Abstract
Despite recent advances in our understanding of how Helicobacter pylori causes disease, the factors that allow this pathogen to persist in the stomach have not yet been fully characterized. To identify new virulence factors in H. pylori, we generated low-infectivity variants of a mouse-colonizing H. pylori strain using the classical technique of in vitro attenuation. The resulting variants and their highly infectious progenitor bacteria were then analyzed by global gene expression profiling. The gene expression levels of five open reading frames (ORFs) were significantly reduced in low-infectivity variants, with the most significant changes observed for ORFs HP1583 and HP1582. These ORFs were annotated as encoding homologs of the Escherichia coli vitamin B6 biosynthesis enzymes PdxA and PdxJ. Functional complementation studies with E. coli confirmed H. pylori PdxA and PdxJ to be bona fide homologs of vitamin B6 biosynthesis enzymes. Importantly, H. pylori PdxA was required for optimal growth in vitro and was shown to be essential for chronic colonization in mice. In addition to having a well-known metabolic role, vitamin B6 is necessary for the synthesis of glycosylated flagella and for flagellum-based motility in H. pylori. Thus, for the first time, we identify vitamin B6 biosynthesis enzymes as novel virulence factors in bacteria. Interestingly, pdxA and pdxJ orthologs are present in a number of human pathogens, but not in mammalian cells. We therefore propose that PdxA/J enzymes may represent ideal candidates for therapeutic targets against bacterial pathogens.
IMPORTANCE
Approximately half of the world’s population is infected with H. pylori, yet how H. pylori bacteria establish chronic infections in human hosts remains elusive. From gene array studies, we identified two genes as representing potentially novel colonization factors for H. pylori. These genes encoded enzymes involved in the synthesis of vitamin B6, an important molecule for many metabolic reactions in living organisms. Little is currently known regarding vitamin B6 biosynthesis in human pathogens. We showed that mutant H. pylori bacteria lacking an enzyme involved in de novo vitamin B6 biosynthesis, PdxA, were unable to synthesize motility appendages (flagella) and were unable to establish chronic colonization in mice. Thus, this work identifies vitamin B6 biosynthesis enzymes as novel virulence factors for bacterial pathogens. Interestingly, a number of human pathogens, but not their mammalian hosts, possess these genes, which suggests that Pdx enzymes may represent ideal candidates for new therapeutic targets.
INTRODUCTION
There are currently only a limited number of antibiotics suitable for the routine treatment of Helicobacter pylori infection; moreover, the efficacies of these compounds have been severely hindered by the rise in resistance among H. pylori isolates (1, 2). It is therefore necessary to develop novel therapeutic and prophylactic strategies for the management of H. pylori-related disease. In an effort to develop such strategies, we need to greatly enhance our understanding of the molecular basis of H. pylori adaptation to the specialized niche of the gastric mucosa (3). In silico analyses have been fruitful in this regard; however, they are not always accurate (4). Thus, several groups have combined genome information and functional data derived from the use of molecular techniques to shed new insights into different facets of H. pylori virulence (5–7). Although these techniques are very powerful, they rely, among other things, on the availability of H. pylori strains that can be readily transformed in vitro. These strains are usually laboratory-adapted isolates that are not able to colonize small rodents. To overcome this problem, workers have used suckling mice, which support the growth of highly transformable laboratory-adapted H. pylori strains that do not colonize adult mice (5). Suckling mice, however, have underdeveloped immune systems and do not possess the normal flora of adult animals, which may bias results toward the identification of factors that are important in the establishment of infection over those required for chronic colonization (5).
Historically, in vitro attenuation has been used as a strategy to study how microorganisms cause disease (8, 9). This strategy has the advantage of being applicable to any microorganism that can be propagated in vitro. Moreover, when combined with modern molecular techniques, such as whole-genome sequencing (10), it has been possible to identify novel virulence factors in otherwise well-characterized bacterial pathogens. The current study describes the use of a dual strategy of in vitro attenuation and gene expression profiling for the identification of novel virulence factors in H. pylori. Using this approach, we identified PdxA, an enzyme involved in de novo vitamin B6 biosynthesis, as being an important factor required for the chronic colonization of mice by H. pylori. Although it is widely accepted that vitamin B6 is a cofactor in a wide array of cellular metabolic reactions, this study is the first to describe a link between this metabolite and bacterial pathogenesis. Specifically, we demonstrated that vitamin B6 is important for flagellar structure, glycosylation, and motility in a human pathogen. Given that mammalian hosts are unable to perform de novo vitamin B6 biosynthesis, we propose that this work opens new avenues for the development of innovative therapeutic targets against bacterial pathogens.
RESULTS
Generation of low-infectivity isolates of H. pylori SS1 by prolonged in vitro passage.
To identify novel colonization factors of H. pylori, we applied the technique of in vitro attenuation to the well-characterized mouse-colonizing SS1 strain (11). We observed a progressive loss in the ability of H. pylori SS1 variants to infect mice, with significant reductions apparent both in the proportion of mice infected and in gastric bacterial levels after 109 passages in vitro (Table 1) (P = 0.0031 and P ≤ 0.0001, respectively). By 179 subcultures, H. pylori SS1 isolates were no longer able to colonize mice. Only minor differences in the DNA profiles were detected between low- and high-infectivity isolates (see Fig. S1A and B in the supplemental material). Furthermore, no differences were observed between isolates with regard to either their abilities to perform urease-mediated protection against acid, essential for H. pylori colonization of the stomach (12), or their in vitro growth properties (Fig. S1C and D).
TABLE 1 .
