Abstract
Invertebrates lack adaptive immune systems homologous to those of vertebrates, yet it is becoming increasingly clear that they can produce diversified antigen recognition molecules. We have previously noted that the snail Biomphalaria glabrata produces a secreted lectin, fibrinogen-related protein 3 (FREP3), unusual among invertebrate defense molecules because it is somatically diversified by gene conversion and point mutation. Here we implicate FREP3 in playing a central role in resistance to a major group of snail pathogens, digenetic trematodes. FREP3 is up-regulated in three models of resistance of B. glabrata to infection with Schistosoma mansoni or Echinostoma paraensei, and functions as an opsonin favoring phagocytosis by hemocytes. Knock-down of FREP3 in resistant snails using siRNA-mediated interference resulted in increased susceptibility to E. paraensei, providing a direct link between a gastropod immune molecule and resistance to trematodes. FREP3 up-regulation is also associated with heightened responsiveness following priming with attenuated digenetic trematodes (acquired resistance) in this model invertebrate immune system.
Keywords: host–parasite interactions, evolution, immunology, parasitology, schistosomiasis
Recent years have witnessed a surge of new information about the immune capabilities of organisms as diverse as plants (1), worms (2), snails (3), flies (4), sea urchins (5), and lampreys (6). One of the insights emerging from these studies is that invertebrates, although they do not have adaptive immune systems in the sense of those possessed by jawed vertebrates, nonetheless do have sophisticated innate immune systems that have revealed an unforeseen ability to produce diversified antigen recognition molecules (3, 4). Possession of expanded families of antigen recognition molecules (5), the ability to engage in extensive diversification of receptors by alternative splicing (4), somatic diversification achieved by gene conversion and point mutation (3), and the presence of complex genomic architecture conducive to the production of diversified antigen receptors (7) have all been shown in invertebrates, potentially blurring some of the traditional distinctions made between innate and adaptive immune systems in the process (8). It is not surprising that invertebrates have immune systems that are not limited to low numbers of pattern recognition receptors, given the diverse suites of pathogens with which they must cope, and the evident success of invertebrates in persisting abundantly over evolutionary time, in both species and numbers. Many invertebrates have life spans longer than those of many of their vertebrate counterparts, further accentuating their need for sophisticated protection systems. Relatively lacking, however, has been further information documenting that diversified invertebrate defense molecules are actually playing a protective role against natural and relevant pathogens.
There has also been a resurgence of attention in the ability of invertebrates to mount specific secondary responses different from and more protective than those mounted following initial exposure to the same pathogens (9, 10). This phenomenon has been observed in two forms that differ in the specificity of the response to secondary challenge. The first state is one in which the immune response is primed by initial exposure, thereby resulting in a generalized heightened immunological state that renders secondary challenges with several pathogens less successful. The second state is one of specificity, in which the initial priming response and successful clearance leave the organism resistant to homologous secondary pathogen challenge. This specific secondary response has been demonstrated in the water flea Daphnia magna (11), the planorbid snail Biomphalaria glabrata (12), and the bumblebee Bombus terrestris (13). The heightened response engendered by exposure of B. glabrata to attenuated digeneans was termed “acquired resistance” by its discoverers (12). In addition to these examples of specific acquired resistance to reinfection, a wealth of observational evidence supporting increases in specific gene expression patterns, survival, and defense-related protein production is available for other invertebrates (10), suggesting that the capacity to acquire resistance is likely to be present in many invertebrate phyla. Unfortunately, to date very few data are available to suggest the mechanisms that underlie these phenomena (14).
