Abstract
Uracil appears in DNA as a result of cytosine deamination and by incorporation from the dUTP pool. As potentially mutagenic and deleterious for cell regulation, uracil must be removed from DNA. The major pathway of its repair is initiated by uracil-DNA glycosylases (UNG), ubiquitously found enzymes that hydrolyze the N-glycosidic bond of deoxyuridine in DNA. This review describes the current understanding of the mechanism of uracil search and recognition by UNG. The structure of UNG proteins from several species has been solved, revealing a specific uracil-binding pocket located in a DNA-binding groove. DNA in the complex with UNG is highly distorted to allow the extrahelical recognition of uracil. Thermodynamic studies suggest that UNG binds with appreciable affinity to any DNA, mainly due to the interactions with the charged backbone. The increase in the affinity for damaged DNA is insufficient to account for the exquisite specificity of UNG for uracil. This specificity is likely to result from multistep lesion recognition process, in which normal bases are rejected at one or several pre-excision stages of enzyme–substrate complex isomerization, and only uracil can proceed to enter the active site in a catalytically competent conformation. Search for the lesion by UNG involves random sliding along DNA alternating with dissociation–association events and partial eversion of undamaged bases for initial sampling.
Keywords: DNA repair, DNA glycosylases, Uracil-DNA glycosylase, Structure, DNA binding, Processivity
1. Introduction: uracil in DNA
Uracil (Ura), a base normally present in RNA, can nevertheless arise in DNA via two main processes: spontaneous deamination of cytosine (Cyt) already present in DNA and utilization of dUTP during DNA synthesis [1]. While the latter gives rise to Ura:Ade pairs and is not pre-mutagenic, Cyt deamination generates Ura:Gua mispairs potentially leading to Cyt → Thy transitions after the next round of replication. The rate of appearance of Ura in diploid human cells through deamination estimated from the chemical rate constant is ~102 per cell per day [2,3], and since deamination occurs much faster in single-stranded (ss) DNA, it may contribute significantly to mutagenesis in actively transcribed genes and replication forks. Incorporation of dUMP by DNA polymerases occurs much more frequently, about one event per 2000–3000 incorporated dNTPs [4]. Although Ura can base-pair adequately with adenine (Ade), a substitution of Ura for Thy can significantly alter DNA interactions with DNA-binding proteins and disrupt specific protein binding. Thus, Ura in DNA presents a serious threat to the informational integrity and regulated expression of the genome, unless it is actively removed.
2. Properties of family I uracil-DNA glycosylases
The discovery of Cyt deamination in DNA prompted a search for enzymes capable of removing Ura from DNA. In 1974, Tomas Lindahl purified “uracil glycosidase” from E. coli, an activity that excised [3H]Ura from DNA substrates leaving AP sites [5]. This enzyme was later renamed uracil-DNA N-glycosylase (Ung1) and was the first DNA glycosylase discovered as such. The human UNG (hUNG) protein possessing the same activity was purified from several tissues and cell types soon thereafter [6], allowing its sequencing and cloning of the human UNG gene [7]. Conserved UNG-like sequences, grouped in the family I of uracil-DNA glycosylases, are found in nearly all cellular organisms as well as in pox-and herpesviruses, attesting to the importance of this enzyme and similarity of its mechanism in all living systems [8]. Generally, these sequences possess a conserved C-terminal part encompassing the catalytic domain, while the N-terminal extensions found in eukaryotic and viral enzymes vary enormously in length and composition and may be involved in regulation, protein–protein interactions, and subcellular localization. Ung from E. coli (Eco-Ung) is a polypeptide 229 amino acids long [9]. Human UNG gene produces two mRNA and two protein products due to alternative transcription initiation sites. The hUNG1 isoform (304 amino acids) bears a mitochondrial leader peptide on its N-terminus and is imported into mitochondria, whereas the hUNG2 isoform (313 amino acids) is nuclear [10–13]. In addition to the ubiquitous family I, several other structural families of uracil-DNA glycosylases are known but their phylogenetic distribution is narrower (reviewed in [14]). UNG enzymes specifically excise Ura bases from both double-stranded (ds) and ssDNA with a slight preference for the latter substrate [15]. No activity has been detected against normal DNA bases or against uracil in RNA. In spite of their high specificity for Ura, UNG enzymes have been reported to remove 5-fluorouracil, an anti-tumor agent [16], and the products of Cyt oxidation, 5-hydroxyuracil, isodialuric acid, and alloxan [17,18].
As for any DNA repair enzyme, the main problem faced by UNG is how to find sparse substrate lesions against a background of normal DNA. In this review, we summarize structural, thermodynamic and kinetic data relevant to this problem, which underlies the essential biological function of UNG. We do not discuss other aspects of UNG work, such as the chemical mechanism of Ura excision, regulation of UNG activity, interactions with other proteins, or the role of UNG in cellular processes, unless relevant to the main focus of the review.
Operationally, it is useful to distinguish search for the lesion and recognition of the lesion by UNG. Search for a molecular target is, in most cases, an unguided process of approaching the target in space, while recognition constitutes a positive identification of the bound molecule as a target. In the case of many enzymes converting low-molecular-weight substrates, search and recognition are fully uncoupled, as described by the Damjanovich–Somogyi kinetic model [19]: search is a diffusion encounter of the enzyme and substrate molecules with the formation of a non-specific complex, which isomerizes to a Michaelis complex if the encountered substrate is a correct one. However, search of a target in DNA, be it a specific sequence or a damaged base, always involves some elements of recognition, as will be described in the following sections. Nevertheless, the processes of search and recognition differ in their objectives and consequently in their optimization criteria. Search optimization should minimize the time needed to reach a lesion in DNA starting from an arbitrary point in space, whereas recognition optimization should maximize the probability of processing the target once reached and minimize the probability of processing bound non-target DNA [20]. The optimization criteria for search and recognition are opposed, with more robust recognition requiring more time for sampling DNA and thus slowing the search. Therefore, the molecular mechanism of target search and recognition by UNG enzymes is a trade-off between the needs to quickly survey long stretches of DNA and to reliably detect Ura bases.
3. Structural aspects of lesion search and recognition by UNG enzymes
Since the first X-ray structures were published for free human [21] and herpes simplex virus 1 [22] UNG, a wealth of structural information on UNG enzymes has been accumulated, allowing a detailed analysis of the process of lesion recognition by these enzymes. At the moment of writing this review, the Protein Data Bank held 43 structures of UNG enzymes from human [21,23–30], Atlantic cod [31,32], Leischmania naiffi (Larson et al., to be published), E. coli [33–38], Mycobacterium tuberculosis [39], Deinococcus radiodurans [40], Vibrio cholerae (Raeder et al., to be published), herpes simplex virus 1 [22,41,42], Epstein–Barr virus [43] and vaccinia virus [44], either free or complexed with Ura, dUrd, various ss- and ds oligodeoxyribonucleotides (ODNs), and inhibitors, both naturally occurring (Ugi protein of PBS bacteriophages) and designed small molecules. Since the general structures of all these proteins and features of their interaction with DNA are quite similar, it makes sense to consider all UNG proteins as minor variations of a single structural system.