Mouse colonization experiments with low- and high-infectivity H. pylori SS1 isolates
| In vitro passage no. of isolatea | Inoculum/mouse (log10 CFU) | No. of mice infected (total)b | Mean ± SD log10 CFU recovered/gc |
|---|---|---|---|
| 58 | 6.3 | 9 (10)d | —e |
| 8 | 5.0 | 9 (9) | — |
| 74 | 6.7 | 5 (5)d | — |
| 9 | 6.7 | 5 (5) | — |
| 109 | 4.6 | 3 (10)f | 3.74 ± 0.50g |
| 15 | 4.6 | 10 (10) | 6.08 ± 0.52 |
| 179 | 5.5 | 0 (10)g | <2.0h |
| 24 | 5.5 | 9 (9) | 6.46 ± 0.8 |
For each experiment, mice were separated into 2 groups, and each group was inoculated with either a low- or high-infectivity H. pylori SS1 isolate.
H. pylori infection was detected by culture and urease activity at 4 or 6 weeks postchallenge.
Bacterial load determinations were performed by quantitative culture. Mean values were calculated from infected mice only (assay sensitivity, 3.0 log CFU/g).
P > 0.99.
—, not determined.
P = 0.0031.
P ≤ 0.0001.
P ≤ 0.0003.
Downregulation of gene expression in low-infectivity H. pylori SS1.
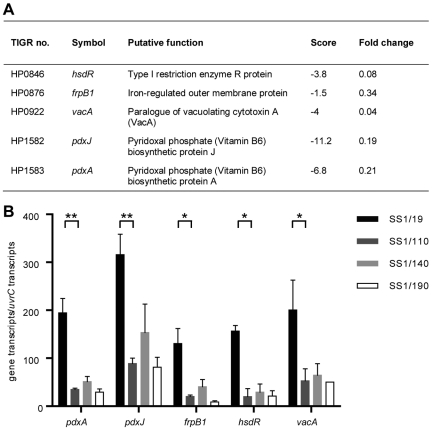
To determine molecular changes correlating with decreased colonization in high-passage-number isolates, we performed global gene expression profiling of high- and low-infectivity isolates. The expression levels of five coding sequences were found to be significantly downregulated in low-infectivity isolates compared to those of high-infectivity bacteria (Fig. 1A). These genes encode proteins involved in vitamin B6 biosynthesis (pdxJ, pdxA), iron metabolism (frpB1), restriction modification (hsdR), and autotransporter (vacA paralog) functions. The expression levels of these genes were confirmed by quantitative reverse transcription-PCR (qRT-PCR) to be significantly reduced in low-infectivity H. pylori SS1 isolates (Fig. 1B). The reduction in the expression levels of these genes correlated with a decrease in infectivity, first observed in H. pylori variants having undergone at least 109 in vitro subcultures, suggesting that the genes may encode factors that contribute to host colonization. The greatest variations in significance analysis of macroarray (SAM) scores were observed for H. pylori pdxJ and pdxA, annotated as encoding homologs of the vitamin B6 biosynthesis proteins PdxJ and PdxA in Escherichia coli. As the role of vitamin B6 has not been extensively studied in a bacterial pathogen, we chose to focus on the functions of the proteins encoded by these genes, including their roles in host colonization by H. pylori.
FIG 1 .
Downregulation of gene expression in low-infectivity H. pylori SS1 isolates. (A) Radioactively labeled cDNA samples from paired low- and high-infectivity H. pylori SS1 isolates were hybridized to macroarrays. TIGR numbers refer to the coding sequence numbers as identified by the J. Craig Venter Institute. Score values represent the relative levels of significance assigned by SAM analysis (34). Fold changes represent the fold changes between low- and high-infectivity isolates (four experiments). (B) qRT-PCR analyses of pdxA, pdxJ, frpB1, hsdR, and vacA paralog mRNA expression in liquid cultures of H. pylori SS1 isolates having undergone different numbers of passages. Gene expression was normalized to the housekeeping gene, uvrC (35). Error bars indicate standard errors of the means (SEM) (three experiments). *, P < 0.05; **, P < 0.01.
PdxJ and PdxA are involved in de novo vitamin B6 biosynthesis in H. pylori.
To determine whether H. pylori pdxJ and pdxA encode bona fide homologs of vitamin B6 biosynthesis enzymes, we constructed isogenic H. pylori pdxA mutants (13) in several laboratory-adapted (251, N6, G27) and mouse-colonizing (SS1 and X47-2AL) H. pylori strains. These mutants could be recovered only when the growth medium was supplemented with pyridoxal-5′-phosphate (PLP), the product of PdxJ/PdxA activities. In contrast, despite numerous attempts and approaches, we were unable to disrupt pdxJ, suggesting that this mutation may be lethal for H. pylori bacteria (data not shown). It is possible that PdxJ may have another vital function(s) besides the synthesis of vitamin B6, as has been reported for certain other H. pylori proteins (14). Alternatively, it is possible that disruption of pdxJ may have downstream effects on pdxA expression. In H. pylori, the pdxJ and pdxA genes are clustered together on the genome, with the stop codon of pdxJ preceding the start codon of pdxA by just 1 nucleotide (Fig. S2A and B). From qRT-PCR analyses, it appears that pdxA is transcribed as part of a longer pdxJ-pdxA cotranscript (Fig. S2C).
