Abstract
The multi-functional histone variant Htz1 (Saccharomyces cerevisiae H2A.Z) is acetylated on up to four N-terminal lysines at positions 3, 8, 10, and 14. It has thus been posited that specific acetylated forms of the histone could regulate distinct roles. Antibodies against Htz1-K8Ac, -K10Ac, and -K14Ac show that all three modifications are added by Esa1 acetyltransferase and removed by Hda1 deacetylase. Completely unacetylatable htz1 alleles exhibit widespread interactions in genome scale genetic screening. However, singly mutated (e.g. htz1-K8R) or singly acetylable (e.g. the triple mutant htz1-K3R/K10R/K14R) alleles show no significant defects in these analyses. This suggests that the N-terminal acetylations on Htz1 are internally redundant. Further supporting this proposal, each acetylation decays with similar kinetics when Htz1 transcription is repressed, and proteomic screening did not find a single condition in which one Htz1Ac was differentially regulated. However, whereas the individual acetylations on Htz1 may be redundant, they are not dispensable. Completely unacetylatable htz1 alleles display genetic interactions and phenotypes in common with and distinct from htz1Δ. In addition, each Htz1 N-terminal lysine is deacetylated by Hda1 in response to benomyl and reacetylated when this agent is removed. Such active regulation suggests that acetylation plays a significant role in Htz1 function.
Keywords: Chromatin Histone Modification, Genome Structure, Histone Acetylase, Histone Deacetylase, Histones, Epistasis Mapping, Genome Stability
Introduction
Chromatin is the highly ordered assembly that packages eukaryotic DNA and regulates nuclear processes such as replication, repair, and transcription. The basic repeating unit of chromatin is the nucleosome core particle: ∼146 bp of DNA wrapped around a core histone octamer composed of a (H3-H4)2 tetramer and two (H2A-H2B) dimers (1). The major histones (H2A, H2B, H3, and H4) are each encoded by multi-copy genes, highly expressed during the S phase, and deposited in chromatin during DNA replication. Histone variants are nonallelic isoforms that are usually (although not exclusively (2)) encoded by single copy genes. In many cases variants can substitute for the major histones in specific nucleosomes through the action of dedicated deposition machineries (3). Both major and variant histones are subject to extensive post-translational modification by the addition of small chemical moieties, including phosphorylation, acetylation, methylation, sumoylation, and ubiquitylation (4). These groups are thought to regulate access to the DNA in the modified nucleosome by various means, including directly modulating the charge on the nucleosome surface or generating sites for the recruitment of regulatory proteins (5, 6).
Histone H2A has one of the largest variant families and includes H2A.Z, a protein that is highly conserved across eukaryotes but differs considerably from the major H2A in each species (7). H2A.Z has been ascribed a large number of roles (8, 9), including most recently suppressing antisense RNAs (10) and stabilizing the association of condensin with mitotic chromosomes (11). We still have a poor understanding of precisely how the variant mediates any specific function, although differential enrichment at specific locations and distinct post-translational modifications are likely to contribute. In all species examined, their respective H2A.Zs are subject to multiple N-terminal tail acetylations (12, 13). This modification is integral to fission yeast H2A.Z function, with completely unacetylatable alleles phenocopying complete deletion of the histone in genome scale transcriptome and genetic analyses (11). Multiple regulatory roles have been assigned to acetylated H2A.Z in Saccharomyces cerevisiae (Sc),2 including heterochromatin restriction (14), transcription (15), and chromosome stability (16). However, many of these ascribed functions were derived from completely unacetylatable alleles, so the importance of any individual acetylation is unclear. Finally, it is also unknown whether the effects of any of these modifications are direct (e.g. steric hindrance or charge modulation) or indirect (e.g. via the recruitment of regulatory proteins).
In this work we examine the regulation and function of four individual acetylations on the Sc H2A.Z (gene name HTZ1) N terminus: K3Ac, K8Ac, K10Ac, and K14Ac. We have raised antibodies to the latter three acetyl forms and show that each is chromatin-associated and primarily regulated by the NuA4 acetyltransferase and Hda1 deacetylase complexes. We have identified a range of novel Htz1Ac regulators, all of which have equivalent effects on K8Ac, K10Ac, and K14Ac. This suggests that each acetylation is redundant, a proposal supported by epistasis mapping analyses of a comprehensive panel of unacetylatable htz1 alleles. However, N-terminal acetylation is important for Htz1 function, and completely unacetylatable htz1 alleles show a large number of genetic interactions in common with and distinct from htz1Δ. Furthermore, each Htz1Ac is actively regulated in response to benomyl, a microtubule destabilizing agent. Thus our results indicate that N-terminal acetylation is important for the Htz1 function, but the cell does not distinguish between individual acetylated lysines on this histone.
EXPERIMENTAL PROCEDURES
Materials
The antibodies and strains used in this study are listed in supplemental Tables S1 and S2.
Antibody Generation
Rabbit polyclonal affinity-purified anti-Sc Htz1-K8Ac, -K10Ac, and -K14Ac were from Millipore, with αK14Ac described previously (16). For each new target (αK3Ac, αK8Ac, and αK10Ac) five rabbits (prescreened for minimal reactivity to Sc proteins) were immunized (peptides as in Fig. 1A coupled to keyhole limpet hemocyanin), and test bleeds/boosts were performed monthly. Initial screening of each bleed was by immunoblotting against whole cell extracts (WCEs) from WT and htz1Δ cells. Attempts to raise αK3Ac failed at this stage because all rabbits failed to raise a detectable immune response (not shown). Higher titer sera (usable at >1/200 dilution) from each rabbit were then tested for specificity against WCEs from unacetylatable htz1 point mutant cells (e.g. htz1-K8R; see Fig. 1B). Specificity was further characterized by Luminex bead assay against a comprehensive peptide panel, including the relative immunogen, its acetylated form, and all other acetyl peptides from the immunization series (supplemental Fig. S1). These analyses confirmed the absence of cross-reaction with an inappropriate Htz1Ac species. To derive preparations for this study, positive bleeds from optimal responders were pooled, affinity-purified on the immunizing acetyl peptide, and subtracted with the relevant unmodified peptide.
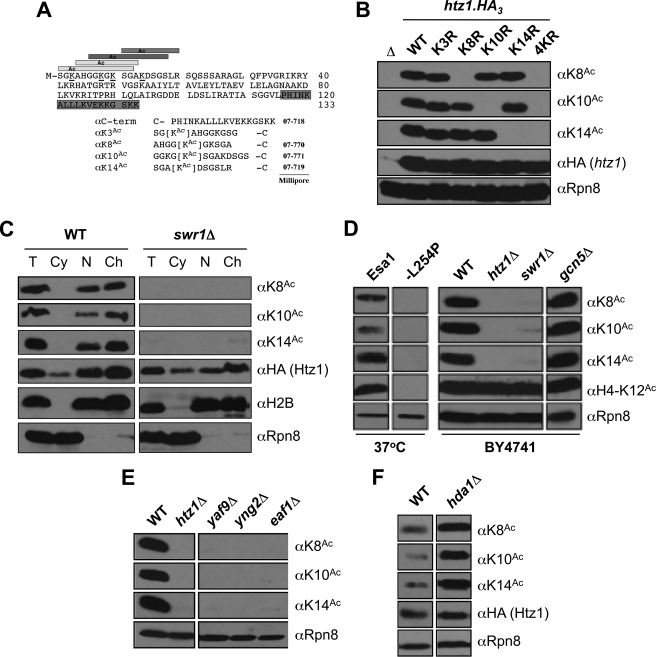
FIGURE 1.
