Abstract
The Mg2+-induced folding of RNA tertiary structures is readily observed via titrations of RNA with MgCl2. Such titrations are commonly analyzed using a site-binding formalism that includes a parameter, the Hill coefficient n, which is sometimes deemed the number of Mg2+ ions bound by the native RNA at specific sites. However, the long-range nature of electrostatic interactions allows ions some distance from the RNA to stabilize an RNA structure. A complete description of all interactions taking place between Mg2+ and an RNA uses a preferential interaction coefficient, Γ2+, which represents the 'excess' Mg2+ neutralizing the RNA charge. The difference between Γ2+ for the native and unfolded RNA forms (ΔΓ2+) is the number of Mg2+ ions ‘taken up’ by an RNA upon folding. Here we ask under what conditions the Hill coefficient n can be equated to the ion uptake ΔΓ2+, and find that two approximations are necessary: (i) the Mg2+ activity coefficient is independent of concentration during a titration, and (ii) the dependence of ΔΓ2+ on Mg2+ concentration is weak. Titration experiments with a Mg2+-binding dye and an adenine-binding riboswitch were designed to test these approximations. Inclusion of a 30-fold excess of KCl over MgCl2 was sufficient to maintain a constant Mg2+ activity coefficient. We also observed that Mg2+ uptake by the RNA varied from near zero to ~2.6 as the Mg2+ concentration increases over ~100-fold range. It is possible to determine ΔΓ2+ from Mg2+ - RNA titrations, but the values are only applicable to a limited range of solution conditions.
For many years, researchers have studied the striking dependence of RNA tertiary structure stability on Mg2+ ions (1–3). The Schimmel laboratory was one of the first to consider the effects of Mg2+ on RNA folding reactions using equations originally derived to describe ligands binding to a fixed number of specific sites on a multisubunit protein (i.e., the Hill equation) (4). Since then, it has become customary to think about ion-RNA interactions as binding events characterized by equilibrium constants and fixed stoichiometries—an approach we refer to as the ‘binding formalism’. In particular, the Hill equation and Hill coefficient (5) have become a standard means for characterizing Mg2+-induced RNA folding reactions and are often interpreted in terms of the binding formalism. But Schimmel himself recognized that “these equations are, essentially, semiempirical and as such provide little insight into the actual mechanism of the binding equilibria. Nevertheless, in order to catalog the information and to compare results of various investigations, they provide a useful common framework” (6). The question remains as to whether the adjustable variables of the Hill equation have meaningful molecular interpretations or should be considered simply empirical parameters.
A thorough consideration of ion effects on RNA folding must take into account the fact that ions interacting with an RNA can experience a variety of different environments, ranging from partially dehydrated ions essentially buried within the RNA to fully hydrated ions some distance from the RNA surface (7). The binding formalism presupposes a model that excludes the possibility of ions interacting via long-range electrostatic interactions, and therefore does not provide a complete description of Mg2+-RNA interactions. Consequently it has been useful to develop a more general formalism for addressing the effects of ions on RNA stability—one that does not specify any particular model of ion-RNA interactions.
A general approach for describing interactions between ions and macromolecules is based on parameters known as preferential interaction coefficients (8). We previously extended this formalism to address the effect of Mg2+ on RNA folding reactions, and derived an equation that simplifies to the form of the Hill equation when two approximations are made (9, 10). Where the approximations are valid, the Hill coefficient n quantifies the 'uptake' of Mg2+ ions that accompany a folding reaction, but has a different molecular interpretation than that attributed to n by the binding formalism.
In the present paper, we experimentally test the two approximations necessary for interpretation of the Hill coefficient. In the first approximation, Mg2+ concentrations are substituted for thermodynamic activities. Because of the strong interactions taking place between ions (e.g. Mg2+ and Cl−), the effective concentration of an ion—known as its activity—is usually very different from its concentration. We present experiments showing how the inclusion of a monovalent salt (such as KCl) can suppress the errors that can arise when Mg2+ concentrations are used in the Hill equation. The second approximation is that the Hill coefficient is a constant, independent of the concentration of Mg2+ present in solution. Using two independent methods for measuring the ion uptake that accompanies folding of a riboswitch RNA, we find that it varies from nearly zero to a maximum of ~2.6 ions per RNA as the Mg2+ concentration required to fold the RNA increases. This strong Mg2+-dependence of the Hill coefficient restricts use of the Hill equation to analysis of folding data over a narrow Mg2+ concentration range, and prevents extrapolation of the derived coefficient to other Mg2+ concentrations.
Background
The empirical Hill equation
The Hill equation was originally proposed as an empirical way to fit titration data. It has the general form
| (1) |
where θ is the extent of a binding reaction, normalized to values between 0 and 1, C is the molar concentration of a ligand, and K and n are empirical parameters (5). If n is fixed at 1, the equation simplifies to a standard isotherm for the binding of a ligand to a single site on a macromolecule. When n is allowed to vary, the equation is able to fit any set of titration data for which the apparent free energy of a macromolecular conformational change has an approximately linear dependence on the log of the titrant concentration (see eqs 9 and 10, below). This condition frequently holds when the effects of salt on nucleic acids are considered (8, 11). In this section, we outline two different approaches that have been used to describe Mg2+-induced RNA folding. Both ultimately yield a relation with the form of eq 1, but the approaches are based on different premises, apply different approximations, and have divergent interpretations of the empirical parameters K and n.
Derivation of the Hill equation from a preferential interaction formalism
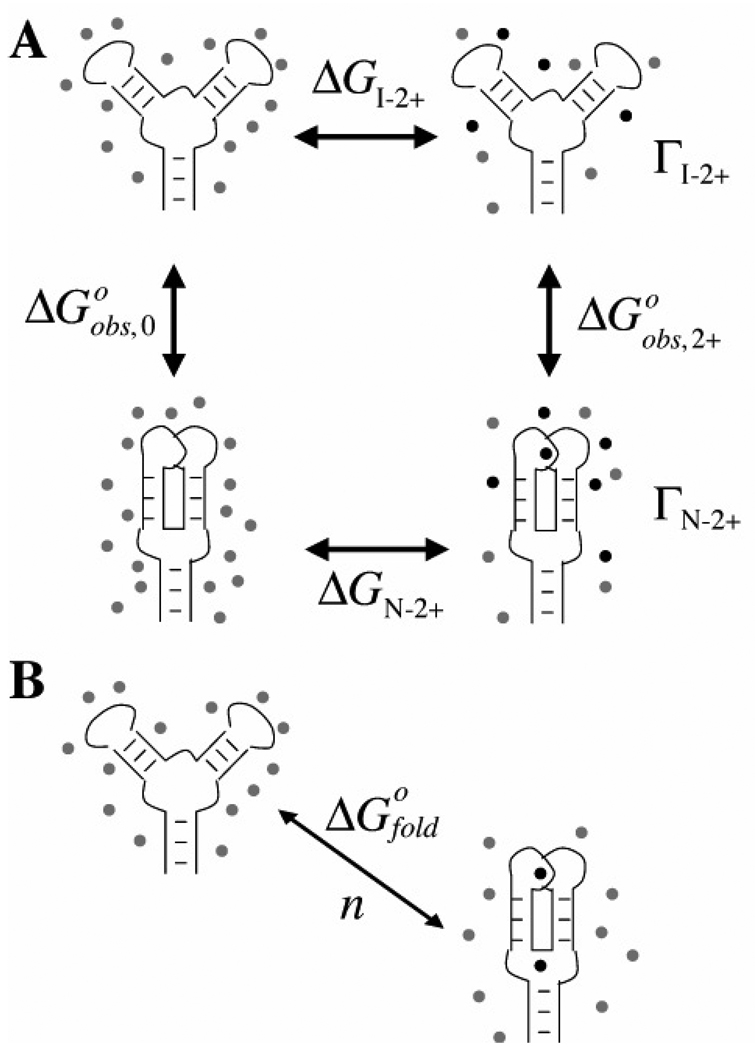
We start with a general scheme that formally distinguishes RNA folding from Mg2+ – RNA interactions. A thermodynamic cycle (Figure 1A) allows the definition of unambiguous free energies for these two aspects of Mg2+-induced RNA folding. Structures in the top row represent partially unfolded forms of the RNA, the so-called “intermediate” or I state, which contains only secondary structure. The vertical arrows represent tertiary contact formation—the folding of the I state to the native structure (N state)—in the presence (right) or absence (left) of Mg2+ ions. The free energies associated with the vertical arrows are the energies typically measured experimentally, based on the ratio of folded and unfolded RNA concentrations (CN and CI respectively) present at a specific concentration of Mg2+:
| (2) |
The horizontal arrows represent the interactions of the I state and the N state with Mg2+. The Γ2+ terms are parameters, sometimes called 'preferential interaction coefficients', that quantitate the accumulation of excess Mg2+ ions by the native or intermediate forms of the RNA, as defined below. From the way the cycle is drawn, nothing is implied about the nature of the interactions between the Mg2+ ions and the RNA.
Figure 1.