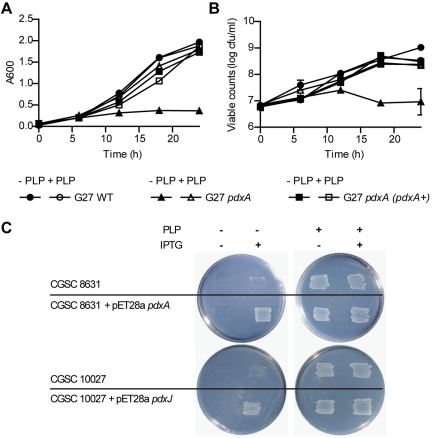
We next investigated the growth properties of the H. pylori pdxA mutants in vitro. In the absence of PLP supplementation, H. pylori pdxA mutants exhibited slower growth in liquid medium than the parent strain (Fig. 2A and B). This defect could, however, be entirely compensated for by the addition of PLP to the media or by complementation using insertion in cis of an intact copy of pdxA into the H. pylori genome (Fig. 2A and B). These data suggest a role for H. pylori PdxA in de novo vitamin B6 biosynthesis. To demonstrate definitively that pdxA and pdxJ encode homologs of vitamin B6 biosynthesis enzymes in H. pylori, we used a cross-complementation strategy with E. coli K-12 ΔpdxA and ΔpdxJ mutants (CGSC8631 and CGSC10027, respectively [15]). For this, we transformed the E. coli mutants with IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression plasmids harboring intact copies of either H. pylori pdxA or pdxJ. The untransformed E. coli mutants were unable to grow on minimal medium without PLP added (Fig. 2C). In contrast, E. coli pdxA and pdxJ mutant bacteria that had been transformed with either H. pylori pdxA- or pdxJ-expressing plasmids, respectively, were able to grow normally on minimal medium containing IPTG without PLP added. Thus, the ability of H. pylori pdxA and pdxJ to cross-complement a PLP deficiency in E. coli provides functional evidence that these genes encode vitamin B6 biosynthesis proteins in H. pylori.
FIG 2 .
In vitro growth of H. pylori pdxA mutants and functional characterization of PdxJ/A. (A and B) In vitro growth properties of H. pylori G27 wild type (WT), pdxA, and pdxA (pdxA+) strains were determined in liquid media supplemented or not supplemented with PLP. At each time point, A600 readings (A) and duplicate viable cell count determinations (B) were performed (three experiments). Data are presented as the log of the mean viable cell count (for each time point) ± SEM. (C) Growth of E. coli CGSC8631 (ΔpdxA) and CGSC10027 (ΔpdxJ) that had been transformed, or not, with pET28a pdxA and pET28a pdxJ, respectively, in the presence or absence of PLP and IPTG.
PdxA is essential for gastric colonization of C57BL/6 mice by H. pylori.
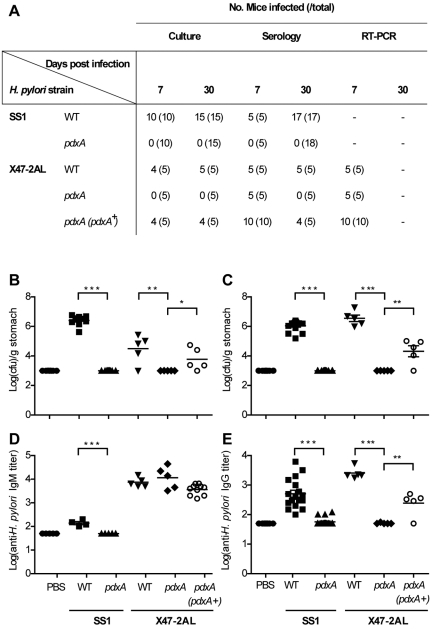
To determine whether vitamin B6 biosynthesis is important for bacterial colonization, we performed mouse infection studies with H. pylori pdxA mutants generated on both the SS1 and X47-2AL genetic backgrounds. In parallel, colonization experiments were performed with mutants in the other putative colonization genes (Fig. 1A). Colonization efficiency was assessed by at least two methods: quantitative culture, serology, and/or RT-PCR (Fig. 3A).
FIG 3 .
PdxA is required for colonization of mice by H. pylori. C57BL/6 mice (five per group) were inoculated with 107 CFU of H. pylori by intragastric gavage. (A) Colonization efficiency was assessed by culture, serology, and/or RT-PCR. −, not determined. (B and C) Quantitative culture assays were performed at 7 (B) and 30 (C) days postinfection. Each point represents the mean of two estimations for a single mouse. Data are pooled from three separate experiments for H. pylori SS1 and two separate experiments for H. pylori X47-2AL. Two independent H. pylori SS1 pdxA clones and two independent H. pylori X47-2AL pdxA (pdxA+) clones were used. Data are presented as log units (CFU/gram stomach). Horizontal bars represent the geometric means of viable cell count determinations for each H. pylori strain. The limit of detection for the assay is 3 log CFU/gram. (D and E) Specific IgM (D) and IgG (E) class antibodies in the sera of H. pylori-infected mice were detected by ELISA at 7 (D) and 30 (E) days postinfection. Data are presented as logs of titers, corresponding to the reciprocal of the serum dilution giving an A595 value that was 5-fold above that obtained for background controls. Horizontal bars represent the geometric means for each H. pylori strain. The limit of detection for the assay is a log titer of 1.7. *, P < 0.05; **, P < 0.01; and ***, P < 0.0001, as determined by the Mann-Whitney U test.
Significantly lower bacterial loads were observed in mice challenged with an H. pylori vacA paralog mutant (P < 0.01), whereas mutations in either frpB1 or hsdR had no significant effects on in vivo colonization by H. pylori (Fig. S3). Interestingly, the most dramatic effects on H. pylori colonization were observed for H. pylori pdxA mutants. Indeed, H. pylori bacteria were not cultured from the gastric biopsy specimens of mice inoculated with pdxA mutants in either H. pylori strain at both 7 and 30 days postinfection (Fig. 3B and C). Similarly, no IgG was detectable in the sera of mice infected with H. pylori pdxA mutant bacteria at 30 days postinfection in either of the strains tested (Fig. 3E). Interestingly, however, low levels of colonization, as indicated by IgM titers, were observed at day 7 postinfection in mice challenged with H. pylori pdxA mutants on the X47-2AL genetic background, whereas no evidence of colonization could be found for H. pylori SS1 pdxA mutants (Fig. 3A and D). In both cases, however, the H. pylori pdxA mutant bacteria were cleared by 30 days.