Htz1 lysines 8, 10, and 14 are acetylated by NuA4 as a component of chromatin. A, peptides used to raise αHtz1Ac antibodies (see “Experimental Procedures”), with the four acetylatable lysines (at positions 3, 8, 10, and 14) on the Htz1 N terminus underlined. The terminal cysteine (–C) on each peptide was used to couple to KLH for immunization or a sulfolink resin for affinity purification. B, αK8Ac, αK10Ac, and αK14Ac are highly specific. C-terminally HA3-tagged forms of the indicated htz1 alleles were individually expressed as the sole source of the histone and WCEs analyzed by immunoblotting. Specific mutation of the appropriate lysine to unacetylatable arginine (e.g. K8R) abolishes recognition by the relevant affinity-purified antibody. Δ is an htz1Δ strain. αHA demonstrates equivalent expression of each htz1.HA3. αRpn8 (19 S proteasome subunit) serves as a loading control. C, each acetylated form of Htz1 is chromatin-associated. WT and swr1Δ cells were spheroblasted (total, T); fractionated into cytoplasm (Cy), nucleus (N), and chromatin (Ch); and immunoblotted as indicated. αHA monitors the localization of Htz1.HA3. Appropriate segregation of histone H2B and Rpn8 indicates efficient fractionation; the former is primarily localized in insoluble chromatin, and the latter is primarily in soluble cytoplasm. D, all of the tested forms of Htz1Ac are Esa1-dependent and Gcn5-independent. Left panel, ESA1 or esa1-L254P strains were incubated at a nonpermissive temperature (37 °C, 2 h), WCEs were harvested and immunoblotted as indicated. Esa1-dependent H4-K12Ac is a positive control. Rpn8 is a loading control. Right panel, BY4741. The indicated WCEs were harvested at 30 °C. E, all of the tested forms of Htz1Ac are dependent on indicated members of the NuA4 acetyltransferase complex. These deletions also reduce H4Ac levels but have no impact on the abundance of Htz1 (16). F, each Htz1Ac is increased on deletion of the Hda1 deacetylase.
Cell Fractionation
Sc cell fractionations were as described (16). The total, cytoplasmic, nuclear, and chromatin fractions were analyzed by SDS-PAGE and immunoblotting.
Creation of htz1 Mutant Strains
Various unacetylatable point mutants at HTZ1 (4KR, K3R, K3Ac*, etc.) were created by a modified Delitto Perfetto approach (17) in a “magic marker” strain compatible with the synthetic genetic array protocol (18) (KFY1069; see supplemental Table S2). In brief, HTZ1 was first replaced by a [KanR/URA3] cassette. The htz1 ORF was then PCR-amplified from Sc genomic DNA (with the desired mutation inserted by megapriming (19)), and the product was transformed into exponentially growing cells. Colonies that replaced [KanR/URA3] with htz1 by homologous recombination were identified on 5-fluoroorotic acid (which counterselects URA3) and confirmed by sequencing. To construct htz1-NΔ, the loxP-KanMX-loxP cassette from pOM10 (20) was used to replace the N terminus of htz1 (residues 3–14, including all four acetylatable lysines; see Fig. 1A) in situ. Cre-induced recombination resulted in cassette removal with retention of the 24-residue loxP element. In the final step a Nourseothricin resistance cassette (NAT) was incorporated immediately downstream of each htz1 to facilitate locus selection during synthetic genetic array (21). An HTZ1::NAT locus-tracking strain was also created as a WT control.
Growth Curve Analysis
Growth curves were monitored with a Bioscreen C (Oy Growth Curves). Seed cultures were grown to mid-log in nonselective medium and diluted to A600 ≤ 0.1 in medium with the appropriate agents, e.g. synthetic complete ± 6-azauracil (6AU) or mycophenolic acid (MPA), YPD ± benomyl, TBZ, or camptothecin. All of the analyses were performed in triplicate, and A600 curves (30 °C, constant agitation, 15-min time points) were monitored for >48 h.
Htz1 Stability Analyses
Translational shut-off: Cycloheximide (final concentration, 35 μg/ml (22)) was added to exponentially growing cultures (A600 = ∼0.5), and 5-ml aliquots were taken at the indicated intervals for WCE isolation (TCA method; see below). Target abundance was estimated by immunoblotting at each time point (see Fig. 3A).
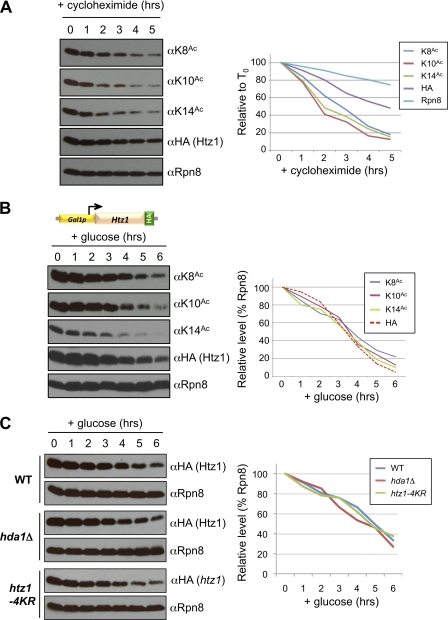
FIGURE 3.
Htz1 acetylation does not regulate turnover of the histone variant. A, cycloheximide was used to repress translation in a HTZ1.HA3 strain, with samples taken for WCE isolation and immunoblotting at times indicated. Quantitation measures each immunoblotted species relative to their abundance at T0 (precycloheximide). B, glucose (final concentration, 2%) was added to repress GAL1 promoter-driven transcription in a GAL1p.HTZ1.HA3 strain, with samples taken for WCE isolation and immunoblotting at times indicated. Quantitation measures each immunoblotted species relative to Rpn8 at the same time point. C, glucose was added to repress transcription from GAL1p.HTZ1.HA3 (WT or hda1Δ backgrounds) or GAL1p.htz1–4KR.HA3, with samples taken for WCE isolation and immunoblotting at times indicated. Quantitation measures each htz1.HA3 relative to Rpn8 at the same time point in the same background.
Transcriptional Shut-off
To create GAL1p.HTZ1.HA3, the GAL1 promoter was integrated by homologous recombination to replace the endogenous promoter upstream of HTZ1.HA3. Immunoblotting (αHA) confirmed that comparable Htz1.HA3 levels are derived from each promoter in YPGR (2% galactose, 1% raffinose) medium (not shown). An exponentially growing GAL1p.HTZ1.HA3 culture (A600 = ∼0.5 in YPGR) was collected by centrifugation, washed in double distilled H2O, and resuspended in YPD (containing 2% glucose to repress transcription from GAL1p). Aliquots were taken at the indicated intervals for WCE isolation. Target abundance was estimated by immunoblotting at each time point (see Fig. 3, B and C).