Two schemes for describing the effect of Mg2+ ions on an RNA folding reaction. In each panel, the RNA in the top row represents the intermediate (I) state containing only secondary structure, and the bottom row diagrams the native (N) state with tertiary structure. Gray and black dots represent excess monovalent and Mg2+ ions, respectively. A) Thermodynamic cycle that distinguishes the free energies of Mg2+-RNA interactions (horizontal arrows) from RNA folding free energies (vertical arrows). The individual free energy (ΔG) and preferential interaction (Γ) terms are defined in the text. The free energy contribution of Mg2+ ion to the RNA folding reaction is defined as: ΔΔG2+ = ΔGN−2+ − ΔGI−2+ = ΔGobs,2+ − ΔGobs,0. B) The “cycle” assumed by the binding formalism (see eqs 8 and 9). Mg2+ ions bind only at specific sites on the RNA, and ΔGofold does not resolve the ion interaction free energy from the intrinsic folding free energy.
A good way to conceptualize the meaning of an interaction coefficient is to consider an equilibrium dialysis experiment. If an RNA solution is dialyzed against a buffer containing MgCl2, at equilibrium some 'excess' Mg2+ ions will accumulate inside the dialysis bag relative to the number of ions in an equivalent volume outside of the bag. (The Mg2+ concentration outside the bag is called the “bulk” concentration, written C2+ here.) There will also be an excess of monovalent cations inside the bag, and because of repulsive interactions, a deficiency of chloride ions. The number of excess cations or excluded anions per RNA is the preferential interaction coefficient for that ion. Because the net charge of a solution must be neutral, the relation
| (3) |
holds. In other words, the total negative charge on the RNA macromolecule, Z, is neutralized by an excess of cations (divalent Γ2+ and/or monovalent Γ+) and the exclusion of anions (note that Γ− is negative) (9). Clearly, Γ2+ cannot exceed 0.5 ions per RNA nucleotide.
Γ 2+ is formally defined as the number of Mg2+ ions that must be added along with an RNA to prevent a change in the chemical potential of Mg2+ in the solution:
| (4) |
where m2+ and mRNA are molal concentrations of Mg2+ and RNA respectively, and μ2+ is the chemical potential of Mg2+. In terms of an equilibrium dialysis experiment, this partial derivative has the following meaning: if the addition of one RNA molecule to the RNA solution is accompanied by Γ2+ ions, there will be no net flow of Mg2+ ions across the membrane (μ2+ is constant). At the relatively low concentrations of ions and RNA used in this work, molar and molal concentration scales are essentially equivalent; although Γ2+ is defined in terms of molal units, we will use molar units throughout this paper.
In principle, Γ2+ can be measured independently for the folded and unfolded forms of an RNA, yielding ΓN−2+ and ΓI−2+ as depicted in Figure 1A. The difference between them is ΔΓ2+, the net ion uptake upon folding. Because less and less Mg2+ accumulates with an RNA as the concentration of Mg2+ decreases, ΓN−2+, ΓI−2+, and ΔΓ2+ all approach zero as the concentration of Mg2+ goes to zero.
Elsewhere we have derived a linkage relationship between ΔΓ2+ and the observed free energy of RNA folding, ΔGobs, 2+ (9, 10):
| (5) |
In the second equality above, the molar bulk concentration of Mg2+ ion (C2+) has been substituted for the thermodynamic activity of MgCl2 (aMgCl2). The conditions under which this approximation is justified will be explored in Results. A summary of the relation between activity and concentration is at the end of this Background section.
Equation 5 is a linkage relation, one of a class of equations that describe the way a macromolecular equilibrium might be shifted by any small molecule, including solvent and ions as well as specific ligands. The application of linkage equations to all varieties of small molecules has been thoroughly considered by Record et al (8), and is based on a general approach first taken by Wyman (see section 6 of reference (12)). Of particular importance to the present work, the derivation makes no assumption about the nature of the interactions involved, whether short-range or long-range—a criterion for discussing electrostatic interactions.
Substitution of eq 2 into eq 5 followed by integration gives a relation with the form of the Hill equation,
| (6) |
where θfold = Kobs/(1+Kobs). The constant K is related to the value of C2+ at the midpoint of the titration curve, . In order to carry out the integration that yields eq 6, ΔΓ2+ must be treated as a constant. As mentioned above, ΔΓ2+ is expected to approach zero at low Mg2+ concentrations; therefore it is important to define the conditions under which the assumption of constant ΔΓ2+ is reasonable. This is the second approximation that will be experimentally tested in the Results.
Another useful equation that follows from eq 5 relates the preferential interaction coefficient Γ2+ to the free energy of Mg2+ – RNA interaction:
| (7) |
where ΔGRNA−2+ may apply to RNA in either the I or N state (Figure 1A) (9, 10). As implied by Figure 1A, ΔGoobs,2+ − ΔGoobs,0 = ΔGN−2+ − ΔGI−2+. Equation 7 includes the approximation that Mg2+ concentration can be substituted for MgCl2 activity. It is important to note that Γ2+ is a Mg2+-dependent variable and, because the lower limit of the integration is at C2+= 0 and Γ2+ = 0, it cannot be factored out of the integration.
Derivation of the Hill equation from a site-binding formalism
Currently, a widely used approach for analyzing a Mg2+-induced RNA folding experiment starts by assuming the equilibrium reaction depicted in Figure 1B:
| (8) |
where Kobs is the molar ratio of N and I state RNAs, as in eq 2. The stoichiometric coefficient n is considered the number of ions “bound” to the folded RNA structure, and C2+ is the concentration of 'free' (unbound) ions. We refer to eq 8 as a binding formalism, because it assumes that all ions can be classified as either bound to the RNA or completely non-interacting (free). (In some formulations an unspecified number of 'non-specific' ions are presumed to interact similarly with the I and N states, and therefore not affect the apparent RNA folding free energy (13).) Equation 8 is an approximation of the MWC model for the linkage of ligand binding to the conformational change of a macromolecule (14, 15), in which ligands are assumed to bind to the macromolecule in an infinitely cooperative, all-or-none fashion. As represented in Figure 1B, the RNA in this model can only adopt either the I state conformation without any associated ions, or the folded N state with a full complement of n bound ions. The model excludes the possibility that changes in long-range electrostatic interactions between the ions and the RNA might contribute to stabilization of the native state.
In using the binding formalism to analyze titrations of RNA with Mg2+, the expression for Kfold (eq 8) is either rearranged to the format of the Hill equation,
| (9) |
or differentiated to yield an expression for the Hill coefficient,
| (10) |
where ΔGobs, 2+ has the same meaning as in eq 2, and θfold is defined by eq 6. In these formulas, the empirical Hill coefficient n is interpreted as either the stoichiometric uptake of Mg2+ ions coupled to RNA folding or a measure of ion-binding cooperativity. The fitted Hill coefficient is then commonly used to extrapolate the free energy of folding to different solution conditions (13, 16, 17).
Equations 5 and 10 are similar in form, but n, interpreted as a stoichiometric coefficient, is assumed to be independent of the Mg2+ concentration, whereas the corresponding quantity ΔΓ2+ in eq 5 must approach zero at low Mg2+ concentrations (see comments on Γ2+ following eq 4). The experiments presented in Results will test how strongly ΔΓ2+ depends on Mg2+ concentration.
Activities vs. concentrations
Thermodynamic equilibrium constants and free energies are properly defined in terms of the activities of the components added to the reaction, rather than their concentrations. The activity of a species, sometimes called its ‘effective concentration,’ is related to its concentration by the activity coefficient,
| (11) |
If the activity coefficient γ is unity, the behavior is ideal and the concentration is equal to the activity. Solutions with moderate concentration of salts do not exhibit ideal behavior primarily because of strong long-range interactions between ions (such as the attraction between Mg2+ and Cl−), though all other sources of attractive and repulsive interactions (e.g. excluded volume, ion-pair formation) are also subsumed into γ. At the concentrations of salts typically encountered in RNA studies, the activity of MgCl2 is lower than its actual concentration due to mutual electrostatic “screening”: a Mg2+ ion surrounded by Cl− ions is less “effective” in solution, and vice versa. The screening becomes more effective as salt concentration increases, a phenomenon that causes the Mg2+ ion activity coefficient to vary strongly with the MgCl2 concentration. Wyman's derivation of linkage relations started with ligand activities and introduced concentrations as an approximation applicable to neutral molecules at low concentrations (12). As pointed out above, the derivation of eqs 5 – 7 includes an approximation in which the MgCl2 activity has been replaced by the molar bulk Mg2+ concentration (C2+) (9). In the Results, we show that this approximation is justified if an excess of a monovalent salt is included in the solution, such that the total Cl− concentration remains approximately constant as MgCl2 is added.