Complementation of H. pylori pdxA mutants with a wild-type copy of pdxA restored infectivity, as demonstrated by a significant increase in both the numbers of mice colonized and the serum IgG levels at day 30 postinfection (Fig. 3B, C, and E) (P < 0.05, P < 0.01, and P < 0.01, respectively). Although the restoration of infectivity for the complemented mutants was incomplete, this can most likely be attributed to the reduced expression levels of pdxJ observed in H. pylori pdxA mutant bacteria (Fig. S4A). Cumulatively, these data identify the essential role of PdxA in H. pylori colonization of the mouse gastric mucosa.
Vitamin B6 contributes to motility and flagellar glycosylation in H. pylori.
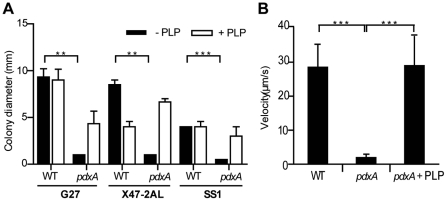
We next wished to investigate the reason(s) for the severe impairment in colonization efficiency of the H. pylori pdxA mutants. Vitamin B6, or its active form, PLP, is classically thought to be a cofactor for many metabolic enzymes and has recently been identified as a cofactor of H. pylori PseC, an aminotransferase involved in flagellar glycosylation (16). Glycosylation of flagella is necessary for the assembly of functional flagellar filaments, motility, and infectivity in H. pylori (17). Therefore, we hypothesized that a defect in flagellar structure or motility may account for the impaired virulence in vivo of the H. pylori pdxA mutants. To test this hypothesis, we examined H. pylori motility in soft agar plates and by live-cell imaging. For each of the H. pylori strains tested (G27, X47-2AL, and SS1), parental bacteria were motile, whereas H. pylori pdxA mutants were nonmotile (Fig. 4A) (P <0.01). Addition of exogenous PLP to the motility media, however, resulted in a diffuse growth pattern for H. pylori pdxA mutants, indicative of motile bacteria (Fig. 4A). Consistent with these findings, in the absence of PLP supplementation, parental H. pylori bacteria exhibited an average velocity of 27.92 ± 1.437 µm/s (see Movie S1 in the supplemental material), whereas H. pylori pdxA mutants moved at an average velocity of only 2.865 ± 0.5626 µm/s (Fig. 4B; Movie S2) (P < 0.0001). Supplementation of broth cultures with PLP restored the motility of H. pylori pdxA mutants to an average velocity of 28.92 ± 3.160 µm/s (Fig. 4B; Movie S3) (P < 0.0001).
FIG 4 .
PdxA is required for motility in H. pylori. (A) The bacterial motility of wild-type and pdxA mutant strains on different H. pylori genetic backgrounds was assayed using 0.4% (wt/vol) semisolid agar plates with (white bars) or without (black bars) the addition of PLP. Bars represent the mean determinations of triplicate measurements of colony diameter ± SEM (three experiments). **, P < 0.01, and ***, P < 0.0001, as determined by Student’s t test. (B) Bacterial motility in 6-h broth cultures was determined by visualization on a Leica AF6000 LX live-cell imager and analyzed using MetaMorph software. Data are presented as means ± SEM derived from over 400 positional data points for each strain (three experiments).
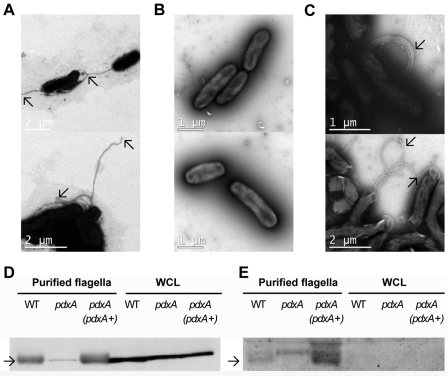
To determine whether the defect in motility of H. pylori pdxA mutants was a result of altered flagellar structure, we examined the flagella from H. pylori G27 parental and pdxA mutant bacteria. Parental H. pylori bacteria and complemented pdxA mutants displayed polar, sheathed flagella, whereas H. pylori pdxA mutants appeared to express no flagella (Fig. 5A to C), indicating a potential impairment in flagellar assembly. To elucidate further the role of vitamin B6 in the production of flagella, we performed Western blot analyses of whole-cell lysates and sheared flagellar preparations using a monoclonal antibody directed against the major H. pylori flagellin, FlaA. Whole-cell lysates prepared from H. pylori X47-2AL pdxA mutants contained amounts of FlaA protein equivalent to those in lysates prepared from parental and complemented H. pylori bacteria (Fig. 5D). Importantly, however, the sheared flagellar preparations from H. pylori pdxA bacteria contained much less FlaA than those prepared from parental bacteria (Fig. 5D). Consistent with previous work (18), exported H. pylori FlaA proteins in the flagellar preparations from parental and complemented H. pylori bacteria were glycosylated, whereas FlaA proteins in the cytoplasm were nonglycosylated (Fig. 5E). Significantly, only the exported FlaA in the flagellar preparations from parental H. pylori bacteria, but not those from the pdxA mutant organisms, were glycosylated (Fig. 5E). The data suggest that the inability of H. pylori pdxA mutants to glycosylate and export functional flagellar filaments was not due to a defect in cellular FlaA production but rather to the absence of a posttranslational modification that requires the PLP-dependent enzyme PseC. Taken together, the data indicate that vitamin B6 biosynthesis is important for flagellar biosynthesis and motility, which are vital for H. pylori colonization of the gastric mucosa.
FIG 5 .