Synthetic Genetic Array Screening
Genetic interactions were determined by the partially automated synthetic genetic array method (21). htz1 alleles in the magic marker background (supplemental Table S2) were mated in quadruplicate to either a mutant library of 1,286 factors involved in chromatin metabolism (23) or two libraries that together cover >98% of yeast genes (i.e. ∼4800 nonessential genes individually deleted with a KanMX cassette (24) or hypomorphic alleles of ∼842 essential genes with KanMX disrupting their polyadenylation site (25)). All of the libraries were arrayed at 1536 colony density/12.5 × 8.5-cm plate and replica plated with a Singer RoToR. The growth of all double-mutant haploid daughters was compared with the respective single-mutant parents to identify and quantify positive or negative genetic interactions (26, 27). For epistasis mapping (23), the genetic profile of each htz1 allele was compared within a set of 2,255 profiles to calculate pair-wise Pearson correlation coefficients.
TCA Cell Extracts for Western Blotting
WCEs were isolated by the TCA method, which efficiently extracts chromatin and preserves labile modifications (28). In brief, ∼10-ml cultures were grown to mid-log (A600 = ∼1.0), and cells were collected by centrifugation and washed with 20% TCA. All further steps were performed on ice with prechilled solutions. Cell pellets were resuspended in 250 μl of 20% TCA and subjected to glass bead lysis. The suspension minus the glass beads was collected, 1 ml of 5% TCA was added, and the precipitate was collected by centrifugation. The pellets were washed with 750 μl of 100% ethanol, and the proteins were solubilized in 50 μl of 1 m Tris, pH 8.0, 100 μl of 2× SDS-PAGE loading buffer (60 mm Tris, pH 6.8, 2% SDS, 10% glycerol, 100 mm DTT, 0.2% bromphenol blue). After 5 min at 95 °C, insoluble material was removed by centrifugation, and the supernatant was analyzed further by immunoblotting.
RESULTS
Htz1-K8Ac, K10Ac, and K14Ac Are Chromatin-associated and Regulated by Esa1 and Hda1
Sc H2A.Z (Htz1) has four acetylatable lysines on its N terminus at positions 3, 8, 10, and 14 (Fig. 1A) (14, 16, 29). Antibodies to each modification would greatly aid in their analysis. We have previously described anti-Htz1-K14Ac (αK14Ac) in detail (16) and have now raised αK8Ac and αK10Ac (Fig. 1B and supplemental Fig. S1). Attempts to raise αK3Ac were unsuccessful (see “Experimental Procedures”). Each reagent (αK8Ac, αK10Ac, and αK14Ac) shows strong specificity for its respective target in immunoblotting, losing recognition if the appropriate lysine is mutated to arginine (e.g. K10R; Fig. 1B). These antibodies also indicate that individual Htz1 N-terminal acetylations show no interdependence (e.g. K10Ac is not impacted by K3R, K8R, or K14R) or compensation (e.g. K10Ac levels do not increase relative to WT if K10 is the sole acetylatable residue on the Htz1 N terminus) (Fig. 1B and supplemental Fig. S2).
The abundance of Htz1 K8Ac, K10Ac, and K14Ac are strongly reduced on deletion of SWR1, the eponymous ATPase subunit of the SWR complex (SWR-C; Fig. 1C). Because Swr1 is required for the insertion of Htz1 into nucleosomes (30, 31), this indicates that all three lysines are acetylated after chromatin assembly. Cell fractionation confirmed that each Htz1Ac is chromatin-associated (Fig. 1C) and further that its acetylation status does not regulate entry to this cellular compartment (supplemental Fig. S3).
The Esa1 and Gcn5 acetyltransferases or Hda1 deacetylase are each reported to regulate the acetylation of Htz1 (15, 16, 32). In direct analyses K8Ac, K10Ac, and K14Ac are abolished in cells containing mutants of NuA4, including a temperature-sensitive allele of Esa1 (esa1-L254P) or deletion of various complex subunits (Fig. 1, D and E). In contrast gcn5Δ has no effect on K8Ac, K10Ac, or K14Ac levels (Fig. 1D), although we note that Gcn5 could still target Htz1-K3Ac; this could not be determined without an antibody to the latter modification. Regarding the deacetylation of Htz1, Hda1 has been shown to target Htz1-K14Ac (32). We have confirmed this and determined that Hda1 also deacetylates K8Ac and K10Ac (Fig. 1F). Thus three distinct Htz1Ac species are chromatin-associated and metabolized by the same enzymes in rapidly growing cells.
Genome Scale Genetic Analyses Indicate That Individual Htz1 Acetylations Are Redundant
To determine whether each Htz1 acetylation could have a specific role, we tested the genetic interactions of a comprehensive range of unacetylatable alleles. A number of synthetic genetic array compatible strains were created, including complete deletion (htz1Δ), N-terminal deletion (NΔ), all four lysines mutated to arginine (4KR) or glutamine (4KQ), each individual lysine mutated to arginine (e.g. K3R) or glutamine (e.g. K3Q), or a single remaining acetylatable residue (e.g. K8R/K10R/K14R = K3Ac*). A locus-marked HTZ1::NAT strain was also created as a WT control. In immunoblotting each mutant was expressed at similar levels to WT with the exception of htz1-NΔ, which is weakly hypomorphic (not shown). Notably this hypomorphism is also seen with a comparable allele of Schizosaccharomyces pombe H2A.Z (pht1-NΔ) (11).
Each strain was individually mated to a mutant library of 1,286 factors involved in chromatin metabolism (23), and double-mutant haploid daughters were isolated. We then derived scores covering each negative (i.e. synthetic sick/lethal (SS/SL)) or positive (i.e. epistasis or suppression) genetic interaction using colony size as a quantitative readout (27, 33). The genetic interaction profile of a particular mutant can be used as a high resolution phenotype, with functionally related factors displaying similar profiles (23, 27). Thus deletions of individual members of the SWR-C are highly correlated with htz1Δ and NΔ (Fig. 2A, green nodes). This is expected because SWR-C is required for the insertion of Htz1 into chromatin (Fig. 1C) (30, 31). The profiles for htz1Δ, NΔ, 4KR, and 4KQ are also highly correlated (Fig. 2A), suggesting that significant functionality of Htz1 resides in its N terminus and furthermore that the primary role of this region is to harbor the acetylations. Indeed the 4KR and 4KQ profiles were most similar to NΔ within those 2,255 profiles collated to date (Fig. 2B) (23, 27, 34). Furthermore the 4KR and 4KQ profiles were themselves very highly correlated (Pearson correlation coefficient (CC) 0.876; Fig. 2C), suggesting that the charge on these residues does not regulate their function (see “Discussion”).
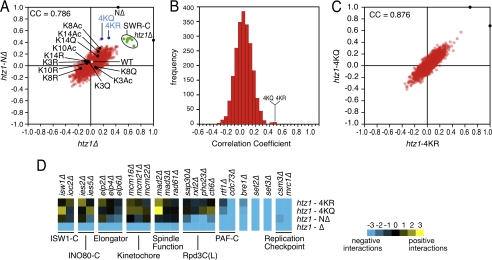
FIGURE 2.