The structural fold of UNG proteins is based on four β-sheets sandwiched between two pairs of α-helices (Fig. 1A). The surface of the protein globule is traversed by a shallow and narrow positively charged groove, where substrate DNA binds. This groove harbors the enzyme’s active site, which in turn contains a deep pocket that accommodates the uracil base (Fig. 1B). Although the protein lacks distinct domains, binding of hUNG to DNA is accompanied by a small but well defined conformational change: three parts of the protein rotate by ~10° relative to each other around hinges located around Asp145–Pro168 and Leu192–Asn215. These flexible regions are juxtaposed in the three-dimensional structure and form the active site and the Ura recognition pocket. The conformational changes in the substrate DNA molecule are much more pronounced. Although the overall structure of free Ura-containing dsDNA is B-DNA [45], when bound to UNG it is kinked by ~45° at the position of dUMP while remaining regular B-form outside the base pair containing the lesion (Fig. 1C and D). The dUrd nucleoside is unstacked from the neighboring bases, everted from the double helix, and inserted into the enzyme’s active site. The eversion is due to changes in the α and ζ torsion angles of the dUrd and is accompanied by a compression of the DNA backbone with the distance between p+1 and p−1 phosphates (Fig. 1C) decreasing from ~13 Å in B-DNA to ~8 Å. The protein binds in the minor groove of dsDNA and inserts a hydrophobic side chain of Leu272 (in hUNG) into a void formed by dUrd eversion. The intercalated residue forms extensive van der Waals contacts with the adjacent bases thus partially restoring stacking in DNA. Hydrogen bonds formed by the protein mostly target phosphates of the damaged DNA strand (Fig. 1C).
Fig. 1.
(A) schematic representation of hUNG structure. Circles indicate α-helices, triangles, β-sheets. The scheme is based on the structure of free hUNG (PDB ID 1AKZ [21]). (B) Structure of hUNG in a complex with DNA containing an uncleavable dUrd analog 2′-deoxypseudouridine (PDB ID 1EMH [25]). Protein is shown as ribbons, DNA, as sticks. (C) scheme of hUNG interactions with DNA after Ura excision (based on the structure of the enzyme–product complex, PDB ID 1SSP [24]). The residues are numbered relative to dUrd0, positive towards 5′, negative towards 3′; indices in the parentheses relate to the complementary strand. Only the central seven base pairs in the structure are shown. Hydrogen bonds are represented by arrows pointing to the acceptor; hydrophobic and van der Waals interactions are shown by direct contacts between the residues. Some hydrophobic/van der Waals contacts with unmodified residues are omitted for clarity. (D) Overlay of DNA structures from the non-specific/early recognition complex (PDB ID 2OXM [29]; light grey) and the specific/late recognition complex (PDB ID 1EMH [25]; dark grey). The void-filling Leu272 residue is also shown.
Five conserved structural motifs can be distinguished in UNG proteins: (1) the loop that activates the catalytic water molecule (144GQDPYH148 in hUNG); (2) the loop compressing the DNA backbone 5′ to the lesion (165PPPPS169), (3) Ura binding motif (199GVLLLN204), (4) the loop compressing the DNA backbone 3′ to the lesion (246GS247), and (5) the loop penetrating the minor groove (268HPSPLS273). Binding of UNG to DNA critically depends on rigid loops 2, 4, and 5, which are rich in Ser, Pro, and Gly residues that allow the polypeptide chain to approach the DNA backbone closely. Loops 2 and 4 compress the DNA backbone and initiate the DNA kink, which reaches its maximum after dUrd eversion and Leu insertion into the void through the minor groove [24,37,46,47].
The highly conserved Ura binding pocket is complementary in its shape and electrostatic potential to the everted Ura base but is too small to accommodate purines. The best match to the pocket can be achieved only after Ura detachment from C1′, thus lowering the energy of the transition state and the enzyme–product complex relative to the enzyme–substrate complex. The structure of hUNG in the complex with DNA containing a 2′-deoxypseudouridine residue reveals a highly distorted conformation of the damaged deoxynucleoside, with the C1′–N1 bond swerving ~50° from the plane of Ura ring [25]. However, Raman spectroscopy [48], computational analysis of dUrd structure [49], inhibitor [50] and mutagenesis studies [37] suggest that this conformation may not hold true for dUrd. Discrimination against Thy and other 5-substituted pyrimidines is performed by the side chain of a Tyr residue (Tyr147 in hUNG) pressing against C5, whereas discrimination against Cyt is due to a set of specific hydrogen bonds with O2, N3 and O4 of Ura. O2 accepts a bond from the main chain amide of a Gln residue in the conserved 143GQ144 motif. The side chain amide of the conserved Asn204, fixed through the interactions with a water cluster near the entrance to the Ura-binding pocket, forms bonds with N3 and O4 that are specific for Ura and cannot occur with Cyt in the pocket. The critical role of complementarity between Ura and the Ura-binding pocket is highlighted by the creation of UNG mutants with the set of bonds optimized for recognition of Cyt or Thy instead of Ura. Such enzymes are able to excise normal pyrimidines from DNA and confer a spontaneous mutator phenotype to overexpressing E. coli cells [51].
Among the published UNG structures, the structure of hUNG in a complex with DNA containing a Thy:4-methylindole pair [29] is the most illuminating for understanding the lesion search process. 4-Methylindole is an adenine isostere incapable of hydrogen bonding, and its pair with Thy is inherently prone to spontaneous opening. In the structure, Thd is flipped out of DNA but not nearly to the same extent as dUrd in other structures (Fig. 1D). The partially everted Thd is bound near the mouth of the Ura-binding site where it forms hydrogen bonds to two conserved His residues, and it is likely that a dUrd residue is initially bound to the same site [29]. A similar “exosite” participating in probing undamaged bases was identified in human 8-oxoguanine-DNA glycosylase [52]. The kink in the axis of Thy:4-methylindole-containing DNA is not yet fully developed in the complex with UNG. Thus, this structure with a non-specific DNA mimic may be also viewed as a structure of an early recognition complex, with Thd later partitioned back to the DNA helix while dUrd proceeding to enter the Ura recognition pocket. It is not known presently whether the exosite of UNG can accommodate other normal bases.
Despite the wealth of information generated by structural approach, it has its shortcomings. Two most important ones are general lack of structures representing important transient enzyme–substrate complexes and problems with correlating a structure with the energetics of interactions in the complex. Thus, the structural information on UNG enzymes must be supplemented with thermodynamic, kinetic, and computational studies to reveal a true picture of Ura recognition and excision. In the following sections of this review, we discuss experimental data that are relevant to the energetic and dynamic aspects of Ura recognition by UNG.
