbols) hisactophilin at pH 6.7 (squares) and pH 8.7 (circles). Dashed lines represent fits of the data to a two-state binomial extrapolation model (12) and fitted values are given in Table S1.
Fig. 3.
Gibbs free energy diagram for myristoylated (- -) and nonmyristoylated (—) hisactophilin. ΔG‡-U and ΔGF-U represent the measured kinetic folding barriers and free energies of folding, respectively (13). The free energy of the unfolded state, U, is proposed to be increased upon myristoylation, as generally occurs upon increased exposure of hydrophobic groups to aqueous solution (47). The free energy of the folded state, F, and transition state, TS, are defined relative to the unfolded state, based on ΔGF-U and ΔG‡-U, respectively. The energies for various states are not drawn to scale, but are consistent with the experimentally determined equilibrium stabilities and kinetic data. Because only relative energy levels can be determined experimentally, the entire profiles for myristoylated and nonmyristoylated hisactophilin may be shifted relative to each other.
Localization of the Myristoyl Group in the Major Hydrophobic Core.
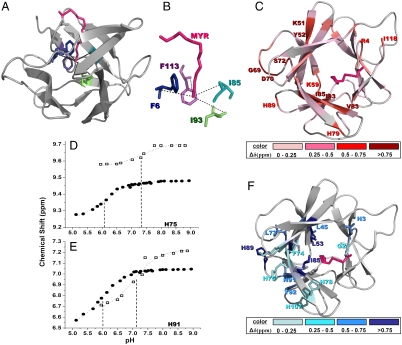
The structure of myristoylated hisactophilin was investigated as a function of pH using NMR, and compared with data obtained for the nonmyristoylated protein. The myristoyl group exhibits just one set of resonances in NMR spectra (Fig. S3), indicating that exchange between the myrseq and myracc states is fast on the NMR timescale. A lower limit for the rate constants of exchange between the myrseq and myracc states was estimated from lineshape analysis to be on the order of ∼1 × 105 s-1 (SI Results). These fast rate constants are consistent with the apparent two-state transitions in denaturant (Figs. 1 and 2). NOE and chemical shift data show that the myristoyl group is buried in the major hydrophobic core of the protein in the myrseq state. The amide protons of F6, I85, I93, and F113 exhibit NOEs with the terminal methyl of the myristoyl moiety (Fig. 4 A and B). Thus, the myristoyl reaches the interface between the hydrophobic bottom layer of the β-barrel (where F6, I85, and F113 are situated) and the upper β-hairpin layer (where I93 is situated). The observation of the largest chemical shift changes upon myristoylation, 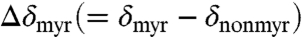 , in this region is consistent with the location of the myristoyl group near these residues, although the NOEs for the residues with perturbed chemical shifts are very similar to those in the nonmyristoylated protein, indicating no major structural reorganization. Nevertheless, myristoylation causes extensive chemical shift changes through the protein structure (Fig. 4C and Fig. S4).
, in this region is consistent with the location of the myristoyl group near these residues, although the NOEs for the residues with perturbed chemical shifts are very similar to those in the nonmyristoylated protein, indicating no major structural reorganization. Nevertheless, myristoylation causes extensive chemical shift changes through the protein structure (Fig. 4C and Fig. S4).
Fig. 4.