The primary function of the Htz1 N terminus is to harbor lysine acetylations. A, Pearson CC plot compares the genetic interaction profiles of htz1Δ or htz1-NΔ (X- or Y-axes) with 2,255 profiles performed to date. Blue and black spots, as labeled; green spots, SWR-C components (swr1Δ, yaf9Δ, vps71Δ, vps72Δ, swc3Δ, swc5Δ, and arp6Δ); white spot, WT control; red spots, all other mutants on array. B, distribution plot of the CCs of NΔ against 2,245 mutant profiles. The most highly correlated profiles include those derived from 4KR and 4KQ. C, a correlation plot of CCs for 2,245 individual genetic screens relative to 4KR or 4KQ. D, as expected from their strong but imperfect pairwise CCs htz1Δ, NΔ, 4KR, and 4KQ show both common and unique genetic interactions. Blue and yellow correspond to negative or positive genetic interactions.
In contrast to the widespread genetic interactions of completely unacetylatable htz1 alleles, single point (e.g. K3R) or singly acetylatable (e.g. K3Ac*) mutants gave few consistent interactions and clustered near the wild-type control (Fig. 2A, white node). Thus the loss of any single Htz1 acetylation does not confer a genotype strong enough to distinguish the allele, suggesting no significant loss-of-function in any case. Because the retention of any single acetylatable lysine has a similar effect, this suggests that the individual Htz1 acetylations are redundant.
Closer inspection of the individual genetic interactions for htz1Δ, NΔ, 4KR, and 4KQ revealed many that are common between these four mutants. This includes SS/SL interactions with components of the PAF complex (RTF1 and CDC73), the replication checkpoint complex (CSM3 and MRC1), and the chromatin regulators BRE1, SET2, and SET3 (Fig. 2D). However, and as expected given their imperfect CCs, htz1Δ also shows many interactions distinct from the completely unacetylatable alleles. This may be related to the loss of the Htz1 core domain or C terminus in the complete deletion (see “Discussion”). For example, we observed SS/SL interactions between htz1Δ and components of the Isw1, Ino80, and Elongator complexes, as well as factors involved in kinetochore (MCM16, MCM21, and MCM22) and spindle (MAD2, MAD3, and RAD61) function. Finally, we also observed SS/SL interactions common to htz1Δ and NΔ but not shared by 4KR or 4KQ, such as with components of the Rpd3L deacetylase complex (Fig. 2D). These last may be related to the hypomorphic nature of the NΔ allele or indicate that the tail contains functional residues other than the acetylatable lysines.
To more comprehensively investigate the various pathways in which Htz1Ac may play a role, we further mated the NΔ or 4KR strains against two libraries that together cover >95% of the entire Sc genome: ∼4800 nonessential deletions (24) and ∼900 essential DAmP hypomorphs (25). As above, NΔ and 4KR shared many genetic interactors, with ∼30% of these conserved with htz1Δ (supplemental Fig. S4). This was a surprise because we had originally predicted that completely unacetylatable htz1 mutants would simply display a subset of the genetic interactions seen in htz1Δ (see “Discussion”). However, gene ontology analysis supports the validity of each group SS/SL with htz1-NΔ, i.e. those in common with htz1Δ or unique to the tail-less mutant. For example, the “telomere maintenance” gene ontology category is over-represented in each group, and this function is known to be mediated by the Htz1 N terminus (14).
Htz1 Is Recycled into Chromatin after Nucleosome Displacement
To further investigate any potential distinction between Htz1-K8Ac, -K10Ac, and -K14Ac, we measured the half-life (t½) of each modification. Htz1 is enriched in “hot” nucleosomes, defined as those with a high turnover rate (35). However, it is not known whether the histone variant is degraded or reused after nucleosome eviction, although the former would likely result in a protein with a relatively short half-life. Furthermore if each Htz1Ac species is enriched at distinct genomic locations, this could lead to a range of decay kinetics.
Two approaches were used: general inhibition of the translational machinery by cycloheximide (Fig. 3A) or the specific transcriptional repression of GAL1p-HTZ1.HA3 by glucose (Fig. 3B). After each shut-off, the abundance of Htz1 and Htz1-K8Ac, -K10Ac, and -K14Ac were monitored for up to 6 h. In each case total Htz1 had a t½ greater than 3 h or approximately two complete cell cycles, strongly suggesting reuse after nucleosome eviction. Each Htz1Ac decayed significantly faster than total Htz1 on cycloheximide (Fig. 3A). However, this is likely due to the translational inhibition of an acetylation regulator (e.g. a subunit of NuA4; Fig. 1, D and E), because each Htz1Ac tracked with total Htz1 when transcription of the histone was repressed (Fig. 3B). Furthermore the observation that Htz1-K8Ac, -K10Ac, and -K14Ac decayed at similar rates on transcriptional shut-off also argues against their differential usage at distinct locations.
To examine whether its acetylation could regulate Htz1 turnover, we compared GAL1p-HTZ1.HA3 to two additional strains: GAL1p-htz1–4KR.HA3 and [GAL1p-HTZ1.HA3/hda1Δ]. The former contains an unacetylatable form of Htz1, the latter is increased for all acetylated species of the histone (Fig. 1F). On transcriptional repression, total Htz1 levels decayed with similar rates in all three backgrounds (Fig. 3C), indicating that its acetylation status has no impact on the turnover of this histone variant.
Each Htz1Ac Is Similarly Regulated in a Range of Mutant Backgrounds
To further investigate the possibility of their differential regulation, we used a proteomic screening approach (36) to identify modifiers of each Htz1Ac. Viable mutants of ∼50 genes, primarily factors related to the gene ontology terms “histone acetylation” or “histone deacetylation,” were isolated from the Sc haploid deletion collection and immunoblotted for Htz1-K8Ac, -K10Ac, and -K14Ac (see supplemental Table S3). This confirmed that all three Htz1Ac forms are dependent on individual subunits of NuA4 (YAF9, YNG2, and EAF1) but independent of SAGA components (see also Fig. 1, D and E). The approach identified many additional regulators of Htz1Ac (e.g. Asf1, Rpd3L, and Elongator) but no differential regulators of any individual acetylation, i.e. K8Ac, K10Ac, and K14Ac tracked together in every background (supplemental Table S3 and data not shown). This includes Bud14 and Clb2, whose deletions were previously identified as reducing K14Ac levels (34); these deletions impact K8Ac and K10Ac to a similar degree (not shown). In many of these deletion backgrounds, the effect on Htz1Ac may be indirect, almost certainly in Rpd3L complex mutants where reduced K8Ac, K10Ac, and K14Ac is an unlikely outcome when inhibiting a deacetylase.