Below we highlight recent studies building on our previous work showing that the freshwater planorbid snail, Biomphalaria glabrata, is capable of producing a unique category of circulating defense molecules called fibrinogen-related proteins (FREPs). FREPs are calcium-dependent lectins with one or two N-terminal Ig superfamily (IgSF) domains and a C-terminal fibrinogen domain. They are known to be up-regulated following infection and to bind to parasite surfaces (15). As one of their most distinctive features, FREPs are diversified somatically by both gene conversion and point mutation (3). Just as mosquitoes have developed into important research models for studying invertebrate immunity because of their role in transmitting malaria, gastropod molluscs such as B. glabrata are of interest because of their role in the transmission of digenetic trematodes that cause schistosomiasis. Many of the more than 18,000 species of digenetic trematodes (digeneans) found worldwide have a significant impact on animal and human health (16). Among them, schistosomes afflict more than 200 million persons, mostly in sub-Saharan Africa (17). Almost all digenean species depend on molluscs, usually snails, for their larval development, a relationship characterized by a high degree of specificity (18). Digenean infections of snails are characterized by an intimate, often life-long association that involves parasite proliferation and castration of the snail host (19). Compatibility between snail and digenean is determined, in part, by immunological interactions believed to be particularly pronounced during the early stages of infection (20): The parasite attempts to suppress the snail immune response (21) or to disguise itself from it (22), thereby allowing the parasite to establish. For its part, the snail host mounts humoral and cellular responses to encapsulate and kill the parasite (20, 23, 24). In some snail–digenean combinations, the snail is resistant to infection and the immune components responsible have been a frequent topic of study (23), in part because of the ramifications for interrupting transmission of schistosomes to humans. Below we provide evidence to implicate FREP3 in resistance of snails to trematode infection, confirm that it is diversified among hemocytes, and that specific knockdown of FREP3 using siRNAs can increase the susceptibility of snails to digenean infection.
Results
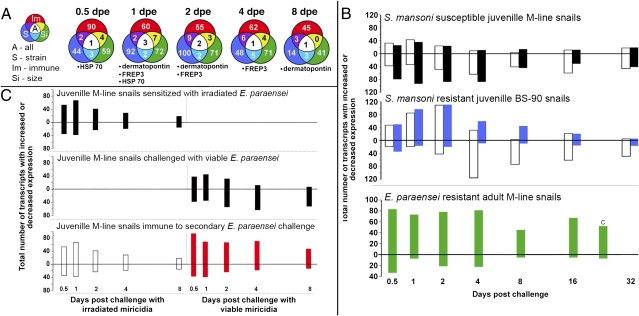
Using a microarray emphasizing known immunological transcripts expressed by the snail B. glabrata (20), we identified FREP3 as one of a small group of transcripts consistently and significantly up-regulated following challenge of resistant snails with digeneans (Fig. 1A). Snails resistant to infection because of their strain, increased age and hence larger size (25), or sensitization by a previous exposure to irradiation-attenuated, homologous trematodes (the latter referred to as “acquired resistance”), all displayed a significant increase in FREP3 transcription as assessed using the microarray, and confirmed using quantitative RT-PCR (Fig. S1; Tables S1–S3). Conversely, snails successfully infected with digeneans exhibit down-regulation of FREP3 expression (21) (Fig. 1 B and C).
Fig. 1.
Microarray studies of responses of B. glabrata to trematode infection, including three different models of resistance. (A) Venn diagrams showing up-regulated transcripts at different days postexposure (dpe) that are shared among the three forms of resistance to trematode infection. Outer colors of the Venn circles correspond with bar colors shown in A and B. Listed below each time point are the transcripts common to all three resistance models (size, strain, and acquired immune resistance. (B) Enumeration of up-regulated (above zero line) and down-regulated array responses (below zero line) over a time course from 0.5 to 32 d postexposure for juvenile M-line strain snails susceptible to both S. mansoni (open bars) and E. paraensei (black bars); BS-90 strain snails susceptible to E. paraensei (open bars) but resistant to S. mansoni (blue bars); and adult M-line snails resistant to E. paraensei (green bars). In this latter experiment, the transcripts represented on the graph are ones that are up-regulated as a result of size resistance, with those up-regulated during challenge of susceptible small M-line snails subtracted. (C) Summary of transcriptional profiles associated with induction of acquired resistance to E. paraensei in M-line snails: sensitized control snails exposed only to irradiated miracidia, which fail to develop; challenge control snails exposed only to normal (viable) miracidia, which establish successful infections; and experimental snails sensitized with irradiated miracidia and challenged 8 d later with viable miracidia that are killed because of the development of acquired resistance (12). That acquired resistance occurred was verified by exposure of snails to determine if they became infected (shed cercariae): 2 of 59 (3%) snails exposed only to irradiated miracidia, 78 of 84 (93%) exposed only to viable miracidia, and 13 of 94 (14%) exposed first to irradiated miracidia and later challenged with viable miracidia became infected.