NMR analysis of structure and localization of ionizable groups involved in controlling switching in myristoylated hisactophilin. The structure of the protein was modeled based on the structure of nonmyristoylated hisactophilin (Protein Data Bank 1HCD) and NOEs observed for the myristoyl group (see SI Results for details). The protein backbone is shown as a ribbon; the myristoyl group (pink) and selected residues are colored (see panel legend) and labeled with single letter code and residue number. (A) The myristoyl group is buried in the hydrophobic core of the protein in the myrseq state. Residues exhibiting NOEs between their amide proton and the terminal methyl of the myristoyl group are shown in stick representation (F6 in blue, I85 in cyan, I93 in green, and F113 in purple). (B) Close-up view of myristoyl group illustrating NOEs (dotted lines) from A. (C) Structure of hisactophilin showing chemical shift changes upon myristoylation, Δδmyr( = δmyr - δnonmyr), calculated using Eq. S1 at pH 8.7. (D and E) pH dependence of chemical shifts for backbone amide 1H resonances of H75 (D) and H91 (E) in nonmyristoylated (□) and myristoylated (●) hisactophilin. Fits of the observed chemical shifts to a single apparent pKa correspond to those determined from fitting the pH dependence of ΔΔGU-F (Fig. S2). Similar pH-dependencies are observed for additional amides in the vicinity of H75 and H91 (Fig. S5), with the magnitude of the chemical shift changes tending to be largest for groups closest to H75 and H91. (F) Structure of hisactophilin color coded according to the magnitude of chemical shift changes associated with a pKa,seq ∼ 6 in myristoylated hisactophilin. The colored amides show a decrease in apparent pKa from ∼7–7.5 to ∼6 for 1H and 15N upon myristoylation. Δδ values were calculated similar to ref. 10 and limiting chemical shift values at low and high pH associated with pKa,seq ∼ 6.
Role of Histidines in Flipping the Myristoyl Switch via Proton Uptake or Release.
Further insight into the molecular mechanism of switching was obtained by analyzing the changes in chemical shifts as a function of pH (Fig. 4 D–F). For many amide (NH) groups, the apparent pKa values for pH-dependent changes in chemical shift change from pH ∼ 7–7.5 in nonmyristoylated hisactophilin to ∼6 in the myristoylated protein (Fig. 4 D and E). These apparent pKa values are very similar to the values of pKa,acc and pKa,seq for the ionizable groups that govern switching obtained from fitting the pH dependence of ΔΔGU-F (Fig. S2). A prominent group of such NHs is clustered on one side of the protein (Fig. 4F) in the vicinity of various ionizable groups, including H75 (Fig. 4D), H91 (Fig. 4E), H78, and H107 (Fig. 4F). It is not possible from the available data to determine the exact contributions of each of these groups to switching (SI Results); however, H75 and H91 are likely to play significant roles because the largest changes in chemical shift occur near these residues. Further inspection of the pattern of chemical shift changes reveals a likely pathway for communication between the ionizable groups and the myristoyl group via hydrophobic residues (L45, L53, F74, I85) that pack near the myristoyl group in the protein core (Fig. 4F). Thus, the combined results from fitting the pH dependence of the switch energetics and the pH dependence of chemical shift changes provides an intriguing model for the molecular basis of the pH dependence of the myristoyl switch controlled by histidine ionizations and propagated by hydrophobic residues.
Discussion
Myristoylation Increases Protein Stability.
Despite the relatively common occurrence of protein myristoylation, there is remarkably little information available concerning its effects on protein folding. Here we show that myristoylation significantly increases the stability of hisactophilin for both the myrseq and the myracc states. The relatively smaller increase in stability for myracc suggests that there are some residual stabilizing interactions and/or burial of the myristoyl group at low pH. Much larger stabilization is observed for the myrseq state when the myristoyl group is buried in the hydrophobic core at high pH. It is well established that, in general, increasing the burial of hydrophobic groups increases protein stability (16). It appears that burial of myristoyl groups may also commonly stabilize other proteins in an analogous fashion. For example, protein melting temperatures are increased upon myristoylation of HIV-1 matrix protein p17 (17), GCAP1 (2, 18), and calcineurin (19). Increased stability upon myristoylation is also implicated by structural data for recoverin, which is well ordered when the myristoyl is sequestered inside the protein but shows increased disorder when the myristoyl is exposed to solvent (3); similar behavior is also observed for GCAP1. It is noteworthy that all of the aforementioned proteins are structurally unrelated to hisactophilin. Recoverin, GCAP1, and calcineurin are highly helical, but the myristoyl group also inserts into the major hydrophobic core of these proteins. Thus, myristoylation may frequently contribute to increasing protein stability via hydrophobic burial in protein cores made by various structural elements.
Myristoylation Increases Global Protein Folding and Unfolding.