We chose Asf1 as a novel Htz1Ac regulator to examine further. Deletion of this H3-H4 chaperone reduces the level of each Htz1Ac without affecting the abundance of the histone (Fig. 4A). Each acetylation takes place after Htz1 is assembled into chromatin (Fig. 1C), so we tested whether asf1Δ impacts the deposition and/or acetylation step(s). In fractionation analyses, the chromatin of asf1Δ cells shows WT levels of total Htz1 but a reduction in each Htz1Ac (Fig. 4B). Thus Asf1 appears to regulate the Htz1 acetylation step. Asf1 is multi-functional and among other roles regulates various acetylations on histones H3 and H4 (H3-K9Ac, K27Ac, K56Ac; H4-K5Ac, K8Ac, K12Ac; see Fig. 3C (37)). We thus tested whether Htz1Ac was dependent on the prior modification of any of these residues using a range of mutant strains with unacetylatable alanine at each position (i.e. H3-K9A, K27A, K56A; H4-K5A, K8A, K12A (38)). In each mutant the abundance of each Htz1Ac was comparable with WT (supplemental Fig. S5).
FIGURE 4.
Htz1Ac is regulated in asf1Δ cells by an indirect mechanism. A, each Htz1Ac (but not total Htz1.HA3) is decreased in asf1Δ cells. Rpn8 serves as a loading control. B, asf1Δ reduces each Htz1Ac but has no effect on the amount of total Htz1 associated with chromatin. WT and asf1Δ cells were spheroblasted (total, T); fractionated into cytoplasm (Cy), nucleus (N), and chromatin (Ch); and immunoblotted as indicated. Appropriate localization of H2B and Rpn8 indicates efficient fractionation (as in Fig. 1C). C, schematic depicts the various roles of Asf1, a histone H3-H4 donor to various acetyltransferase complexes (substrates indicated), CAF/proliferating cell nuclear antigen (PCNA) at replication forks (for replication-dependent deposition (RDD)), and HIR in actively transcribed regions (for replication-independent deposition (RID)). D, glucose (final concentration, 2%) was added to repress GAL1p-driven transcription in GAL1p.ASF1.HA3 or GAL1p.ASF1.HA3.PEST strains, with samples taken for WCE isolation and immunoblotting at times indicated. Rpn8 serves as a loading control. E, GAL1p.ASF1.HA3 or GAL1p.ASF1.HA3.PEST strains were maintained in glucose (G, 2%) or galactose/raffinose (GR, 2%/1%) containing medium for >24 h before the samples were collected for WCE isolation and immunoblotting. Rpn8 serves as a loading control.
Given the multi-functionality of Asf1, it could prove difficult to identify whether a specific role is required for efficient Htz1Ac. We thus took an approach where Asf1 was rapidly removed from cells, and each Htz1Ac was monitored over a succeeding time course. We used two shut-off strains, namely GAL1p.ASF1.HA3 and GAL1p.ASF1.HA3.PEST as the only source of the factor, with ASF1 transcription in each repressed by glucose addition (39). The Cln2 C-terminal PEST domain confers a rapid, cell cycle-independent turnover on heterologous proteins (40), and its addition to Asf1.HA3 significantly shortens protein half-life (t½ Asf1.HA3 > 60 min; t½ Asf1.HA3.PEST < 15 min; Fig. 4D) (39). However, the abundance of each Htz1Ac (K8Ac, K10Ac, or K14Ac) remained unchanged over each 6-h time course (i.e. more than three complete cell cycles under experimental conditions) rather than reducing as expected (Fig. 4D). This is not a strain background issue because their prolonged (> 24 h) incubation in glucose-containing medium recapitulates the original observation of reduced Htz1Ac (Fig. 4E). Thus the reduced Htz1Ac in asf1Δ cells may not be due to a direct regulation by Asf1 of Htz1 acetylation but rather may involve some adaptation to the chronic absence of the histone chaperone. This also raises a cautionary note about overinterpreting the relationship of those additional Htz1 regulators identified by the proteome screening approach (supplemental Table S3).
Htz1 Acetylation Regulates the Response to Microtubule Depolymerizing Agents
The deletion of H2A.Z in S. cerevisiae or S. pombe leads to genome instability, synthetic genetic interactions with components of the kinetochore and spindle checkpoint machineries, and sensitivity to benomyl, a microtubule-depolymerizing agent (Figs. 2D and 5A) (11, 16, 41). H2A.Z acetylation contributes to the genome stability role in each species, with strains containing unacetylatable alleles showing increased rates of chromosome loss (11, 16).
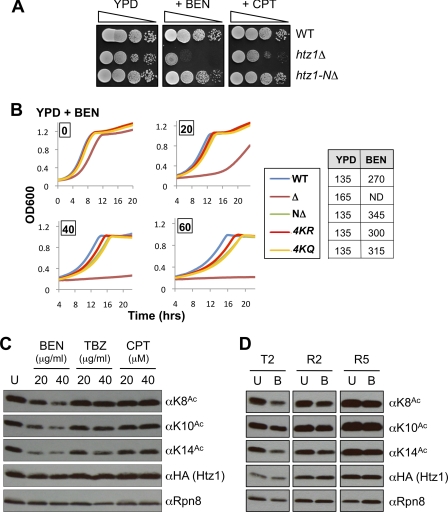
FIGURE 5.
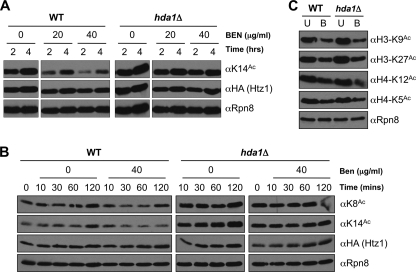
Htz1Ac is reduced in response to benomyl. A, htz1Δ (and to a lesser degree NΔ) cells are sensitive to benomyl (BEN, 15 μg/ml). In contrast htz1Δ but not NΔ cells are sensitive to camptothecin (CPT, 20 μm). Spots are 10-fold serial dilutions on YPD plates ± each agent after 2 days at 30 °C. B, unacetylatable htz1 strains (NΔ, 4KR, or 4KQ) are weakly sensitive to benomyl. Growth curves were analyzed on a Bioscreen (see “Experimental Procedures”) during culture in YPD + benomyl (0–60 μg/ml as indicated). The table indicates the time taken (in minutes) for each strain to traverse A600 0.45 to 0.9 (i.e. exponential part of growth curve) in YPD ± benomyl (60 μg/ml). ND, not determined. C, each Htz1Ac is reduced on benomyl treatment. WT cells were grown in YPD to A600 ∼0.5 and benomyl, TBZ, or camptothecin was added (concentrations indicated) before growth for another 2 h. WCEs were harvested and immunoblotted as indicated. Rpn8 is a loading control. U, untreated. D, the decrease of each Htz1Ac in response to benomyl is reversible. WT cells were grown in YPD to A600 ∼0.5, and benomyl was added (40 μg/ml) for 2 h (T2). The cells were then washed, resuspended in YPD, and allowed to recover for 2 (R2) or 5 (R5) h. WCEs were harvested and immunoblotted as indicated. Rpn8 is a loading control. U, untreated; B, benomyl.