PdxA is required for functional flagellar assembly and flagellar glycosylation in H. pylori. (A to C) TEM micrographs of negatively stained H. pylori G27 wild-type (A), G27 pdxA (B), and G27 pdxA (pdxA+) strains (C). Arrows indicate the ends of the flagella in each strain. (D) Western blot of whole-cell lysates (WCL) and sheared flagellar preparations, detected with monoclonal anti-H. pylori 54-kDa flagellin antibodies. (E) Detection of glycosylated proteins in WCL and sheared flagellar preparations of H. pylori, oxidized with periodate and visualized with Pro-Q Emerald 488 reagent. Arrows indicate the bands corresponding to FlaA.
DISCUSSION
This study reports the identification of novel virulence factors in a pathogenic bacterium using the classical approach of in vitro attenuation in combination with global gene expression profiling. Although some of the factors (i.e., HsdR and FrpB1) were not essential for colonization, we suggest that the cumulative effects of lowered gene expression levels for these and other factors, including PdxA and the VacA paralog, are likely to have contributed to the difference in colonization levels between the low- and high-passage-number isolates of H. pylori SS1. Most importantly, however, this approach has allowed us to demonstrate, for the first time, the importance of de novo vitamin B6 biosynthesis for host colonization by a human pathogen. Vitamin B6 is an enzyme cofactor in a plethora of cellular reactions, including transamination, decarboxylation, and racemization (reviewed in reference 19). PdxA and PdxJ catalyze two sequential reactions in the DXP-dependent de novo vitamin B6 biosynthesis pathway (20); the by-product of these reactions is pyridoxine-5′-phosphate (PNP) (21). In turn, PNP is oxidized to form PLP, the bioactive vitamer of vitamin B6 (22). The DXP-dependent pathway is found predominantly in a small subset of gammaproteobacteria (23–25) but has been extensively studied only in E. coli (20–23). In H. pylori, only the genes pdxJ and pdxA are annotated as belonging to the pdx family and are clustered together on the H. pylori genome (Fig. S2A). Interestingly, the genetic organization of pdxJ and pdxA appears to be conserved for all Helicobacter and Campylobacter spp., as well as for several other related organisms (Fig. S2A). Conversely, these genes are spatially separated in all other sequenced bacterial genomes containing the pdxJ and pdxA genes, including E. coli. Thus, there appears to have been divergent evolution of bacterial vitamin B6 biosynthesis genes. We suggest that this further highlights the differences between the physiology of the model bacterium E. coli and those of other bacteria and emphasizes the need to study the physiology of pathogenic bacterial species.
From the data presented here, it appears that H. pylori pdxJ and pdxA are transcriptionally coregulated. This finding is consistent with reports that both pdxA and pdxJ are repressed in the stationary phase of growth (26) but that both genes are induced by low pH (pH 4.5) (27). Furthermore, these findings are in agreement with a recent study describing the primary transcriptome of H. pylori, where pdxJ and pdxA were found to be transcribed as an operon (28). This further validates PdxJ and PdxA as colonization factors for H. pylori, as low pH is encountered by H. pylori in its preferred ecological niche (27). No differences in the upstream DNA regions of pdxJ and pdxA were identified between low- and high-infectivity variants of H. pylori SS1 (Fig. S2B), suggesting that transcriptional repression by a proximal cis-acting sequence is unlikely to be responsible for altered gene expression in the low-infectivity variants. Recently, a new layer of complexity in H. pylori gene expression and regulation, which is encoded by antisense and/or trans-acting regulatory short RNAs, was uncovered (28) and may be the mechanism of regulation of pdxJ and pdxA expression. The reason for the reduced levels of gene expression in the low-infectivity variants requires further investigation.
In this study, we identified H. pylori PdxA (HP1583) as being essential for in vivo colonization of mice. We therefore reasoned that the colonization defect of H. pylori pdxA mutants may be the result of the loss of a PLP-dependent function involved in colonization. One such function is the glycosylation of flagella, which is necessary for motility and colonization in H. pylori (16–18). H. pylori glycosylates its flagellins with a pseudaminic acid (Pse) moiety, synthesized via a PLP-dependent reaction mediated by PseC (16). We therefore hypothesized that the absence of PLP in H. pylori pdxA mutants may result in defective flagellar synthesis. Indeed, H. pylori pdxA mutants were nonmotile and expressed no flagella. Furthermore, these mutants accumulated nonglycosylated flagellins of lower molecular weights within the cytoplasm, indicative of an impairment in flagellar glycosylation (Fig. 5A to E). Hence impairment of PseC activity, due to a lack of available PLP, is likely to be one factor contributing to the colonization defect of H. pylori pdxA mutants.
It is widely accepted that vitamin B6 plays an important cellular role as an enzyme cofactor. To our knowledge, however, this study is the first to describe a link between this vitamin and bacterial pathogenesis. This finding is consistent with emerging evidence supporting the importance of vitamin B6 in cellular processes involved in the biosynthesis of surface-exposed structures that are likely to contribute to adhesion and colonization in some pathogenic bacteria (16, 29, 30). Indeed, PLP-dependent enzymes are involved in the synthesis of surface-exposed glycosyl moieties, such as flagella and lipopolysaccharide (LPS), in Salmonella enterica serovar Typhimurium (29), Campylobacter jejuni (16), Vibrio cholerae (30), and E. coli O157:H7 (30); LPS is necessary for efficient bovine colonization by O157:H7 (30). It is worth noting that in addition to the bacteria named above, the PdxJ/A-dependent pathway of vitamin B6 biosynthesis is present in several other human pathogens, including Neisseria spp. and Yersinia pestis, but is absent in mammals. Thus, this work identifies PdxJ and PdxA as novel potential therapeutic targets for the treatment of infections by microbial pathogens. Indeed, various PLP-dependent enzymes have already been identified as drug targets for the treatment of a diverse range of diseases (20–22). We suggest that the disruption of de novo PLP biosynthesis per se, via the inhibition of PdxJ/A, offers the added advantage of selectively inhibiting microbial growth without nonspecifically affecting vital host cell functions. Thus, this study opens new avenues for the treatment of microbial infections.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table S1 in the supplemental material. H. pylori isolates were routinely subcultured as previously described (31). For in vitro growth studies, H. pylori bacteria were grown for up to 24 h in 10 ml brain heart infusion (BHI; Oxoid) broth supplemented or not supplemented with 10 mg/liter PLP (Sigma). Viable cell counts and optical density (A600) determinations were performed at the selected time points. E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar (containing 1.5% [wt/vol] agar) supplemented with kanamycin (20 mg/liter), ampicillin (100 mg/liter), or chloramphenicol (25 mg/liter), as appropriate.