Materials and Methods
Materials and solution preparation
All solutions were prepared using distilled, deionized water at18.3 MΩ resistivity. High purity (> 99%) KCl, KOH, and MOPS were purchased from Fluka. 8-hydroxy-5-quinolinic acid (HQS1) was purchased from Sigma and recrystallized before use as described (18). Buffers used in the experiments were made accounting for the K+ present in the KOH used to adjust the buffer pH to 6.8 (1.4 mM KOH for 5 mM MOPS). Thus, all the Cl− concentrations were 1.4 mM less than the noted K+ concentrations in HQS titrations. KMOPS buffer was 5 mM MOPS, pH 6.8 for HQS titrations, and 20 mM MOPS, pH 6.8 for UV melting experiments and RNA-HQS titrations. Buffer solutions all contained 2 µM EDTA to scavenge heavy metals. At this concentration and pH, a negligible fraction of the added Mg2+ is bound to EDTA (K ~ 105 M−1), while transition metals are stoichiometrically bound (K ~ 1014 - 1024 M−1) (19). Riboswitch ligands (2,6-diaminopurine, 2-aminopurine, adenine, and purine) were purchased from Sigma and dissolved in 1% (v/v) HCl solutions (5 or 10 mM stocks depending on solubility). MgCl2 in hexahydrate form was purchased from Sigma. Due to the hygroscopic nature of this compound, MgCl2 solution concentrations were determined by stoichiometric titration of EDTA as described (18).
The A-riboswitch RNA used in the experiments was obtained by in vitro transcriptions with T7 phage RNA polymerase from a plasmid DNA template. The transcribed sequence was that of the add riboswitch from V. vulnificus (20) with a P1 stem sequence modified to enhance transcription efficiency and allow run-off transcription after plasmid cleavage with Sma I restriction endonuclease. The desired sequence was cloned into a pUC18 plasmid construct (pLL2) that has a T7 RNA polymerase promoter followed by a Stu I restriction site (15). The integrity of the cloned sequence was confirmed by DNA sequencing. Run-off transcriptions were purified on denaturing 12% polyacrylamide gels (18). Bands corresponding to RNA were excised and subject to electroelution to recover the sample, which was concentrated and extensively exchanged into the buffer of choice using Millipore MW3 Centricon filter units (Amicon). Before being used in titration experiments, RNA samples were renatured in KMOPS buffer that included the specified K+ and ligand concentrations, by heating to 65°C for 5 minutes and then sitting at room temperature for 15 minutes.
Spectroscopic titrations
Automated Mg2+ titrations of the fluorescent indicator dye HQS (50 µM) were carried out at 20 °C in an Aviv ATF-105 fluorimeter equipped with two computer-controlled Hamilton titrators dispensing the Mg2+ titrant. Samples were prepared in 1 cm × 1 cm cells and stirred continuously over the course of the titration. Binding curves were collected in standard 5 mM KMOPS buffer at various salt concentrations as specified in the figure legends. Titration data were fit to different binding isotherms, as specified, after selecting evenly spaced points on a log scale to avoid weighting different parts of the curve unequally. The data were then normalized by dividing by the maximum fluorescence returned by fitting of the isotherm. Residuals of fits of the data to the Hill equation were calculated in Kaleidagraph. Bootstrap analysis, as implemented by Regress+ 2.3 (causaScientia.org), revealed no systematic correlations in the data. The reported Hill coefficient errors are the standard deviations of at least three repeated experiments.
Similar automatic titrations of A-riboswitch RNA with MgCl2 were monitored by UV absorption in a Cary 400 spectrophotometer interfaced with a Hamilton titrator. Samples were assembled in 1 cm path length cuvettes, kept at 20 °C, and stirred continuously during experiments. The instrument was used in double beam mode, with a non-titrated reference cuvette containing the appropriate monovalent salt and ligand concentration. The MgCl2 titrants included exactly the same salt and ligand concentrations as the titrated sample. Absorbance data were collected at 260, 280, and 295 nm; for some ligand conditions, the change in the 280 nm signal was too small for reliable analysis. In the case of titrations performed in 2-aminopurine (2-AP), the substantial absorbance of this ligand at longer wavelengths precluded collection of the 295 nm signal. Ligand concentrations were chosen so as to be in sufficient excess over the RNA to minimize changes in the free ligand concentration over the course of the titration experiment. The titration data were fit to either of two formulas based on the Hill equation that included a linear baseline term, mbl, as well as initial and final absorbances, Ii and If, respectively:
| (12a) |
| (12b) |
In either equation, the term in brackets is θfold as defined by eq 6. Equation 12b was used when a significant fraction of the RNA was present in the native conformation before addition of the MgCl2 titrant. Kobs,0 is the equilibrium constant for folding in the absence of Mg2+ (eq 2), and the equivalent of the Hill coefficient is calculated from the fitted parameters as the derivative
| (13) |
Insufficient data could be collected prior to the folding transition to accurately determine an initial (I state) baseline. In eq 12 we assume that the same slope characterizes the folded and unfolded state baselines, but similar results are obtained for these data sets if the I state baseline is assigned a slope of zero. Omission of the baseline term had little or no effect (<4%) on the Hill coefficients obtained for data collected at 260 or 280 nm, but the term was essential for analysis of data collected at 295 nm.
The accumulation of Mg2+ by A-riboswitch RNA, known as the preferential interaction coefficient Γ2+ (eq 3), was measured via parallel fluorescence Mg2+-titrations of HQS in the presence and absence of RNA, as described in detail elsewhere (18). These titrations were carried out in the Aviv ATF-105 fluorimeter at 20 °C with the buffers specified in the legend to Figure 6. RNA concentrations were in the range 16 – 32 µM (1 – 2 mg/mL). Four data sets were collected for titrations performed in both the presence or absence of DAP ligand. The presence of DAP has no effect on the affinity of HQS for Mg2+ ion, as measured in control titrations. To average data from different titrations, Γ2+ values were linearly interpolated at the same regular concentration intervals in each data set, using about the same number of points (~200) as present in the original data set. The average and standard deviation for the four sets were then calculated at each of the interpolated concentrations.
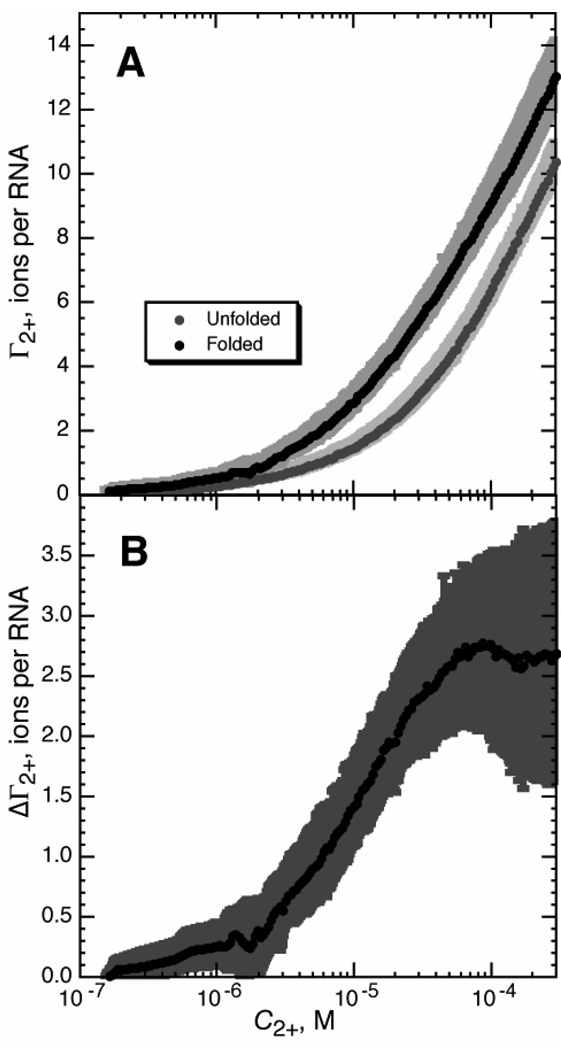
Figure 6.
Preferential interaction coefficients (Γ2+) for the A-riboswitch, as calculated from RNA-HQS titration experiments. Titrations were carried out at 20 °C in buffer containing 20 mM MOPS, pH 6.8, and 50mM K+. A) Γ2+ measured in the presence (black circles) or absence (gray circles) of 250 µM DAP. Error bars are standard deviations calculated from four interpolated data sets. B) ΔΓ2+, the difference between Γ2+ for folded and unfolded A-riboswitch RNA (panel A), as a function of Mg2+ concentration.
UV melting curves
Thermal denaturation of A-riboswitch RNA was carried out in the Cary 400 spectrophotometer using the following program of temperature changes: a renaturation stage (5 minutes at 65 °C followed by 1 deg/min cooling to 5 °C), then heating at 0.66 deg/min from 5 °C to 95 °C. 1 cm or 2 mm pathlength stoppered cuvettes were used, as appropriate, and data were collected at 260, 280, and 295 nm if possible. For ease of visualization and data analysis, melting data are plotted as the first derivative of the absorbance at each wavelength with respect to temperature. Transitions are revealed by peaks in this melting profile. These peaks can be treated as sequential two-state transitions, each characterized by an enthalpy (related to the width of the peak) and a Tm (the position of the apex of the peak). Global fits of data obtained at two wavelengths retrieve the relevant Tms and enthalpies for the first transition (which reports formation of tertiary structure), which were then used to calculate RNA folding free energies from the formula ΔGo = (ΔHo)(T0/Tm − 1), where R is the gas constant and T0 is 293 K. Reported errors are the result of bootstrap analysis on individual melting profiles. Software for taking the derivative of the absorbance data, fitting multiple transitions to data sets, and bootstrap analysis of errors has been described (21).