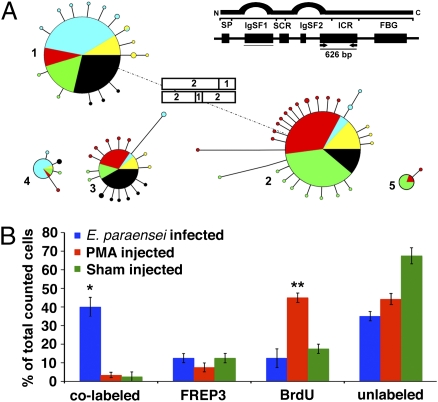
Characterization of bacterial artificial chromosome (BAC) sequences revealed a cluster arrangement of both multiple B. glabrata FREP3 genes and partial sequences. This spatial configuration and the high level of sequence similarity among these sequences are conducive for, and consistent with, gene conversion (26) (Fig. S2). The association that we noted between FREP3 and resistance to trematode infection, as well as its remarkable capacity for diversification, provided strong justification for further investigation of FREP3. To examine FREP3 diversification, we chose to focus on exon 5 because it is convenient to amplify, encompasses parts of both the second IgSF domain and the interceding region of FREP3, and represents a part of the FREP3 molecule that has not previously been surveyed. We examined four subsets of 20–40 hemocytes taken from each of five individual snails (Fig. 2A). As expected (3), this analysis yielded a limited number of frequently recovered FREP3 “source sequences” in common to all hemocyte subsets from a given snail (five for each snail investigated). Source sequences are the unaltered FREP3 alleles present in the snail's germline. Previous studies based on Southern blot analysis have shown there to be no more than 2–5 FREP3 loci present in B. glabrata (27) a number possibly inflated by cross-reactivity of our FREP3 cDNA probe with IgSF-encoding portions of fragmented or other incomplete FREP genes. Subsequent analysis of a FREP3-containing BAC clone revealed no more than two to three intact FREP3 loci. Assuming some heterozygosity at these loci, this smaller number of loci is consistent with the presence of five source sequences per snail that we noted in the present study. These results again emphasize the point that the number of source sequences is very limited relative to the diversity of FREP3 sequences recovered from each snail in our analyses (3) (Fig. S3). In addition, unique FREP3 genomic sequences were found in each hemocyte subset (between four and 11 variants/hemocyte subset per snail), reflecting involvement of both point mutation and gene conversion in generation of greater FREP3 sequence diversity than that encompassed by the source sequences (Fig. S3), and consistent with the occurrence of random diversification events unique within hemocytes comprising each subset.
Fig. 2.
(A) Presence of variant FREP3 genomic sequences in the indicated 626-bp portion of the molecule was ascertained among multiple subsets of 20–40 hemocytes taken from each of five snails (identified by different colors). The five large circles (diameter indicating frequency of recovery) represent the presumptive FREP3 source sequences. Note that the five snails had mostly the same source sequences. The attached small satellite circles (diameter representing recovery rate from 1–3) represent relatively rare FREP3 variants recovered from particular hemocyte subsets that were derived from a source sequence by point mutation (length of connecting lines represents number of differing nucleotides, range 1–6). One variant sequence of source sequence 1 was recovered from two individual snails (satellite circle with two colors). Rectangles indicate additional novel sequences generated by gene conversion, with the contributing source sequences indicated by numbers. The observation of two occurrences does not imply that gene conversion involving FREP3 sequences is rare, as no more than 800 hemocytes were sampled. Our data are consistent with the contribution of gene conversion (additional to point mutations) to extensive somatic diversification of FREP genes as recorded from whole body tissues of B. glabrata (3). (B) Circulating hemocytes taken from snails with 8-d E. paraensei infections were more likely to exhibit both BrdU incorporation (indicative of recent origin) and expression of FREP3 transcripts than hemocytes from snails either injected with PMA or sham injected. *Significant differences from sham-injected controls (P < 0.05, one-way ANOVA). Bars indicate SE.