4. Thermodynamics of UNG interactions with undamaged and damaged substrates
It is currently believed that reactions catalyzed by most nucleic acid (NA)-dependent enzymes proceed in at least three stages [53]:
After the encounter E·NA complex is formed, both components undergo one or several conformational changes characterized by the overall equilibrium constant Keq(2). The third stage is the catalysis of breakage or creation of covalent bonds, characterized by the rate constant (kcat). The overall reaction rate depends not only on this stage but also on how easily the conformation of the encounter complex can be adjusted to the conformation optimal for catalysis.
The energy of primary binding can be conveniently estimated and analyzed using the stepwise increase in ligand complexity (SILC) approach. This method, as applied to DNA-dependent enzymes (reviewed in [54,55]), is based on analysis of the affinities of an enzyme for a series of ligands including orthophosphate (Pi), deoxyribose or mononucleotides (often the minimal ligands of DNA-dependent enzymes), ss non-specific homo-ODNs, ss non-specific mixed-sequence ODNs, ss specific mixed-sequence ODNs, ds non-specific homo-ODNs, ds non-specific mixed-sequence ODNs, ds specific mixed-sequence ODNs, and specific long DNA. Appropriately selected ligands allow one to quantitatively dissect contributions of individual elements of DNA structure (bases, phosphates, deoxyribose; each deoxynucleotide; each DNA strand) into the binding affinity.
The Gibbs free energy characterizing the enzyme–ligand complex formation in most cases can be presented as the sum of ΔG° values for the individual contacts [53]:
with ΔG° i = −RT ln Kdi, where Kdi is the contribution of an individual contact (normalized for 1 M and hence dimensionless). Therefore, the overall Kd value characterizing the formation of the complex can be formally expressed as a product of the Kd values for individual contacts:
and
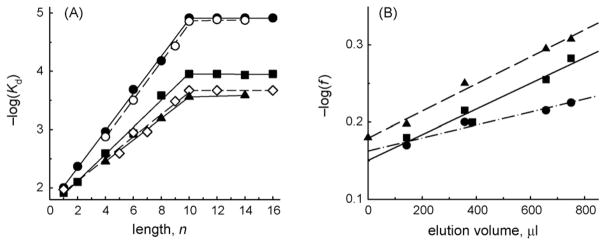
SILC was successfully applied to dissect the energetics of DNA binding by hUNG [56–59]. An example of SILC analysis is shown in Fig. 2. To test for the basic SILC assumption of the additivity of energies of DNA interaction with the enzyme, the log Kd values for d(pN)n were plotted as the logarithmic against the ligand length, n (0 ≤ n ≤ 16, n = 0 corresponds to Pi). The plots were linear to n = 9–10, indicating that, within this range, the addition of one nucleotide contributes the same amount of binding energy, and therefore the binding energy is additive with respect to the ODN length. hUNG binds dUMP approximately one order of magnitude more efficiently than TMP or other dNMPs, and the same difference in affinity is observed between d(pU)n and d(pT)n, or between heteroduplexes either containing a single dUrd or not, indicating that hUNG possesses only one Ura-specific binding site [56]. Dependences of log Kd on n abruptly plateau at n = 9–10, indicating that hUNG covers no more than 9–10 DNA units (Fig. 2) [56,58,59]. The affinity of UNG for non-specific homo-ODNs depends on their composition. Values of f, the factor defined as the increase in affinity of the enzyme for DNA per unit increase in DNA length, can be found from the slopes of the ascending parts of the plots: 1.46 (C units), 1.65 (T units), 1.82 (G units), 1.92 (A units) (Fig. 2). Interestingly, the slope of the plot for d(pR)n, the oligomers containing only the sugar–phosphate backbone, is comparable with that for d(pC)n containing the least hydrophobic of all DNA bases (Fig. 2) [56,58,59].
Fig. 2.
(A) Dependences of hUNG affinity for ss and ds ODNs of various composition on their length (n). (●) d(pA)n; (○) d(pA)n + d(pT)n; (■) d(pT)n; (◇) d(pC)n; (▲) d(pR)n (an oligomer consisting of n units of (3-hydroxytetrahydrofuran-2-yl)methyl phosphate, a deoxynucleotide analog lacking the nucleobase). The data are from Ref. [59]. (B) Determination of the electrostatic factor e reflecting an increase in the affinity due to one internucleotide phosphate group of d(pN)n using log dependence of factor f (an increase in the affinity for various d(pN)n per unit increase in length) on the relative hydrophobicity of mononucleotides. The relative hydrophobicities of mononucleotides were estimated by isocratic reverse-phase chromatography [98] and are expressed as the volume of retention on the column. Dependences for hUNG (■, e = 1.35), AP endonuclease APEX1 (●, e = 1.45) and DNA polymerase α (▲, e = 1.52) are shown; e factors are the intercepts of the linear dependences. The data are from Refs. [59,61,99].
We have previously shown that the interaction of sequence- or structure-specific DNA enzymes (repair enzymes, topoisomerases, restriction endonucleases, integrases) with each nucleotide unit of non-specific ss- or dsDNA is usually a sum of weak electrostatic and hydrophobic or van der Waals interactions with the individual structural elements of DNA [60–64]. The interaction of all investigated enzymes with non-specific DNA can be described by the following equation:
where Kd[(Pi)] is the Kd for the minimal orthophosphate ligand, e is an “electrostatic factor” reflecting an increase in the affinity due to one internucleotide phosphate group, hN are factors of increase in the affinity due to hydrophobic and/or van der Waals interactions of enzymes with one of the bases (A, C, G, and T), the numbers of which in d(pN)n are a, c, g, and t, respectively [55]. The f factor mentioned above is equal to h × e in the case of additive interactions. Only the values of e and hN usually change from one enzyme to another (Fig. 2B). The affinities of some DNA-dependent enzymes for non-specific d(pN)n do not depend on the relative hydrophobicity of the bases, reflecting preferential interactions with the phosphodiester backbone. However, if the enzyme interacts with the bases, the increase in affinity for such ODNs usually follows the same order as the increase in the relative hydrophobicity of the bases: C < T < G < A [55].
In the case of hUNG, the e factor of 1.35 was found from the dependence of log(f) on the relative hydrophobicity of the bases (Fig. 2B). Assuming additive contribution of the phosphate groups and the nucleobases to the hUNG affinity for DNA, the hN factors were calculated: hC = 1.08, hT = 1.22, hG = 1.35, and hA = 1.41 [59].
As evident from the values of e and hN factors for UDG, the contribution of hydrophobic interactions grows with the increase in the hydrophobicity of the bases and becomes comparable with that of electrostatic interactions only for the most hydrophobic bases, Gua and Ade. Thus, on average, internucleoside phosphates contribute most to the UNG affinity for DNA. This conclusion agrees with the results of X-ray structural studies of UNG (see above).