Quite unexpectedly, despite increasing protein stability, myristoylation of hisactophilin also increases protein dynamics, as is evident in the markedly increased rates of global folding and unfolding. These increased dynamics may be linked to and promote the rapid interconversion between myrseq and myracc states, which is revealed by the apparent two-state transitions in denaturant (Fig. 1A) and averaged resonances in NMR experiments. The increased dynamics with increased stability may initially seem counterintuitive; however, there is precedence for similar effects from Φ-values involving hydrophobic amino acids in other proteins. The global kinetic and thermodynamic data for hisactophilin reveal that myristoylation has a larger stabilizing effect on the transition state of folding relative to the ground (native and denatured) states, manifested as a nonclassical Φ-value. Such Φ-values, although not common, have been reported in a number of experimental (20–22) and theoretical studies (23). In various wild-type proteins, removal of hydrophobic groups (analogous to removal of the myristoyl moiety) has analogous effects of decreasing stability and decreasing folding and unfolding rates, e.g., I34A in src SH3 (21), V21T in CheY (20), and I23V in ADA2h (22); in other proteins, the rates are decreased but stability is increased or changes little (24). Similar effects also occur in designed proteins where stability, and folding and unfolding rates are all increased when the hydrophobicity of the protein core is increased (comparable to addition of the myristoyl group). Examples include src SH3 best 4 and best 5 (25), Src SH3 A39V/V55I (24), and acylphosphatase with a redesigned hydrophobic core (16). More dramatic increases (several orders of magnitude) in folding and unfolding rates resulted from redesigning the hydrophobic core of Rop, with slight decreases in stability (26). Together these results suggest that hydrophobic groups, including myristoyl, can accelerate global protein dynamics. Increased dynamics may be important for facilitating folding in general and for facilitating conformational changes associated with function.
Complex Energy Changes upon Myristoylation and Mechanisms for Increased Dynamics.
We propose that the mechanism for the increased stability and dynamics in myristoylated hisactophilin involves the following: (i) destabilization of the denatured state due to increased exposure of the hydrophobic myristoyl group; (ii) strain in the native myrseq state upon burial of the myristoyl group in the protein core; and (iii) nonnative interactions and/or relief of strain in the transition state (Fig. 3). This mechanism is based on and includes elements of different mechanisms proposed previously to explain nonclassical Φ-values including denatured state effects (27), overpacking of hydrophobic groups (25), and nonnative interactions of hydrophobic groups in the transition state (23). In general, folding rates are favored by classical hydrophobic burial effects (14, 23), which would also apply to burial of the myristoyl group in going from the unfolded to the transition state. The effects of hydrophobic groups on unfolding tend to be less pronounced, perhaps due to a different mechanism involving rate limiting disruption of tight native packing (23). However, the myristoyl group in hisactophilin appears not to be tightly packed, as suggested by the rapid interconversion between myrseq and myracc states. The increased dynamics in myristoylated hisactophilin suggest overpacking and strain in the folded state. This strain may be relieved in the transition state in which, based on the Tanford βT(βT = mf/meq) (13) of ∼0.7, there is ∼30% exposure of hydrophobic surface. In addition, there may be some nonnative interactions in the transition state. Nonspecific nonnative interactions have been proposed for mutations of solvent exposed Tyr to Phe mutations in SH3, where the increased hydrophobicity of Phe accelerates both folding and unfolding rates with little effect on stability (28). There is also evidence for position-specific nonnative interactions of hydrophobic groups increasing rates for Fyn SH3 (23). Considering that a myristoyl group is relatively long and flexible compared to natural hydrophobic amino acids, it has a high potential in general for making nonspecific and/or specific nonnative interactions in the transition state and thereby facilitating structural transitions.
Decreased Folding Frustration upon Myristoylation.