A common result of trematode infection in snails is that hematopoiesis is stimulated (25, 28). This observation led us to hypothesize that the increased FREP3 expression associated with resistance is linked to the production of new hemocytes. Using in situ hybridization (Fig. S4), we noted expression of FREP3 transcripts in newly produced hemocytes (as assessed by BrdU incorporation) was greater than in the original hemocyte population, or in PMA-stimulated proliferation controls (Fig. 2B and Fig. S4), supporting our hypothesis that FREP3 is somatically diversified in hemocytes.
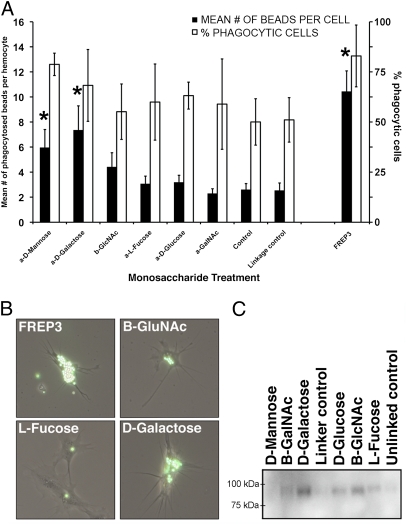
FREP3 binding targets were identified using monosaccharides that have been shown to be associated with surface glycoproteins of larval digeneans (29). We reasoned that FREP3 would have lectin binding properties because FREP4 had previously been shown to precipitate digenean secretory/excretory products in a monosaccharide-inhibitable manner (15). When conjugated to fluorescent microspheres and injected into snails, α-d-mannose and α-d-galactose induced higher rates and proportions of phagocytosis in the resident population of hemocytes than the other monosaccharides tested (Fig. 3 A and B). To confirm that FREP3 contributed to the increase in phagocytosis observed using α-d-galactose, it was purified from snail hemolymph and conjugated to microspheres that were then injected into snails. FREP3-coated beads were phagocytosed at significantly higher rates and proportions than control beads, or beads conjugated to any of the monosaccharides (Fig. 3 A and B). Finally, by incubating monosaccharide-conjugated microspheres with cell-free snail plasma we demonstrated that FREP3 bound a range of monosaccharides including d-glucose, GlcNAc and most strongly, α-d-galactose (Fig. 3C). Because α-d-mannose was not bound by FREP3 in this study, it is likely the phagocytosis induced by α-d-mannose was because it was bound by mannose binding lectin, which has been identified in other invertebrates (30) and has been shown to be involved in the recognition and phagocytosis of pathogens (31, 32). A C-type lectin-like sequence from B. glabrata with significant similarity to macrophage mannose receptor (20), array feature BGC04266), is frequently up-regulated following exposure to digenean infection (Tables S1–S3). These assays indicate FREP3 has lectin activity and opsonic properties for hemocytes which are known to be involved in attacking and phagocytosing portions of the tegument of digenean sporocysts as part of the encapsulation response in resistant snails (33). A number of other phagocytosis-related transcripts were also shown to be up-regulated in resistant snails by our transcriptional analysis, underscoring the importance of this process in resistance to digenean infection (Table S1). Of particular interest is feature BGC0426, which, because of sequence similarities to dermatopontin (an extracellular matrix protein) and hemagglutinin/amebocyte aggregation factor (20), may attract hemocytes and regulate cellular adherence (Fig. 1A).
Fig. 3.
FREP3 is involved in detection of monosaccharides and is able to enhance phagocytosis. (A) Streptavidin beads conjugated to different monosaccharides or to purified FREP3 were injected into snails; 2 h later, hemocytes were removed and assayed for presence of beads. Graph shows mean number of beads phagocytosed per hemocyte for 100 hemocytes counted from each snail (n = 10) (filled bars, left axis). Also shown is the percentage of cells counted that had phagocytosed beads (open bars, right axis). *Significant difference from corresponding control (P < 0.05, one-way ANOVA). Bars indicate SE. (B) Hemocytes observed during the phagocytosis experiment with varying numbers of beads within. Beads were conjugated with the substance indicated on each figure. (C) Monosaccharide-conjugated beads were incubated with cell-free snail plasma. Polypeptides were then solubilized from the beads, run on an SDS/PAGE gel, and transferred to nitrocellulose. The Western blot was probed with anti-FREP3 antibody to show which sugars were bound by FREP3.