Results
Concentration vs. activity in MgCl2 titrations
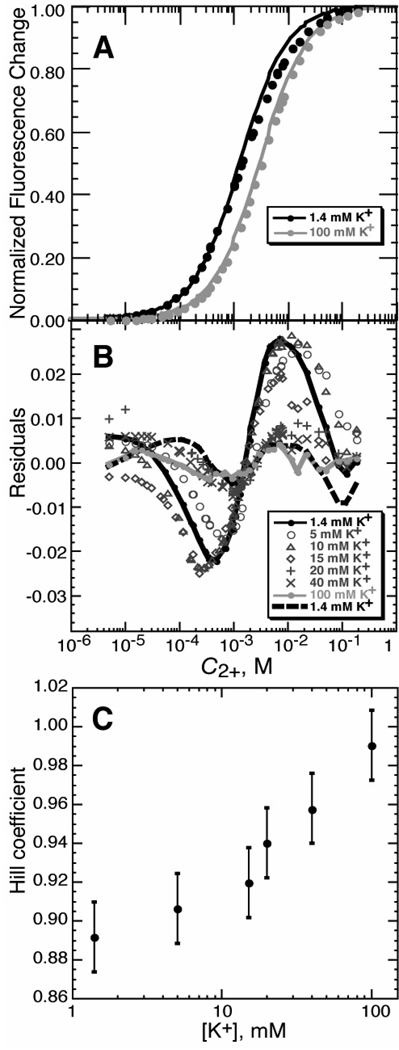
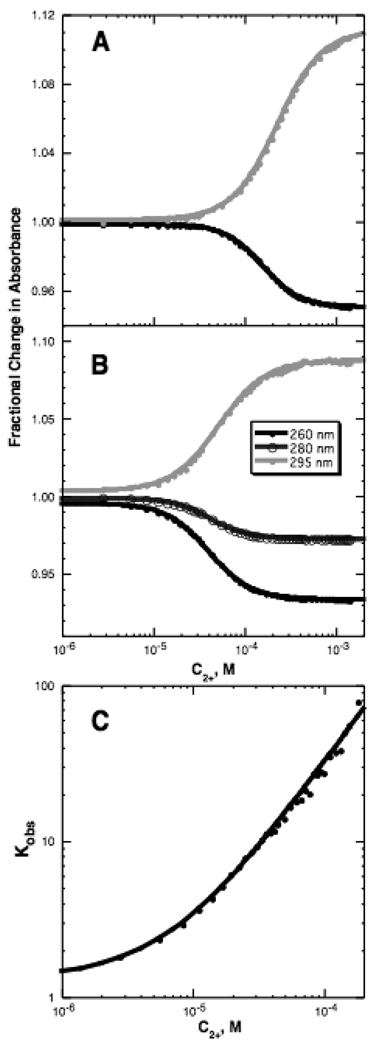
We first examine the question of the potential errors that are introduced in the analysis of a titration experiment when Mg2+ concentrations are used instead of activities. A sensitive way to look at this question is by use of a fluorescent dye, 8-hydroxy-5-quinoline sulfonic acid (HQS1), which chelates Mg2+ ion (22). The stoichiometry of the complex is 1:1 (18); therefore titrations of the dye with Mg2+ should fit a single-site binding isotherm (eq 9 with n set to 1). Because Mg2+ binding enhances the HQS fluorescence signal by a large factor (~40 fold), it is possible to detect small deviations of the titrations from the expected isotherm. Data obtained in titrating solutions of HQS and buffer with MgCl2 do in fact systematically deviate from a 1:1 binding curve (1.4 mM K+ curves in Figure 2A and B). We attribute the deviations to the fact that Mg2+ activity is decreasing as MgCl2 concentration increases; thus, more MgCl2 must be added to achieve the same effective concentration as the titration progresses, and the curve is broader than expected.
Figure 2.
Mg2+ - HQS binding isotherms. A) Titrations of HQS with MgCl2 in either buffer alone (black symbols; 5 mM KMOPS buffer, 1.4 mM K+, pH 6.8) or the same buffer with added KCl (100 mM total K+; gray symbols). The curves are least squares best fits of a single-site binding isotherm (eq 9, with n = 1). Fluorescence intensity data were collected and normalized as described in Materials and Methods. B) Residuals of single-site binding isotherms fit to HQS titrations carried out with various concentrations of added KCl; total K+ concentrations, which include 1.4 mM contributed by the buffer, are indicated in the legend (see Materials and Methods for further details). The dashed black line indicates the residuals of a fit of eq 9 to the titration in 1.4mM K+, for which the Hill coefficient n was allowed to float; the same data fit with n = 1 are indicated with a solid black line. The gray solid line indicates the quality of fit for the single-site binding isotherm for the data collected in 100 mM K+. C) The HQS-Mg2+ titrations were fit to eq 9, treating the Hill coefficient n as a variable. n approaches 1 as the concentration of KCl increases. The K+ concentration is equal to the initial anion concentration (the sum of the chloride and MOPS anion concentrations). Error bars are from bootstrap analysis (see Materials and Methods).
Mg2+ activity is largely determined by its interactions with anions, both Cl− and ionized MOPS in the Figure 2 titrations. Over the course of a titration with MgCl2, the concentration of Cl− changes dramatically (from none to > 100 mM, compared to 1.4 mM MOPS anion), and the activity of Mg2+ progressively decreases. If a change in Mg2+ activity was the reason for the broadened titration curve, then the effect should be suppressed by including a large amount of monovalent salt in the titration conditions—a high initial concentration of Cl− will minimize the effect of further screening of Mg2+ by chloride ions from added MgCl2. Indeed, with the addition of 98.6 mM KCl (see gray curve in Figure 2A) a single-site isotherm gave an excellent fit to the titration data, as indicated by the dramatic decrease in the systematic deviations between the fitted curve and data points (cf. residuals in Figure 2B). Thus, in order to minimize changes to Mg2+ activity during a titration, experiments are best performed in a sufficient excess of chloride to keep the total anion concentration fairly constant.
The titration data from Figures 2A and B were reanalyzed with the Hill equation (eq 9). All fits were excellent, regardless of KCl concentration (e.g., dashed line in Figure 2B), as long as the empirical Hill coefficient was allowed to deviate from n = 1. The best-fit values of n were significantly less than 1 in low salt conditions, but approached the true binding stoichiometry at higher salt concentrations, when the Mg2+ activity coefficient was nearly invariant during the titration (Figure 2C).
Equilibrium folding of the A-riboswitch
In the titration of an RNA with MgCl2, the exponent in the Hill equation (n or ΔΓ2+, eqs 6 and 9) is expected to increase if the folding transition midpoint is shifted to higher Mg2+ concentration. How strong is this dependence? An RNA that lends itself to such a study is the adenine-binding riboswitch (A-riboswitch), a regulatory RNA found in the 5’ UTRs of several bacterial mRNAs encoding proteins involved in purine metabolism (23). Upon binding adenine, the aptamer portion of the switch (Figure 3A) adopts a specific tertiary structure that affects the expression of the gene under regulation. The advantage of the A-riboswitch for our purposes is that the RNA folds in different Mg2+ concentration ranges depending on the affinity and concentration of the ligand (adenine, or an adenine derivative) that is present.
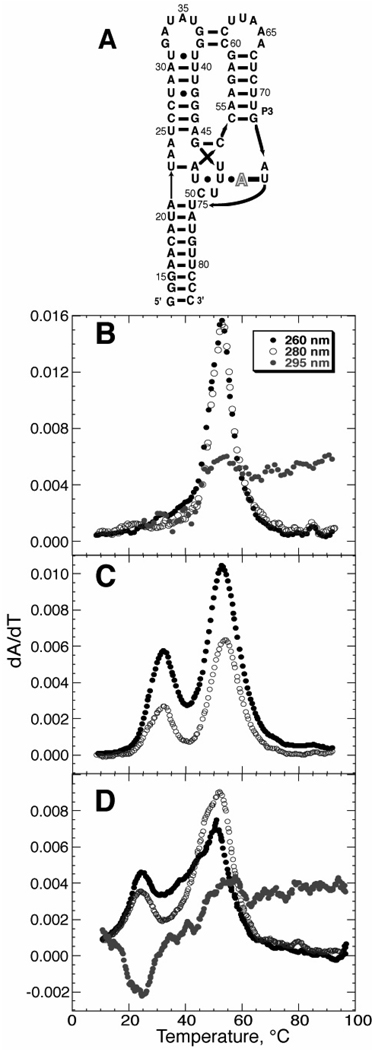
Figure 3.