To further examine how individual acetylations on Htz1 may regulate benomyl resistance, we tested how the full range of unacetylatable alleles (htz1Δ, NΔ, 4KR, K3R, K3Ac*, etc.) responded to this agent. Rather than spotting onto plates, growth curves were monitored in liquid culture because the latter approach can more easily distinguish the reduced fitness of htz1Δ relative to WT on rich media (YPD; Fig. 5, compare A and B). In this manner completely unacetylatable htz1 alleles (NΔ, 4KR or 4KQ) show a dose-dependent sensitivity to benomyl, although not to the same degree as htz1Δ (Fig. 5B). We (and others) had previously reported an htz1-K14R strain as sensitive to benomyl (16, 32). However, we have been unable to consistently repeat this phenotype in current analyses using any single unacetylatable (e.g. K14R) or acetylatable (e.g. K14Ac*) htz1 allele (not shown). This may be related to strain differences or some other as yet uncharacterized factor.
Because Htz1Ac contributes to benomyl resistance, we next asked whether the acetylations are themselves regulated by the agent. In parallel analyses, cells were treated with thiabendazole (TBZ, another microtubule depolymerizer) or camptothecin (a topoisomerase I inhibitor). These were chosen for comparison because htz1Δ (and to a lesser degree NΔ, 4KR and 4KQ) strains are sensitive to thiabendazole, whereas htz1Δ shows sensitivity to camptothecin under some conditions (i.e. on plates but not in liquid culture: Fig. 5A and supplemental Fig. S6). The cells were exposed to each agent for 2 h, and the abundance of Htz1 and each acetylated form (K8Ac, K10Ac, K14Ac) was determined. Under these conditions each Htz1Ac was reduced in a dose-dependent manner by benomyl but unchanged in response to thiabendazole or camptothecin (Fig. 5C). The reduction in Htz1Ac by benomyl is post-translational, because it is not accompanied by a change in Htz1 levels (Fig. 5C). It is also reversible, largely returning to normal 2 h after removal of the agent (Fig. 5D).
Benomyl could modulate Htz1Ac levels by various means, including impacting Htz1 deposition (by Swr1; Fig. 1C), acetylation (by Esa1; Fig. 1D), or deacetylation (by Hda1; Fig. 1F). We thus exposed WT or hda1Δ cells to benomyl as above and determined that those lacking the deacetylase (or its accessory subunits Hda2 and Hda3) show no reduction in any Htz1Ac (Fig. 6A, supplemental Fig. S7A, and data not shown). Benomyl has no effect on the abundance of any members of the HDA complex (Hda1, Hda2, or Hda3; supplemental Fig. S7, B and C), so the agent is likely to regulate either its activity or localization. In addition, the response is rapid, with Htz1Ac levels visibly reduced within 10 min of exposure to benomyl and bottoming out by 30 min (Fig. 6B).
FIGURE 6.
The benomyl-induced decrease of each Htz1Ac is Hda1-dependent. A, cultures were grown in YPD to A600 ∼0.5, benomyl was added (40 μg/ml) and incubated as indicated before WCEs were harvested and immunoblotted. B, Htz1Ac is reduced within 30 min of benomyl treatment. The cultures were grown in YPD to A600 ∼0.5 before benomyl addition (40 μg/ml). WCEs were harvested and immunoblotted as indicated. C, acetylations on histones H3 (K9Ac and K27Ac) and H4 (K5Ac and K12Ac) are reduced by benomyl but independently of Hda1. The cultures were grown in YPD to A600 ∼0.5 before benomyl addition (40 μg/ml, 2 h). WCEs were harvested and immunoblotted as indicated. U, untreated; B, benomyl.
Finally we determined whether the benomyl-induced deacetylation of Htz1 is the sole chromatin change in response to this agent. Of four acetylations tested (H3-K9Ac, H3-K27Ac, H4-K5Ac, and H4-K12Ac), all are reduced in response to benomyl (Fig. 6C). However, each is independent of Hda1 (Fig. 6C), even although H3-K9Ac and K27Ac are substrates of this deacetylase at Tup1-repressed promoters (41). Thus benomyl induces widespread changes in histone acetylation and likely via multiple mediators. The benomyl sensitivity of strains containing unacetylatable htz1 alleles suggests that deacetylation of this histone variant is one component of the effective cellular response to this agent.
DISCUSSION
Histones are subject to a bewildering array of covalent modifications (42). In this work we set out to test whether each acetylation on a single histone tail could mediate a specific function. We examined the acetylations of H2A.Z (S. cerevisiae Htz1) rather than of a major histone because the variant provides a number of advantages. It is encoded by a single, nonessential gene, and the protein has a dedicated deposition machinery (the SWR-C) and limited genomic distribution (e.g. highly enriched in the nucleosomes flanking the nucleosome-free region over promoters (7, 9)). In addition Htz1 is multiply acetylated and multi-functional. It has thus been posited that specific acetylated forms could regulate distinct roles (14–16). We find that this is not the case: rather the modifications are redundant, with any single lysine on the Htz1 tail sufficient to mediate function. However, we also show that the acetylation of Htz1 is highly regulated and required for full functionality of this histone variant.
N-terminal Acetylation Is a Widespread Modification of H2A.Z Orthologs
The H2A.Z N terminus is acetylated in S. cerevisiae (14–16), S. pombe (11), Tetrahymena thermophila (43, 44), and metazoans (13, 45, 46). In each organism multiple acetyl-lysines are possible: up to four in budding and fission yeasts, five in metazoans, and six in Tetrahymena. However, these acetylations are unequally distributed across all potential lysine substrates in each species. For example, in Sc K3Ac, K8Ac, K10Ac, and K14Ac are ∼3, 7, 14, and 38% of total Htz1, respectively (15). In contrast the histone is ∼90% unmodified in chicken erythrocytes, with the remainder primarily mono-acetylated on the more N-terminal lysines (K4Ac ∼ K7Ac > K11Ac > K13Ac ≫ K15Ac) (2, 13). Thus the yeast and chicken H2A.Z N-terminal tails differ in both the abundance and precise location of their acetylations. This, coupled with the fact that the N terminus is one of the most variable regions across H2A.Z orthologs, would tend to suggest differences in how these modifications are regulated. Despite this, the respective Kat5 family acetyltransferase appears to be the primary H2A.Z modifier in all tested species, i.e. S. pombe Mst1 (11), Drosophila Tip60 (47), or Caenorhabditis elegans MYS-1 (48). This is certainly the case in Sc, with Htz1-K8Ac, -K10Ac, and -K14Ac each dependent on Esa1 (the budding yeast Kat5 homolog) and multiple members of the Esa1-containing NuA4 complex (Fig. 1, D and E) (14, 16). Esa1 thus shows great promiscuity in its ability to acetylate each lysine on the Htz1 N terminus (MSGKAHGGKGKSGAKD). However, the acetyltransferase is not infinitely capable, with an N-terminal HA3-tagged form of Htz1 being very poorly acetylated (not shown). N-terminal GFP tagging of Drosophila H2A.Z also disrupts its acetylation and compromises the ability of the variant to regulate the heat shock response (45). Alternatively, because the Htz1 N terminus (residues 1–53) regulates nuclear localization (49), N-terminal tagging of the histone may simply interfere with nuclear import and thus availability for nucleosome assembly and subsequent acetylation.