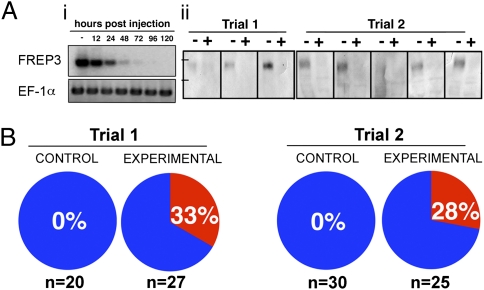
To further test the role played by FREP3 in antidigenean responses in B. glabrata, we developed an alternative method to effect RNA interference mediated knock-down of FREP3 expression using a combination of siRNAs rather than a single large dsRNA construct as used previously for RNAi in this snail (34). Significant knock-down of FREP3 was confirmed at both transcript and protein level in experimental snails whereas control (pre–knock-down), or siGFP-specific knock-down snails showed no alteration in FREP3 expression (Fig. 4A). The effect of FREP3 knock-down on susceptibility to E. paraensei was assessed in adult M-line B. glabrata (10- to 15-mm shell diameter), which are typically resistant to E. paraensei infection (25) (Fig. 4B). Knock-down of FREP3 in these large snails resulted in an alteration of the resistance phenotype such that 16 of 52 (31%) of these snails became infected with E. paraensei that had developed to the redial stage, as compared with 0 of 50 control snails that received GFP-targeted siRNA oligos. The phenotype of susceptibility to digenean infection has thus been uniquely changed in a snail as a consequence of experimental alteration of the expression of a specific immune gene product. Furthermore, although these data suggest that FREP3 is not the only defense factor involved in digenean resistance, it does imply that it is one of importance.
Fig. 4.
RNAi-mediated knock-down of FREP3. (A) Confirmation of transcriptional knock-down (i) using representative results of RT-PCR assays on samples taken from unexposed control snails between 0 and 120 h postinjection. FREP3 knockdown specificity was confirmed using an elongation factor 1-α (EF-1α) endogenous control. Protein level knock-down (ii) shown for two trials, each a Western blot analysis loading 100 μg plasma protein from eight individual snails either before (−) or 4 d after (+) injection of FREP3 specific siRNA. Tick marks represent standard size markers of 75 kDa (Lower) and 100 kDa (Upper). (B) Results of two trials comparing the effects of injection of either GFP oligos (control) or four 27mer FREP3 oligos (experimental). The percentage of snails in each group found to contain rediae at 12–14 dpe is shown and indicated by the red pie segments. The number of snails injected for each group is shown at the bottom of the figure.
Discussion
The focus of this study was to identify transcripts that are important to defense of B. glabrata to trematode infection and to then functionally characterize these transcripts in relation to their role in snail defense. Microarray analysis of the transcriptional profiles associated with size, strain, and acquired resistance models identified FREP3 as a transcript that is commonly up-regulated in resistant snails. Conversely, previous studies have demonstrated that FREP3 is a transcript that is targeted by the parasites for suppression during successful infections (21).
FREP3 is a member of a diverse group of B. glabrata hemolymph lectins that possess one or two IgSF domains coupled to a fibrinogen domain (3). Immune-related molecules containing fibrinogen domains have been identified in a steadily increasing number of invertebrate phyla (15, 35–39), however gastropod FREPs are structurally unique and, thus far, have not been found outside the Phylum Mollusca. FREPs change in abundance in snail hemolymph following digenean infection (15, 35, 36) and can precipitate secretory/excretory products of sporocysts implying they play a role in response to infections (15). FREPs have recently been shown to form complexes with highly polymorphic mucin molecules produced by S. mansoni sporocysts, complexes that also include thioester-containing proteins believed to favor phagocytosis and encapsulation responses (40). Previous analysis of genomic sequence variants of the first IgSF domain of FREP3 (exon 2 underlined in Fig. 2A) revealed that a limited number of germline alleles are somatically diversified by both gene conversion and point mutation, and that correspondingly diverse populations of FREP3 mRNAs are produced (3). High levels of variability in another subfamily of FREPs (FREP2) have also recently been independently reported (40).