The speed at which a protein folds is often explained in terms of its energy landscape: proteins with smooth landscapes tend to fold quickly, whereas those with rugged or frustrated landscapes tend to fold slowly and populate partly folded intermediate states (29). A key finding here is that myristoylation decreases frustration in the energy landscape of hisactophilin. Decreased frustration is supported by the increased folding and unfolding rates, and good fits of equilibrium and kinetic data to a two-state transition, with no detectable population of partly folded intermediates (Figs. 1 and 2, and Tables S1 and S2). In contrast, previous studies on nonmyristoylated hisactophilin showed population of a folding intermediate, evidenced by rollover and double exponential folding kinetics (30). The differences suggest the myristoyl group makes specific nonnative interactions that compete with other nonnative interactions favoring intermediate formation. Similarly, specific nonnative interactions by certain hydrophobic residues in SH3 were found to accelerate folding and unfolding, whereas other hydrophobic residues had the opposite effect (23). The much faster folding of R15 compared the R16 and R17 α-spectrins may also be related to key transition state interactions of residues in the cores of these proteins, with the core of R15 notably containing more hydrophobic residues (31). In general, nonnative interactions may act to favor or disfavor folding (32). Folding simulations have identified frustration in the folding pathway of β-trefoil proteins, including nonmyristoylated hisactophilin and interleukin-1β, manifested as formation of intermediates and backtracking during folding (29, 33, 34). In interleukin-1β, the frustration is particularly pronounced due to nonnative interactions made by a hydrophobic loop (not found in hisactophilin) which is required for receptor binding. The authors concluded that the flux through multiple pathways on the β-trefoil folding landscapes may differ as a result of different functional requirements of the various trefoil proteins. It is very interesting that the naturally occurring hydrophobic myristoyl group on hisactophilin has a critical role in function and does not hinder but rather dramatically enhances folding.
Switching Facilitated by Hydrophobic Interactions and Regulated by Ligand Binding.
The increased dynamics in hisactophilin upon myristoylation may illustrate a general mechanism whereby hydrophobic groups facilitate conformational changes and switching. Another striking example is the switching of a repacked hydrophobic core mutant of Rop, which folds and unfolds 2 and 4 orders of magnitude faster, respectively, than the wild-type protein (26), and undergoes a switch between active and inactive folded states (35). Also, nonclassical Φ-values are indicative of increased dynamics (16) and have been observed in various proteins that undergo switches, such as CheY (36) and ADA2h (22). The residues exhibiting nonclassical Φ-values are often hydrophobic, again implicating a key role for hydrophobic groups in facilitating conformational dynamics. Thus, the presence of strain and/or nonnative effects seen in the global dynamics and Φ-values may be linked to switching in these proteins as well.
Another noteworthy finding here is that the thermodynamic and ligand (H+) binding characteristics of the myristoyl switch in hisactophilin are nicely tuned to enable high-sensitivity signaling. The dynamic range and detection limit for switching controlled by ligand binding is determined by the thermodynamics of switching (i.e., the equilibrium constant between the two switching states) (37–39). In hisactophilin, the ΔGswitch of 2.03 kcal·mol-1 centered around a pKswitch of 6.95 allows for a large signal (i.e., change in populations of the myracc and myrseq states) upon reversible H+ binding (37). These characteristics measured in vitro provide an explanation for in vivo observations of large changes in membrane binding by hisactophilin with changes in cellular pH (40). Switching from myrseq to myracc in hisactophilin is accompanied by the binding of ∼1.5 protons, due to an increase in the apparent pKa of ionizable groups. It is not possible from the available data to precisely define the number and identity of the ionizable groups that control switching. However, fitting of the pH dependence of the switch energetics (Fig. S2, and SI Results) combined with the pH dependence of NMR chemical shifts (Fig. 4 D–F, Figs. S5–S7, and SI Results) suggest that a small number of ionizable groups make a major contribution to switching. The lower apparent pKa in the sequestered state, which disfavors proton binding, may be a consequence of various effects, in particular, increased hydrophobic environment of the ionizable groups or their closer proximity to other positively charged groups (Fig. 4F). An analogous switching mechanism applies to recoverin, where the binding of 2 Ca2+ ions also results in altered interactions of hydrophobic groups and myristoyl switching to the accessible state (3). Similar mechanisms may also occur in the maltose binding protein (39) and N-terminal domain of calmodulin (38), where hydrophobic moieties tune a switch through alteration of ligand (maltose and Ca2+, respectively) binding affinity. Thus, the results presented here reveal a general mechanism of switching based on cooperativity between hydrophobic groups and ligand binding.
Conclusions.