Folding of the adenine-binding riboswitch (A-riboswitch) tertiary structure. A) Schematic of the aptamer domain of the A-riboswitch. Arrows denote 5’-3’ backbone connectivity, horizontal black bars represent canonical Watson-Crick base pairing, black dots represent non-canonical pairs, and gray bars represent tertiary interactions. The outlined A denotes the ligand. Panels B, C, and D are representative melting profiles of the A-riboswitch in the presence or absence of purine ligand and MgCl2. Data were collected at three wavelengths: 260 nm (black circles), 280 nm (open circles), and 295 nm (gray circles). B) Representative melting profile of the A-riboswitch secondary structure in the absence of either MgCl2 or purine ligand (20 mM KMOPS buffer with 3 µM RNA, 50 mM K+). C) A new A-riboswitch unfolding transition appears (~ 30 °C) in the presence of DAP ligand and absence of MgCl2 (20 mM KMOPS buffer with 8 µM RNA, 50 mM K+, 200 µM DAP). The melt was performed in a 2 mm path length cuvette because of the high absorbance with DAP present; the change at 295 nm was too small to detect. D) A-riboswitch tertiary structure forms in the absence of ligand if MgCl2 is present (20 mM KMOPS buffer with 3 µM RNA, 50 mM K+, 2 mM MgCl2). The signal at 295 nm is hypochromic upon unfolding, and reports on the same tertiary transition detected by the hyperchromic changes at 260 nm and 280 nm.
The stability of the A-riboswitch tertiary structure is sensitive to both Mg2+ and purine ligand. In their absence, a single peak in the UV melting profile represents disruption of the secondary structures present in the partially unfolded (I state) RNA (Figure 3B). Folding of the riboswitch tertiary structure can then be induced by adding either a purine ligand (2,6-diaminopurine, DAP, in Figure 3C) or Mg2+ (Figure 3D), as indicated by the appearance of a new, low-temperature peak in the profile. This peak is identified with the formation of tertiary structure because its Tm depends on the concentration of DAP present (data not shown). The unfolding transitions can be observed by monitoring three different wavelengths: the hyperchromic change in 260 nm and 280 nm have different sensitivities to base stacking, and the more unusual hypochromic change observed at 295 nm reports on an entirely different interaction between bases (24). Observation of the transition at 295 nm is convenient since this change in absorbance is unique to tertiary structure formation in this RNA. These melting experiments and others established the solution conditions under which the riboswitch is folded at 20 °C, information that was needed for the design of subsequent titration experiments.
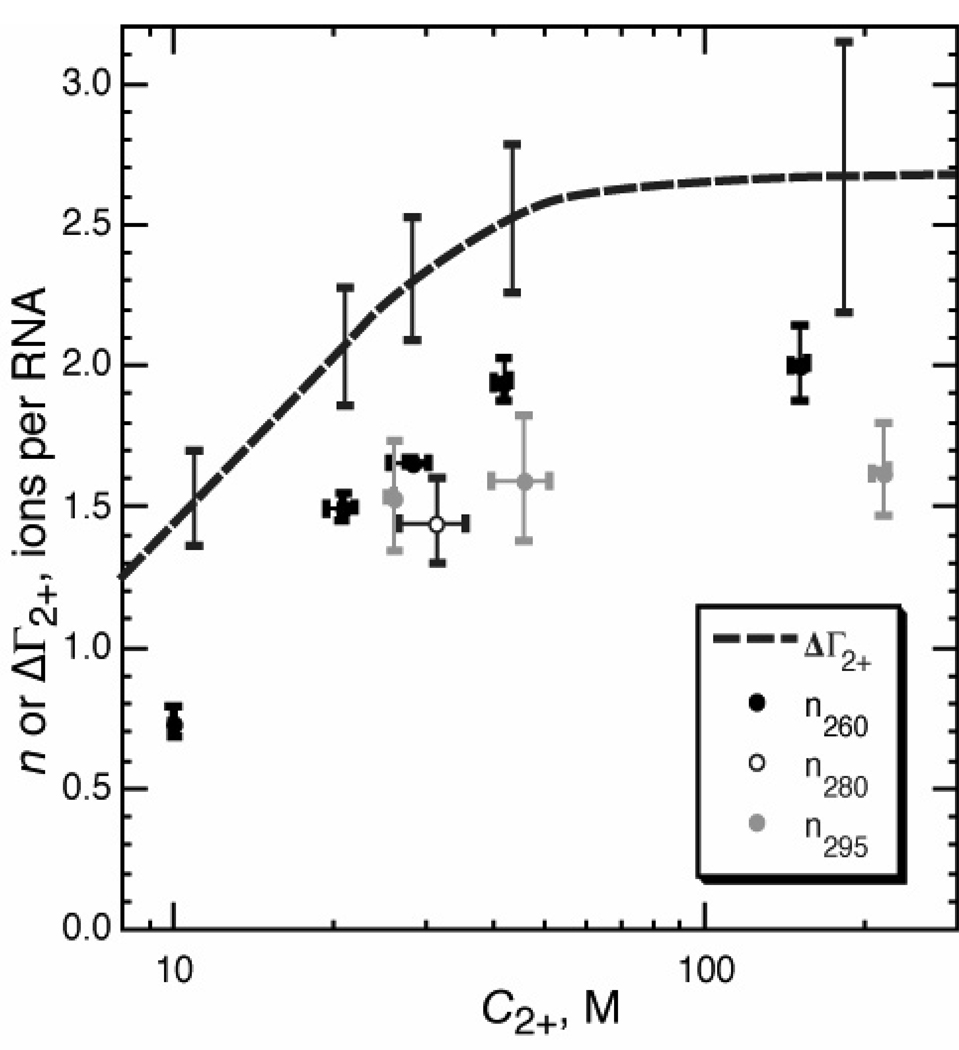
A series of isothermal Mg2+-titrations with the riboswitch in the presence of different ligands was performed. The ligands used in these experiments (in order of increasing affinity) are purine, adenine, 2-aminopurine (2-AP), and DAP. In total, the four ligands in five conditions were used to access folding reactions with a range of folding midpoints between 10 and 200 µM Mg2+ (Table 1). Data collected at each of three wavelengths were independently fit to a modified Hill equation that included baseline corrections to account for changes in extinction coefficient (eq 12, Materials and Methods). (In some conditions, the change in the signal at one wavelength was too small to be reliable, and thus was omitted.) If folding is two-state, analysis of all three signals should be self-consistent. Midpoints determined at different wavelengths were the same within error with the exception of the titration in the presence of purine; in addition, a wavelength-dependent variation in n suggests deviation from two-state behavior in the case of 11 µM adenine. There is a trend towards larger n as the midpoint of the titration shifts to higher Mg2+ concentrations (Figure 5 and Discussion).
Table 1.
Measurements of the Hill coefficient n and ΔΓ2+ for A-riboswitch foldinga
| UV titrations | RNA-HQS titrations | |||
|---|---|---|---|---|
| Condition | Wavelength (nm) | Midpoint (µM) | Avg n | ΔΓMg+2 |
| 11 µM purine | 260 | 152 ± 5 | 2.00 ± 0.13 | 2.65 ± 0.48 |
| 295 | 217 ± 8 | 1.62 ± 0.16 | ||
| 11 µM adenine | 260 | 41.9 ± 1.4 | 1.94 ± 0.08 | 2.51 ± 0.26 |
| 295 | 45.9 ± 5.9 | 1.59 ± 0.22 | ||
| 82 µM adenine | 260 | 28.1 ± 2.4 | 1.65 ± 0.02 | 2.30 ± 0.22 |
| 280 | 31.2 ± 4.5 | 1.44 ± 0.15 | ||
| 295 | 25.9 ± 0.4 | 1.53 ± 0.20 | ||
| 82 µM 2-AP | 260 | 20.9 ± 1.2 | 1.49 ± 0.05 | 2.06 ± 0.21 |
| 11 µM DAPb | 260 | 10.0 | 0.73 ± 0.05 | 1.52 ± 0.17 |
All titrations with MgCl2 were performed in 20 mM KMOPS buffer, 50 mM K+, at 20 °C. The ligand identity and concentration was the only variable among the data sets, as listed in the first column of the Table. RNA folding was monitored by absorbance changes at one to three different wavelengths (see Materials and Methods for details). The midpoints and Hill coefficients of the titration curves are the parameters (1/K)1/n and n, respectively, obtained from independent fits of eq 12a to each data set. The errors reported are the standard deviation of values obtained from three repetitions of each experiment. ΔΓ2+ values were calculated at the average of the titration midpoints for each condition from the data shown in Figure 7 (ΔΓ2+ = ΓN−2−ΓI−2+). The reported errors are standard deviations of four repeated experiments.
Figure 5.
Comparison of the empirical Hill coefficient n for A-riboswitch folding with a direct measurement of ΔΓ2+. (circles), plot of Hill coefficients n vs. titration midpoints (C2+) taken from Table 1. Subscripts of n refer to the wavelength at which titration data were collected. (dashed line), ΔΓ2+ calculated from measurements of Γ2+ for folded and partially unfolded RNA (Figure 6). Error bars are shown at selected C2+ for comparison with n values.
For one set of titration conditions (11 µM DAP, Figure 4C), melting experiments under the same conditions show that the RNA tertiary structure has a Tm of 20.7 °C, corresponding to Kobs = 1.32 at 20 °C (data not shown). Because eq 6 assumes that θfold varies from 0 to 1 over the course of the titration, we used a modified equation which takes on a value of θfold = Kobs/(1+Kobs) when C2+ = 0 (see Materials and Methods, eq 12b). The fit of this equation to the data set is shown in Figure 4C, which is graphed in a way similar to a standard 'Hill plot' (12) in that the slope of this plot at a particular value of C2+ is ΔΓ2+ (cf. eq 5). The slope clearly approaches zero at low C2+, and increases steadily over the range of accessible C2+.