Our ability to link FREP3 expression patterns within B. glabrata to newly generated hemocytes responding to E. paraensei infection, coupled with our confirmation of diversification of the FREP3 molecule implies that B. glabrata hemocytes are not all of the same and, in aggregate, produce a considerable diversity of FREP3 sequences (between 4 and 11 variants/hemocyte subsets per snail). In addition, confirmation of sequence diversification in the second Ig domain provides a more complete picture of the potential variability of the final FREP3 protein. We now know that both IgSF domains and the ICR are diversified by point mutation and gene conversion events providing the potential for differing FREP3 molecules to recognize different pathogen-associated targets.
Our RNAi data imply an important role for FREP3 in the B. glabrata response to the digenean E. paraensei, and suggest this will be a useful approach for testing several other promising factors also shown by our microarray studies to be involved in snail defense against digeneans. As FREP3 transcripts are also up-regulated in the BS-90 strain B. glabrata, which are resistant to S. mansoni, our results suggest it is also an important molecule in defense against this important human-infecting digenean as well. Identification of specific factors associated with resistance to schistosome infection opens the door for further study of snail susceptibility to schistosomes in areas endemic for S. mansoni and how this attribute might be manipulated to assist with schistosomiasis control.
Finally, we note that in the model of acquired resistance discussed above, snails previously sensitized by exposure to attenuated digeneans respond to challenge with homologous viable parasites with a strongly up-regulated FREP3 response. This makes FREP3 one of a small group (41–43) of invertebrate immune-associated molecules that has the potential to increase antigen recognition capability during a distinctive period of heightened immune responsiveness that, in this case, confers meaningful protection against parasite infection. Similar activation of defenses might occur in field situations when snails are exposed to miracidia either of digenean species that they normally do not host or of normally compatible digeneans that have been attenuated naturally (as, for example, by age), resulting in an increase in the ability of the snail to reject medically relevant digeneans such as S. mansoni. Our studies also point out a role for somatic diversification in invertebrate immune defense and help to explain how heightened levels of responsiveness could be achieved in organisms lacking an adaptive immune system homologous to that found in the vertebrates (44).
Materials and Methods
Microarray Scanning and Analysis.
Microarrays used to assess transcriptional differences between controls and snails with size, strain, and acquired resistance were probed in triplicate, with each probe derived from a separate pool of cDNA from five snails. Analysis was performed using Genepix and Acuity software (Molecular Devices). Statistical analysis was assessed using SAM software (45), and a 1.5-fold change in expression was imposed on all data.
Assessment of FREP3 Sequence Diversity.
Snail hemolymph was isolated and diluted using sterile snail saline to a concentration of 20–40 hemocytes per 10 μL. Cells were lysed for 10 min at 94 °C and then used as a template for PCR amplification of FREP3. The resulting amplicons were cloned and sequenced, and then analyzed to identify sequence differences.
Generation of an Anti-FREP3 Antibody and Detection of Native FREP3 in Snail Plasma.
The polyclonal antibody to FREP3 was generated against a recombinant fragment of the FREP3 protein representing the first IgSF domain and joining region between the two IgSF domains. This antibody was used to purify FREP3 from snail plasma as well as to detect FREP3 on Western blots.
BrdU Labeling and Detection of FREP3 Using in Situ Hybridization.
Approximately 5–15 μL (depending on snail body size) of the undiluted BrdU labeling reagent was injected into the hemocoel of the snail. Snails were bled 24 h after injection, and hemolymph was placed on microscope slides to allow hemocytes to adhere. BrdU incorporation into the DNA of the hemocytes was assessed following the FLUOS protocol (Roche). FREP3 expression in hemocytes was assessed using DIG-labeled RNA probes specific for FREP3. The presence of the DIG-labeled probe was detected using anti-DIG antibody.
Assessment of FREP3 Binding and Function Using Streptavidin-Conjugated Microspheres.
FREP3 and monosaccharides were biotinylated and conjugated to streptavidin-coated latex beads (Bang Laboratories). Beads were injected through the shell into the hemocoel of the snail directly beside the heart. Injected snails were bled by head–foot retraction 2 h after injection, and hemolymph was placed on microscope slides to allow cells to adhere before assaying for phagocytosis.