Fig. 2 demonstrates that the second DNA strand does not contribute detectably to the UNG affinity for 10–15-mer DNA duplexes. In this respect, UNG is different from many other DNA-dependent enzymes that require the second strand for the formation of the stable enzyme–DNA complex [55]. The positive electrostatic potential of the DNA-binding groove, in principle, allows it to form electrostatic interactions with both strands of dsDNA. In the structures of UNG bound to specific dsDNA, most of the apparent electrostatic or hydrogen-bonding interactions occur with the damaged strand. However, in the structure of the early recognition complex [29], contacts with the second DNA strand are more extensive. Lack of the structure of UNG complex with true non-specific DNA precludes the analysis of the electrostatic interactions of the enzyme with two undamaged strands but from our SILC data it is likely that interactions with the second strand may be gained only with a compensating loss of interactions with the first strand.
Interactions of UNG with the bases in dsDNA, as evident from the structures, are limited to the interactions of the everted base either with the base recognition pocket in the late recognition complex or with the transient exosite in the early recognition complex, and of the Pro271–Leu272 plug with the bases adjacent to the sampled base and its pairing partner. As UNG likely samples each base pair with partial eversion [29,65–67], these hydrophobic interactions most probably also contribute in the affinity of the enzyme for undamaged DNA. Again, no structural information is available on the interactions of UNG with nucleobases in ssDNA.
The increase in affinity on going from non-specific to specific dsDNA depends to some extent on the ODN sequence. The affinity of UNG for specific ds or ss homo- and hetero-ODNs is only ~10–30-fold higher than that for the best-binding non-specific ODNs [59]. Taking into account all types of non-specific contacts, the complete thermodynamic model of interaction of UNG with non-specific and specific DNA can be constructed (Fig. 3A and B). Specific interactions of UNG with a dUrd residue, namely five hydrogen bonds to the Ura base, hydrophobic and van der Waals interactions, provide only ~1 order of affinity (ΔG° ~ −1.4 to −1.8 kcal/mol), while non-specific interactions provide ~5 orders of the affinity (ΔG° ~ −5.9 kcal/mol) [56–59,68]. A later thermodynamic analysis of contribution of Ura using Eco-Ung and ODN analogs containing 2′-fluoro-2′-deoxyuridine confirms that the eversion of Ura and its interaction with UNG is characterized by ΔG° ~ −2 kcal/mol [69]. Therefore, Michaelis complex formation cannot provide the major part of the UNG specificity. The catalytic substrate specificity of UNG rather lies in the kcat term; the reaction rate is increased by at least five orders of magnitude by the transition from non-specific to specific ODNs [59].
Fig. 3.
Thermodynamic model for interaction of hUNG with non-specific d(pA)n:d(pT)n duplex (A) and specific dsDNA containing a dU deoxynucleotide (B). *Weak electrostatic interactions with internucleotide phosphate groups. Adenine bases forming weak hydrophobic interactions and/or van der Waals contacts with the enzyme are shadowed.
The conformational parameters of the DNA sequence may be of some importance for UNG binding to DNA. The adjustment of some dsODNs to the catalytically active conformation may proceed more easily than with ssODNs [56–59,68]. For example, while the affinity of UNG for homo-ssODNs is comparable with that for the corresponding dsODNs, the affinity for homo-ssODNs containing one “atypical” nucleotide, e.g., d[(pT)n(pC)(pT)m], is ~4–50-fold lower than for completely homogeneous ssODNs. The affinity of UNG for such ODNs does not depend on the type of the intervening base, and the effect is also observed when the intervening residue is dUrd. Notably, neither dUrd-containing mixed-sequence ssODN nor any dsODNs show such anomalous interaction with UNG. Consequently, the efficiency of conformational adjustment of the UNG–DNA complex partly depends on the sequence of ssDNA, and the complementary strand can promote the adjustment. Computational studies suggest that the activity of UNG is higher in more flexible dsDNA sequences, which are easily distorted by the enzyme [70,71].
The active site of UNG imposes stricter constraints on the DNA structure than the other subsites participating in DNA binding (Fig. 3). For example, the affinities of UNG for ss-ribo-(pN)n, dsRNA and RNA–DNA duplexes are on average an order of magnitude lower than for corresponding ODNs of the same length [58,59]. However, this corresponds to a decrease in the affinity of a single residue, the one bound in the active site, to the level of other subsites. The only RNA–DNA duplex bound by UNG with the same affinity as its DNA–DNA analog is (pA)n:d(pT)n; its higher affinity for UNG is most probably due to its ability, unique for RNA–DNA duplexes, to adopt B-form [72]. Thus, it is quite likely that the tighter interactions of the enzyme with a single damaged or undamaged dN in the lesion-binding site are due to the formation of defined bonds, while the rest of DNA interacts with the binding groove through less defined electrostatic contacts, perhaps similar to the attraction of two oppositely charged surfaces, and through general dehydration of the binding interface.
The efficiency of excision of regular bases by UNG is at least five orders of magnitude lower than that of Ura [59]. The catalytic stage appears to be significantly more sensitive to the DNA structure than the stage of DNA binding. In the case of Eco-Ung, the minimal reported substrate is dUrd, but the efficiency of cleavage of the optimal substrate, d(ApUpApA) is 3.6 × 109-fold higher [73]. As mentioned above, contribution of specific interactions to the total UNG affinity for DNA is relatively low (1–2 orders of magnitude), and it is obvious that such interactions cannot ensure the substrate discrimination at the stage of DNA binding. Ultimately, all structural changes of the enzyme–substrate complex are necessary for orbital steering, a precise (to ~10°) alignment of the reacting orbitals [53]. Only then effective catalysis is possible. Even minimal modifications of dU, including those that barely influence the properties of glycosidic bond but likely perturb the conformation of dU in the active site (for example, introduction of a Br atom into position 5 of Ura base or reversion of C3′ carbon configuration) may cause a drastic drop in UNG activity while not affecting ligand affinity [57]. Similar several orders of magnitude decrease in kcat with non-specific DNA is seen in many sequence-specific enzymes [55].
5. Mechanism of lesion search by uracil-DNA glycosylase
Search of specific targets in DNA has been a matter of intense theoretical and experimental scrutiny over several past decades. The current concepts are based on the model originally developed for site-specific DNA-binding proteins such as LacI [74–76]. According to this model, the proteins increase the rate of target search through the reduction of the search dimensionality, substituting one-dimensional diffusion (“scanning”) along the DNA contour for three-dimensional diffusion with random association events. Mechanistically, one-dimensional diffusion may involve sliding (movement of a protein molecule along DNA without microscopic dissociation of the complex) and short-range hopping (dissociation of the protein followed by its instant re-binding in the immediate vicinity of the previous position); also, inter-segmental transfer may occur in the statistical DNA coils when the protein dissociates and rapidly re-binds DNA in a position close to the previous one in space but remote along the DNA contour [74]. As the one-dimensional random walk does not change the mean coordinate, the proteins that rely entirely on passive scanning without energy expenditure should alternate between one-dimensional diffusion, three-dimensional diffusion and inter-segmental transfer to maximize the contour length surveyed per unit time [74]. The one-dimensional diffusion model has been confirmed for many DNA-dependent proteins, including several DNA glycosylases (reviewed in [20]).