We have shown here that myristoylation can simultaneously favor protein stability, folding, and function. Increases in stability and folding rates resulting from myristoylation may be advantageous for generating and maintaining proteins in vivo, whereas increases in global dynamics upon myristoylation may facilitate switching and regulation of function. It will be of great interest to determine how general these effects are for myristoylation and for other posttranslational modifications, for which there is currently very little quantitative data (41). Myristoyl and other lipid modification-based switches in a wide range of proteins can be modulated by binding of various ligands, including H+, Ca2+, GTP, and even regulatory proteins, which favor extrusion of the lipid group from a binding pocket within the protein (1). Changing intracellular pH is a common mechanism for regulating protein function and often acts in cooperation with other binding interactions (42). Regulation of the interactions of various hydrophobic moieties via ligand binding is not yet well understood in terms of mechanisms and energetics, although it occurs in many types of switching proteins, such as recoverin, maltose binding protein, calmodulin, calbindin (43), and the low-density lipoprotein receptor (44) and is of tremendous biological significance. The methodology for analyzing the energetics of switching presented herein is general and can be used to gain valuable insights into many other switches.
Materials and Methods
Recombinant Hisactophilin Expression and Purification.
Wild-type hisactophilin was expressed as described (11), after performing site-directed mutagenesis (QuickChange, Stratagene) to remove extraneous N-terminal Gly-Glu-Phe-Gly residues (45). In addition, human N-myristoyltransferase 1 was coexpressed using pHV738 (46). Myristoylated hisactophilin was purified as described previously (11) with an additional RP-HPLC purification step (see SI Materials and Methods).
Urea Denaturation Curves and Kinetic Measurements.
Equilibrium denaturation curves and kinetics of folding–unfolding were determined as described (11, 12) (SI Materials and Methods). To determine pKswitch and ΔGswitch( = ΔΔGU-F,low pH - ΔΔGU-F,high pH), the dependence of ΔΔGU-F on pH was fit to a general titration equation:
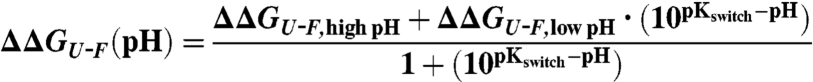 |
[1] |
NMR Analysis.
All NMR spectra were acquired at 298.15 K on a Bruker AMX 600 MHz spectrometer (SI Materials and Methods).
Supplementary Material
Acknowledgments.
The authors thank Joe Gaspar, Jessica Rumfeldt, and Daša Lipovček for their stimulating input and support, and Dr. Richard Kahn (Emory University) for the N-myristoyltransferase 1 expression plasmids. This research was funded by Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008026107/-/DCSupplemental.
References
- 1.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 2.Orban T, et al. Conformational changes in guanylate cyclase-activating protein 1 induced by Ca2+ and N-terminal fatty acid acylation. Structure. 2010;18:116–126. doi: 10.1016/j.str.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames JB, et al. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 4.Kahn RA, et al. The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport. J Biol Chem. 1992;267:13039–13046. [PubMed] [Google Scholar]
- 5.Resh MD. A myristoyl switch regulates membrane binding of HIV-1 Gag. Proc Natl Acad Sci USA. 2004;101:417–418. doi: 10.1073/pnas.0308043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim S, Peshenko I, Dizhoor A, Ames JB. Effects of Ca2+, Mg2+, and myristoylation on guanylyl cyclase activating protein 1 structure and stability. Biochemistry. 2009;48:850–862. doi: 10.1021/bi801897p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randazzo PA, et al. The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch. J Biol Chem. 1995;270:14809–14815. doi: 10.1074/jbc.270.24.14809. [DOI] [PubMed] [Google Scholar]
- 8.Hanakam F, Gerisch G, Lotz S, Alt T, Seelig A. Binding of hisactophilin I and II to lipid membranes is controlled by a pH-dependent myristoyl-histidine switch. Biochemistry. 1996;35:11036–11044. doi: 10.1021/bi960789j. [DOI] [PubMed] [Google Scholar]