Knock-Down of FREP3 in E. paraensei-Resistant Snails.
siRNA 27mer oligos were synthesized by Integrated DNA Technologies (IDT) using different conserved locations along the FREP3 transcript. Knock-down was confirmed by RT-PCR and Western blot analysis. For RNAi experiments, snails were injected with FREP3 RNAi oligos and infected with E. paraensei on the same day. Analysis of altered resistance phenotypes was by dissection and searching for rediae, performed 12–14 d after exposure.
Detailed Materials and Methods.
A detailed description of the materials and methods used in this study is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. M. Madrid, C. M. Lun, J. Gauntt, and S. D. Stansbury, who provided technical assistance with sequencing of BAC DNA and FREP clones, and microarrays. Primary support for this project came from the National Institutes of Health (NIH) AI24340 (to E.S.L.), with secondary support from the NIH National Center for Research Resources P20RR18754 (to E.S.L.), NIH AI052363 (to C.M.A.), and NIH AI067686 (to S.-M.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The microarray data reported in this paper have been deposited in the GEO database (accession nos. GSE21878, GSE21880, GSE21881). BAC sequencing reads have been deposited in the NCBI trace archive (accession nos. gnl|ti|2278068446–2278071234).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011242107/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 4.Watson FL, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 5.Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley KM, Terwilliger DP, Smith LC. Sequence variations in 185/333 messages from the purple sea urchin suggest posttranscriptional modifications to increase immune diversity. J Immunol. 2008;181:8585–8594. doi: 10.4049/jimmunol.181.12.8585. [DOI] [PubMed] [Google Scholar]
- 8.Litman GW, Dishaw LJ, Cannon JP, Haire RN, Rast JP. Alternative mechanisms of immune receptor diversity. Curr Opin Immunol. 2007;19:526–534. doi: 10.1016/j.coi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Little TJ, Hultmark D, Read AF. Invertebrate immunity and the limits of mechanistic immunology. Nat Immunol. 2005;6:651–654. doi: 10.1038/ni1219. [DOI] [PubMed] [Google Scholar]
- 10.Kurtz J. Specific memory within innate immune systems. Trends Immunol. 2005;26:186–192. doi: 10.1016/j.it.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Carius HJ, Little TJ, Ebert D. Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution. 2001;55:1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 12.Lie KJ, Heyneman D. Acquired resistance to echinostomes in four Biomphalaria glabrata strains. Int J Parasitol. 1979;9:533–537. doi: 10.1016/0020-7519(79)90009-2. [DOI] [PubMed] [Google Scholar]
- 13.Schmid-Hempel P, Puhr K, Kruger N, Reber C, Schmid-Hempel R. Dynamic and genetic consequences of variation in horizontal transmission for a microparasitic infection. Evolution. 1999;53:426–434. doi: 10.1111/j.1558-5646.1999.tb03778.x. [DOI] [PubMed] [Google Scholar]
- 14.Hauton C, Smith VJ. Adaptive immunity in invertebrates: A straw house without a mechanistic foundation. Bioessays. 2007;29:1138–1146. doi: 10.1002/bies.20650. [DOI] [PubMed] [Google Scholar]
- 15.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooker S, et al. An updated atlas of human helminth infections: The example of East Africa. Int J Health Geogr. 2009;8:42. doi: 10.1186/1476-072X-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chitsulo L, Loverde P, Engels D. Schistosomiasis. Nat Rev Microbiol. 2004;2:12–13. doi: 10.1038/nrmicro801. [DOI] [PubMed] [Google Scholar]
- 18.Cribb TH, Bray RA, Olson PD, Littlewood DT. Life cycle evolution in the digenea: A new perspective from phylogeny. Adv Parasitol. 2003;54:197–254. doi: 10.1016/s0065-308x(03)54004-0. [DOI] [PubMed] [Google Scholar]
- 19.Lockyer AE, Jones CS, Noble LR, Rollinson D. Trematodes and snails: An intimate association. Can J Zool. 2004;82:251–269. [Google Scholar]