Figure 4.
Representative titrations of A-riboswitch RNA with Mg2+ in the presence of different ligands. The spectroscopic signals at 260 (black circles), 280 (open circles), and 295 nm (gray 37 circles) were collected and analyzed. Solid lines are least squares fits of two-state transitions to the data; for clarity, linear baselines have been subtracted (see Materials and Methods, eqs 12a and 12b). All titrations were carried out in buffer containing 1 µM RNA, 20 mM KMOPS, 50 mM K+, and the indicated concentration of ligand. A) Titration of RNA in 11 µM purine. Folding in the presence of purine shows the largest discrepancy in transition midpoints (Table 1). The change in absorbance at 280 nm was too small to be recorded. B) Titration of RNA in 11 µM adenine. C) Titration of RNA in the presence of 11 µM DAP. The graph is in the form of a 'Hill plot', the slope of which corresponds to ΔΓ2+.
Direct measurement of excess Mg2+ ions (Γ2+)
A direct way to measure Mg2+ accumulation by an RNA is to use the aforementioned fluorescent dye HQS as a sensor of Mg2+ activity in solution. In the absence of RNA, HQS binds Mg2+ as predicted by a single-site binding isotherm, reported by an increase in fluorescence (Figure 2A). In the presence of RNA, the titrated Mg2+ interacts with both the HQS dye and the RNA, so a higher Mg2+ concentration is needed to achieve the same level of HQS fluorescence. It is not necessary for Mg2+ ions to be in direct contact with the RNA in order for the dye to detect a change in ion activity; long-range electrostatic interactions between Mg2+ and RNA will also reduce the apparent Mg2+-HQS binding affinity. The difference between the two titration curves obtained in the presence and absence of RNA is related to the preferential interaction coefficient (Γ2+) (9).
With the A-riboswitch, RNA-HQS titrations can be performed with either folded or unfolded RNA. Γ2+ for the unfolded form is obtained in the absence of ligand, and data for the folded form are gathered in the presence of a large excess of DAP, the most stabilizing of the ligands. Melt analysis (Figure 3C) as well as small-angle x-ray scattering data (data not shown) reveal that the RNA is completely folded at 20 °C in the absence of Mg2+ ion when 250 µM DAP and 50mM K+ are included; if DAP is omitted, the RNA tertiary structure does not form at the highest Mg2+ concentrations accessed in these experiments (~ 300 µM). The titrations were all performed in excess monovalent salt (50 mM K+) to avoid the Mg2+ activity problem discussed previously.
Excess Mg2+ curves for the folded and unfolded A-riboswitch are shown in Figure 6A. Of note is the substantial accumulation of Mg2+ by the unfolded (I state) form of the RNA. The difference between the two curves in Figure 6A is the uptake of Mg2+ (ΔΓ2+) during the folding reaction, as a function of Mg2+ concentration (Figure 6B). An important outcome of these measurements is the variation seen in ΔΓ2+, which approaches zero at very low Mg2+ concentrations and reaches a maximum value at ~0.1 mM Mg2+ (see Discussion).
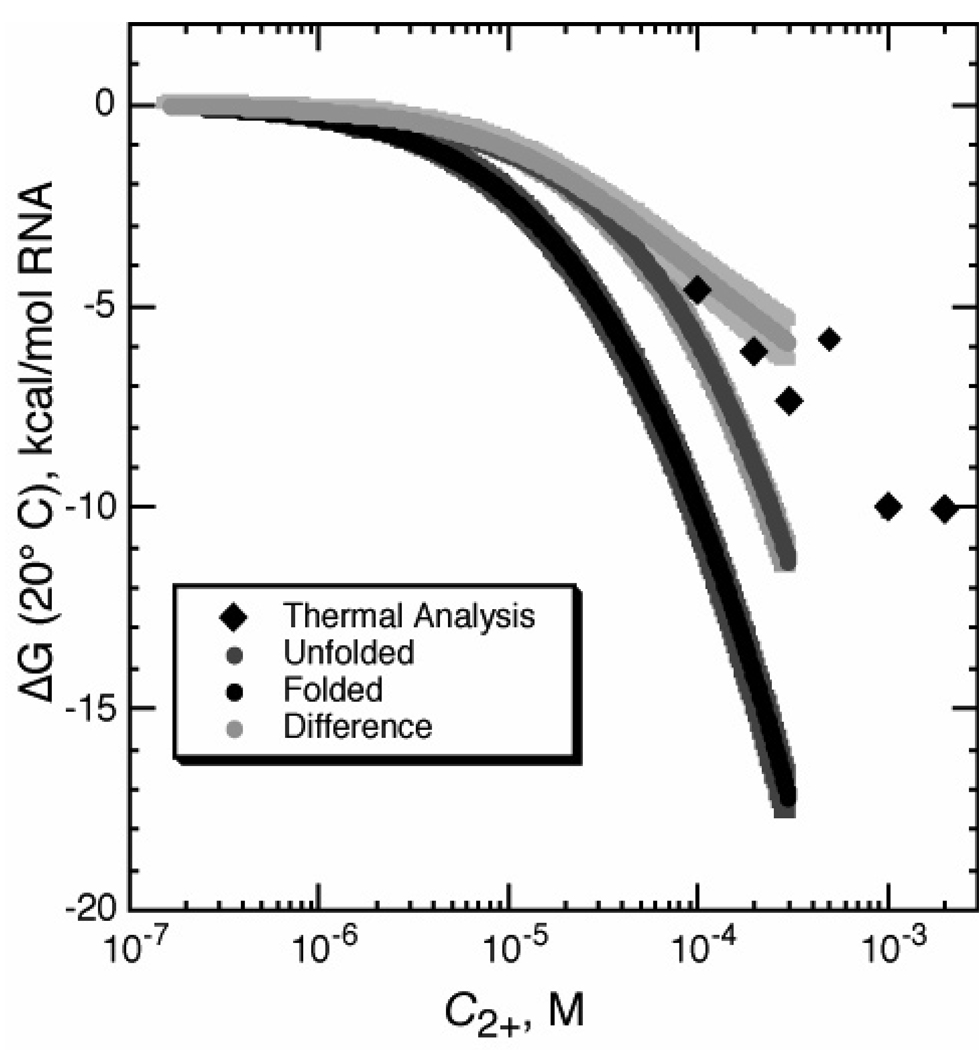
Integration of the Γ2+ data sets in Figure 6A yields the free energy of Mg2+ interaction with either of the forms of the RNA (Figure 7). These free energies, ΔGI−2+ and ΔGN−2+, are associated with the horizontal arrows of the thermodynamic cycle in Figure 1. Clearly the magnitude of ΔGI−2+ makes it a substantial consideration in the overall thermodynamics of the Mg2+-induced folding reaction (see Discussion). The difference between the free energy curves for the folded and unfolded RNA states is the free energy contribution of Mg2+ to the RNA folding reaction (ΔΔG2+, Figures 1 and 7). This free energy difference, obtained by direct measurements of Mg2+ - RNA interactions, can be compared with the same quantity obtained by an independent method, the stabilization of the RNA by Mg2+ in thermal melting experiments (Figure 7). Where the two data sets overlap, the agreement is very good. Unlike the thermal melting analysis, the Γ2+ measurements reveal the individual contributions from the native and unfolded forms to the overall stabilization free energy.
Figure 7.
Free energy changes upon addition of MgCl2 to folded (black curve, in the presence of ligand) and unfolded (gray curve, absence of ligand) A-riboswitch, as calculated by integration of data in Figure 6. The difference between the curves (ΔΔG2+) is also plotted (light gray). Other data points (black diamonds) were calculated from thermal melting experiments (pH 6.8 20 mM MOPS, 50 mM K+, 20 µM DAP), from the increment in Tm of the tertiary unfolding transition upon addition of Mg2+ (see Materials and Methods).
Discussion
Riboswitches are potentially useful systems for studying the influence of Mg2+ on RNA folding reactions, since added ligand can shift the folding equilibrium and allow a wide range of salt concentrations to be explored. The A-riboswitch is a particularly good candidate for such studies because the purine ligand is a natural RNA base; therefore no interactions take place in the ligand complex that are not typical of folded RNA molecules. In this paper, we use the A-riboswitch to investigate the Mg2+ – dependent thermodynamics of an RNA folding equilibrium.
In the Background, we discussed two accounts of Mg2+ ion interactions with RNA: a simplified, all-or-none ligand-binding model (Figure 1B, eq 9) and a thermodynamic cycle that is developed in terms of preferential interaction factors and a Wyman linkage equation (Figure 1A). The two accounts lead to equations with the form of the empirical Hill equation (eqs 6 and 9) even though very different assumptions underlie the derivations. The formalism using interaction coefficients is a more rigorous and general method to treat ions, since Γ2+ describes the overall Mg2+ - RNA interactions in a way that includes long-range electrostatics as well as any specific site-binding. The binding formalism, in contrast, starts with the very restrictive assumption that only a stoichiometric number of n Mg2+ ions, bound to specific sites in the native RNA, are thermodynamically important. The primary goal of this work is to determine the experimental conditions under which it is valid to use the Hill equation in data analysis and interpret the Hill coefficient as having physical significance. To derive eqs 5 and 6, which can be used to extract ΔΓ2+ from experimental data, it was necessary to make two approximations: (i) Mg2+ concentration can be substituted for MgCl2 activity, and (ii) the data being analyzed cover a narrow enough range in Mg2+ concentration that ΔΓ2+ can be treated as a constant. The next sections discuss experiments that quantitatively establish the conditions under which (i) and (ii) are good approximations.