Infection of mice with H. pylori.
Animal handling and experimentation were performed in accordance with national and institutional guidelines (French Laws 87 to 848 and Monash University AEC no. BAM/M2005/44, respectively). H. pylori suspensions for mouse inoculation were prepared directly in broth (31). Six- to eight-week-old female specific-pathogen-free mice were each intragastrically administered a single 100-µl aliquot of the inoculating suspension (107 CFU/mouse) using polyethylene catheters (31).
Assessment of H. pylori infection in mice.
The presence of H. pylori infection in mice was determined by quantitative culture, detection of anti-H. pylori antibodies in sera, and/or RT-PCR on RNA isolated from gastric biopsy specimens, as previously described (31).
Molecular biology techniques.
Genomic and plasmid DNA were isolated using the Masterpure DNA purification (Epicentre) and PureLink plasmid midi kits (Invitrogen), respectively. Restriction endonuclease digestions, ligations, and other enzymatic reactions were conducted by standard procedures as described by Sambrook et al. (32).
Macroarray analyses.
Macroarrays (Eurogentec, SA, Belgium), consisting of the complete set of genes from the H. pylori 26695 strain, were hybridized to [α-33P]dCTP-labeled cDNA which had been reverse transcribed from RNA isolated from paired liquid cultures of high (17 to 23 in vitro passages)- or low (180 to 192 passages)-infectivity H. pylori SS1 variants (33). The intensities of hybridizing spots from four independent biological replicates were determined using the XdotsReader program COSE (33). Significant differences in gene expression were determined using the program significance analysis of macroarrrays (SAM) (34).
RNA isolation.
Mouse gastric biopsy specimens, stored in RNA later (Ambion), were placed in TRIzol reagent (Invitrogen) and homogenized on ice using a PRO-200 homogenizer (PRO Scientific, CT). RNA was purified from gastric biopsy specimens or harvested H. pylori bacteria (107 CFU) using TRIzol reagent and Phase Lock gel tubes (Eppendorf) and then DNase treated using RQ1 DNase (Promega). RNA was subsequently purified using the RNeasy minikit (Qiagen).
qRT-PCR.
RNA (200 ng to 2 µg) was reverse transcribed using random hexamers and SuperScript III (Invitrogen). Primers (Table S2) were designed based on the H. pylori 26695 genomic DNA sequence (GenBank accession no. AE000511) using the Primer Express primer design software (Applied Biosystems). qRT-PCR was performed as previously described (35). cDNA concentrations of the target genes for each sample were normalized to expression of the housekeeping gene, uvrC (26).
Construction of H. pylori mutants by allelic exchange.
PCR fragments, amplified using the primers listed in Table S2 in the supplemental material, were purified and then ligated into pILL570 or pGEM-T Easy (Promega). Constructs were amplified by inverse PCR (Failsafe PCR System; Epicentre) using primers SL7 and SL8, SL3 and SL4, or SL12 and SL13, respectively. The digested PCR products were ligated with a BamHI/KpnI-digested nonpolar kanamycin resistance cassette (aphA3) (13). The construct used to inactivate the vacA paralog gene was generated by PCR with the 3T primer pool (3T-PCR), using the oligonucleotides H206, H207, H208, and H209 (Table S2). Isogenic H. pylori mutants were created by either electroporation (36) or natural transformation (37) and selected on horse blood agar (HBA) plates containing kanamycin (20 mg/liter). PLP was added to media used to culture H. pylori pdxA mutant strains. Kanamycin-resistant transformants in each strain of H. pylori were verified by sequencing and PCR analyses (primer H17 or H50) (Table S2).
Complementation of H. pylori pdxA mutants.
A 150-bp fragment containing the H. pylori 26695 ureA promoter sequence was amplified by PCR from the pUGO plasmid (M. Kaparakis-Liaskos, unpublished data) using primers AG14F and AG14R (Table S2). H. pylori pdxA was amplified from H. pylori 26695 chromosomal DNA using primers AG15F and AG5R. The partially complementary PCR products (1 ng) were used as templates in splicing by overlap extension (SOE) PCR, consisting of five cycles of 94°C for 1 min, 35°C for 1 min, and 72°C for 2 min without the addition of primers, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min in the presence of 20 pmol of each of the primers AG14F and AG5R. The resultant PCR products were cloned into the BamHI/EcoRI sites of the linearized pRdxA vector (38). Complemented H. pylori pdxA mutants were selected on HBA plates supplemented with metronidazole (8 mg/liter) in the absence of PLP. These mutants were verified by PCR sequencing using the primers AG1F, AG1R, AG5F, and AG5R.
Detection of antibodies by ELISA.
H. pylori antigen-specific antibodies in mouse serum samples were detected by enzyme-linked immunosorbent assay (ELISA) (39). Bound H. pylori-specific immunoglobulins were detected with biotinylated goat anti-mouse antibodies (IgM µ-chain [Millipore] or IgG [Jackson ImmunoResearch Laboratories, Inc.]) and streptavidin-peroxidase conjugate (Chemicon).