Coregulation and Genetics Indicate Minimal Specialization of Each Htz1Ac
Disparate analyses demonstrate that the individual N-terminal acetylations on Htz1 are coregulated. Thus K8Ac, K10Ac, and K14Ac are added by NuA4 acetyltransferase after Htz1 is inserted into nucleosomes by the SWR-C (Fig. 1, C–E) and subsequently removed by the Hda1 deacetylase complex (Fig. 1F and supplemental Fig. S7). All three acetylations decay with similar kinetics after Htz1 transcription is repressed (Fig. 3). All three are similarly affected in every mutant background tested (Fig. 4 and supplemental Table S3). Finally, all three acetylations are similarly effected by a diverse range of agents, including 6-azauracil, benomyl, camptothecin, mycophenolic acid, and TBZ (Fig. 5C and supplemental Fig. S8).
Their coregulation strongly suggests that individual Htz1 N-terminal acetylations are redundant, a proposal supported by comparing the genetic profiles of a comprehensive range of htz1 alleles (Fig. 2). If the cell utilizes each individual acetylation for a specific role, we would expect to see distinct defects (and thus unique genetic interactions) associated with the mutation of these residues. However, completely unacetylatable mutants (NΔ, 4KR, and 4KQ) are highly correlated with htz1Δ, whereas singly mutated (e.g. K3R) or singly acetylatable (e.g. K3Ac*) alleles cluster closer to HTZ1 (Fig. 2A). This suggests that a significant degree of the function of Htz1 resides in its N terminus or more specifically in the acetylatable lysines of this region. Furthermore it is strong evidence against an individual function for each N-terminal acetylation.
How Does Acetylation Regulate Htz1 Function?
Previous analyses have identified some role for Htz1 acetylation in the maintenance of euchromatin-heterochromatin boundaries (14), transcription (15), and chromosome transmission (16). Gene ontology categories covering these functions are among the most highly represented when the genetic interactions of completely unacetylatable htz1 alleles are examined (supplemental Fig. S4). In addition, and consistent with a role in chromosome stability, unacetylatable htz1 alleles are sensitive to the microtubule destabilizing agents benomyl and TBZ (Fig. 5, A and B, and supplemental Fig. S6, D and E). One possibility is that Htz1Ac somehow regulates the response to spindle stress; this is supported by the observation that each acetylation is regulated by the HDA complex on exposure to benomyl (Fig. 5C). This could in turn suggest how this deacetylase complex regulates normal chromosome segregation (50).
Approximately 30% of the genetic interactions of htz1Δ are conserved with NΔ or 4KR (Fig. 2 and supplemental Fig. S4). However, the list of interactors with unacetylatable htz1 alleles are not simply a subset of those with the complete deletion. The htz1Δ-specific group may be due to a loss of the Htz1 core domain or C terminus. Alternatively it may be caused by the inappropriate activity of SWR-C in the absence of Htz1 (51, 52). However, the genetic interactors unique to unacetylatable htz1 alleles are more difficult to explain. H2A.Z acetylation does not regulate the incorporation of the histone into chromatin in either Sc (supplemental Fig. S3A) or S. pombe (11). In addition the acetylation status of Htz1 does not impact nucleosome turnover (Fig. 3C). It is possible that nucleosomes containing unacetylatable htz1 are unable to use an H2AAc-dependent compensation pathway. This might indicate that cells differentiate certain histone tails as one of two states, acetylated or not, and make little distinction between the particular modified residue. This would appear to be a retrograde step in our understanding of chromatin function. However, we note that NuA4 acetylates Htz1 (Fig. 1, D and E), H2A, and H4 (53), and mutant combinations that reduce the total number of NuA4-dependent acetylations (e.g. [htz1–4KR, eaf1Δ] or [htz1–4KR, hhf2-K5/8/12R]) are invariably synthetic (14). In addition the Esa1-targeted H4 N terminus also shows significant redundancy, such that the loss of all four acetylatable lysines leads to slow growth and camptothecin sensitivity, but the retention of any single lysine complements both phenotypes (54). Furthermore an ectopic lysine in an otherwise unacetylatable H4 N terminus is acetylated (likely by Esa1) and also repairs the slow growth and camptothecin sensitivity (54).
As noted above, the simultaneous loss of all N-terminal acetylation on Htz1 has widespread effects. Thus the modifications regulate the functions of this histone variant. However, is this by charge modulation or effector recruitment? Arginine and glutamine are respectively considered charge mimics for unacetylated and acetylated lysine (13), although there is no evidence that they mimic the unmodified/modified state as it relates to recognition by acetyl-binding proteins. We note that the htz1–4KR and 4KQ mutants are indistinguishable throughout this work, an observation also made with comparable mutants in fission yeast (11). This strongly argues against the acetylation of Htz1 acting via charge modulation. However, no chromatin effector has yet been identified that is so selectively promiscuous in its binding (i.e. only to KAcs on a H2A.Z tail). One possibility is that the Htz1 tail functions by dynamic charge modulation; i.e. the acetylations must be added and then removed (constitutively one or the other will not work). If this is indeed the case, our current belief that the simple presence of many histone marks is enough may need some revision.
Supplementary Material
Acknowledgments
We thank Steve Buratowski (Harvard Medical School), Jeffrey Fillingham (Ryerson University), Dan Finley (Harvard Medical School), Jack Greenblatt (University of Toronto), Jessica Tyler (University of Colorado, Denver), Fred Winston (Harvard Medical School), and Ali Shilatifard (Stowers Institute) for the generous supply of antibodies and yeast strains (detailed in supplemental Tables S1 and S2). We also thank Brian Strahl (University of North Carolina, Chapel Hill), Joseph Wade (Wadsworth Center), Jon Warner (AECOM), and members of the Krogan and Keogh labs for advice and comments.
This work was supported, in whole or in part, by the National Institutes of Health. This work was also supported by the Searle and Keck Foundations, by National Cancer Institute Cancer Centre Grant 2P30CA013330 to Albert Einstein College of Medicine, and by the Speaker's Fund for Biomedical Research: Toward the Science of Patient Care awarded by the City of New York.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S8.
- Sc
- S. cerevisiae
- SWR-C
- SWR complex
- WCE
- whole cell extract
- TBZ
- thiabendazole
- CC
- correlation coefficient
- SS/SL
- synthetic sick/lethal.