- 9.Pintsch T, Zischka H, Schuster SC. Hisactophilin is involved in osmoprotection in Dictyostelium. BMC Biochem. 2002;3:10–17. doi: 10.1186/1471-2091-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond MS, Houliston RS, Meiering EM. Two-dimensional 1H and 15N NMR titration studies of hisactophilin. Biochem Cell Biol. 1998;76:294–301. doi: 10.1139/bcb-76-2-3-294. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, et al. Thermodynamics of denaturation of hisactophilin, a beta-trefoil protein. Biochemistry. 2001;40:3817–3827. doi: 10.1021/bi002609i. [DOI] [PubMed] [Google Scholar]
- 12.Wong HJ, Stathopulos PB, Bonner JM, Sawyer M, Meiering EM. Non-linear effects of temperature and urea on the thermodynamics and kinetics of folding and unfolding of hisactophilin. J Mol Biol. 2004;344:1089–1107. doi: 10.1016/j.jmb.2004.09.091. [DOI] [PubMed] [Google Scholar]
- 13.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. pp. 508–614. [Google Scholar]
- 14.Cho JH, Sato S, Raleigh DP. Thermodynamics and kinetics of non-native interactions in protein folding: A single point mutant significantly stabilizes the N-terminal domain of L9 by modulating non-native interactions in the denatured state. J Mol Biol. 2004;338:827–837. doi: 10.1016/j.jmb.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Mirny LA, Shakhnovich EI. Kinetics, thermodynamics and evolution of non-native interactions in a protein folding nucleus. Nat Struct Biol. 2000;7:336–342. doi: 10.1038/74111. [DOI] [PubMed] [Google Scholar]
- 16.Kuhlman B, Baker D. Exploring folding free energy landscapes using computational protein design. Curr Opin Struct Biol. 2004;14:89–95. doi: 10.1016/j.sbi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, et al. Total chemical synthesis of N-myristoylated HIV-1 matrix protein p17: Structural and mechanistic implications of p17 myristoylation. Proc Natl Acad Sci USA. 2004;101:11587–11592. doi: 10.1073/pnas.0404649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dell’orco D., Behnen P, Linse S, Koch KW. Calcium binding, structural stability and guanylate cyclase activation in GCAP1 variants associated with human cone dystrophy. Cell Mol Life Sci. 2010;67:973–984. doi: 10.1007/s00018-009-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy MT, Brockman H, Rusnak F. Contributions of myristoylation to calcineurin structure/function. J Biol Chem. 1996;271:26517–26521. doi: 10.1074/jbc.271.43.26517. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Hernandez E, Serrano L. Structure of the transition state for folding of the 129 aa protein CheY resembles that of a smaller protein, CI-2. Folding Des. 1996;1:43–55. [PubMed] [Google Scholar]
- 21.Riddle DS, et al. Experiment and theory highlight role of native state topology in SH3 folding. Nat Struct Biol. 1999;6:1016–1024. doi: 10.1038/14901. [DOI] [PubMed] [Google Scholar]
- 22.Villegas V, Martinez JC, Aviles FX, Serrano L. Structure of the transition state in the folding process of human procarboxypeptidase A2 activation domain. J Mol Biol. 1998;283:1027–1036. doi: 10.1006/jmbi.1998.2158. [DOI] [PubMed] [Google Scholar]
- 23.Zarrine-Afsar A, et al. Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding. Proc Natl Acad Sci USA. 2008;105:9999–10004. doi: 10.1073/pnas.0801874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northey JG, Maxwell KL, Davidson AR. Protein folding kinetics beyond the phi value: Using multiple amino acid substitutions to investigate the structure of the SH3 domain folding transition state. J Mol Biol. 2002;320:389–402. doi: 10.1016/S0022-2836(02)00445-X. [DOI] [PubMed] [Google Scholar]
- 25.Ventura S, et al. Conformational strain in the hydrophobic core and its implications for protein folding and design. Nat Struct Biol. 2002;9:485–493. doi: 10.1038/nsb799. [DOI] [PubMed] [Google Scholar]
- 26.Munson M, Anderson KS, Regan L. Speeding up protein folding: mutations that increase the rate at which Rop folds and unfolds by over four orders of magnitude. Folding Des. 1997;2:77–87. doi: 10.1016/S1359-0278(97)00008-4. [DOI] [PubMed] [Google Scholar]
- 27.Cho JH, Raleigh DP. Denatured state effects and the origin of nonclassical phi values in protein folding. J Am Chem Soc. 2006;128:16492–16493. doi: 10.1021/ja0669878. [DOI] [PubMed] [Google Scholar]