Cross-complementation in E. coli.
pdxA and pdxJ were amplified from H. pylori 26695 genomic DNA using the primers AG5F and AG5R and AG6F and AG6R, respectively, and ligated into the BamHI/EcoRI-linearized pET28a vector (Novagen). These plasmids were transformed by electroporation (40) into E. coli CGSC8631 (pdxA) and CGSC10027 (pdxJ) and selected on Glu B1 56/2 minimal medium (41), supplemented or not supplemented with PLP and IPTG (1 mM).
Motility testing.
Bacterial motility was determined using semisolid agar plates (17). The velocities of individual bacteria were determined by differential interference contrast (DIC) imaging using an Integrated Leica AF6000 LX live-cell workstation (Leica Microsystems). Aliquots (10 µl) of H. pylori (5 × 107 CFU/ml), which had been harvested from 6-h liquid broth cultures supplemented or not with PLP, were prepared in 0.4% methylcellulose in a FluoroDish (World Precision Instruments Inc.). Image analysis was performed using MetaMorph software (Molecular Devices). For each experiment, positional data for 20 bacteria over a minimum of 20 frames each were used to determine average velocity.
Electron microscopy.
Bacteria were applied to Formvar-carbon-coated copper grids (400 mesh) and negatively stained for 5 to 10 s with 1% phosphotungstic acid (pH 6.8). Grids were examined using a Hitachi H-7500 transmission electron microscope.
Flagellin preparation.
Flagella were isolated according to a modified method of Reeves et al. (42). Briefly, bacteria were harvested from 500-ml liquid cultures, resuspended in phosphate-buffered saline (PBS), and sheared for 1 min with an MSE homogenizer (MSE Ltd.). Cell debris was removed by centrifugation, and flagella were concentrated from the supernatants by ultracentrifugation using a TH-641 rotor in the Sorvall RC-90 ultracentrifuge (285,000 × g for 1 h at 10°C). Pellets containing flagella were resuspended in 100 µl Milli-Q H2O. Protein concentrations were determined by the Bradford (Bio-Rad) or Qubit (Invitrogen) assay.
SDS-PAGE and Western blotting.
Samples were separated in 10 to 12.5% (vol/vol) acrylamide gels and transferred to nitrocellulose. Flagellin was detected using mouse monoclonal antibodies to the H. pylori 54-kDa flagellin antigen (Bioscientific), followed by biotinylated secondary goat anti-mouse IgG antibodies (Chemicon) and streptavidin-horseradish peroxidase (HRP) (Chemicon). Antigen-antibody complexes were detected using ECL detection reagent (GE Healthcare). Glycosylated proteins were detected using the Pro-Q Emerald 488 glycoprotein gel and blot stain kit (Invitrogen).
Statistics.
Differences in colonization levels between low- and high-infectivity H. pylori SS1 variants were determined using contingency tables (Fisher’s exact test). Differences in the bacterial loads or antibody responses between animal groups were determined by the Mann-Whitney U test (two-sided). Differences in expression levels determined by qRT-PCR were analyzed using the Student t test. Differences were considered significant when P was ≤0.05.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
This work was funded by an NHMRC project grant (no. 384232). R.L.F. thanks the Embassy of France in Australia for providing him with a travel grant. A.G. was supported by a Dora Lush Postgraduate Biomedical Scholarship. M.T. was supported by the Fondation pour la Recherche Médicale. The work performed at MIMR was supported by the Victorian Government’s Operational Infrastructure Support Program.
We thank J. Clark and Sarah Walker for their assistance with transmission electron microscopy (TEM) and B. Wanner for his generosity in providing us with E. coli mutant strains.
Footnotes
Citation Grubman, A., A. Phillips, M. Thibonnier, M. Kaparakis-Liaskos, C. Johnson, et al. 2010. Vitamin B6 is required for full motility and virulence in Helicobacter pylori. mBio 1(3):e00112-10. doi:10.1128/mBio.00112-10.
REFERENCES
- 1. Katelaris P. H., Adamthwaite D., Midolo P., Yeomans N. D., Davidson G., Lambert J. 2000. Randomized trial of omeprazole and metronidazole with amoxycillin or clarithromycin for Helicobacter pylori eradication, in a region of high primary metronidazole resistance: the HERO study. Aliment. Pharmacol. Ther. 14:751–758 [DOI] [PubMed] [Google Scholar]
- 2. Mollison L. C., Stingemore N., Wake R. A., Cullen D. J., McGechie D. B. 2000. Antibiotic resistance in Helicobacter pylori. Med. J. Aust. 173:521–523 [DOI] [PubMed] [Google Scholar]
- 3. Ferrero R. L., Labigne A. 2001. Helicobacter pylori vaccine development in the post-genomic era: can in silico translate to in vivo. Scand. J. Immunol. 53:443–448 [DOI] [PubMed] [Google Scholar]
- 4. Marais A., Mendz G. L., Hazell S. L., Megraud F. 1999. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol. Mol. Biol. Rev. 63:642–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo B. P., Mekalanos J. J. 2002. Rapid genetic analysis of Helicobacter pylori gastric mucosal colonization in suckling mice. Proc. Natl. Acad. Sci. U. S. A. 99:8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kavermann H., Burns B. P., Angermuller K., Odenbreit S., Fischer W., Melchers K., Haas R. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salama N. R., Shepherd B., Falkow S. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steenken J. W.1935. Lysis of tubercle bacilli in vitro. Proc. Soc. Exp. Biol. Med. 33:253–255 [Google Scholar]
- 9. Behr M. A., Wilson M. A., Gill W. P., Salamon H., Schoolnik G. K., Rane S., Small P. M. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523 [DOI] [PubMed] [Google Scholar]
- 10. Brosch R., Gordon S. V., Buchrieser C., Pym A. S., Garnier T., Cole S. T. 2000. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee A., O’Rourke J., De Ungria M. C., Robertson B., Daskalopoulos G., Dixon M. F. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386–1397 [DOI] [PubMed] [Google Scholar]