Substitution of Mg2+ concentration for activity
Mg2+ concentrations can be substituted for MgCl2 activity in the linkage eq 5 when the activity coefficient for MgCl2 (γMgCl2, eq 11) is independent of Mg2+ concentration. The activity of a Mg2+ ion is affected by the presence of its counterion, Cl−, due to screening: in the concentration ranges considered here, the more concentrated Cl− ion becomes, the less 'effective' Mg2+ will be. In titrations of a metal-chelating dye with MgCl2, the broadened titrations in low-salt solutions (Figure 2A) suggest that the 'effectiveness' of Mg2+ in binding the dye decreases as higher Mg2+ concentrations accumulate. This behavior is consistent with the expected decrease in Mg2+ activity coefficient as the total Cl− concentration increases. The titration curves approached the expected shape of a single-site binding isotherm when a large excess of monovalent salt was included, as anticipated when the Cl− concentration remained nearly constant during the titration. The deviation of n from the expected value of one was fairly small in the HQS - Mg2+ complex considered here, only ~10% when Mg2+ and K+ concentrations were in similar ranges. However, a KCl excess over added MgCl2 of more than 10-fold was needed to bring n within experimental error of one (Figure 1C). We therefore expect that low concentrations of monovalent salt could introduce some bias in extracting ΔΓ2+ from Mg2+ - RNA titrations. Though somewhat counterintuitive, the way to minimize the contribution of chloride ion is to have it present in excess so the activity coefficient of Mg2+ stays constant throughout the titration experiment.
Early folding studies of tRNA generally used a large excess of monovalent over divalent salts (2, 25). Subsequent studies have shifted to using concentrations of monovalent salt, including buffer ion, comparable in magnitude to the MgCl2 concentration (26–28). These are low enough concentrations to make interpretation of the Hill coefficient problematic. An additional benefit of using an excess of KCl over MgCl2 is that the physiological K+ and Mg2+ activities are about 0.15 M and 0.5–1 mM respectively, so experiments with excess monovalent salt are actually more relevant to in vivo folding conditions (29–31).
Dependence of ΔΓ2+ on Mg2+ concentration
In regard to the dependence of ΔΓ2+ on Mg2+ concentration, we find that ΔΓ2+ (as calculated from independent measurements of ΓN−2+ and ΓI−2+) approaches zero at low Mg2+ concentrations, as expected, and increases monotonically to a maximum value at approximately 0.1 mM Mg2+ (Figure 6B). For the purpose of data analysis using the linkage eq 5 or the Hill eq 6, ΔΓ2+ can be considered a constant if the range of Mg2+ concentrations being analyzed is either narrow or in the region where ΔΓ2+ has reached a maximum value. When titration curves were simulated with ΔΓ2+ varying approximately in the way seen in Figure 6A, fits of the Hill equation to the simulated curves gave Hill coefficients that only slightly differ from the actual value of ΔΓ2+ at the midpoint of the curve. In fitting the Hill equation to a data set, the Hill coefficient tends to be heavily biased by the slope of the data at the midpoint of the titration curve, and relatively insensitive to the shape of the curve at values of θfold > 0.9 or < 0.1. For this reason, eq 5 works well in assigning a correct value of ΔΓ2+ to a data set, but the value only applies to the Mg2+ concentraton at the titration midpoint.
A different way to approach the calculation of ΔΓ2+ is to re-cast the titration data as a plot of ln(Kobs) as a function of ln(C2+). The slope of such a plot taken at any one value of C2+ is ΔΓ2+, as is clear from the linkage eq 5. In principle, the Mg2+-dependence of ΔΓ2+ should cause the plot to appear curved. In most cases, accurate values of ln(Kobs) cannot be obtained over a wide enough range of Mg2+ concentrations to reliably observe the expected curvature. An exception is the titration shown in Figure 4C, in which the RNA is partly folded in the absence of Mg2+. Because there is an independent measure of Kobs before MgCl2 is added, the Mg2+ - dependence of ΔΓ2+ can be observed from the lowest Mg2+ concentrations. It is apparent that ΔΓ2+ approaches zero at low C2+, and increases sharply over the ~100 fold range of C2+ for which ln(Kobs) is available, consistent with the variation of ΔΓ2+ with C2+ seen by direct measurements (Figure 6B).
Contrasting interpretation of the Hill exponent in the binding and preferential interaction formalisms
To assess the cooperativity of ligand binding to allosteric proteins, titration data are sometimes graphed as a 'Hill plot', ln(θL/(1−θL)) vs. ln(CL), where θL is the fractional saturation of binding sites by ligand and CL is the ligand activity (32, 33). The slope of this plot, at times referred to as the "coefficient of cooperativity" or the Hill coefficient nH, reaches a maximum at the midpoint of the titration curve where it takes on the value of n in the empirical Hill equation (eq 9). The maximum of nH cannot exceed the number of ligand binding sites in the protein. The minimum value of nH is unity, which is approached at high and low ligand concentrations. The Hill plot for ligand binding data is analogous to Figure 4C, which substitutes Kobs = θfold/(1−θfold) for θL/(1−θL) and has a slope of ΔΓ2+. The contrasting range of slopes (and values of nH or ΔΓ2+) in the two kinds of plots illustrates a fundamental difference between ligand - protein interactions and ion - nucleic acid interactions.
For ligand-binding proteins, the allowed values of nH are constrained by binding stoichiometries: the minimum value is unity because the minimum binding unit is a single ligand molecule, and the maximum possible value is the ligand:protein stoichiometry at saturation. In contrast, the presence of strong, long-range electrostatic interactions between ions and an RNA means that Γ2+ cannot be identified with a specific set of ions, and is not a binding stoichiometry. (An exception is extremely high salt buffers that minimize long-range interactions, where Γ2+ may approach a stoichiometric ratio (34, 35).) The increase in charge density that accompanies RNA folding causes a global re-configuration of long- and short-range interactions with all ions present in solution (36); thus ΔΓ2+ is not restricted to integral numbers, and in particular may be less that one. It is frequently assumed that the minimum number of Mg2+ ions that may be taken up in an RNA folding reaction is one, or that a Hill coefficient near one indicates 'non-cooperative' binding of Mg2+ ions to an RNA (37, 38). These assumptions are based on an allosteric protein - ligand binding model which is an inappropriate representation of RNA - Mg2+ interactions.
ΔΓ2+ obtained by direct methods vs. linkage equations
Between the two methods discussed for measuring the ion uptake, application of the linkage eq 5 to spectroscopic titrations always gave smaller values of ΔΓ2+ than those obtained from RNA-HQS titrations (Figure 5 and Table 1). Perhaps this should be unsurprising, given the fundamental differences between the techniques. In each of the RNA-HQS titrations, the populations of RNA are either entirely folded or entirely unfolded (Figure 6A); taken together, they measure the ion uptake upon folding (ΔΓ2+) as a function of bulk Mg2+ concentration. In contrast, interpretation of the spectroscopic folding experiments depends on the validity of the two-state approximation implicit in eq 5. If there is a population of RNAs at the transition midpoint that is mostly folded but not quite native in structure, then the titration curve would be broadened and the calculated value of ΔΓ2+ will be smaller than that calculated from the difference between ΓN−2+ and ΓI−2. Data in Table 1 suggest that A-riboswitch folding is not always two-state; for the weakest-interacting ligand (purine), the global measure of folding (260 nm signal) exhibits a lower midpoint than the local, tertiary-specific measure (295 nm signal). These results are consistent with single molecule studies of a similar A-riboswitch (pbuE from B. subtilis), which have detected an intermediate, partially folded RNA at Mg2+ concentrations near the transition midpoint (39).
For the folding of a 58mer fragment of rRNA, ΔΓ2+ derived from linkage analysis is within error of ΔΓ2+ found by HQS-RNA titrations (9), which suggests that the HQS-RNA titration method itself does not introduce systematic errors in the determination of ΔΓ2+. We therefore attribute the differences between direct and linkage - based measurements of ΔΓ2+ in the A-riboswitch to deviations from strictly two-state behavior and suggest that this kind of comparison could be an informative way to assess the cooperativity with which an RNA folds.