- 20.Adema CM, et al. Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes) Mol Immunol. 2010;47:849–860. doi: 10.1016/j.molimm.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanington PC, Lun CM, Adema CM, Loker ES. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int J Parasitol. 2010;40:819–831. doi: 10.1016/j.ijpara.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu XJ, et al. Proteomic analysis of Schistosoma mansoni proteins released during in vitro miracidium-to-sporocyst transformation. Mol Biochem Parasitol. 2009;164:32–44. doi: 10.1016/j.molbiopara.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayne CJ. Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: A 2009 assessment. Mol Biochem Parasitol. 2009;165:8–18. doi: 10.1016/j.molbiopara.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller AN, Raghavan N, FitzGerald PC, Lewis FA, Knight M. Differential gene expression in haemocytes of the snail Biomphalaria glabrata: Effects of Schistosoma mansoni infection. Int J Parasitol. 2001;31:687–696. doi: 10.1016/s0020-7519(01)00133-3. [DOI] [PubMed] [Google Scholar]
- 25.Loker ES, Cimino DF, Stryker GA, Hertel LA. The effect of size of M line Biomphalaria glabrata on the course of development of Echinostoma paraensei. J Parasitol. 1987;73:1090–1098. [PubMed] [Google Scholar]
- 26.Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. Gene conversion: Mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 27.Zhang SM, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–266. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mounkassa JB, Jourdane J. Dynamics of the leukocytic response of Biomphalaria glabrata during the larval development of Schistosoma mansoni and Echinostoma liei. J Invertebr Pathol. 1990;55:306–311. doi: 10.1016/0022-2011(90)90071-d. [DOI] [PubMed] [Google Scholar]
- 29.Peterson NA, Hokke CH, Deelder AM, Yoshino TP. Glycotope analysis in miracidia and primary sporocysts of Schistosoma mansoni: Differential expression during the miracidium-to-sporocyst transformation. Int J Parasitol. 2009;39:1331–1344. doi: 10.1016/j.ijpara.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kvennefors EC, Leggat W, Hoegh-Guldberg O, Degnan BM, Barnes AC. An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev Comp Immunol. 2008;32:1582–1592. doi: 10.1016/j.dci.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Gruden-Movsesijan A, Milosavljevic LjS. The involvement of the macrophage mannose receptor in the innate immune response to infection with parasite Trichinella spiralis. Vet Immunol Immunopathol. 2006;109:57–67. doi: 10.1016/j.vetimm.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 32.Kawabata S, Tsuda R. Molecular basis of non-self recognition by the horseshoe crab tachylectins. Biochim Biophys Acta. 2002;1572:414–421. doi: 10.1016/s0304-4165(02)00322-7. [DOI] [PubMed] [Google Scholar]
- 33.Ataev GL, Coustau C. Cellular response to Echinostoma caproni infection in Biomphalaria glabrata strains selected for susceptibility/resistance. Dev Comp Immunol. 1999;23:187–198. doi: 10.1016/s0145-305x(99)00023-3. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y, Loker ES, Zhang SM. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Comp Immunol. 2006;30:855–866. doi: 10.1016/j.dci.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev Comp Immunol. 2005;29:295–303. doi: 10.1016/j.dci.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SM, Zeng Y, Loker ES. Expression profiling and binding properties of fibrinogen-related proteins (FREPs), plasma proteins from the schistosome snail host Biomphalaria glabrata. Innate Immun. 2008;14:175–189. doi: 10.1177/1753425908093800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems—not homogeneous, not simple, not well understood. Immunol Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, et al. A fibrinogen-related protein from bay scallop Argopecten irradians involved in innate immunity as pattern recognition receptor. Fish Shellfish Immunol. 2009;26:56–64. doi: 10.1016/j.fsi.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Middha S, Wang X. Evolution and potential function of fibrinogen-like domains across twelve Drosophila species. BMC Genomics. 2008;9:260. doi: 10.1186/1471-2164-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moné Y, et al. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litman GW, Cannon JP, Rast JP. New insights into alternative mechanisms of immune receptor diversification. Adv Immunol. 2005;87:209–236. doi: 10.1016/S0065-2776(05)87006-3. [DOI] [PubMed] [Google Scholar]
- 43.Kurtz J, Armitage SA. Alternative adaptive immunity in invertebrates. Trends Immunol. 2006;27:493–496. doi: 10.1016/j.it.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Tusher V, Tibshirani R, Chu C. Significance analysis of microarrays applied to ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.