- 12. Eaton K. A., Brooks C. L., Morgan D. R., Krakowka S. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skouloubris S., Thiberge J. M., Labigne A., De Reuse H. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 66:4517–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cover T. L., Blanke S. R. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320–332 [DOI] [PubMed] [Google Scholar]
- 15. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:20060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schoenhofen I. C., McNally D. J., Vinogradov E., Whitfield D., Young N. M., Dick S., Wakarchuk W. W., Brisson J. R., Logan S. M. 2006. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J. Biol. Chem. 281:723–732 [DOI] [PubMed] [Google Scholar]
- 17. Schirm M., Soo E. C., Aubry A. J., Austin J., Thibault P., Logan S. M. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579–1592 [DOI] [PubMed] [Google Scholar]
- 18. Josenhans C., Vossebein L., Friedrich S., Suerbaum S. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210:165–172 [DOI] [PubMed] [Google Scholar]
- 19. Fitzpatrick T. B., Amrhein N., Kappes B., Macheroux P., Tews I., Raschle T. 2007. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem. J. 407:1–13 [DOI] [PubMed] [Google Scholar]
- 20. Banks J., Cane D. E. 2004. Biosynthesis of vitamin B6: direct identification of the product of the PdxA-catalyzed oxidation of 4-hydroxy-l-threonine-4-phosphate using electrospray ionization mass spectrometry. Bioorg. Med. Chem. Lett. 14:1633–1636 [DOI] [PubMed] [Google Scholar]
- 21. Laber B., Maurer W., Scharf S., Stepusin K., Schmidt F. S. 1999. Vitamin B6 biosynthesis: formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-l-threonine and 1-deoxy-d-xylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett. 449:45–48 [DOI] [PubMed] [Google Scholar]
- 22. Zhao G., Winkler M. E. 1995. Kinetic limitation and cellular amount of pyridoxine (pyridoxamine) 5′-phosphate oxidase of Escherichia coli K-12. J. Bacteriol. 177:883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ehrenshaft M., Bilski P., Li M. Y., Chignell C. F., Daub M. E. 1999. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 96:9374–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mittenhuber G.2001. Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J. Mol. Microbiol. Biotechnol. 3:1–20 [PubMed] [Google Scholar]
- 25. Tanaka T., Tateno Y., Gojobori T. 2005. Evolution of vitamin B6 (pyridoxine) metabolism by gain and loss of genes. Mol. Biol. Evol. 22:243–250 [DOI] [PubMed] [Google Scholar]
- 26. Thompson L. J., Merrell D. S., Neilan B. A., Mitchell H., Lee A., Falkow S. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wen Y., Marcus E. A., Matrubutham U., Gleeson M. A., Scott D. R., Sachs G. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma C. M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermuller J., Reinhardt R., Stadler P. F., Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 29. Noland B. W., Newman J. M., Hendle J., Badger J., Christopher J. A., Tresser J., Buchanan M. D., Wright T. A., Rutter M. E., Sanderson W. E., Muller-Dieckmann H. J., Gajiwala K. S., Buchanan S. G. 2002. Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: a 4-amino-4-deoxy-l-arabinose lipopolysaccharide-modifying enzyme. Structure 10:1569–1580 [DOI] [PubMed] [Google Scholar]
- 30. Sheng H., Lim J. Y., Watkins M. K., Minnich S. A., Hovde C. J. 2008. Characterization of an Escherichia coli O157:H7 O-antigen deletion mutant and effect of the deletion on bacterial persistence in the mouse intestine and colonization at the bovine terminal rectal mucosa. Appl. Environ. Microbiol. 74:5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferrero R. L., Thiberge J. M., Huerre M., Labigne A. 1998. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect. Immun. 66:1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Philpott D. J., Belaid D., Troubadour P., Thiberge J. M., Tankovic J., Labigne A., Ferrero R. L. 2002. Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylori isolates. Cell. Microbiol. 4:285–296 [DOI] [PubMed] [Google Scholar]
- 34. Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grubman A., Kaparakis M., Viala J., Allison C., Badea L., Karrar A., Boneca I. G., Le Bourhis L., Reeve S., Smith I. A., Hartland E. L., Philpott D. J., Ferrero R. L. 2010. The innate immune molecule, NOD1, regulates direct killing of Helicobacter pylori by antimicrobial peptides. Cell. Microbiol. 12:626–639 [DOI] [PubMed] [Google Scholar]
- 36. Ferrero R. L., Cussac V., Courcoux P., Labigne A. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 174:4212–4217 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y., Roos K. P., Taylor D. E. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485–2493 [DOI] [PubMed] [Google Scholar]
- 38. Smeets L. C., Bijlsma J. J., Boomkens S. Y., Vandenbroucke-Grauls C. M., Kusters J. G. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferrero R. L., Thiberge J. M., Kansau I., Wuscher N., Huerre M., Labigne A. 1995. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc. Natl. Acad. Sci. U. S. A. 92:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morrison S. L.2001. Transformation of E. coli by electroporation. Curr. Protoc. Immunol. [DOI] [PubMed] [Google Scholar]
- 41. Monod J., Cohen-Bazire G., Cohn M. 1951. The biosynthesis of beta-galactosidase (lactase) in Escherichia coli; the specificity of induction. Biochim. Biophys. Acta 7:585–599 [DOI] [PubMed] [Google Scholar]
- 42. Reeves E. P., Ali T., Leonard P., Hearty S., O’Kennedy R., May F. E., Westley B. R., Josenhans C., Rust M., Suerbaum S., Smith A., Drumm B., Clyne M. 2008. Helicobacter pylori lipopolysaccharide interacts with TFF1 in a pH-dependent manner. Gastroenterology 135:2043–2054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.