Implications of the dependence of ΔΓ2+ on Mg2+ concentration
The dependence of ΔΓ2+ on Mg2+ concentration has not been generally recognized in RNA studies; the Hill coefficient is usually thought to remain constant over a very wide range of Mg2+ concentrations. For instance, the Hill coefficient determined for an RNA at one Mg2+ concentration has been used to extrapolate the expected free energy change for the RNA at a different Mg2+ concentration, according to the formula
| (13) |
where ΔΔGMg2+ is the difference in folding free energy for two related RNAs whose titration curves have midpoints at C2+,1 and C2+,2; the same coefficient n is assumed to apply to both RNAs (16). This extrapolation is reasonable for ratios of C2+,1 to C2+,2 near one, but could become inaccurate as the difference in the titration midpoints increases. Another potential interpretive problem arises when folding of the same RNA domain is considered in two different contexts. The isolated P4–P6 domain shows a larger value of the Hill coefficient than the same region incorporated into the full-length Tetrahymena ribozyme (28), but the midpoint of the transition also occurs at a higher Mg2+ concentration in the isolated domain. Without knowing the detailed dependence of ΔΓ2+ on Mg2+ concentration, it is not possible to say whether the difference in Hill coefficient reflects any fundamental difference in RNA-ion interactions or folding mechanism in the two contexts.
Conclusion
The main focus of this work has been on the question of whether the Hill coefficient, an empirical factor frequently used as a measure of the Mg2+-dependence of RNA folding, can ever be related to the Figure 1A thermodynamic cycle. We find that the empirical Hill coefficient n can have physical meaning under the right set of conditions: if the ratio of monovalent to divalent salt is high enough and if a narrow range of Mg2+ concentrations is considered. In these cases n corresponds to ΔΓ2+, a well-defined thermodynamic parameter. The importance of the relatively strong interactions of Mg2+ with the partially unfolded (I state) form of an RNA in determining the magnitude of ΔΓ2+ has been pointed out for other RNAs (9, 40, 41) and is further supported by measurements of the A-riboswitch presented here (Figures 6A and 7). Finally, it is important to note that ΔΓ2+ describes a global reconfiguration of ions that accompanies the folding reaction and includes ions interacting at long ranges as well as any at discrete sites.
Footnotes
This work was supported by National Institutes of Health Grants GM58545 (to D.E.D.) and T32 GM008403 (Program in Molecular Biophysics).
Abbreviations used: HQS, 8-hydroxy-quinoline-5-sulfonic acid; 2-AP, 2-aminopurine; DAP, 2,6 diaminopurine; A-riboswitch, the adenine-binding domain of an adenine riboswitch.
References
- 1.Cole PE, Yang SK, Crothers DM. Conformational Changes of Transfer Ribonucleic Acid. Equilibrium Phase Diagrams. Biochemistry. 1972;11:4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- 2.Stein A, Crothers DM. Conformational changes of transfer RNA. The role of magnesium(II) Biochemistry. 1976;15:160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 3.Latham JA, Cech TR. Defining the Inside and Outside of a Catalytic RNA Molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 4.Schreier AA, Schimmel PR. Interaction of Manganese with Fragments, Complementary Fragment Recombinations, and Whole Molecules of Yeast Phenylalanine Specific Transfer RNA. J. Mol. Biol. 1974;86:601–620. doi: 10.1016/0022-2836(74)90183-1. [DOI] [PubMed] [Google Scholar]
- 5.Hill AV. The possible effects of the aggregation of the molecules of hemoglobin on its dissociation curves. J Physiol (Suppl) 1910;40:4–7. [Google Scholar]
- 6.Schimmel PR, Redfield AG. Transfer RNA in Solution: Selected Topics. Annu Rev Biophys Bioeng. 1980;9:181–221. doi: 10.1146/annurev.bb.09.060180.001145. [DOI] [PubMed] [Google Scholar]
- 7.Draper DE, Grilley D, Soto A. Ions and RNA Folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 8.Record MT, Courtenay ES, Cayley S, Guttman HJ. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem Sci. 1998;23:143–148. doi: 10.1016/s0968-0004(98)01196-7. [DOI] [PubMed] [Google Scholar]
- 9.Grilley D, Soto A, Draper DE. Mg2+–RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc Natl Acad Sci USA. 2006;103:14003–14008. doi: 10.1073/pnas.0606409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leipply D, Lambert D, Draper DE. Ion - RNA interactions: thermodynamic analysis of the effects of mono- and divalent ions on RNA conformational equilibria. Methods in Enzymol. 2009;469:427–457. doi: 10.1016/S0076-6879(09)69021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dove WF, Davidson N. Cation Effects on the Denaturation of DNA. J Mol Biol. 1962;5:467–478. [Google Scholar]
- 12.Wyman JJ. Linked functions and reciprocal effects in hemoglobin: a second look. Adv Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 13.Fang X, Pan T, Sosnick TR. A Thermodynamic Framework and Cooperativity in the Tertiary Folding of a Mg2+-Dependent Ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- 14.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 15.Laing LG, Gluick TC, Draper DE. Stabilization of RNA Structure by Mg Ions: Specific and Non-specific Effects. J Mol Biol. 1994;237:577–587. doi: 10.1006/jmbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- 16.Silverman SK, Cech TR. Energetics and Cooperativity of Tertiary Hydrogen Bonds in RNA Structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- 17.Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR. Structure of a Folding Intermediate Reveals the Interplay Between Core and Peripheral Elements in RNA Folding. J Mol Biol. 2005;352:712–722. doi: 10.1016/j.jmb.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Grilley D, Soto A, Draper DE. Direct Quantitation of Mg2+-RNA Interactions by Use of a Fluorescent Dye. Methods Enzymol. 2009;455:71–94. doi: 10.1016/S0076-6879(08)04203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sillen LG, Martell AE. Stability Constants of Metal-Ion Complexes. London: The Chemical Society; 1964. [Google Scholar]
- 20.Serganov A, Yuan Y, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draper DE, Bukhman YV, Gluick TC. Thermal Methods for the Analysis of RNA Folding Pathways. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Current Protocols in Nucleic Acid Chemistry. New York: John Wiley & Sons; 2000. pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 22.Bishop JA. Complex Formation and Fluorescence Part I: Complexes of 8-Hydroxyquinoline-5-sulfonic acid. Analytica Chimica Acta. 1963;29:172–177. [Google Scholar]
- 23.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 24.Testa SM, Gilham PT. Analysis of oligonucleotide structure using hyperchromism measurements at long wavelengths. Nucleic Acids Res. 1993;21:3907–3908. doi: 10.1093/nar/21.16.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch DC, Schimmel PR. Cooperative Binding of Magnesium to Transfer Ribonucleic Acid Studied by a Fluorescent Probe. Biochemistry. 1974;13:1841–1852. doi: 10.1021/bi00706a012. [DOI] [PubMed] [Google Scholar]
- 26.Celander DW, Cech TR. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991;251:401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- 27.Zarrinkar PP, Williamson JR. Kinetic Intermediates in RNA Folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 28.Uchida T, He Q, Ralston CY, Brenowitz M, Chance MR. Linkage of Monovalent and Divalent Ion Binding in the Folding of the P4–P6 Domain of the Tetrahymena Ribozyme. Biochemistry. 2002;41:5799–5806. doi: 10.1021/bi020042v. [DOI] [PubMed] [Google Scholar]
- 29.Cayley S, Lewis BA, Guttman HJ, Record MT. Characterization of the Cytoplasm of Escherichia coli K-12as a Function of External Osmolarity: Implications for Protein-DNA Interactions in Vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 30.London RE. Methods for Measurement of Intracellular Magnesium: NMR and Fluorescence. Annu Rev Physiol. 1991;53:241–258. doi: 10.1146/annurev.ph.53.030191.001325. [DOI] [PubMed] [Google Scholar]
- 31.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. Fluorescence measurements of free [Mg+2] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Wyman JJ. Allosteric Effects in Hemoglobin. Cold Spr Harb Symp Quant Biol. 1963;28:483–489. [Google Scholar]
- 33.Wyman JJ, Gill SJ. Binding and Linkage: Functional Chemistry of Biological Macromolecules. Mill Valley, CA: University Science Books; 1990. [Google Scholar]
- 34.Bukhman YV, Draper DE. Affinities and selectivities of divalent cation binding sites within an RNA tertiary structure. J Mol Biol. 1997;273:1020–1031. doi: 10.1006/jmbi.1997.1383. [DOI] [PubMed] [Google Scholar]
- 35.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J Am Chem Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J Mol Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 37.Downey CD, Fiore JL, Stoddard CD, Hodak JH, Nesbitt DJ, Pardi A. Metal Ion Dependence, Thermodynamics, and Kinetics for Intramolecular Docking of a GAAA Tetraloop and Receptor Connected by a Flexible Linker. Biochemistry. 2006;45:3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley D. The Complete VS Ribozyme in Solution Studied by Small-Angle X-Ray Scattering. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemay JPJ, Tremblay R, Lilley D, Lafontaine DA. Folding of the Adenine Riboswitch. Chem Biol. 2006;13:847–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Soto AM, Misra V, Draper DE. Tertiary Structure of an RNA Pseudoknot Is Stabilized by “Diffuse” Mg2+ Ions. Biochemistry. 2007;46:2973–2983. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- 41.Grilley D, Misra V, Caliskan G, Draper DE. Importance of Partially Unfolded Conformations for Mg2+-Induced Folding of RNA Tertiary Structure: Structural Models and Free Energies of Mg2+ Interactions. Biochemistry. 2007;46:10266–10278. doi: 10.1021/bi062284r. [DOI] [PubMed] [Google Scholar]