- 28.Viguera AR, Vega C, Serrano L. Unspecific hydrophobic stabilization of folding transition states. Proc Natl Acad Sci USA. 2002;99:5349–5354. doi: 10.1073/pnas.072387799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosavi S, Chavez LL, Jennings PA, Onuchic JN. Topological frustration and the folding of interleukin-1 beta. J Mol Biol. 2006;357:986–996. doi: 10.1016/j.jmb.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Gaspar JA, Wong HJ, Meiering EM. Conserved and nonconserved features of the folding pathway of hisactophilin, a beta-trefoil protein. Protein Sci. 2002;11:669–679. doi: 10.1110/ps.31702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wensley BG, et al. Experimental evidence for a frustrated energy landscape in a three-helix-bundle protein family. Nature. 2010;463:685–688. doi: 10.1038/nature08743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton VL, Friel CT, Allen LR, Paci E, Radford SE. The effect of increasing the stability of non-native interactions on the folding landscape of the bacterial immunity protein Im9. J Mol Biol. 2007;371:554–568. doi: 10.1016/j.jmb.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Chavez LL, Gosavi S, Jennings PA, Onuchic JN. Multiple routes lead to the native state in the energy landscape of the beta-trefoil family. Proc Natl Acad Sci USA. 2006;103:10254–10258. doi: 10.1073/pnas.0510110103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capraro DT, Roy M, Onuchic JN, Jennings PA. Backtracking on the folding landscape of the beta-trefoil protein interleukin-1beta? Proc Natl Acad Sci USA. 2008;105:14844–14848. doi: 10.1073/pnas.0807812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gambin Y, et al. Direct single-molecule observation of a protein living in two opposed native structures. Proc Natl Acad Sci USA. 2009;106:10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sola M, et al. Towards understanding a molecular switch mechanism: Thermodynamic and crystallographic studies of the signal transduction protein CheY. J Mol Biol. 2000;303:213–225. doi: 10.1006/jmbi.2000.4507. [DOI] [PubMed] [Google Scholar]
- 37.Vallee-Belisle A, Ricci F, Plaxco KW. Thermodynamic basis for the optimization of binding-induced biomolecular switches and structure-switching biosensors. Proc Natl Acad Sci USA. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ababou A, Shenvi RA, Desjarlais JR. Long-range effects on calcium binding and conformational change in the N-domain of calmodulin. Biochemistry. 2001;40:12719–12726. doi: 10.1021/bi010405b. [DOI] [PubMed] [Google Scholar]
- 39.Marvin JS, Hellinga HW. Manipulation of ligand binding affinity by exploitation of conformational coupling. Nat Struct Biol. 2001;8:795–798. doi: 10.1038/nsb0901-795. [DOI] [PubMed] [Google Scholar]
- 40.Hanakam F, Albrecht R, Eckerskorn C, Matzner M, Gerisch G. Myristoylated and non-myristoylated forms of the pH sensor protein hisactophilin II: Intracellular shuttling to plasma membrane and nucleus monitored in real time by a fusion with green fluorescent protein. EMBO J. 1996;15:2935–2943. [PMC free article] [PubMed] [Google Scholar]
- 41.Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci USA. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuccitelli R, Deamer DW. Intracellular pH: Its Measurement, Regulation, and Utilization in Cellular Functions. New York: Liss; 1982. [Google Scholar]
- 43.Stratton MM, Mitrea DM, Loh SN. A Ca2+-sensing molecular switch based on alternate frame protein folding. ACS Chem Biol. 2008;3:723–732. doi: 10.1021/cb800177f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto T, Chen HC, Guigard E, Kay CM, Ryan RO. Molecular studies of pH-dependent ligand interactions with the low-density lipoprotein receptor. Biochemistry. 2008;47:11647–11652. doi: 10.1021/bi801117t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang HW. Ontario, Canada: Univ of Waterloo; 2002. The Role of Temperature, Osmolytes and Myristoylation on the Stability and Folding of Hisactophilin. MSc thesis. [Google Scholar]
- 46.Duronio RJ, et al. Protein N-myristoylation in Escherichia coli: Reconstitution of a eukaryotic protein modification in bacteria. Proc Natl Acad Sci USA. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.