REFERENCES
- 1.Luger K. (2003) Curr. Opin. Genet. Dev. 13, 127–135 [DOI] [PubMed] [Google Scholar]
- 2.Dryhurst D., Ishibashi T., Rose K. L., Eirín-López J. M., McDonald D., Silva-Moreno B., Veldhoen N., Helbing C. C., Hendzel M. J., Shabanowitz J., Hunt D. F., Ausió J. (2009) BMC Biol. 7, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henikoff S., Furuyama T., Ahmad K. (2004) Trends Genet 20, 320–326 [DOI] [PubMed] [Google Scholar]
- 4.Latham J. A., Dent S. Y. (2007) Nat. Struct. Mol. Biol. 14, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 5.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caterino T. L., Hayes J. J. (2007) Nat. Struct. Mol. Biol. 14, 1056–1058 [DOI] [PubMed] [Google Scholar]
- 7.Mehta M., Kim H. S., Keogh M. C. (2010) J. Biol. 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raisner R. M., Madhani H. D. (2006) Curr. Opin. Gen Dev 16, 119–124 [DOI] [PubMed] [Google Scholar]
- 9.Zlatanova J., Thakar A. (2008) Structure 16, 166–179 [DOI] [PubMed] [Google Scholar]
- 10.Zofall M., Fischer T., Zhang K., Zhou M., Cui B., Veenstra T. D., Grewal S. I. (2009) Nature 461, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H. S., Vanoosthuyse V., Fillingham J., Roguev A., Watt S., Kislinger T., Treyer A., Carpenter L. R., Bennett C. S., Emili A., Greenblatt J. F., Hardwick K. G., Krogan N. J., Bähler J., Keogh M. C. (2009) Nat. Struct. Mol. Biol. 16, 1286–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thambirajah A. A., Li A., Ishibashi T., Ausió J. (2009) Biochem. Cell Biol. 87, 7–17 [DOI] [PubMed] [Google Scholar]
- 13.Ishibashi T., Dryhurst D., Rose K. L., Shabanowitz J., Hunt D. F., Ausió J. (2009) Biochemistry 48, 5007–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babiarz J. E., Halley J. E., Rine J. (2006) Genes Dev. 20, 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millar C. B., Xu F., Zhang K., Grunstein M. (2006) Genes Dev. 20, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keogh M. C., Mennella T. A., Sawa C., Berthelet S., Krogan N. J., Wolek A., Podolny V., Carpenter L. R., Greenblatt J. F., Baetz K., Buratowski S. (2006) Genes Dev. 20, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storici F., Resnick M. A. (2006) Methods Enzymol. 409, 329–345 [DOI] [PubMed] [Google Scholar]
- 18.Tong A., Boone C. (2006) Methods Mol. Biol. 313, 171–192 [DOI] [PubMed] [Google Scholar]
- 19.Keogh M. C., Cho E. J., Podolny V., Buratowski S. (2002) Mol. Cell. Biol. 22, 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauss R., Trautwein M., Sommer T., Spang A. (2005) Yeast 22, 1–12 [DOI] [PubMed] [Google Scholar]
- 21.Tong A. H., Boone C. (2006) Methods Mol. Biol. 313, 171–192 [DOI] [PubMed] [Google Scholar]
- 22.Fillingham J., Recht J., Silva A. C., Suter B., Emili A., Stagljar I., Krogan N. J., Allis C. D., Keogh M. C., Greenblatt J. F. (2008) Mol. Cell. Biol. 28, 4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 24.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 25.Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., Newman H. W., Braun S., Madhani H. D., Krogan N. J., Weissman J. S. (2008) Nat. Methods 5, 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins S. R., Schuldiner M., Krogan N. J., Weissman J. S. (2006) Genome Biol. 7, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., Krogan N. J. (2005) Cell 123, 507–519 [DOI] [PubMed] [Google Scholar]
- 28.Keogh M. C., Kim J. A., Downey M., Fillingham J., Chowdhury D., Harrison J. C., Onishi M., Datta N., Galicia S., Emili A., Lieberman J., Shen X., Buratowski S., Haber J. E., Durocher D., Greenblatt J. F., Krogan N. J. (2006) Nature 439, 497–501 [DOI] [PubMed] [Google Scholar]
- 29.Millar C. B., Grunstein M. (2006) Nat. Rev. Mol. Cell Biol. 7, 657–666 [DOI] [PubMed] [Google Scholar]
- 30.Krogan N. J., Keogh M. C., Datta N., Sawa C., Ryan O. W., Ding H., Haw R. A., Pootoolal J., Tong A., Canadien V., Richards D. P., Wu X., Emili A., Hughes T. R., Buratowski S., Greenblatt J. F. (2003) Mol. Cell 12, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 31.Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. (2004) Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 32.Lin Y. Y., Qi Y., Lu J. Y., Pan X., Yuan D. S., Zhao Y., Bader J. S., Boeke J. D. (2008) Genes Dev. 22, 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuldiner M., Collins S. R., Weissman J. S., Krogan N. J. (2006) Methods 40, 344–352 [DOI] [PubMed] [Google Scholar]
- 34.Fiedler D., Braberg H., Mehta M., Chechik G., Cagney G., Mukherjee P., Silva A. C., Shales M., Collins S. R., van Wageningen S., Kemmeren F., Holstege F. C., Weissman J. S., Keogh M. C., Koller D., Shokat K. M., Krogan N. J. (2009) Cell 136, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dion M. F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O. J. (2007) Science 315, 1405–1408 [DOI] [PubMed] [Google Scholar]
- 36.Schneider J., Dover J., Johnston M., Shilatifard A. (2004) Methods Enzymol. 377, 227–234 [DOI] [PubMed] [Google Scholar]
- 37.Ransom M., Dennehey B. K., Tyler J. K. (2010) Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakanishi S., Sanderson B. W., Delventhal K. M., Bradford W. D., Staehling-Hampton K., Shilatifard A. (2008) Nat. Struct. Mol. Biol. 15, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zabaronick S. R., Tyler J. K. (2005) Mol. Cell. Biol. 25, 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mateus C., Avery S. V. (2000) Yeast 16, 1313–1323 [DOI] [PubMed] [Google Scholar]
- 41.Wu J., Suka N., Carlson M., Grunstein M. (2001) Mol. Cell 7, 117–126 [DOI] [PubMed] [Google Scholar]
- 42.Lall S. (2007) Nat. Struct. Mol. Biol. 14, 1110–1115 [DOI] [PubMed] [Google Scholar]
- 43.Ren Q., Gorovsky M. A. (2001) Mol. Cell 7, 1329–1335 [DOI] [PubMed] [Google Scholar]
- 44.Ren Q., Gorovsky M. A. (2003) Mol. Cell. Biol. 23, 2778–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanabe M., Kouzmenko A. P., Ito S., Sawatsubashi S., Suzuki E., Fujiyama S., Yamagata K., Zhao Y., Kimura S., Ueda T., Murata T., Matsukawa H., Takeyama K., Kato S. (2008) Genes Cells 13, 1279–1288 [DOI] [PubMed] [Google Scholar]
- 46.Bruce K., Myers F. A., Mantouvalou E., Lefevre P., Greaves I., Bonifer C., Tremethick D. J., Thorne A. W., Crane-Robinson C. (2005) Nucleic Acids Res. 33, 5633–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., Yates J. R., 3rd, Abmayr S. M., Washburn M. P., Workman J. L. (2004) Science 306, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 48.Updike D. L., Mango S. E. (2006) PLoS Genetics 2, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straube K., Blackwell J. S., Jr., Pemberton L. F. (2010) Traffic 11, 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanta H., Laprade L., Almutairi A., Pinto I. (2006) Genetics 173, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morillo-Huesca M., Clemente-Ruiz M., Andújar E., Prado F. (2010) PLoS One 5, e12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halley J. E., Kaplan T., Wang A. Y., Kobor M. S., Rine J. (2010) PLoS Biol. 8, e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M. (2001) Mol. Cell 8, 473–479 [DOI] [PubMed] [Google Scholar]
- 54.Bird A. W., Yu D. Y., Pray-Grant M. G., Qiu Q., Harmon K. E., Megee P. C., Grant P. A., Smith M. M., Christman M. F. (2002) Nature 419, 411–415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.