With a single exception [77], all studies of kinetics of lesion search by DNA glycosylases have so far relied on the phenomenon of processive cleavage, first described for phage T4 pyrimidine dimer endonuclease [78]. Processive cleavage requires two conditions: first, lesion search must proceed by one-dimensional scanning; second, the enzyme must not dissociate from DNA after base excision and strand nicking. Only if both conditions are satisfied, after cleavage at one target site, the enzyme has a higher probability to cleave at the second target site near the first one. Opposite to processive cleavage is distributive cleavage, in which the sites of two consecutive catalytic acts by an enzyme molecule are totally uncorrelated.
Historically, two assays have been used to study the processivity of DNA glycosylases. In the first one, randomly damaged plasmids are used as substrates, and the processive cleavage shows itself as preferential accumulation of linear products, which appear when two lesions happen to occur close to each other (within ~16 bp) in opposite strands [78]. The second assay uses a concatemer substrate ligated from a number of identical dsODN units, with the processive cleavage evident as preferential accumulation of unit-length products [79]. Eco-Ung has been studied using both the plasmid assay [80] and the concatemer assay [79,81], with the former one demonstrating processivity over the range of 1.5–2 kb and the latter one suggesting either the distributive [79] or processive [81] mode of action. Rat liver mitochondrial UNG was reported to be processive in the concatemer assay [81]. The use of differently constructed substrates and the lack of a standard quantitative measure of processivity likely account for conflicts in these reports.
Recently, we [82,83] and others [84,85] have independently developed a new approach to study the processive cleavage. The assay employs defined ODN substrates (Fig. 4) and provides a quantitative measure of processivity, the probability of correlated cleavage [82], which could be used to estimate the diffusion constant. The substrate structure can be easily varied, allowing one to address the influence of DNA sequence and local conformation on the processive cleavage. The ODN assay have been used to study Eco-Ung [82,84] and hUNG (D.O.Z. and G.V.M., paper in preparation). In both cases, it is shown that UNG can slide along dsDNA over relatively short distances (e.g., the probability of correlated cleavage was at the best ~0.4 for 20 nt separation between two Ura bases), and that it uses both sliding and short-distance hopping for translocation along DNA. The processivity of ssDNA cleavage is lower than for dsDNA. With hUNG, we have found that its processivity is the same for the full-length protein and the catalytic fragment lacking the first 84 residues (D.O.Z. and G.V.M., paper in preparation).
Fig. 4.

Oligonucleotide-based assay for studying processive cleavage by DNA glycosylases. The structure of the substrate is shown on the left. In the reaction scheme, P1 and EP1 refer to the substrate cleaved at any one of the Ura residues and to the respective enzyme–product complex; EP2, the complex with the substrate cleaved at one Ura and the enzyme translocated to the other Ura site; P2, the substrate cleaved at both Ura sites. Pcc, probability of correlated cleavage, i.e., the cleavage at the second Ura after the cleavage of the first Ura by the same enzyme molecule [82]. The initial rates of accumulation of 32-, 27-, and 19-mer products under the conditions of large substrate excess are denoted ν32, ν27, and ν19, respectively.
The relevance of in vitro processive cleavage to the mechanisms of in vivo search is always an issue in the studies of DNA target location, because ion concentrations typical of the cell efficiently screen out electrostatic DNA–protein interactions [74]. With ionic strength increasing to near-physiological levels, the processivity of UNG sharply decreases, and it is also notably lower in cell extracts rich in proteins competing with UNG for DNA binding [82]. However, in the two cases when the processivity of a DNA glycosylase [86] or a restriction endonuclease [87] was studied in vivo, the evidence for processivity was reported. In a highly crowded intracellular environment, a considerable fraction of ions is bound to proteins, perhaps relieving the unfavorable screening effects [88]. Interestingly, we have found no significant effect of polyamines and crowding agents (polyethylene glycols) on the processivity of UNG. In addition, the concentration of DNA in the bacterial cell or the eukaryotic nucleus is sufficiently high for the short-range hopping to turn into intersegment transfer [74], which is not processive by definition but still maintains one-dimensional search mechanism.
Irrespective of the mean DNA length surveyed per association event, the primary binding of UNG will almost inevitably be with undamaged DNA. After that, the enzyme will engage in scanning until it either finds a lesion or falls off DNA. Structurally, the scanning likely involves UNG–DNA complexes similar to the sampling complex with the T:4-methylindole base pair [29]; it would be interesting to use molecular modeling to get an insight into the movement of the enzyme from one sampled position in DNA to the adjacent one. NMR studies suggest that binding undamaged DNA makes the UNG molecule conformationally mobile, easing the commitment to further recognition if the damaged base is encountered [89].
UNG may also use a completely different mechanism of lesion search coupled with replication. Incorporation of dUMP from dUTP is a major source of Ura in DNA [4,90]. hUNG binds replication protein A and the proliferating cell nuclear antigen [91–93]. Structurally, this binding involves the N-terminal part of hUNG, which does not participate in DNA binding or catalysis and probably exists as either separately folded domain or a poorly folded fragment [26]. Thus, UNG may travel with the replication fork removing misincorporated Ura bases without a need for extensive search. However, this mechanism may operate in S-phase only, and the repair under other circumstances would still require lesion search by diffusion.
The early stages of lesion recognition are actively investigated at present. Direct NMR imino proton exchange measurements of the rates of spontaneous base flipping in the presence and in the absence of Eco-Ung suggest that the enzyme may stabilize dUrd that has spontaneously flipped out during DNA breathing, rather than actively promote its eversion [65,67]. On the contrary, stopped-flow studies of the reaction catalyzed by Eco-Ung [47] or herpes simplex virus 1 UNG [94] employing fluorescent reporters incorporated next to Ura indicate that UNG does enhance the rate of dUrd eversion. Given the similarity of UNG enzymes, it is unlikely that they use different mechanisms of lesion search; obviously, this apparent discrepancy has to be resolved. As the equilibrium constant for spontaneous Ura flipping is ~3 × 10−6 [65], relying entirely on the capture of extrahelical Ura appears to be quite an inefficient strategy of lesion search. It should be emphasized that both NMR and stopped-flow experiments mentioned above were performed with short substrates with a single Ura base, which translates to the lesion concentration at least two orders of magnitude higher than can ever be found in normal DNA. Under such conditions, many association events will place the damaged base directly to the lesion-binding subsite of the enzyme’s DNA-binding groove, and the recognition process in this case and in the case of sideways sliding may involve different conformational events [95].
Stopped-flow and site-directed mutagenesis studies led to an establishment of a multistep reaction scheme for UNG [46,47]. At the first stage, the enzyme is bound to DNA non-specifically, and then dUrd is flipped, either actively or passively. The void-plugging Leu is inserted in a kinetically separate step, followed by the N-glycoside bond breakage. The Leu residue is then retracted, and the enzyme–product complex dissociates. Such multistep schemes have an inherent advantage of minimizing errors during lesion recognition [20].
6. Conclusions
Based on the structural, thermodynamic, and kinetic data, the mechanism of lesion search and recognition by UNG can be summarized as follows. UNG binds in a random place in DNA and starts scanning it by one-dimensional diffusion, with a partial eversion of the base sampled at the moment. Non-specific contacts with DNA outside of the sampled base hold the enzyme in place but are not strong enough to prevent the diffusion-driven translocation along DNA. After surveying a short stretch of DNA, the enzyme may dissociate from it. However, if a Ura base is found, it can be drawn into the Ura-binding pocket, increasing the time of residence at this position and allowing for more precise conformational adjustment of the enzyme–substrate complex. Only dUrd and other UNG substrates, but not the normal deoxynucleotides, may adopt a correct conformation in the enzyme’s active site and proceed to breakage of the N-glycosidic bond.
Important questions regarding the UNG search mechanism remain to be answered. The structures of UNG in a complex with undamaged DNA sampling various base pairs will be needed to draw a detailed picture of early recognition. A technique of disulfide cross-linking, which have proved tremendously useful with some other DNA glycosylases [52,96] may be applied to obtain such complexes. The controversy whether UNG actively opens the sampled base pair or relies on its spontaneous opening needs to be reconciliated. Mechanistic details of the movement of UNG along DNA are mostly obscure; the studies to determine the diffusion constant, edge effects, influence of the reaction environment, and the relationship between sliding and hopping mechanisms are in progress in our laboratory. Given the recent emergence of UNG proteins as promising therapeutic targets [97], detailed knowledge of their mechanisms may be valuable for development of new antiviral, antibacterial, and anticancer drugs.
Acknowledgments
This research was supported by the Presidium of the Russian Academy of Sciences (22.7, 22.14) and by Russian Foundation for Basic Research (07-04-00395, 08-04-00596).
Abbreviations
- ds
double stranded
- ODN(s)
oligodeoxyribonucleotide(s)
- Eco-Ung
uracil-DNA glycosylase from Escherichia coli
- hUNG
human uracil-DNA glycosylase
- ss
single-stranded
- UNG
uracil-DNA N-glycosylase
Footnotes
In the literature, family I uracil-DNA glycosylases are often abbreviated Udg or UDG. However, this abbreviation is also often used for UDP-glucose 6-dehydrogenase. Hence, we will refer to bacterial uracil-DNA glycosylase as Ung, and to eukaryotic uracil-DNA glycosylase isoform 2 as UNG, in accordance with the standard genetic nomenclature for E. coli (cgsc.biology.yale.edu) and the Human Genome Organization Nomenclature Committee convention (www.genenames.org). We will use the term UNG when describing family I uracil-DNA glycosylases in general.
Conflict of interest
This research was supported by the Presidium of the Russian Academy of Sciences and by Russian Foundation for Basic Research. The study sponsors were not involved in the study design, collection, analysis and interpretation of data, the writing of the manuscript, or the decision to submit the manuscript for publication.
Contributor Information
Dmitry O. Zharkov, Email: dzharkov@niboch.nsc.ru.
Georgy A. Nevinsky, Email: nevinsky@niboch.nsc.ru.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 3. ASM Press; Washington, DC: 2006. [Google Scholar]
- 2.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 3.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 4.Tye BK, Chien J, Lehman IR, Duncan BK, Warner HR. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1978;75:233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krokan H, Wittwer CU. Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981;9:2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen LC, Aasland R, Wittwer CU, Krokan HE, Helland DE. Molecular cloning of human uracil DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989;8:3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L, Koonin EV. The α/β fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1:research0007. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varshney U, Hutcheon T, van de Sande JH. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung) J Biol Chem. 1988;263:7776–7784. [PubMed] [Google Scholar]
- 10.Slupphaug G, Markussen FH, Olsen LC, Aasland R, Aarsæther N, Bakke O, Krokan HE, Helland DE. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haug T, Skorpen F, Lund H, Krokan HE. Structure of the gene for human uracil-DNA glycosylase and analysis of the promoter function. FEBS Lett. 1994;353:180–184. doi: 10.1016/0014-5793(94)01042-0. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visnes T, Doseth B, Pettersen HS, Hagen L, Sousa MML, Akbari M, Otterlei M, Kavli B, Slupphaug G, Krokan HE. Uracil in DNA and its processing by different DNA glycosylases. Philos Trans R Soc Lond B: Biol Sci. 2009;364:563–568. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 16.Warner HR, Rockstroh PA. Incorporation and excision of 5-fluorouracil from deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1980;141:680–686. doi: 10.1128/jb.141.2.680-686.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes: 5-hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 18.Dizdaroglu M, Karakaya A, Jaruga P, Slupphaug G, Krokan HE. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keleti T. Basic Enzyme Kinetics. Akadémiai Kiadó; Budapest: 1986. [Google Scholar]
- 20.Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 22.Savva R, McAuley-Hecht K, Brown T, Pearl L. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 23.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 24.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA glycosylase–DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 27.Bianchet MA, Seiple LA, Jiang YL, Ichikawa Y, Amzel LM, Stivers JT. Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase. Biochemistry. 2003;42:12455–12460. doi: 10.1021/bi035372+. [DOI] [PubMed] [Google Scholar]
- 28.Krosky DJ, Bianchet MA, Seiple L, Chung S, Amzel LM, Stivers JT. Mimicking damaged DNA with a small molecule inhibitor of human UNG2. Nucleic Acids Res. 2006;34:5872–5879. doi: 10.1093/nar/gkl747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung S, Parker JB, Bianchet M, Amzel LM, Stivers JT. Impact of linker strain and flexibility in the design of a fragment-based inhibitor. Nat Chem Biol. 2009;5:407–413. doi: 10.1038/nchembio.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leiros I, Moe E, Lanes O, Smalas AO, Willassen NP. The structure of uracil-DNA glycosylase from Atlantic cod (Gadus morhua) reveals cold-adaptation features. Acta Crystallogr. 2003;D59:1357–1365. doi: 10.1107/s0907444903011144. [DOI] [PubMed] [Google Scholar]
- 32.Moe E, Leiros I, Riise EK, Olufsen M, Lanes O, Smalås AO, Willassen NP. Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG) from atlantic cod (Gadus morhua) J Mol Biol. 2004;343:1221–1230. doi: 10.1016/j.jmb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Ravishankar R, Sagar MB, Roy S, Purnapatre K, Handa P, Varshney U, Vijayan M. X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (Ec UDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG. Nucleic Acids Res. 1998;26:4880–4887. doi: 10.1093/nar/26.21.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao G, Tordova M, Jagadeesh J, Drohat AC, Stivers JT, Gilliland GL. Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins. 1999;35:13–24. [PubMed] [Google Scholar]
- 35.Drohat AC, Xiao G, Tordova M, Jagadeesh J, Pankiewicz KW, Watanabe KA, Gilliland GL, Stivers JT. Heteronuclear, NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187. Biochemistry. 1999;38:11876–11886. doi: 10.1021/bi9910880. [DOI] [PubMed] [Google Scholar]
- 36.Putnam CD, Shroyer MJN, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J Mol Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 37.Werner RM, Jiang YL, Gordley RG, Jagadeesh GJ, Ladner JE, Xiao G, Tordova M, Gilliland GL, Stivers JT. Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase. Biochemistry. 2000;39:12585–12594. doi: 10.1021/bi001532v. [DOI] [PubMed] [Google Scholar]
- 38.Saikrishnan K, Bidya Sagar M, Ravishankar R, Roy S, Purnapatre K, Handa P, Varshney U, Vijayan M. Domain closure and action of uracil DNA glycosylase (Udg): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving Udg. Acta Crystal-logr. 2002;D58:1269–1276. doi: 10.1107/s0907444902009599. [DOI] [PubMed] [Google Scholar]
- 39.Kaushal PS, Talawar RK, Krishna PDV, Varshney U, Vijayan M. Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources. Acta Crystallogr. 2008;D64:551–560. doi: 10.1107/S090744490800512X. [DOI] [PubMed] [Google Scholar]
- 40.Leiros I, Moe E, Smalås AO, McSweeney S. Structure of the uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans. Acta Crystallogr. 2005;D61:1049–1056. doi: 10.1107/S090744490501382X. [DOI] [PubMed] [Google Scholar]
- 41.Savva R, Pearl LH. Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase-uracil glycosylase inhibitor protein complex. Nat Struct Biol. 1995;2:752–757. doi: 10.1038/nsb0995-752. [DOI] [PubMed] [Google Scholar]
- 42.Krusong K, Carpenter EP, Bellamy SRW, Savva R, Baldwin GS. A comparative study of uracil-DNA glycosylases from human and herpes simplex virus type 1. J Biol Chem. 2006;281:4983–4992. doi: 10.1074/jbc.M509137200. [DOI] [PubMed] [Google Scholar]
- 43.Géoui T, Buisson M, Tarbouriech N, Burmeister WP. New insights on the role of the γ-herpesvirus uracil-DNA glycosylase leucine loop revealed by the structure of the Epstein–Barr virus enzyme in complex with an inhibitor protein. J Mol Biol. 2007;366:117–131. doi: 10.1016/j.jmb.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Schormann N, Grigorian A, Samal A, Krishnan R, DeLucas L, Chattopadhyay D. Crystal structure of vaccinia virus uracil-DNA glycosylase reveals dimeric assembly. BMC Struct Biol. 2007;7:45. doi: 10.1186/1472-6807-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carbonnaux C, Fazakerley GV, Sowers LC. An NMR structural study of deaminated base pairs in DNA. Nucleic Acids Res. 1990;18:4075–4081. doi: 10.1093/nar/18.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang YL, Stivers JT. Mutational analysis of the base-flipping mechanism of uracil DNA glycosylase. Biochemistry. 2002;41:11236–11247. doi: 10.1021/bi026226r. [DOI] [PubMed] [Google Scholar]
- 47.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteadystate analysis of a single catalytic turnover by Escherichia coli uracil-DNA glycosylase reveals a “pinch-pull-push” mechanism. J Biol Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 48.Dong J, Drohat AC, Stivers JT, Pankiewicz KW, Carey PR. Raman spectroscopy of uracil DNA glycosylase-DNA complexes: insights into DNA damage recognition and catalysis. Biochemistry. 2000;39:13241–13250. doi: 10.1021/bi001437m. [DOI] [PubMed] [Google Scholar]
- 49.Berti PJ, Tanaka KSE. Transition state analysis using multiple kinetic isotope effects: mechanisms of enzymatic and non-enzymatic glycoside hydrolysis and transfer. Adv Phys Org Chem. 2002;37:239–314. [Google Scholar]
- 50.Jiang YL, Cao C, Stivers JT, Song F, Ichikawa Y. The merits of bipartite transition-state mimics for inhibition of uracil DNA glycosylase. Bioorg Chem. 2004;32:244–262. doi: 10.1016/j.bioorg.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Kavli B, Slupphaug G, Mol CD, Arvai AS, Petersen SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 53.Fersht A. Enzyme Structure and Mechanism. W.H. Freeman & Co; New York: p. 1985. [Google Scholar]
- 54.Zharkov DO, Ishchenko AA, Douglas KT, Nevinsky GA. Recognition of damaged DNA by Escherichia coli Fpg protein: insights from structural and kinetic data. Mutat Res. 2003;531:141–156. doi: 10.1016/j.mrfmmm.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Nevinsky GA. The role of weak specific and nonspecific interactions in enzymatic recognition and conversion of long DNAs. Mol Biol (Mosk) 2004;38:636–662. [PubMed] [Google Scholar]
- 56.Vasilenko NL, Bulychev NV, Gorn VV, Levina AS, Nevinskii GA. Recognition of uracil in DNA by uracil-DNA-glycosylase from human placenta. Mol Biol (Mosk) 1994;28:450–460. [PubMed] [Google Scholar]
- 57.Kubareva EA, Volkov EM, Vinogradova NL, Kanevsky IA, Oretskaya TS, Kuznetsova SA, Brevnov MG, Gromova ES, Nevinsky GA, Shabarova ZA. Modified substrates as probes for studying uracil-DNA glycosylase. Gene. 1995;157:167–171. doi: 10.1016/0378-1119(94)00771-j. [DOI] [PubMed] [Google Scholar]
- 58.Vinogradova NL, Yamkovoi VI, Tsvetkov IV, Nevinskii GA. Interaction of human placental uracil DNA glycosylase with single-stranded oligonucleotides and their duplexes. Mol Biol (Mosk) 1996;30:130–135. [PubMed] [Google Scholar]
- 59.Vinogradova NL, Bulychev NV, Maksakova GA, Johnson F, Nevinskii GA. Uracil DNA glycosylase: interpretation of X-ray data in the light of kinetic and thermodynamic studies. Mol Biol (Mosk) 1998;32:400–409. [PubMed] [Google Scholar]
- 60.Ishchenko AA, Vasilenko NL, Sinitsina OI, Yamkovoy VI, Fedorova OS, Douglas KT, Nevinsky GA. Thermodynamic, kinetic, and structural basis for recognition and repair of 8-oxoguanine in DNA by Fpg protein from Escherichia coli. Biochemistry. 2002;41:7540–7548. doi: 10.1021/bi0121297. [DOI] [PubMed] [Google Scholar]
- 61.Beloglazova NG, Kirpota OO, Starostin KV, Ishchenko AA, Yamkovoy VI, Zharkov DO, Douglas KT, Nevinsky GA. Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta. Nucleic Acids Res. 2004;32:5134–5146. doi: 10.1093/nar/gkh846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolocheva TI, Maksakova GA, Bugreev DV, Nevinsky GA. Interaction of endonuclease EcoRI with short specific and nonspecific oligonucleotides. IUBMB Life. 2001;51:189–195. doi: 10.1080/152165401753544269. [DOI] [PubMed] [Google Scholar]
- 63.Bugreev DV, Baranova S, Zakharova OD, Parissi V, Desjobert C, Sottofattori E, Balbi A, Litvak S, Tarrago-Litvak L, Nevinsky GA. Dynamic, thermodynamic, and kinetic basis for recognition and transformation of DNA by human immunodeficiency virus type 1 integrase. Biochemistry. 2003;42:9235–9247. doi: 10.1021/bi0300480. [DOI] [PubMed] [Google Scholar]
- 64.Bugreeva IP, Bugreev DV, Nevinsky GA. Formation of nucleoprotein RecA filament on single-stranded DNA: analysis by stepwise increase in ligand complexity. FEBS J. 2005;272:2734–2745. doi: 10.1111/j.1742-4658.2005.04693.x. [DOI] [PubMed] [Google Scholar]
- 65.Cao C, Jiang YL, Stivers JT, Song F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat Struct Mol Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 66.Krosky DJ, Song F, Stivers JT. The origins of high-affinity enzyme binding to an extrahelical DNA base. Biochemistry. 2005;44:5949–5959. doi: 10.1021/bi050084u. [DOI] [PubMed] [Google Scholar]
- 67.Cao C, Jiang YL, Krosky DJ, Stivers JT. The catalytic power of uracil DNA glycosylase in the opening of thymine base pairs. J Am Chem Soc. 2006;128:13034–13035. doi: 10.1021/ja062978n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kubareva EA, Vasilenko NL, Vorobjeva OV, Volkov EM, Oretskaya TS, Korshunova GA, Nevinsky GA. Role of DNA definite structural elements in interaction with repair enzyme uracil-DNA glycosylase. Biochem Mol Biol Int. 1998;46:597–606. doi: 10.1080/15216549800204122. [DOI] [PubMed] [Google Scholar]
- 69.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 70.Fuxreiter M, Luo N, Jedlovszky P, Simon I, Osman R. Role of base flipping in specific recognition of damaged DNA by repair enzymes. J Mol Biol. 2002;323:823–834. doi: 10.1016/s0022-2836(02)00999-3. [DOI] [PubMed] [Google Scholar]
- 71.Seibert E, Ross JBA, Osman R. Role of DNA flexibility in sequence-dependent activity of uracil DNA glycosylase. Biochemistry. 2002;41:10976–10984. doi: 10.1021/bi026121o. [DOI] [PubMed] [Google Scholar]
- 72.Zimmerman SB, Pheiffer BH. A RNA·DNA hybrid that can adopt two conformations: an X-ray diffraction study of poly(rA)·poly(dT) in concentrated solution or in fibers. Proc Natl Acad Sci USA. 1981;78:78–82. doi: 10.1073/pnas.78.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang YL, Stivers JT. Reconstructing the substrate for uracil DNA glycosylase: tracking the transmission of binding energy in catalysis. Biochemistry. 2001;40:7710–7719. doi: 10.1021/bi010622c. [DOI] [PubMed] [Google Scholar]
- 74.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 75.Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor–operator interaction: equilibrium measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 76.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 77.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lloyd RS, Hanawalt PC, Dodson ML. Processive action of T4 endonuclease V on ultraviolet-irradiated DNA. Nucleic Acids Res. 1980;8:5113–5127. doi: 10.1093/nar/8.21.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purmal AA, Lampman GW, Pourmal EI, Melamede RJ, Wallace SS, Kow YW. Uracil DNA N-glycosylase distributively interacts with duplex polynucleotides containing repeating units of either TGGCCAAGCU or TGGC-CAAGCTTGGCCAAGCU. J Biol Chem. 1994;269:22046–22053. [PubMed] [Google Scholar]
- 80.Higley M, Lloyd RS. Processivity of uracil DNA glycosylase. Mutat Res. 1993;294:109–116. doi: 10.1016/0921-8777(93)90019-d. [DOI] [PubMed] [Google Scholar]
- 81.Bennett SE, Sanderson RJ, Mosbaugh DW. Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- 82.Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Correlated cleavage of single- and double-stranded substrates by uracil-DNA glycosylase. FEBS Lett. 2008;582:410–414. doi: 10.1016/j.febslet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 83.Sidorenko VS, Zharkov DO. Correlated cleavage of damaged DNA by bacterial and human 8-oxoguanine-DNA glycosylases. Biochemistry. 2008;47:8970–8976. doi: 10.1021/bi800569e. [DOI] [PubMed] [Google Scholar]
- 84.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci USA. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hedglin M, O’Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gruskin EA, Lloyd RS. Molecular analysis of plasmid DNA repair within ultraviolet-irradiated Escherichia coli. I. T4 endonuclease V-initiated excision repair. J Biol Chem. 1988;263:12728–12737. [PubMed] [Google Scholar]
- 87.Jeltsch A, Wenz C, Stahl F, Pingoud A. Linear diffusion of the restriction endonuclease EcoRV on DNA is essential for the in vivo function of the enzyme. EMBO J. 1996;15:5104–5111. [PMC free article] [PubMed] [Google Scholar]
- 88.Völker J, Breslauer KJ. Communication between noncontacting macromolecules. Annu Rev Biophys Biomol Struct. 2005;34:21–42. doi: 10.1146/annurev.biophys.33.110502.133332. [DOI] [PubMed] [Google Scholar]
- 89.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andersen S, Heine T, Sneve R, König I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 91.Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R, Krokan HE. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J Biol Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 92.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hagen L, Kavli B, Sousa MML, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, Otterlei M, Hørning O, Jensen ON, Krokan HE, Slupphaug G. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bellamy SRW, Krusong K, Baldwin GS. A rapid reaction analysis of uracil DNA glycosylase indicates an active mechanism of base flipping. Nucleic Acids Res. 2007;35:1478–1487. doi: 10.1093/nar/gkm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zaika EI, Perlow RA, Matz E, Broyde S, Gilboa R, Grollman AP, Zharkov DO. Substrate discrimination by formamidopyrimidine-DNA glycosylase: a mutational analysis. J Biol Chem. 2004;279:4849–4861. doi: 10.1074/jbc.M310262200. [DOI] [PubMed] [Google Scholar]
- 96.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 97.Sousa MML, Krokan HE, Slupphaug G. DNA-uracil and human pathology. Mol Aspects Med. 2007;28:276–306. doi: 10.1016/j.mam.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 98.Doronin SV, Lavrik OI, Nevinsky GA, Podust VN. The efficiency of dNTP complex formation with human placenta DNA polymerase α as demonstrated by affinity modification. FEBS Lett. 1987;216:221–224. doi: 10.1016/0014-5793(87)80693-2. [DOI] [PubMed] [Google Scholar]
- 99.Kolocheva TI, Maksakova GA, Zakharova OD, Nevinsky GA. The algorithm of estimation of the Km values for primers in DNA synthesis catalyzed by human DNA polymerase α. FEBS Lett. 1996;399:113–116. doi: 10.1016/s0014-5793(96)01298-7. [DOI] [PubMed] [Google Scholar]